Abstract
Endocytosis, and the subsequent trafficking of endosomes, requires dynamic physical alterations in membrane shape that are mediated in part by endophilin proteins. The endophilin B family of proteins contains an N-terminal Bin/amphiphysin/Rvs (N-BAR) domain that induces membrane curvature to regulate intracellular membrane dynamics. Whereas endophilin B1 (SH3GLB1/Bif-1) is known to be involved in a number of cellular processes, including apoptosis, autophagy, and endocytosis, the cellular function of endophilin B2 (SH3GLB2) is not well understood. In this study, we used genetic approaches that revealed that endophilin B2 is not required for embryonic development in vivo but that endophilin B2 deficiency impairs endosomal trafficking in vitro, as evidenced by suppressed endosome acidification, EGFR degradation, autophagic flux, and influenza A viral RNA nuclear entry and replication. Mechanistically, although the loss of endophilin B2 did not affect endocytic internalization and lysosomal function, endophilin B2 appeared to regulate the trafficking of endocytic vesicles and autophagosomes to late endosomes or lysosomes. Moreover, we also found that despite having an intracellular localization and tissue distribution similar to endophilin B1, endophilin B2 is dispensable for mitochondrial apoptosis. Taken together, our findings suggest that endophilin B2 positively regulates the endocytic pathway in response to growth factor signaling, autophagy induction, and viral entry.
Keywords: autophagy, endocytosis, influenza, intracellular trafficking, virus entry, EGFR degradation, Endophilin B2, SH3GLB2, endosome maturation
Introduction
Endocytosis, and the subsequent trafficking of endosomes, requires dynamic physical alterations in membrane shape (1) that are mediated in part by endophilin proteins and changes in lipid composition (2, 3). The endocytic pathway is tightly regulated to maintain cellular homeostasis (4, 5). For example, endocytic trafficking is essential for the processing and silencing of receptors for extracellular growth factor signaling, the clearance of exogenous pathogens, and autophagy (4–9). During autophagy, damaged organelles and cytosolic components are enclosed in a double-membrane structure known as the autophagosome and delivered to the lysosome for degradation. Interestingly, autophagy has extensive cross-talk with the endocytic pathway, because autophagosomes obtain membrane from endosomes during biogenesis (10, 11), utilize endosomal effectors for trafficking (8, 12), and fuse with late endosomes and lysosomes for cargo degradation (13).
The endophilin B family of proteins, comprising endophilin B1 (SH3GLB1/Bif-1) and endophilin B2 (SH3GLB2), contains an N-terminal Bin/amphiphysin/Rvs (N-BAR)3 domain and a C-terminal Src homology 3 (SH3) domain that are responsible for inducing membrane curvature and mediating interactions with proteins containing proline-rich regions, respectively (1, 14). In contrast to the endophilin A family proteins that act predominantly at the plasma membrane, the endophilin B family proteins appear to regulate intracellular membrane dynamics (1). Endophilin B1 was originally identified as a pro-apoptotic BCL-2-associated X protein (BAX)-interacting factor that regulates mitochondrial morphogenesis and apoptosis (15–18). Subsequent studies, however, revealed that endophilin B1 also interacts with the class III phosphatidylinositol 3-kinase complex, dynamin 2, and the HIV-1 Tat interactive protein 2, 30 kDa (TIP30) complex to regulate autophagy (19–23) and the endocytic trafficking of growth factor receptors (24–26). Endophilin B1 deficiency in mice promotes obesity (27), Alzheimer's disease pathology (28), and tumor development (19, 21), indicating the importance of endophilin B1 in the maintenance of tissue homeostasis. Endophilin B2 was discovered by a yeast two-hybrid screen using endophilin B1 as a bait and exhibits 58% homology in amino acids and 67% homology in nucleotide sequence to endophilin B1 in humans (15). Despite its high sequence and structural similarity to endophilin B1 (29), the cellular function of endophilin B2 remains unclear.
In this study, we utilized genetic approaches to demonstrate that, whereas endophilin B2 is dispensable for embryonic development and mitochondrial apoptosis, loss of endophilin B2 impairs endosome acidification, leading to suppressed EGF-stimulated EGF receptor (EGFR) degradation, autophagic flux, and influenza A virus entry.
Results
Endophilin B2 is dispensable for embryonic development
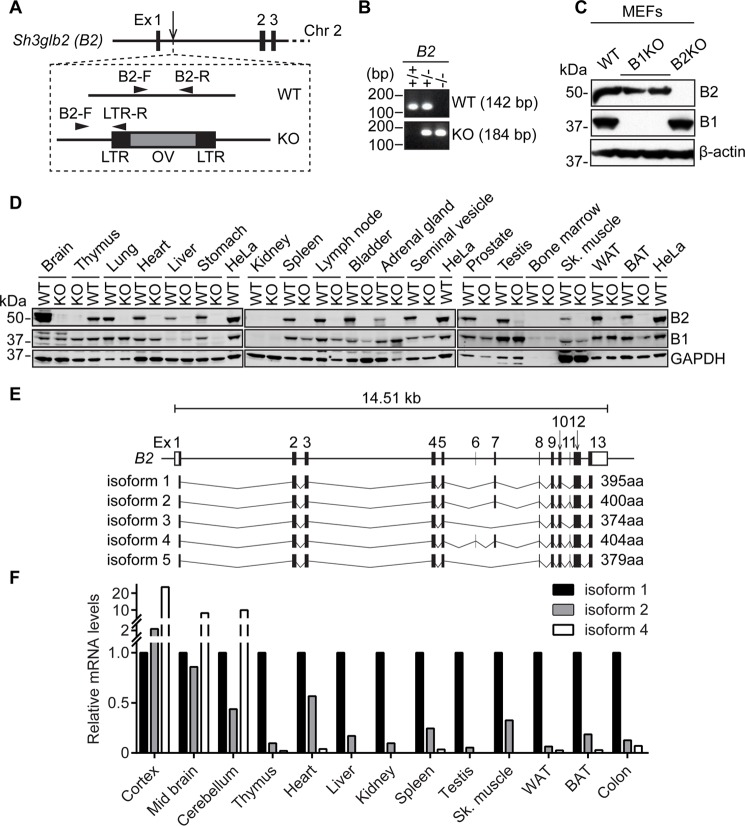
To determine the importance of endophilin B2 in vivo, endophilin B2-deficient mice were generated from an embryonic stem cell clone containing a gene trap cassette in the first intron of the Sh3glb2 gene (Fig. 1A). Genotyping of the resultant mice demonstrated successful insertion of the cassette, which disrupted endophilin B2 protein expression in isolated mouse embryonic fibroblasts (MEFs) (Fig. 1, B and C). Similar to endophilin B1 (18), loss of endophilin B2 does not affect embryonic development, because pups were born with normal ratios of Mendelian inheritance and were indistinguishable from wild-type littermates. Moreover, endophilin B2 demonstrated a similar tissue expression profile to endophilin B1, as indicated by co-expression of endophilins B1 and B2 in the majority of tissues examined (Fig. 1D). Interestingly, because endophilin B2 assembles heterodimers with endophilin B1 (15), we found that loss of endophilin B2 suppressed endophilin B1 expression in several tissues, including the heart, stomach, spleen, lymph node, bladder, skeletal muscle, and brown adipose tissue, indicating that an epistatic interaction between endophilin B1 and B2 genes may regulate the stability of endophilin B1 in these tissues.
Figure 1.
Endophilin B1 and B2 share similar expression profiles in mice. A, schematic of gene trap cassette insertion into endophilin B2 (B2) gene intron, genotyping primer design (described under “Experimental Procedures”), and cassette scheme (LTR, long terminal repeat; OV, Omnibank vector). B, genomic DNA prepared from mouse tail was subjected to PCR analysis using the primer sets shown in A. C, total cell lysates were prepared from MEFs with the indicated genotypes and subjected to immunoblotting using the indicated antibodies. B1, endophilin B1; B2, endophilin B2. D, tissue homogenates were prepared from 10-week-old endophilin B2KO and WT mice and subjected to immunoblotting using the indicated antibodies. Sk. muscle, skeletal muscle; WAT, white adipose tissue; BAT, brown adipose tissue. E, schematic of predicted murine endophilin B2 isoforms with predicted splicing and amino acid length. F, the mRNA expression profile for each endophilin B2 isoform was determined by semi-quantitative RT-PCR as described in supplemental Fig. S3. Relative expression of each isoform in each tissue was normalized to isoform 1 expression.
Furthermore, several tissue-specific isoforms of endophilin B1 have been implicated in alternative cellular functions in the brain and testis (30, 31). Because at least three distinct bands for endophilin B2 were detected in brain tissue lysates (Fig. 1D and supplemental Fig. S1), we sought to identify and characterize the isoforms of endophilin B2 in mice. The mouse endophilin B2 gene is located on chromosome 2 and contains 13 exons (32) that are predicted by the Ensembl database to encode five isoforms varying from 374 to 404 amino acids in length (Fig. 1E and supplemental Fig. S2). To assess the tissue expression profile of the endophilin B2 isoforms, we performed semi-quantitative RT-PCR (supplemental Fig. S3 and Table S2). We observed that isoform 1 of endophilin B2 was the most ubiquitously expressed, whereas isoforms 2 and 4 were increased in the brain, and isoforms 3 and 5 were undetectable in the tissues that we examined (Fig. 1F and supplemental Fig. S3).
Collectively, these results demonstrate that endophilin B2 is dispensable for murine embryonic development, shares a similar tissue distribution profile with endophilin B1, and exists as three predominant isoforms with isoform 1 ubiquitously expressed and isoforms 2 and 4 enriched in the brain.
Endophilin B2 is dispensable for mitochondrial apoptosis
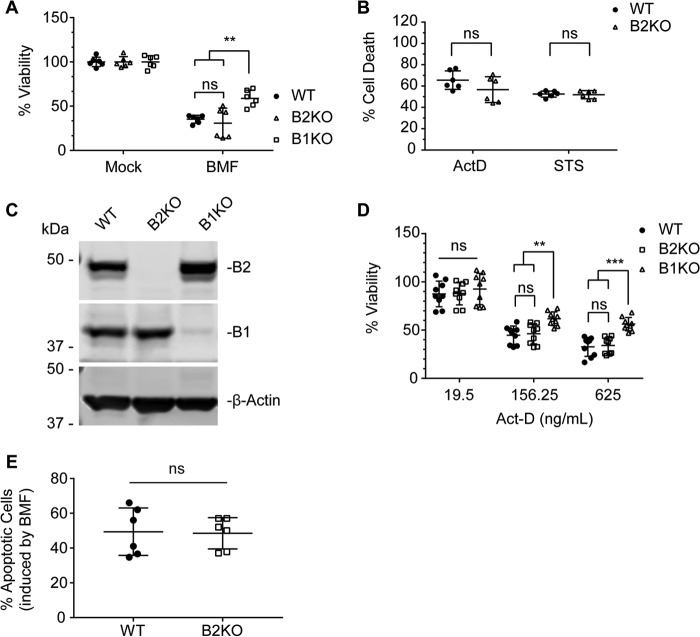
Endophilin B1 was discovered as a pro-apoptotic BAX-interacting protein that promotes mitochondrial apoptosis (15, 16, 18). Because endophilin B1 and endophilin B2 display a similar tissue expression profile (Fig. 1D), are highly similar in protein structure, and heterodimerize (15, 33), we first aimed to examine whether endophilin B2 also plays an important role in mitochondrial apoptosis. To this end, WT MEFs or MEFs lacking endophilin B1 (B1KO) or endophilin B2 (B2KO) were transduced with lentivirus encoding the BH3-only protein BCL-2-modifying factor (BMF) at an equal multiplicity of infection (MOI). BMF sensitizes cells to apoptosis by directly antagonizing anti-apoptotic BCL-2 family proteins (BCL-2 (B-cell lymphoma 2), BCL-XL (B-cell lymphoma extra large), and MCL-1 (myeloid cell leukemia 1)) to allow for BAX and BAK (BCL-2 antagonist/killer) activation (34). Unexpectedly, unlike endophilin B1 deficiency, the loss of endophilin B2 did not enhance cell viability in MEFs transduced with BMF lentivirus (Fig. 2A; BMF-treated: WT to B2KO, p = 0.9578; WT to B1KO, p = 0.0015; B2KO to B1KO, p = 0.0001). Likewise, no statistical difference in cell death was detected between WT and endophilin B2-deficient MEFs exposed to the potent apoptosis inducers actinomycin D (ActD) and staurosporine (STS) (Fig. 2B). To further confirm the results, we generated endophilin B1- and B2-deficient HeLa cells using the CRISPR-Cas9 gene editing system (Fig. 2C). Notably, as observed in MEFs (Fig. 1C), expression of endophilin B1 was not altered by the loss of endophilin B2 in HeLa cells (Fig. 2C). Consistently, loss of endophilin B2 did not affect the induction of mitochondrial apoptosis in response to BMF or ActD, whereas endophilin B1 depletion partially restored cell viability during ActD treatments (Fig. 2, D and E; 156.25-ng/ml dose: WT to B2KO, p = 0.9602; WT to B1KO, p = 0.0035; B2KO to B1KO, p = 0.0078; 625-ng/ml dose: WT to B2KO, p = 0.9585; WT to B1KO, p < 0.0001; B1KO to B2KO, p = 0.0001). Taken together, we conclude that, unlike endophilin B1, endophilin B2 does not regulate intrinsic apoptosis.
Figure 2.
Endophilin B2 is dispensable for mitochondrial apoptosis. A, WT, endophilin B1 knock-out (B1KO), and endophilin B2 knock-out (B2KO) MEFs were transduced with lentiviruses encoding BMF at an MOI of 3 for 48 h and subjected to a sulforhodamine B (SRB) cell viability assay. B, WT and B2KO MEFs were treated with 5 μg/ml ActD or 1 μm STS for 48 h and subjected to an MTS cell viability assay. Data from two independent experiments performed in triplicate are shown in A and B. C, WT, B2KO, and B1KO HeLa cells were subjected to immunoblotting using the indicated antibodies. D, HeLa cells were treated with the indicated doses of ActD for 24 h and subjected to a PrestoBlue cell viability assay. Data represent three independent experiments performed in triplicate. E, HeLa cells were transduced with lentiviruses encoding BMF at an MOI of 3 for 24 h, stained with annexin V, and analyzed by flow cytometry. Data represent three independent experiments performed in duplicate. Statistical significance was determined by unpaired two-way ANOVA with Tukey's multiple comparison test (A, B, and D) and unpaired Student's t test with Holm-Sidak multiple-comparison correction (E). All values are mean ± S.D. ns, not significant; **, p ≤ 0.01; ***, p ≤ 0.001.
Endophilin B2 plays a role in endosome maturation
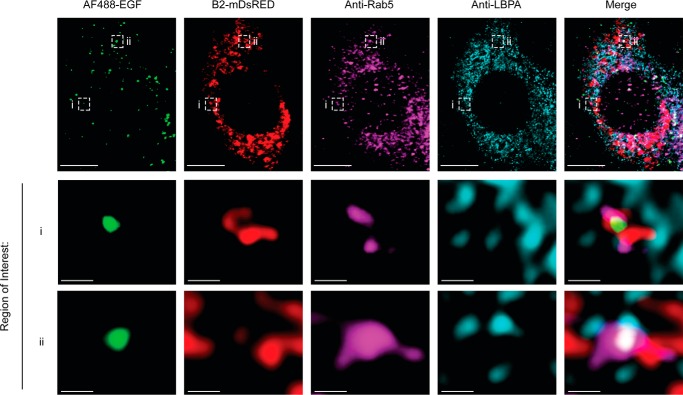
To further explore the cellular function of endophilin B2, we examined the subcellular localization of endophilin B2. Immunofluorescence microscopy revealed that endophilin B2 resides on cytoplasmic structures, where it partially colocalizes with endophilin B1 (supplemental Fig. S4A). Subcellular fractionation analysis further revealed an enrichment of endophilin B2 and endophilin B1 in early (Rab5) and late (Rab7) endosomal fractions as well as in the microtubule-associated protein 1A/1B light chain 3 (LC3)-II-enriched autophagosomal fractions (supplemental Fig. S4B). To confirm endophilin B2 localization to endosomes, HeLa cells expressing C-terminal tagged monomeric DsRed endophilin B2 were treated with fluorescence-tagged EGF to stimulate endosomal trafficking and analyzed by immunofluorescence microscopy. Indeed, we observed that endophilin B2 localizes to EGF-positive, Rab5-positive early endosomes and lysobisphosphatidic acid (LBPA)-positive late endosome structures in response to EGF stimulation (Fig. 3 and supplemental Fig. S6). Because endophilin B1 promotes endosomal acidification and trafficking (24, 25, 35), we hypothesized that endophilin B2 may have a similar role in endocytic processes.
Figure 3.
Endophilin B2 localizes to early/late endosome compartments in response to EGF stimulation. HeLa cells expressing a c-terminally tagged monomeric DsRED endophilin B2 (B2-mDsRED) were serum-starved for 16 h and stimulated with 1 μg/ml Alexa Fluor 488-conjugated EGF (AF488-EGF) for 15 min. Cells were then immunostained with the indicated antibodies and subjected to confocal microscopy. Scale bars, 10 and 0.5 μm in magnified images of boxed areas. Images are representative of two independent experiments.
To determine whether endophilin B2 regulates fluid-phase endocytosis, cells were pulsed with FITC-labeled dextran, and fluid-phase endocytosis was quantified by measuring the relative fluorescence intensity (36). In contrast to 5-(N-ethyl-N-isopropyl)-amiloride (EIPA), a selective inhibitor of sodium-hydrogen exchange that impairs fluid-phase endocytosis (37, 38) (supplemental Fig. S5), we did not observe any difference in the uptake of FITC-dextran between WT and endophilin B1- or B2-deficient HeLa cells (Fig. 4A). Thus, in contrast to endophilin A proteins, the endophilin B family of proteins do not appear to be required for plasma membrane internalization during fluid-phase endocytosis. Next, to assess whether endophilin B2 regulates the endocytic trafficking of internalized vesicles, we utilized dextran conjugated to the pH-sensitive pHrodo-Red dye. As the fluorescence intensity of this dye increases in response to decreasing pH, pHrodo-Red dextran monitors the acidification and maturation of endosomes (5). As expected, a time-dependent increase in the fluorescence intensity of pHrodo-Red dextran was observed upon pulse-chase of WT HeLa cells (Fig. 4B), indicating the progressive acidification of endosomes that reaches a plateau ∼5 min after the addition of the dye. Interestingly, whereas pulse-chase of endophilin B1- and B2-deficient cells with pHrodo-Red dextran resulted in a time-dependent increase in fluorescence intensity, the maximal fluorescence intensity was significantly reduced compared with WT cells (Fig. 4C; WT to B2KO, p = 0.0134; WT to B1KO, p = 0.0459). Similarly, endophilin B2 depletion in MEFs did not affect the uptake of FITC-dextran (Fig. 4D) but significantly reduced the maximal fluorescence intensity of pHrodo-Red dextran (Fig. 4, E and F; p = 0.0322 (F)). Taken together, these data suggest that endophilin B2 functions in a similar manner to endophilin B1 to promote the acidification of endosomes.
Figure 4.
Endophilin B2 deficiency impairs endosomal acidification. HeLa cells (A–C) and MEFs (D–F) were incubated with 50 μg/ml FITC-dextran (A and D) or 50 μg/ml pHrodo-Red dextran (B, C, E, and F) for 5 min and analyzed by spectrofluorometry. A and D, relative fluorescence intensity (RFI) values were normalized to the median of WT cells and are a result of three independent experiments performed in triplicate. B and E, fluorescence intensities were measured at 30-s intervals for the indicated periods of time. Shown are representative curves from one of three independent experiments performed in triplicate. C and F, maximum signal intensities in B and E were determined by curve fit analysis. Independent assays were normalized to WT signal, and then WT signals from the three independent assays were normalized to mean WT signal. Statistical significance was determined by unpaired one-way ANOVA with Tukey's multiple-comparison test (A and C) or unpaired Student's t test with Holm-Sidak multiple comparison correction (D and F). All values are mean ± S.D. (error bars). ns, not significant; *, p < 0.05.
Because endosomal acidification is critical for receptor degradation upon receptor-mediated endocytosis, we sought to determine the effect of endophilin B2 on the lysosomal degradation of EGFR. To this end, cells were serum-starved overnight to accumulate EGFR at the plasma membrane. Upon stimulation with EGF, EGFR is internalized (supplemental Fig. S6) and trafficked to the lysosome for degradation with minimal receptor recycling to the plasma membrane (36, 39). Using fluorescently tagged-EGF to stimulate and track EGFR (40), we observed that the loss of endophilin B2 significantly impairs the delivery of internalized EGFR to lysosomal-associated membrane protein 1 (Lamp1)-positive lysosomal structures (Fig. 5, A and B; p < 0.0001). Importantly, and in accordance with our results for fluid-phase endocytosis, the plasma membrane internalization of EGFR is independent of endophilin B2 (Fig. 5A).
Figure 5.
Endophilin B2 deficiency impairs the lysosomal delivery of internalized EGF. A, HeLa cells were serum-starved for 16 h, incubated with 1 μg/ml Alexa Fluor 488-conjugated EGF (AF488-EGF) for the indicated periods of time, immunostained for Lamp1, and subjected to confocal microscopy. Images are representative of two independent experiments. Magnified images of boxed areas are shown on the right. AF488-EGF and Lamp1 colocalization was determined as described under “Experimental procedures.” Scale bars, 10 μm. B, Mander's overlap coefficient for AF488-EGF with Lamp1 in A (n > 20 from two independent experiments). Statistical significance was determined by two-way ANOVA with Tukey's multiple-comparison test. All values are mean ± S.D. (error bars). ****, p < 0.0001.
Acidification is a critical part of endosomal maturation that allows for the activation of acid hydrolases along the endosomal-lysosomal pathway. Our results reveal that the loss of either endophilin B1 or B2 significantly delayed the degradation of EGFR upon EGF stimulation (Fig. 6, A and B; WT to B2KO, p < 0.0001; WT to B1KO, p = 0.0002). Lysosomes are dynamic organelles that are re-formed after fusion events and require an acidic lumen for the acid hydrolases to properly degrade material (41). To determine whether endophilin B2 is important for the biogenesis and/or acidification of lysosomes, HeLa cells were stained with the cell-permeable lysosomal dye LysoTracker and the pH-responsive lysosomal dye LysoSensor, respectively. Whereas LysoTracker signals were not significantly altered by the loss of endophilin B2, LysoSensor signals were surprisingly slightly enhanced in endophilin B2-deficient cells to indicate that the compartments are more acidic compared with WT (Fig. 6C; p = 0.0001). Collectively, these results suggest that suppressed EGFR degradation observed in endophilin B2 deficient cells is not indirectly caused by lysosome impairment.
Figure 6.

Loss of endophilin B2 suppresses EGFR degradation. A, HeLa cells were serum-starved for 16 h, incubated with 100 ng/ml EGF for the indicated durations, and subjected to immunoblotting using the indicated antibodies. Representative immunoblot from one of three independent experiments is shown. B, EGFR levels in A were quantified and normalized to β-actin. C, HeLa cells were co-stained with 50 nm LysoTracker Deep Red and 1 μm LysoSensor Green DND-189 for 30 min and analyzed by flow cytometry (n = 12; three independent experiments performed in quadruplicate). Statistical significance was determined by two-way ANOVA with Tukey's multiple-comparison test. All values are mean ± S.D. (error bars). ns, not significant; ***, p ≤ 0.01.
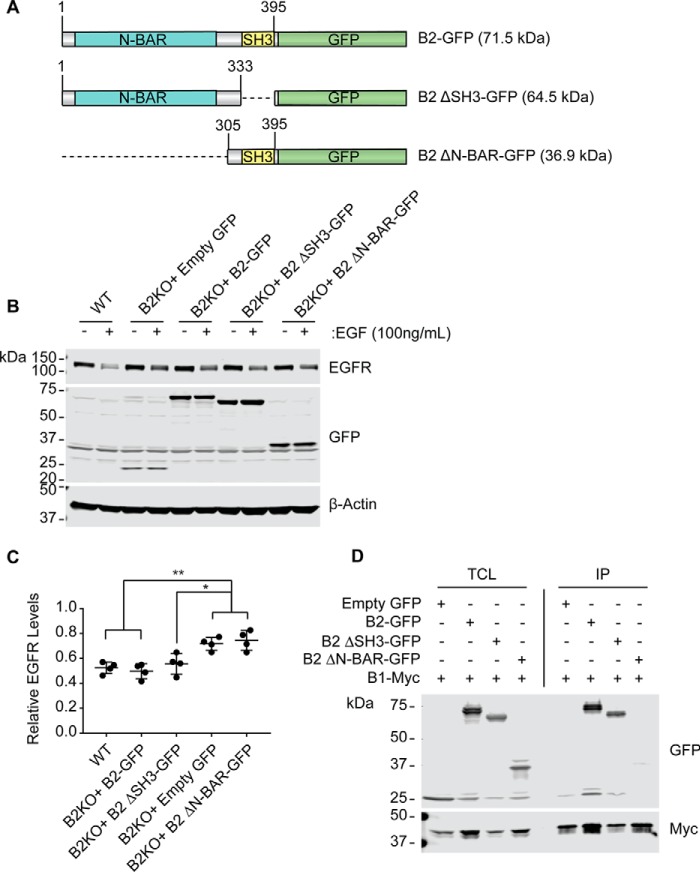
Importantly, re-expression of endophilin B2-GFP but not GFP control was able to rescue EGFR degradation in endophilin B2-deficient cells (Fig. 7, A–C). To identify the domain(s) of endophilin B2 that regulate EGFR degradation, cells were also restored with expression of endophilin B2 mutants lacking the SH3 domain or the N-BAR domain (Fig. 7, A–C). Interestingly, we found that the N-BAR domain of endophilin B2 is required to rescue EGFR degradation in endophilin B2-deficient cells, whereas the SH3 domain appears to be dispensable (Fig. 7, B and C; B2KO + GFP to B2KO + B2-GFP, p = 0.0030; B2KO + GFP to B2KO + B2-ΔSH3-GFP, p = 0.0345; B2KO + GFP to B2KO + B2-ΔN-BAR-GFP, p = 0.9897). Using co-immunoprecipitation, we confirmed that endophilin B2, indeed, heterodimerizes with endophilin B1 (15) and that heterodimization requires the N-BAR domain (Fig. 7D). These data suggest that the membrane curvature-inducing activity of endophilin B2 and/or heterodimerization of endophilin B2 and endophilin B1 are important for the endocytic degradation of EGFR.
Figure 7.
The N-BAR domain of endophilin B2 is essential for EGFR trafficking. A, schematic of endophilin B2 domain mutants. B, HeLa cells were transduced with the indicated lentiviruses at an MOI of 1. 72 h post-transduction, cells were serum-starved for 16 h, incubated with 100 ng/ml EGF for 30 min, and subjected to immunoblotting using the indicated antibodies. Representative immunoblot from one of four independent experiments is shown. C, EGFR levels in B were quantified and normalized to β-actin and presented as values relative to untreated controls. Statistical significance was determined by two-way ANOVA with Tukey's multiple comparison test. All values are mean ± S.D. (error bars). *, p ≤ 0.05; **, p ≤ 0.01. D, HEK293T/17 cells were co-transfected with plasmids containing endophilin B1-Myc and the indicated endophilin B2 constructs. After 16 h of transfection, cells were subjected to immunoprecipitation using anti-Myc-coated beads and subjected to immunoblotting with the indicated antibodies. Immunoblotting data represent one of two independent experiments. TCL, total cell lysates; IP, immunoprecipitation.
Taken together, these results indicate that endophilin B2 facilitates endosome maturation in both fluid-phase and receptor-mediated endocytic pathways. Further investigation is under way to dissect the precise role of endophilin B family proteins in endocytic trafficking.
Endophilin B2 deficiency decreases autophagic flux upon nutrient starvation
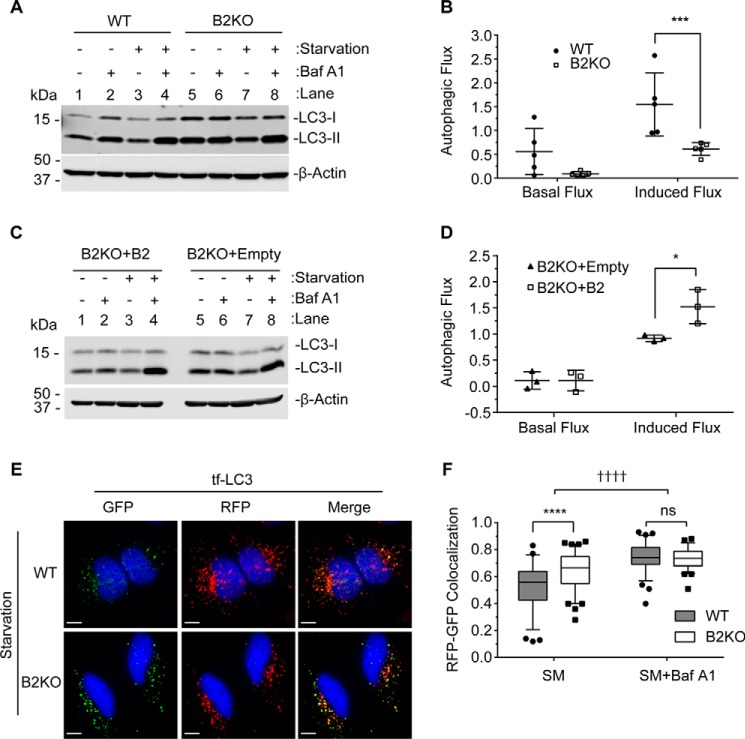
During autophagy, autophagosomes mature into degradative autophagic vacuoles by fusing with late endosomes and/or lysosomes (reviewed in Refs. 7 and 8). To determine whether the impairment in endosome maturation observed in endophilin B2-deficient cells affects autophagy, we performed an autophagic flux assay. During autophagic stimulation, such as nutrient starvation, cytosolic LC3-I is lipidated to form LC3-II bound to the autophagosome membrane (11). Autophagosome maturation occurs upon fusion with lysosomes and degradation of luminal contents; thus, measurement of the lysosomal turnover of LC3-II has been used as a reliable method to monitor autophagic flux (42, 43). Because increases in LC3-II can indicate either an increase in autophagosome biogenesis or a block in lysosomal degradation, autophagic flux assays were performed in the presence or absence of the lysosomal inhibitor, bafilomycin A1 (Baf A1). We found that the starvation-induced lysosomal turnover of LC3-II was significantly suppressed in endophilin B2-deficient HeLa cells (Fig. 8A, lane 8 minus lane 7) compared with WT (lane 4 minus lane 3). Quantification analysis further revealed that, indeed, starvation-induced autophagic flux is significantly suppressed by loss of endophilin B2 (Fig. 8B; p = 0.0007). Although endophilin B2-deficient cells also appear to have a decrease in basal autophagic flux under normal culture conditions (Fig. 8A; compare lane 6 minus lane 5 versus lane 2 minus lane 1), statistical significance was not obtained (Fig. 8B). Similar experiments were performed in MEFs. Consistently, we observed that lack of endophilin B2 expression significantly suppressed starvation-induced autophagic flux (Fig. 8, C and D; p = 0.0317).
Figure 8.
Loss of endophilin B2 suppresses autophagic flux. HeLa cells (A and B) and MEFs (C and D) were incubated in starvation medium (SM) or complete medium (CM) in the presence or absence of 100 nm Baf A1 for 2 h. A and C, total cell lysates were subjected to immunoblotting using the indicated antibodies. Representative immunoblots from five (A) and three (B) independent experiments are shown. B and D, autophagic flux under non-starved (basal flux) and starved (induced flux) conditions in A and C were calculated as described under “Experimental procedures.” All values are mean ± S.D. (error bars). E, HeLa cells were transduced with lentiviruses encoding mRFP-GFP-LC3 (tf-LC3) for 72 h, starved for 2 h, and analyzed by fluorescence deconvolution microscopy. Nuclei were stained with DAPI. Scale bars, 10 μm. F, Pearson's correlation coefficient for mRFP and GFP in E (n > 70; duplicates from two independent experiments). The lines, boxes, and error bars represent median values, 25th to 75th percentiles, and 5th to 95th percentiles, respectively. Statistical significance was determined by two-way ANOVA with Tukey's multiple-comparison test. *, p ≤ 0.05; ***, p ≤ 0.001; **** and ††††, p < 0.0001; ns, not significant.
To further demonstrate the importance of endophilin B2 in autophagic flux, we performed the tandem-fluorescent LC3 (tf-LC3; mRFP-GFP-LC3) assay. The tf-LC3 assay discriminates between autophagosomes from autolysosomes (autophagosomes fused with lysosomes) due to the differences in pKa values between RFP and GFP. Upon autophagosome and lysosome fusion, the GFP moiety of tf-LC3 becomes quenched by the acidic environment to result in RFP+GFP− puncta (44). We found that nutrient starvation of HeLa cells increased RFP+GFP+ puncta (autophagosomes) regardless of endophilin B2 expression, indicating that endophilin B2 is not required for autophagosome formation (Fig. 8E). In contrast, the generation of RFP+GFP− puncta (autolysosomes) was reduced in nutrient-starved endophilin B2-deficient cells compared with WT (Fig. 8E), suggesting impaired delivery of autophagosomes to acidified late endosomes and/or lysosomes. Indeed, RFP+GFP+ signals significantly accumulated in endophilin B2-deficient cells during nutrient starvation compared with WT (Fig. 8F; p < 0.0001). Because Baf A1 slightly but significantly enhanced the abundance of RFP+GFP+ puncta in nutrient-starved endophilin B2-deficient cells (Fig. 8F; ††††, p < 0.0001), a small degree of autophagic flux is maintained in the absence of endophilin B2. Nonetheless, these results clearly demonstrate an important role of endophilin B2 in autophagosome maturation.
Loss of endophilin B2 delays nuclear translocation and replication of influenza A viruses
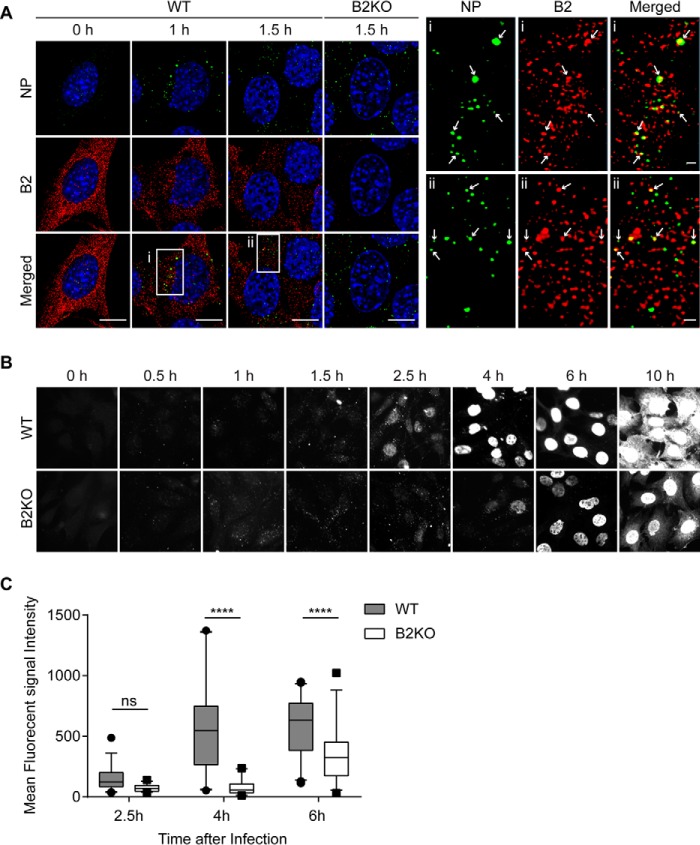
To address the biological significance of the above results, we monitored influenza A virus infection in WT and endophilin B2-deficient cells. Influenza A virus is a member of the Orthomyxoviridiae family of enveloped viruses. Influenza A virus is internalized through receptor-mediated endocytosis and trafficked to late endosomes. The acidic environment (pH ∼5.0) of late endosomes triggers conformational changes in the viral envelope hemagglutinin that result in the fusion of the endosomal and viral membranes, release and transport of viral ribonucleoprotein particle to the nucleus, and replication of the viral RNA genome (vRNA) in the nucleus (45–50). Notably, influenza A is a pathogen that requires both endosomal acidification and deployment of autophagy to efficiently replicate in host cells (51). Because endophilin B2-deficient cells were observed to have a defect in endosome acidification, we hypothesized that endophilin B2 may regulate viral trafficking. To this end, we monitored the subcellular localization of endophilin B2 in MEFs during influenza A virus infection. Indeed, endophilin B2 is observed to colocalize with the influenza A vRNA-binding nucleoprotein (NP) in the cytoplasm at ∼1–1.5 h post-infection (Fig. 9A), suggesting a role for endophilin B2 in viral trafficking. We next determined the effect of endophilin B2 depletion on vRNA trafficking and replication. In WT cells, we observed a time-dependent increase of NP in the nucleus and then in the cytoplasm, indicating replication of the viral genome and generation of new virions (Fig. 9B). Similar to fluid-phase and receptor-mediated endocytosis, endophilin B2 was dispensable for the internalization of influenza A virus into host cells (Fig. 9B). However, both the nuclear and cytoplasmic accumulation of NP were significantly delayed by loss of endophilin B2 (Fig. 9C; p < 0.0001), suggesting that endophilin B2 is important for the efficient trafficking of vRNAs to the nucleus. Taken together, these data suggest that endophilin B2 deficiency effectively impairs the pH-dependent escape of influenza A virus from endosomes to suppress vRNA translocation into the nucleus for replication, a result consistent with impaired endosomal acidification upon the loss of endophilin B2.
Figure 9.
Endophilin B2 deficiency attenuates nuclear trafficking of influenza A viral nucleoprotein. A–C, WT or endophilin B2 KO MEFs were infected with influenza A virus H1N1 PR8 strain at an MOI of 5, incubated for the indicated durations and co-stained for viral nucleoprotein (NP) and endophilin B2 (A) or stained for NP alone (B and C). A, fluorescence images were analyzed by confocal microscopy. Magnified images of boxed areas are shown on the right. The arrows indicate colocalization of endophilin B2 with NP. Scale bars, represent 10 μm. B, cells were analyzed by fluorescence microscopy. C, mean nuclear NP fluorescence signal intensity at the indicated time points in B was quantified using Slidebook version 5.0 software (n > 30 from two independent experiments). The lines, boxes, and error bars represent median values, 25th to 75th percentiles, and 5th to 95th percentiles, respectively. Statistical significance was determined by two-way ANOVA with Tukey's multiple-comparison test. ns, not significant; ****, p < 0.0001.
Discussion
In this study, we demonstrate a novel role for endophilin B2 in endocytic membrane trafficking upon fluid-phase endocytosis, receptor-mediated endocytosis, starvation-induced autophagy, and influenza A viral infection. Notably, the loss of endophilin B2 appears to impair endosomal maturation, as supported by suppressed endosomal acidification during fluid-phase endocytosis, decreased lysosomal degradation of internalized EGFR, impaired starvation-induced autophagic flux, and delayed nuclear translocation of influenza A vRNA. Furthermore, our studies reveal that loss of endophilin B2 fails to efficiently deliver EGFR to Lamp1-positive compartments upon receptor-mediated endocytosis or newly generated autophagosomes to acidified late endosomes and/or lysosomes. Because the loss of endophilin B2 does not impair internalization from the plasma membrane or lysosomal function, endophilin B2 appears to regulate endocytic membrane trafficking along the endosomal-lysosomal pathway.
In contrast, endophilin B2 has recently been reported to be indispensable for the internalization of ligand-stimulated glutamate receptors in cultured neuronal cells (52). We propose that the discrepancy in results may be due to the differential expression of endophilin B2 isoforms in neuronal and non-neuronal cells. Indeed, we have identified and characterized the murine endophilin B2 isoforms and reveal a tissue-specific increase of isoforms 2 and 4 in neuronal tissues compared with non-neuronal tissues that predominantly express isoform 1. Interestingly, whereas endophilin B1 promotes apoptosis in non-neuronal cells, brain-specific isoforms of endophilin B1 have been reported to have anti-apoptotic functions during ischemic injury (30).
Although we observed that endophilin B1 and endophilin B2 can assemble heterodimers and have a similar subcellular localization in endothelial and fibroblast cells, our studies provide insight into unique functions of each family member. Most notably, we revealed that endophilin B2 is dispensable for mitochondrial apoptosis. Although this result is not entirely surprising, given that endophilin B2 is not a direct interactor of BAX (15), our results suggest that endophilin B1 homodimers may be critical for its pro-apoptotic function. In contrast, the novel roles of endophilin B2 in endocytic trafficking, endosomal maturation, and autophagy are similar to previously defined functions of endophilin B1 in EGFR trafficking and degradation (24) and autophagic flux (19, 21), suggesting that heterodimers of endophilin B proteins may regulate endocytic membrane dynamics. In support of this hypothesis, we observed the co-fractionation of endophilin B2 with endophilin B1 in endosome- and autophagosome-enriched fractions and demonstrated that the N-BAR domain, but not the SH3 domain, is necessary for the heterodimerization of endophilin B1 and B2 as well as EGFR degradation. Moreover, loss of endophilin B2 leads to impairment of starvation-induced autophagic flux. Consistently, a similar effect on autophagic flux is observed in endophilin B1-deficient models (19, 21), and a recent report has shown that endophilin B2 promotes the selective degradation of mitochondria by autophagy (mitophagy) through dimerization with endophilin B1 (33). Moreover, this finding is in agreement with the importance of endosome maturation for autophagic flux (7). Further studies are warranted to elucidate the interaction between endophilin B family homodimers and heterodimers in the regulation of intracellular membrane dynamics.
The precise molecular mechanism behind the regulation of endosome maturation by endophilin B2 is yet to be determined. As discussed above, endophilin B2 heterodimerization with endophilin B1 may promote endosome maturation through the activation of phosphatidylinositol 3-kinase Vps34 and/or the recruitment of the UVRAG (UV radiation resistance-associated gene)-Hops complex (homotypic fusion and protein sorting complex) to early endosomes (19, 53). In fact, our preliminary data show that UVRAG co-immunoprecipitates with endophilin B1 and endophilin B2 (data not shown). In addition, our data indicated that the N-BAR domain of endophilin B2 is necessary for endocytic degradative trafficking, whereas the SH3 domain is dispensable. This suggests that the membrane curvature functions and dimerization of endophilin B2 are necessary for endocytic trafficking. Alternatively, or in addition, we cannot eliminate the possibility that endophilin B2 may regulate endosomal maturation through its effects on the cytoskeleton. Recently, endophilin B2 has been identified as a regulator of the vimentin cytoskeletal network via its SH3-mediated interaction with the cytoskeletal linker protein plectin 1 (54). Because the vimentin network controls late endosome and lysosome positioning as well as the acidification of endosomes and lysosomes (55), we propose that this interaction may be critical for the phenotype reported here. Moreover, vimentin has also been identified as a vital regulator for the trafficking and endosomal escape of influenza A viral ribonucleoprotein particle (56). Because the phenotype of vimentin-null cells infected with influenza A virus is similar to that observed in endophilin B2-deficient cells, we hypothesize that endophilin B2 may cooperate with vimentin to regulate endosome acidification and/or the positioning of endosomes during maturation.
Because endophilin B2-deficient mice are born at the expected Mendelian ratios and are indistinguishable from wild-type littermates, evidence of endophilin B2 in the pathogenesis of disease is currently lacking. However, high expression of endophilin B2 has been reported in aggressive human prostate tumors as well as primary tumors from a transgenic mouse model of prostate cancer to suggest that endophilin B2 could serve as a potential biomarker (57–59). In direct opposition, decreased endophilin B1 expression is associated with cancer development and progression (60–63). The opposite changes in expression observed for the endophilin B proteins in cancer have generated much speculation regarding the function of endophilin B2. On one hand, endophilin B2 has been proposed to have pro-apoptotic functions like endophilin B1 but to persist in an up-regulated state for unknown reasons. Alternatively, endophilin B2 has been suggested to be anti-apoptotic, such that its expression is positively selected for during tumor development. Because loss of endophilin B2 did not further sensitize cells to mitochondrial apoptosis in our studies, endophilin B2 may not be a direct suppressor of apoptosis (1). Interestingly, the identification of endophilin B2 as a novel promoter of endocytic trafficking may provide insight into its role in tumorigenesis, because dysregulated growth factor signaling and autophagy are hallmarks of cancer (64). Additionally, our findings implicate endophilin B2 in influenza A viral infection to provide a novel target for future investigation of viruses requiring an acidic pH for infectivity. Collectively, our studies provide a foundation to understand the physiological and pathological roles of endophilin B2 in human health and disease.
Experimental procedures
Antibodies and plasmids
The following antibodies were used for immunoblotting (IB) and immunofluorescence (IF): mouse anti-β-actin monoclonal (Sigma, catalog no. A5441; 1:20,000 for IB); rabbit anti-calnexin polyclonal (Thermo Fisher Scientific, catalog no. MA3027; 1:2,000 for IB); rabbit anti-endophilin B2 polyclonal (Proteintech Group, catalog no. 15897-1-AP; 1:5,000 for IB, 1:1,000 for IF); rabbit anti-TGN46 polyclonal (Novus, catalog no. NBP1-49643; 1:400 for IB); mouse anti-endophilin B1 monoclonal (Novus, catalog no. NBP2-24733; 1:500 for IB, 1:800 for IF); goat anti-endophilin B1 polyclonal (GeneTex, catalog no. GTX89961; 1:500 for IB); rabbit anti-LC3 polyconal (Novus, catalog no. NB100-2220; 1:5,000 for IB); rabbit anti-LC3 monoclonal (Cell Signaling Technology, catalog no. 3868S; 1:2,000 for IB); rabbit anti-EGF receptor monoclonal (Cell Signaling Technology, catalog no. 4267S; 1:1,000 for IB); rabbit anti-Rab5 monoclonal (Cell Signaling Technology, catalog no. 3547; 1:700 for IB); rabbit anti-Rab7 monoclonal (Cell Signaling Technology, catalog no. 9367; 1:700 for IB); mouse anti-LBPA monoclonal (Echelon Biosciences, catalog no. Z-PLBPA; 1:5,000 for IF); mouse anti-human c-Myc monoclonal (BD Biosciences, catalog no. 551101; 1:1,000 for IB); rabbit anti-GFP polyclonal (Abcam, catalog no. ab6556; 1:2,000 for IB); mouse anti-human-CD107A (Lamp1) monoclonal (BD Biosciences, catalog no. 555798; 1:200 for IF); guinea pig anti-P62 polyclonal (American Research Products, catalog no. 03-GP62-C; 1:5,000 for IB); rabbit anti-Tom20 polyclonal (Santa Cruz Biotechnology, catalog no. SC-11415; 1:500 for IB); mouse anti-influenza A-nucleoprotein monoclonal (EMD Millipore, catalog no. MAB825; 1:1,000 for IF).
The pCDH1–3xFLAG-IRES-GFP plasmid was generated by subcloning PCR-amplified 3xFLAG cDNA (XbaI-EcoRI site) from p3xFLAG-CMV-10 (Sigma, catalog no. E7658) and the IRES-GFP sequence (EcoRI-SalI site) from pBMN-I-GFP (Addgene, catalog no. 1736) into pCDH1-MCS1-EF1-Puro vector (System Biosciences, catalog no. CD510A-1). The cDNA encoding human BMF (Addgene, catalog no. 24264) was amplified by PCR and subcloned into the EcoRI-XhoI site of pCDH1–3xFLAG-BMF-IRES-GFP. The human endophilin B2 was obtained from Invitrogen (catalog no. FL1002), amplified by PCR, and subcloned into the XbaI-EcoRI site of pCDH1-MCS1-EF1-Puro vector. The endophilin B2 insert was subsequently mutated to make it resistant to recognition by CRISPR-Cas9 using a site-directed mutagenesis kit (Agilent Technologies, catalog no. 200521) with the following primer: 5′-caacatgaagaagctggcatcagatgcaggcatcttcttcacccgg-3′. CRISPR-resistant endophilin B2 domain deletion mutants were PCR-amplified using the primer sets outlined in supplemental Table S1 and subcloned using the XbaI and BamHI sites into the pCDH1-AcGFP-N1-Puro vector. pCDH1-AcGFP-N1-Puro vector was created using the Ac-GFP N1 cloning site from pAcGFP-N1 Clontech (catalog no. 632469) by subcloning using NheI-NotI into the pCDH1-MCS1-EF1 plasmid. The pCDH1-endophilin B2-mDsRED vector was created by subcloning the DsRED-monomer-N1 from Clontech (catalog no. 632465) into the pCDH1 plasmid using NheI and NotI restriction sites and subsequently cloning the endophilin B2 insert using the XbaI and BamHI sites. The primer sequences used to amplify 3xFLAG, human BMF, and human endophilin B2 cDNAs were as follows: 3xFLAG, 5′-GCTCTAGAGCCACCATGGACTACAAAGACCATGAC-3′ and 5′-GGAATTCCTTGTCATCGTCATCCTTG-3′; human BMF, 5′-GGGAATTCGAGCCATCTCAGTGTGT-3′ and 5′-GGTGTCGACTCACCTAGGGCCTGCC-3′; human endophilin B2, 5′-CCGCCAGTTCTAGAGCCACCATGGACTTCAA-3′ and 5′-ATGCATGGAATTCCTAGCTGAGCAGTTC-3′. The guide sequence targeting exon 1 of human endophilin B2 (5′-AAGAAGCTGGCGTCGGACGC-3′) and endophilin B1 (5′-AAGAAGCTGGCGGCCGACGC-3′ was designed and subcloned into lentiCRISPRv2 plasmid (Addgene, catalog no. 52961) according to Dr. Feng Zhang's laboratory protocol (65). The mRFP-GFP-LC3 (tf-LC3) cDNA was obtained from Addgene (catalog no. 21074) and subcloned into the NheI-EcoRI site of pCDH1-MCS1-EF1-Puro vector.
Generation of endophilin B2 knock-out mice
Endophilin B2-deficient mice with a 129/SvEv and C57BL/6 mixed genetic background were generated by the Texas A&M Institute for Genomic Medicine (Houston, TX) by the gene trap strategy using the OMNIBANK ES cell clone OST224737. This gene trap cassette includes a splice adaptor at the 5′ end with neomycin selection and poly(A) termination signals followed by another ES cell-only promotor that expresses the first exon of the Bruton's tyrosine kinase gene with multiple termination codons to prevent expression of downstream fusion transcripts. The resultant mice were back-crossed with C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) more than 10 times to obtain endophilin B2KO mice with the genetic background used in this study. Genotyping was performed using standard PCR methods with the following primers: genomic forward primer (B2-F), 5′-GTTGGTGCTAATGGTTGCATCC-3′; genomic reverse primer (B2-R), 5′-AGATCCTAGCCTTCTGACATCC-3′; and LTR-reverse (LTR-R), 5′-ATAAACCCTCTTGCAGTTGCATC-3′. All mice were maintained in accordance with federal guidelines, and studies were approved by the Pennsylvania State University Animal Care and Use Committee.
Semi-quantitative RT-PCR
Total RNA was isolated from tissues using TRIzol reagent (Thermo Fisher Scientific, catalog no. 15596026) according to the manufacturer's protocol. Quality of RNA was tested using the BioAnalyzer RNA 6000 nanochip (Agilent Technologies). cDNA was created from RNA using the High Capacity cDNA Reverse Transcriptase Kit (Thermo Fisher Scientific, catalog no. 4368814) according to the manufacturer's protocol. PCR amplification of endophilin B2 isoforms was achieved by using 2.5 ng of each cDNA using the primer pairs found in supplemental Table S1. Semi-quantitative PCR was performed using the GoTaq Colorless Master Mix (Promega, catalog no. M713) according to the manufacturer's protocol, using the scheme and protocol outlined in supplemental Fig. S2A and Table S2, respectively. Isoforms of endophilin B2 were quantified by the BioAnalyzer High Sensitivity chip (Agilent Technologies), and relative mRNA values of each isoform were calculated based on the ratios and differences of the PCR1 and -2 products.
Cell culture, viral transduction, and generation of stable cell lines
HeLa (CCL-2) and 293T/17 (HEK293T/17) cells were obtained from ATCC. SV40 large T antigen-immortalized MEFs were generated as described previously (21). All cells were maintained in DMEM supplemented with 10% FBS and 1% antibiotic-antimycotic solution (Corning, catalog no. 30-004-CI). Recombinant lentiviruses were produced and transduced to targeted cells as described previously (66). Lentiviral MOI was calculated from infectious viral units/ml and determined using the Lenti-X quantitative RT-PCR kit (Clontech, catalog no. 631235). To generate endophilin B2 knock-out HeLa cells, cells transduced with lentiviruses encoding Cas9 and endophilin B2 single-guide RNA were selected with 1.5 μg/ml puromycin for 5 days followed by single-clone isolation by serial dilution. The resultant single clones (three individual clones) were pooled together and used for the experiments.
Immunoblotting
Tissue homogenates and total cell lysates were prepared in radioimmunoprecipitation assay buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mm EDTA, pH 8.0) containing protease and phosphatase inhibitors and subjected to immunoblotting as described previously (21, 22). The signals were measured and quantified using a LI-COR Odyssey CLx image scanner with the Image Studio version 5 software (LI-COR Biotechnology).
Fluorescence microscopy
Cells were fixed in 4% paraformaldehyde-PBS for 10 min, permeabilized with 100 μg/ml digitonin for 10 min, and stained for the indicated antibodies unless otherwise noted. Fluorescence images were obtained using an Olympus IX81 deconvolution microscope (×60 PLAN S-APO oil immersion objective, numeric aperture = 1.35; ×40 PLAN FLUORITE oil immersion objective, numeric aperture = 1.30) equipped with a Hamamatsu ORCA-R2 Digital CCD camera (catalog no. C10600-10B) or a Leica AOBS SP8 laser-scanning confocal microscope (×40 PLAN-APOCHROMAT, numeric aperture = 1.3 oil immersion objective), deconvolved using SlideBook software (Intelligent Imaging Innovations) or Huygens deconvolution software (Scientific Volume Imaging), and analyzed using SlideBook software and/or Imaris software (Bitplane).
Cell viability and apoptosis assays
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Promega, catalog no. G3582), sulforhodamine B (SRB) (Sigma, catalog no. S1402), and PrestoBlue (Thermo Fisher Scientific, catalog no. A-13261) assays according to the manufacturer's protocols. Spectrophotometry and fluorometry was performed using a CLARIOstar plate reader with MARS data management software (BMG Labtech). To determine apoptosis by flow cytometry, cells were stained with Annexin V-PE (BD Biosciences, catalog no. 559763) and analyzed by flow cytometry using a FACSCanto II flow cytometer (BD Biosciences) and FlowJo version 10 data analysis software (Tree Star).
Endocytosis assays
To measure fluid-phase endocytosis and endosome acidification, cells were incubated in Live Cell Imaging Solution (Thermo Fisher Scientific, catalog no. A14291DJ) containing 50 μg/ml FITC-dextran (10,000 Da; Sigma, catalog no. FD10S-100MG) or pHrodo Red-dextran (10,000 Da; Thermo-Fisher Scientific, catalog no. P10361), respectively, at 37 °C for 5 min, rinsed twice with Live Cell Imaging Solution, and analyzed by spectrofluorometry using a CLARIOstar plate reader with MARS data management software (BMG Labtech). Fluorescence signals were normalized according to cell viability determined by PrestoBlue staining as described above. For Alexa Fluor 488-dextran (Thermo-Fisher Scientific, catalog no. D22910) treated with EIPA (Enzo Life Sciences, catalog no. ALX-550-266-M005), cells were either preincubated with 80 mm EIPA for 30 min or not and then incubated with 50 μg/ml Alexa Fluor 488-dextran for 30 min with or without EIPA before fixation. Cell images were captured using the Olympus IX-81 microscope and processed using Slidebook version 6.0 software. EGF receptor-mediated endocytosis and endocytic trafficking were determined as described previously (24). Briefly, cells were serum-starved for 16 h; incubated in DMEM containing 20 mm Hepes (pH 7.5), 0.1% BSA (EMD Millipore, catalog no. 126575), and 100 ng/ml EGF (Thermo Fisher Scientific, catalog no. PHG0311) at 37 °C for the indicated periods of time; and subjected to immunoblotting to measure EGFR degradation. To monitor receptor-mediated endocytosis, serum-starved cells were rinsed once with ice-cold PBS and then incubated in uptake medium (DMEM, 2% BSA, 20 mm Hepes, pH 7.5) containing 1 μg/ml Alexa Fluor 488-EGF (Thermo Fisher Scientific, catalog no. E13345) on ice for 1 h. After washing three times with ice-cold PBS to remove unbound ligands, cells were incubated at 37 °C for the indicated periods of time, fixed in 4% PFA-PBS for 10 min, and subjected to immunofluorescence microscopy. Colocalization reported as Manders' overlap coefficient was determined using the colocalization module in the Imaris version 8.1 software by manual thresholding.
Immunoprecipitation assay
HEK293T/17 cells were co-transfected by calcium phosphate co-precipitation with either empty GFP or one of the endophilin B2 mutant plasmids together with an endophilin B1-Myc tag vector. Upon overnight incubation post-transfection, cells were harvested and lysed in 1% Triton X-100 with 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 20 mm Tris-HCl (pH 7.5) containing protease and phosphatase inhibitors. Cell lysates (400 μg of total protein) were immunoprecipitated using Myc-Trap_A beads (ChromoTek, catalog no. yta-10) and processed according to the manufacturer's protocol.
Autophagic flux assays
To determine lysosomal turnover of LC3-II, cells were incubated in serum- and amino acid-free Dulbecco's modified Eagle's medium (starvation medium; SM) (custom-ordered from Invitrogen) or control complete medium (CM) in the presence or absence of 100 nm Baf A1 (Enzo Life Sciences, catalog no. BML-CM110-0100) for 2 h and subjected to immunoblotting as described above. The levels of LC3-II were normalized to β-actin, and autophagic flux of LC3-II was calculated as follows: basal flux = (CM with Baf A1) − CM; induced flux = (SM with Baf A1) − SM, as defined previously (42).
For the mRFP-GFP-LC3 (tf-LC3) assay, cells were transduced with lentiviruses encoding tf-LC3 for 3 days, starved for 2 h, fixed in 4% PFA for 10 min, and subjected to fluorescence microscopy. Pearson's correlation coefficient was determined using SlideBook software.
Virus entry assay
The mouse-adapted influenza A virus H1N1 strain A/PR 8/34 (PR8) was produced in 10-day-old embryonated chicken eggs, purified by sucrose density gradient centrifugation, and quantitated by a fluorescence focus assay as described previously (67). To monitor viral entry, trafficking, and replication, MEFs seeded on coverslips at 150,000 cells/well of a 24-well plate were rinsed once with PBS and incubated in serum-free DMEM containing PR8 viruses for 1.5 h at 4 °C. After rinsing twice with PBS, cells were further incubated in DMEM supplemented with 5% FBS at 37 °C for the indicated periods of time, rinsed twice with PBS, fixed in 4% paraformaldehyde-PBS containing 0.1% Triton X-100 for 15 min, blocked in 1% normal goat serum for 60 min, stained for endophilin B2 and/or NP, and subjected to fluorescence deconvolution or confocal microscopy. Nuclei were stained with DAPI to generate nuclear masks, and NP fluorescence signals in the nucleus were quantified using SlideBook software.
Statistical analyses
Multiple independent experiments were tested for statistical significance using GraphPad Prism version 7.0. Statistical tests performed include Student's t test or ANOVA as outlined in the figure legends with appropriate post-hoc tests when necessary. The threshold for statistical significance for each test was set at 95% confidence (p < 0.05).
Author contributions
J. M. S., Y. T., L. L., and Z. Z. performed the experiments. Y. I. K., Y. L., Z. T., N. T., L. Y., J. M. A., and M. M. Y. assisted with the experiments and manuscript preparation. J. M. S., Y. T., Z. C. C., and H.-G. W. designed the experiments, analyzed data, prepared the figures, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Neelam Desai and Longgui Chen for technical support. We thank Nate Sheaffer, Joseph Bednarzyk, and Jade Vogel (Pennsylvania State College of Medicine Flow Cytometry Core Facility) for assistance with flow cytometry analysis. We also thank Dr. Thomas Abraham and Wade Edris of (Pennsylvania State College of Medicine imaging core) for assistance with the confocal microscope.
This work was supported by National Institutes of Health Grants CA82197 and CA129682 (to H. G. W.) and HL128746 (to Z. C.) and by the Lois High Berstler Endowment Fund and the Four Diamonds Fund of the Pennsylvania State University College of Medicine. Core facility services and instruments used in this project were funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1 and S2 and Figs. S1–S6.
- N-BAR
- N-terminal Bin/amphiphysin/Rvs
- SH3
- Src homology 3
- EGFR
- epidermal growth factor receptor
- SM
- starvation medium
- CM
- complete medium
- MEF
- mouse embryo fibroblast
- BMF
- BCL-2-modifying factor
- MOI
- multiplicity of infection
- ActD
- actinomycin D
- STS
- staurosporine
- LBPA
- lysobisphosphatidic acid
- EIPA
- 5-(N-ethyl-N-isopropyl)-amiloride
- Baf A1
- bafilomycin A1
- tf-LC3
- tandem-fluorescent LC3
- vRNA
- viral RNA genome
- NP
- nucleoprotein
- IB
- immunoblotting
- IF
- immunofluorescence
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- ANOVA
- analysis of variance.
References
- 1. Kjaerulff O., Brodin L., and Jung A. (2011) The structure and function of endophilin proteins. Cell Biochem. Biophys. 60, 137–154 [DOI] [PubMed] [Google Scholar]
- 2. Hullin-Matsuda F., Taguchi T., Greimel P., and Kobayashi T. (2014) Lipid compartmentalization in the endosome system. Semin. Cell Dev. Biol. 31, 48–56 [DOI] [PubMed] [Google Scholar]
- 3. Lindmo K., and Stenmark H. (2006) Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 4. Scott C. C., Vacca F., and Gruenberg J. (2014) Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 31, 2–10 [DOI] [PubMed] [Google Scholar]
- 5. Huotari J., and Helenius A. (2011) Endosome maturation. EMBO J. 30, 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tooze S. A., Abada A., and Elazar Z. (2014) Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 6, a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hyttinen J. M. T., Niittykoski M., Salminen A., and Kaarniranta K. (2013) Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim. Biophys. Acta 1833, 503–510 [DOI] [PubMed] [Google Scholar]
- 8. Lamb C. A., Dooley H. C., and Tooze S. A. (2013) Endocytosis and autophagy: shared machinery for degradation. BioEssays 35, 34–45 [DOI] [PubMed] [Google Scholar]
- 9. Goh L. K., and Sorkin A. (2013) Endocytosis of receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 5, a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lamb C. A., Yoshimori T., and Tooze S. A. (2013) The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 [DOI] [PubMed] [Google Scholar]
- 11. Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 12. Ao X., Zou L., and Wu Y. (2014) Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 21, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berg T. O., Fengsrud M., Strømhaug P. E., Berg T., and Seglen P. O. (1998) Isolation and characterization of rat liver amphisomes: evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273, 21883–21892 [DOI] [PubMed] [Google Scholar]
- 14. Mim C., Cui H., Gawronski-Salerno J. A., Frost A., Lyman E., Voth G. A., and Unger V. M. (2012) Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pierrat B., Simonen M., Cueto M., Mestan J., Ferrigno P., and Heim J. (2001) SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics 71, 222–234 [DOI] [PubMed] [Google Scholar]
- 16. Cuddeback S. M., Yamaguchi H., Komatsu K., Miyashita T., Yamada M., Wu C., Singh S., and Wang H.-G. (2001) Molecular cloning and characterization of Bif-1. J. Biol. Chem. 276, 20559–20565 [DOI] [PubMed] [Google Scholar]
- 17. Karbowski M., Jeong S.-Y., and Youle R. J. (2004) Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 166, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi Y., Karbowski M., Yamaguchi H., Kazi A., Wu J., Sebti S. M., Youle R. J., and Wang H.-G. (2005) Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol. Cell Biol. 25, 9369–9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mulé J. J., Pledger W. J., and Wang H.-G. (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9, 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi Y., Meyerkord C. L., Hori T., Runkle K., Fox T. E., Kester M., Loughran T. P., and Wang H.-G. (2011) Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy 7, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takahashi Y., Hori T., Cooper T. K., Liao J., Desai N., Serfass J. M., Young M. M., Park S., Izu Y., and Wang H.-G. (2013) Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood 121, 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi Y., Tsotakos N., Liu Y., Young M. M., Serfass J., Tang Z., Abraham T., and Wang H.-G. (2016) The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget 7, 20855–20868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong A. S., Lee R. H., Cheung A. Y., Yeung P. K., Chung S. K., Cheung Z. H., and Ip N. Y. (2011) Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat. Cell Biol. 13, 568–579 [DOI] [PubMed] [Google Scholar]
- 24. Runkle K. B., Meyerkord C. L., Desai N. V., Takahashi Y., and Wang H.-G. (2012) Bif-1 suppresses breast cancer cell migration by promoting EGFR endocytic degradation. Cancer Biol. Ther. 13, 956–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang C., Li A., Zhang X., and Xiao H. (2011) A novel TIP30 protein complex regulates EGF receptor signaling and endocytic degradation. J. Biol. Chem. 286, 9373–9381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan J., Cheung A. Y., Fu W.-Y., Wu C., Zhang M., Mobley W. C., Cheung Z. H., and Ip N. Y. (2008) Endophilin B1 as a novel regulator of nerve growth factor/TrkA trafficking and neurite outgrowth. J. Neurosci. 28, 9002–9012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y., Takahashi Y., Desai N., Zhang J., Serfass J. M., Shi Y.-G., Lynch C. J., and Wang H.-G. (2016) Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci. Rep. 6, 20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang D. B., Kinoshita Y., Kinoshita C., Uo T., Sopher B. L., Cudaback E., Keene C. D., Bilousova T., Gylys K., Case A., Jayadev S., Wang H.-G., Garden G. A., and Morrison R. S. (2015) Loss of endophilin-B1 exacerbates Alzheimer's disease pathology. Brain 138, 2005–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takahashi Y., Meyerkord C. L., and Wang H. G. (2009) Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 16, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang D. B., Uo T., Kinoshita C., Sopher B. L., Lee R. J., Murphy S. P., Kinoshita Y., Garden G. A., Wang H. G., and Morrison R. S. (2014) Bax interacting factor-1 promotes survival and mitochondrial elongation in neurons. J. Neurosci. 34, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hrabchak C., Henderson H., and Varmuza S. (2007) A testis specific isoform of endophilin B1, endophilin B1t, interacts specifically with protein phosphatase-1c γ2 in mouse testis and is abnormally expressed in PP1c γ null mice. Biochemistry 46, 4635–4644 [DOI] [PubMed] [Google Scholar]
- 32. Yates A., Akanni W., Amode M. R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hunt S. E., Janacek S. H., et al. (2016) Ensembl 2016. Nucleic Acids Res. 44, D710–D716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y.-H., Wang J.-Q., Wang Q., Wang Y., Guo C., Chen Q., Chai T., and Tang T.-S. (2016) Endophilin B2 promotes inner mitochondrial membrane degradation by forming heterodimers with endophilin B1 during mitophagy. Sci. Rep. 6, 25153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piñon J. D., Labi V., Egle A., and Villunger A. (2008) Bim and Bmf in tissue homeostasis and malignant disease. Oncogene 27, S41–S52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thoresen S. B., Pedersen N. M., Liestøl K., and Stenmark H. (2010) A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 316, 3368–3378 [DOI] [PubMed] [Google Scholar]
- 36. Kerr M., and Teasdale R. D. (2014) Live imaging of endosome dynamics. Semin. Cell Dev. Biol. 31, 11–19 [DOI] [PubMed] [Google Scholar]
- 37. Koivusalo M., Welch C., Hayashi H., Scott C. C., Kim M., Alexander T., Touret N., Hahn K. M., and Grinstein S. (2010) Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188, 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dutta D., and Donaldson J. G. (2012) Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell. Logist. 2, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doherty G. J., and McMahon H. T. (2009) Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 [DOI] [PubMed] [Google Scholar]
- 40. Pinilla-Macua I., and Sorkin A. (2015) Methods to study endocytic trafficking of the EGF receptor. Methods Cell Biol. 130, 347–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luzio J. P., Hackmann Y., Dieckmann N. M. G., and Griffiths G. M. (2014) The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6, a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tooze S., Dooley H., Jefferies H. J., Joachim J., Judith D., Lamb C., Razi M., and Wirth M. (2015) Assessing mammalian autophagy. in Membrane Trafficking (Tang B. L., ed) pp. 155–165, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 43. Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C. M., Adams P. D., Adeli K., Adhihetty P. J., Adler S. G., Agam G., Agarwal R., Aghi M. K., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kimura S., Noda T., and Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 45. Edinger T. O., Pohl M. O., and Stertz S. (2014) Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 95, 263–277 [DOI] [PubMed] [Google Scholar]
- 46. Sieczkarski S. B., and Whittaker G. R. (2003) Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4, 333–343 [DOI] [PubMed] [Google Scholar]
- 47. Daniels R. S., Downie J. C., Hay A. J., Knossow M., Skehel J. J., Wang M. L., and Wiley D. C. (1985) Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40, 431–439 [DOI] [PubMed] [Google Scholar]
- 48. Maeda T., and Ohnishi S. (1980) Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 122, 283–287 [DOI] [PubMed] [Google Scholar]
- 49. White J. M., and Wilson I. A. (1987) Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 105, 2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshimura A., and Ohnishi S. (1984) Uncoating of influenza virus in endosomes. J. Virol. 51, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hahn D. R., Na C. L., and Weaver T. E. (2014) Reserve autophagic capacity in alveolar epithelia provides a replicative niche for influenza A virus. Am. J. Respir. Cell Mol. Biol. 51, 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Loebrich S., Benoit M. R., Konopka J. A., Cottrell J. R., Gibson J., and Nedivi E. (2016) CPG2 recruits endophilin B2 to the cytoskeleton for activity-dependent endocytosis of synaptic glutamate receptors. Curr. Biol. 26, 296–308 [DOI] [PubMed] [Google Scholar]
- 53. Liang C., Lee J.-S., Inn K.-S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., and Jung J. U. (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vannier C., Pesty A., San-Roman M. J., and Schmidt A. A. (2013) The Bin/Amphiphysin/Rvs (BAR) domain protein endophilin B2 interacts with plectin and controls perinuclear cytoskeletal architecture. J. Biol. Chem. 288, 27619–27637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Styers M. L., Salazar G., Love R., Peden A. A., Kowalczyk A. P., and Faundez V. (2004) The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol. Biol. Cell 15, 5369–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu W., and Panté N. (2016) Vimentin plays a role in the release of the influenza A viral genome from endosomes. Virology 497, 41–52 [DOI] [PubMed] [Google Scholar]
- 57. Lapointe J., Li C., Higgins J. P., van de Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Ekman P., DeMarzo A. M., Tibshirani R., Botstein D., Brown P. O., et al. (2004) Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 101, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fassò M., Waitz R., Hou Y., Rim T., Greenberg N. M., Shastri N., Fong L., and Allison J. P. (2008) SPAS-1 (stimulator of prostatic adenocarcinoma-specific T cells)/SH3GLB2: a prostate tumor antigen identified by CTLA-4 blockade. Proc. Natl. Acad. Sci. 105, 3509–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bainbridge M. N., Warren R. L., Hirst M., Romanuik T., Zeng T., Go A., Delaney A., Griffith M., Hickenbotham M., Magrini V., Mardis E. R., Sadar M. D., Siddiqui A. S., Marra M. A., and Jones S. J. (2006) Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics 7, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Coppola D., Khalil F., Eschrich S. A., Boulware D., Yeatman T., and Wang H. G. (2008) Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer 113, 2665–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coppola D., Oliveri C., Sayegh Z., Boulware D., Takahashi Y., Pow-Sang J., Djeu J. Y., and Wang H. G. (2008) Bax-interacting factor-1 expression in prostate cancer. Clin. Genitourin. Cancer 6, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim S. Y., Oh Y. L., Kim K. M., Jeong E. G., Kim M. S., Yoo N. J., and Lee S. H. (2008) Decreased expression of Bax-interacting factor-1 (Bif-1) in invasive urinary bladder and gallbladder cancers. Pathology 40, 553–557 [DOI] [PubMed] [Google Scholar]
- 63. Lee J. W., Jeong E. G., Soung Y. H., Nam S. W., Lee J. Y., Yoo N. J., and Lee S. H. (2006) Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas. Pathology 38, 312–315 [DOI] [PubMed] [Google Scholar]
- 64. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 65. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Young M. M., Takahashi Y., Khan O., Park S., Hori T., Yun J., Sharma A. K., Amin S., Hu C.-D., Zhang J., Kester M., and Wang H.-G. (2012) Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 287, 12455–12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sever-Chroneos Z., Murthy A., Davis J., Florence J. M., Kurdowska A., Krupa A., Tichelaar J. W., White M. R., Hartshorn K. L., Kobzik L., Whitsett J. A., and Chroneos Z. C. (2011) GM-CSF modulates pulmonary resistance to influenza A infection. Antiviral Res. 92, 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.