Abstract
Contributions of metabolic changes to cancer development and maintenance have received increasing attention in recent years. Although many human cancers share similar metabolic alterations, it remains unclear whether oncogene-specific metabolic alterations are required for tumor development. Using an RNAi-based screen targeting the majority of the known metabolic proteins, we recently found that oncogenic BRAFV600E up-regulates HMG-CoA lyase (HMGCL), which converts HMG-CoA to acetyl-CoA and a ketone body, acetoacetate, that selectively enhances BRAFV600E-dependent MEK1 activation in human cancer. Here, we identified HMG-CoA synthase 1 (HMGCS1), the upstream ketogenic enzyme of HMGCL, as an additional “synthetic lethal” partner of BRAFV600E. Although HMGCS1 expression did not correlate with BRAFV600E mutation in human melanoma cells, HMGCS1 was selectively important for proliferation of BRAFV600E-positive melanoma and colon cancer cells but not control cells harboring active N/KRAS mutants, and stable knockdown of HMGCS1 only attenuated colony formation and tumor growth potential of BRAFV600E melanoma cells. Moreover, cytosolic HMGCS1 that co-localized with HMGCL and BRAFV600E was more important than the mitochondrial HMGCS2 isoform in BRAFV600E-expressing cancer cells in terms of acetoacetate production. Interestingly, HMGCL knockdown did not affect HMGCS1 expression levels, whereas HMGCS1 knockdown caused a compensating increase in HMGCL protein level because of attenuated protein degradation. However, this increase did not reverse the reduced ketogenesis in HMGCS1 knockdown cells. Mechanistically, HMGCS1 inhibition decreased intracellular acetoacetate levels, leading to reduced BRAFV600E-MEK1 binding and consequent MEK1 activation. We conclude that the ketogenic HMGCS1-HMGCL-acetoacetate axis may represent a promising therapeutic target for managing BRAFV600E-positive human cancers.
Keywords: acetoacetate, cancer biology, colon cancer, melanoma, metabolism, oncogenic BRAFV600E, HMG-CoA synthase 1, HMGCS1, ketogenesis, cancer metabolism, synthetic lethal
Introduction
The importance of metabolic alterations in cancer has been increasingly recognized over the past decade. Cancer cells appear to coordinate bioenergetics (aerobic glycolysis), anabolic biosynthesis, and appropriate redox status to promote cancer cell proliferation and tumor growth (1, 2). Interestingly, although increasing evidence suggests that different human cancers may share common metabolic properties, such as the Warburg effect, it is unclear whether distinct oncogene mutations including oncogenes and tumor suppressor genes (TSGs) may regulate and require distinct metabolic alterations for tumor development.
We approached this question by identifying a unique metabolic vulnerability required by distinct oncogenic mutations (3). We chose oncogenic BRAFV600E-positive melanoma as a platform. Melanoma is one of the most common human cancers. BRAFV600E mutation is found in ≥ 60% of malignant melanomas (4–6), as well as in 10% of colorectal cancer (7, 8), 100% of hairy cell leukemia (HCL) (9), and 5% of multiple myeloma (10). The pathogenic importance of BRAFV600E in diverse human cancers makes it a promising therapeutic target in cancer treatment. However, although BRAF-mutant and MEK inhibitors have been demonstrated to be clinically effective in treating BRAFV600E-positive melanoma patients, resistance eventually develops that leads to cancer relapse of treated patients (4–6). Thus, identification of co-dependent targets in BRAFV600E-positive melanomas may inform more successful long-term treatment strategies.
We designed a novel shRNA library that targets the majority of metabolism-related proteins in the human genome (3). Using this library, we identified synthetic lethal (SL) metabolic partners of oncogenic BRAFV600E in human melanoma cells, attenuation of which only inhibited BRAFV600E-expressing melanoma cells but not cells expressing NRAS mutant or wild type (WT) BRAF and NRAS (3). Top candidates include two key enzymes in the ketogenic pathway, HMG-CoA lyase (HMGCL)3 and HMG-CoA synthase 1 (HMGCS1) (3).
Ketogenesis mainly occurs in the mitochondria of liver cells, which normally produces ketone bodies as a result of fatty acid breakdown to generate energy when glucose levels in the blood are low (11, 12). β-oxidation breaks down fatty acids to form acetyl-CoA, which, under normal conditions, is further oxidized in the TCA cycle. However, if TCA cycle activity is low, or the acetyl-CoA generation rate of β-oxidation exceeds the capacity of the TCA cycle, ketogenesis will be activated to convert acetyl-CoA to ketone bodies via HMG-CoA. HMGCS converts acetyl-coA to HMG-CoA, whereas HMGCL converts HMG-CoA to acetyl-CoA and a ketone body, acetoacetate (AA), which can be further converted to two other ketone bodies, including d-β-hydroxybutyrate (3HB) and acetone. Ketone bodies can be transported from the liver to other tissues, where acetoacetate and 3HB but not acetone will be further oxidized via the TCA cycle to produce acetyl-CoA for energy production. Organs, including the heart and brain, can use acetoacetate and 3HB for energy. Acetoacetate, if not used for energy, will be decarboxylated to acetone that is removed as waste (13, 14).
We found that BRAFV600E activates a transcription factor, Oct-1, leading to HMGCL up-regulation in BRAFV600E-expressing human melanoma and hairy cell leukemia cells (3). HMGCL increases intracellular levels of acetoacetate that selectively promotes BRAFV600E (but not WT) binding to MEK1 and subsequent MEK1 activation, which represents a new “wiring” that only occurs in cancer cells expressing BRAFV600E but not normal cells (3). Moreover, we recently reported that a high-fat ketogenic diet resulted in increased serum levels of acetoacetate, leading to enhanced tumor growth potential of BRAFV600E-expressing human melanoma cells in xenograft mice. Treatment with hypolipidemic agents to lower circulating acetoacetate levels or an inhibitory homologue of acetoacetate, dehydroacetic acid (DHAA), to antagonize acetoacetate-BRAFV600E binding attenuated BRAFV600E tumor growth. These findings together reveal a signaling basis underlying a pathogenic role of dietary fat-fueled ketogenesis in BRAFV600E-expressing human cancer (15).
Here we demonstrate that HMGCS1 as a key ketogenic enzyme also functions as a synthetic lethal partner of BRAFV600E in human melanoma cells. There are two isoforms of HMGCS, including cytosolic HMGCS1 and mitochondrial HMGCS2 (16). Although mitochondrial HMGCS2 has been suggested to be the main regulatory site in ketogenesis, our results suggest that cytosolic HMGCS1 is important to regulate intracellular acetoacetate levels that enhance BRAFV600E-dependent MEK1 activation and consequent cell proliferation and tumor growth potential in BRAFV600E-positive melanoma.
Results
HMGCS1 is a synthetic lethal partner of BRAFV600E in human melanoma cells
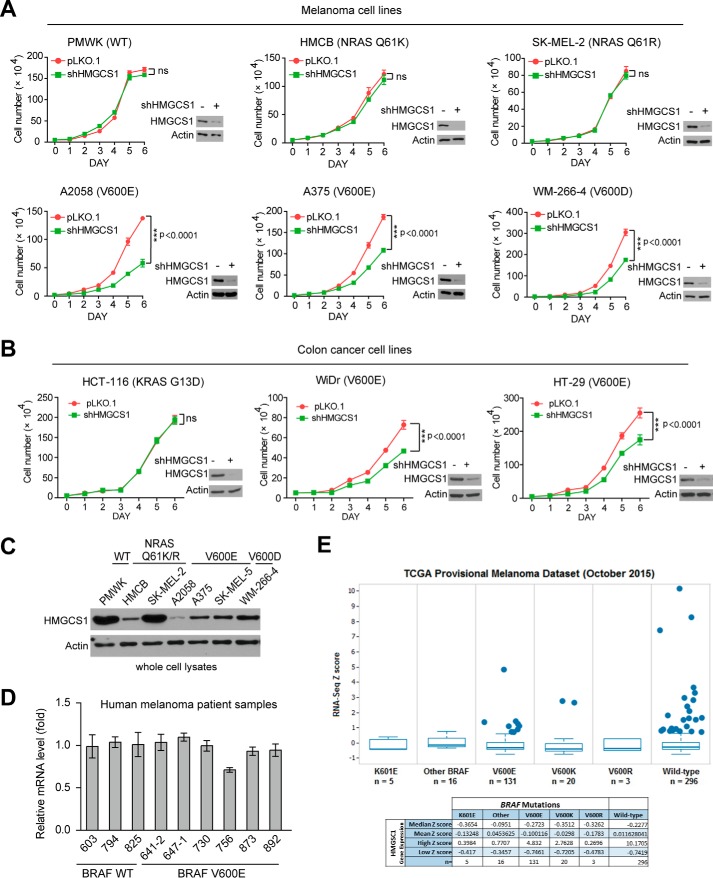
We previously identified HMGCS1 as a top synthetic lethal partner candidate of BRAFV600E, a systematic RNAi screen using human melanoma cells expressing BRAFV600E mutant and control BRAF WT-expressing melanoma cells (3). Future cell-based analyses revealed that stable knockdown of HMGCS1 resulted in more attenuated cell proliferation rates in BRAFV600D/E-expressing melanoma cells including A2058, A375, and WM-266-4 cells compared with control PMWK cells expressing BRAF WT as well as HMCB and SK-ME-2 cells expressing active NRAS Q61K or Q61R mutant, respectively, suggesting selective importance of HMGCS1 in BRAFV600E-induced melanoma transformation (Fig. 1A). Consistent with this finding, we found that stable knockdown of HMGCS1 in BRAFV600E-expressing human colon cancer WiDr and HT-29 cells also resulted in reduced cell proliferation rates, but not in control colon cancer HCT-116 cells (Fig. 1B).
Figure 1.
HMGCS1 knockdown selectively decreases cell proliferation potential in BRAFV600E-expressing melanoma cells. A and B, distinct human melanoma (A) or colon cancer (B) cells with HMGCS1 knockdown were tested for cell proliferation rates by daily cell counting. Immunoblotting results show HMGCS1 knockdown efficiency in corresponding cells. p values were obtained by two-tailed Student's t test. Error bars indicate ± S.D.; n = 3 each. C, immunoblotting shows HMGCS1 protein level in diverse human melanoma cells expressing BRAFV600D/E (A2058, A375, SK-MEL-5, WM-266-4) or BRAF WT (PMWK, HMCB, SK-MEL-2). D, results of quantitative PCR show HMGCS1 gene expression in human primary melanoma tissue samples expressing BRAFV600E compared with samples expressing BRAF WT. E, TCGA provisional melanoma dataset analysis shows HMGCS1 expression (mRNA level) in diverse clinical samples harvesting V600E, V600K, V600R, K601E or other mutations compared with BRAF WT. The statistic result was analyzed by population-based assessment Z scores (S.D. scores). n means sample size of each subgroup.
Surprisingly, unlike HMGCL that is up-regulated by BRAFV600E in human melanoma cells, we found that HMGCS1 protein expression levels did not correlate with BRAFV600E mutational status in diverse human melanoma cell lines (Fig. 1C). In addition, BRAFV600E mutation did not affect mRNA levels of HMGCS1 in primary human tumor tissue samples from a group of BRAFV600E-positive and -negative melanoma patients (Fig. 1D). Consistent with these findings, gene expression levels of HMGCS1 were not up-regulated in human melanoma patients harboring distinct mutations at V600 including V600E, V600K, and V600R compared with patients expressing BRAF WT as assessed using TCGA Provisional melanoma dataset (Fig. 1E). These data suggest that the physiological levels of HMGCS1 in human melanoma cells are sufficient to fulfill the elevated ketogenic flux and consequent acetoacetate production because of increased HMGCL expression that is up-regulated by BRAFV600E.
HMGCS1 signals through acetoacetate to promote cell proliferation and BRAFV600E-dependent MEK1 activation in BRAFV600E-expressing cells
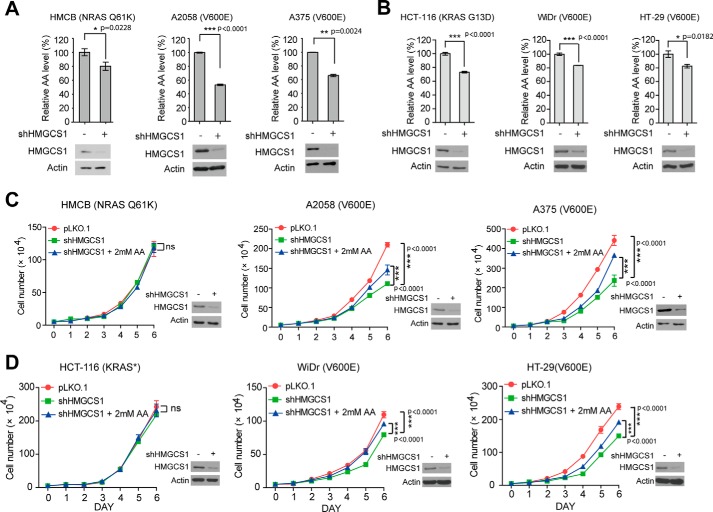
We next sought to explore the underlying molecular mechanism. Previously we demonstrated that HMGCL product acetoacetate selectively promotes BRAFV600E but not BRAF WT binding to MEK1 and consequent MEK1 phosphorylation and activation (3). Similar as observed in HMGCL knockdown cells (3), silencing HMGCS1 by shRNA resulted in significantly decreased intracellular concentration of acetoacetate in diverse human melanoma (Fig. 2A) and colon cancer cells (Fig. 2B), despite BRAFV600E status. However, stable knockdown of HMGCS1 selectively reduced cell proliferation rates in BRAFV600E-expressing melanoma A2058 and A375 cells but not in control NRAS Q61K-expressing HMCB cells (Fig. 2C), as well as in BRAFV600E-expressing colon cancer WiDr and HT-29 cells but not in control KRAS G13D-expressing HCT-116 cells (Fig. 2D). Addition of AA in the culture media significantly reversed the decreased cell proliferation rates only in BRAFV600E-expressing cells with stable HMGCS1 knockdown (Fig. 2, C and D).
Figure 2.
HMGCS1 controls production of ketone body acetoacetate to selectively promote cell proliferation of BRAFV600E-expressing melanoma cells. A–D, intracellular concentrations of acetoacetate (AA) (A and B) and rescue effect of adding acetoacetate in culture media on cell proliferation rates (C and D) were examined in melanoma (A and C) and colon cancer (B and D) cells upon HMGCS1 knockdown. Mean ± S.D. Cell proliferation rates were determined by daily cell counting. Error bars indicate ± S.D.; n = 3 each. p values were obtained by two-tailed Student's t test. Western blot shows knockdown efficiency of HMGCS1 in corresponding cells.
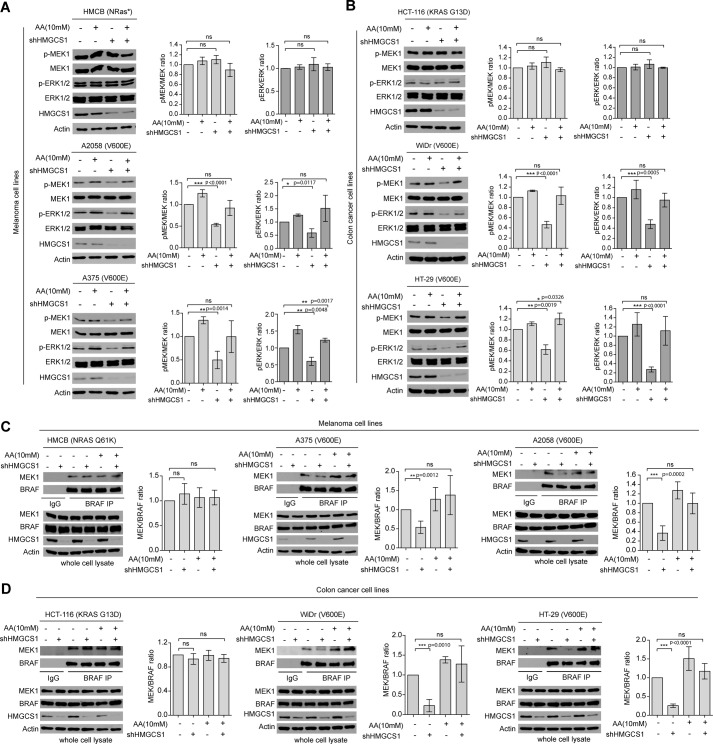
In addition, stable knockdown of HMGCS1 selectively attenuated phosphorylation of MEK1 and ERK1/2 only in BRAFV600E-expressing A2058 and A375 melanoma cells but not control HMCB cells, whereas AA treatment reversed such decreased phosphorylation and activation of MEK-ERK pathway because of HMGCS1 knockdown only in BRAFV600E-expressing melanoma (Fig. 3A) and colon cancer cells (Fig. 3B). Moreover, AA treatment resulted in increased MEK1 binding to BRAFV600E in A2058 and A375 cells, but not to BRAF WT in control HMCB cells, which was reversed by treatment with AA (Fig. 3C). Similar results were obtained using colon cancer cells with HMGCS1 knockdown (Fig. 3D). These data together suggest that HMGCS1 plays an important role in ketogenesis-fueled BRAFV600E transformation in human melanoma and colon cancer cells through regulation of downstream HMGCL product acetoacetate to promote a BRAFV600E-activated MEK-ERK signaling cascade.
Figure 3.
HMGCS1 is selectively important to regulate acetoacetate that specifically promotes BRAFV600E-MEK1 binding and consequent MEK-ERK activation in BRAFV600E-expressing cells. A–D, effect of adding acetoacetate in culture media on phosphorylation levels of MEK1 and ERK1/2 (A and B) and BRAF-MEK1 interaction (C and D) in distinct melanoma (A and C) and colon cancer (B and D) cells and cells with stable HMGCS1 knockdown. IP, immunoprecipitate. Quantification error bars along with each Western blot indicate ± S.D.; p values were obtained by two-tailed Student's t test. Western blots are from a representative experiment for which n = 3 for A, B, and D and n = 4 for C.
Cytosolic HMGCS1 but not mitochondrial HMGCS2 is selectively important for BRAFV600E cancer cells
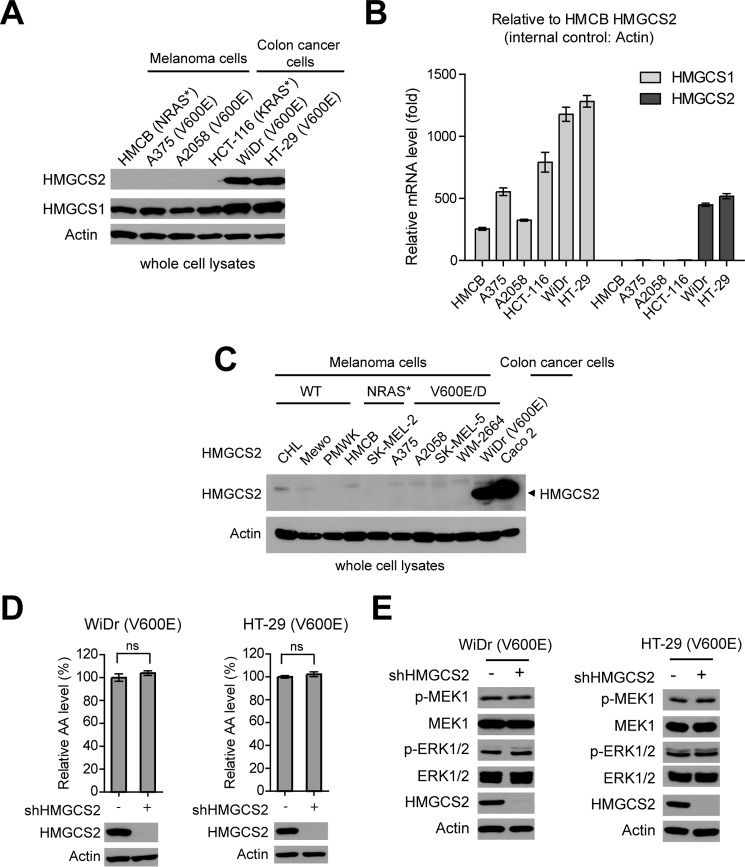
To distinguish the different roles of the two HMGCS isoforms, cytosolic HMGCS1 and mitochondrial HMGCS2, we examined the expression levels of HMGCS1 and HMGCS2 in a group of human cancer cells. We found that HMGCS1 protein expression levels are comparable in all the tested melanoma and colon cancer cells despite BRAFV600E mutational status (Fig. 3A). In contrast, HMGCS2 was only detected in BRAFV600E-expressing colon cancer WiDr and HT-29 cells but not in KRAS G13D-expressing HCT-116 colon cancer cells or NRAS mutant-expressing HMCB and BRAFV600E-expressing A375 and A2058 melanoma cells (Fig. 4A). In addition, we found that the relative HMGCS2 mRNA levels were approximately > 450-fold higher in colon cancer WiDr and HT-29 cells compared with HCT-116 cells and melanoma HMCB, A375, and A2058 cells, whereas the relative HMGCS1 mRNA levels were detectable in all of the tested cell lines (Fig. 4B). Consistent with these results, mitochondrial HMGCS2 protein expression was detected in WiDr cells and a control colon cancer Caco 2 cell line (Abcam), whereas the protein expression levels of HMGCS2 were much lower and almost undetectable in Western blot experiments using cell lysates from diverse human melanoma cell lines despite BRAF and NRAS mutational states (Fig. 4C).
Figure 4.
Cytosolic HMGCS1 but not mitochondrial HMGCS2 is selectively important for BRAFV600E cancer cells. A and B, protein expression (A) and relative mRNA (B) levels of HMGCS1 and HMGCS2 in diverse human melanoma and colon cancer cells were examined. Error bars indicate ± S.D.; n = 3 each. p values were obtained by two-tailed Student's t test. C, results of Western blot detecting mitochondrial HMGCS2 in diverse melanoma cells. D and E, effect of stable knockdown of HMGCS2 on intracellular acetoacetate (D) and phosphorylation levels of MEK1 and ERK1/2 (E) in BRAFV600E-expressing human colon cancer cells.
We thus next examined the effect of stable knockdown of HMGCS2 in the two BRAFV600E-expressing colon cancer cell lines including WiDr and HT-29, which have relatively high levels of HMGCS2 protein expression. In contrast to HMGCS1 knockdown (Figs. 2 and 3), we found that HMGCS2 knockdown did not affect the intracellular levels of acetoacetate (Fig. 4D) or phosphorylation levels of MEK1 and ERK1/2 (Fig. 4E) in BRAFV600E-expressing human colon cancer cells.
HMGCS1 co-localizes with HMGCL and BRAFV600E in cytosol and regulates HMGCL protein stability
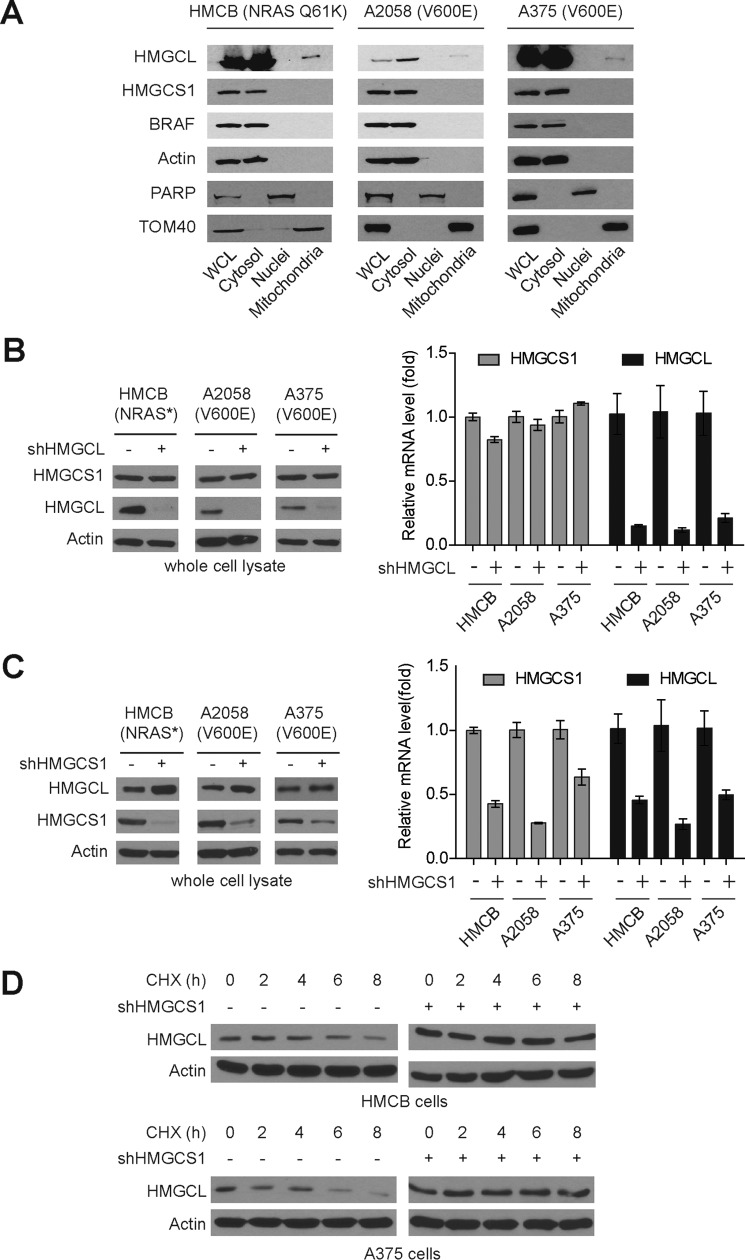
Consistent with these results, we found that HMGCS1 co-localized with HMGCL and BRAF in cytosolic fractions in human melanoma cells expressing BRAF WT or V600E (Fig. 5A). To determine the potential interplay between HMGCS1 and HMGCL in BRAFV600E-expressing cancer cells, we examined the effect of stable knockdown of one protein on the other. We found that stable knockdown of HMGCL did not affect the protein expression and mRNA levels of HMGCS1 in HMCB, A375, and A2058 melanoma cells despite BRAFV600E mutational status (Fig. 5B, left and right, respectively). Interestingly, stable knockdown of HMGCS1 resulted in increased protein levels of HMGCL (Fig. 5C, left) but with decreased HMGCL mRNA levels (Fig. 5C, right). These results suggest that attenuation of HMGCS1 leads to a compensating increase in HMGCL protein levels, which is likely involving alteration in protein stability rather than gene expression. Indeed, we found that treatment with cycloheximide (CHX) induced protein degradation of HMGCL in HMCB and A375 cells but not in cells with stable knockdown of HMGCS1 (Fig. 5D). Together, these data suggest that HMGCS1 knockdown caused a compensating increase in downstream HMGCL protein level because of attenuated protein degradation, which, however, could not reverse the reduced ketogenesis in cells with HMGCS1 knockdown.
Figure 5.
HMGCS1 co-localizes with HMGCL and BRAFV600E in cytosol and regulates HMGCL protein stability. A, subcellular localization of HMGCS1, BRAF, and HMGCL in cytoplasm, nucleus, and mitochondria in BRAF WT cells HMCB and BRAFV600E-positive cells A2058 and A375. Actin, PARP, and TOM40 were included as markers for cytoplasm, nucleus, and mitochondria, respectively. B and C, effect of stable knockdown of HMGCL (B) or HMGCS1 (C) on HMGCS1 or HMGCL protein expression (left panels) and mRNA levels (right panels), respectively. D, results of Western blot showing effect of cycloheximide (CHX) treatment on HMCGL1 in the melanoma cells in the presence or absence of stable knockdown of HMGCS1.
HMGCS1 is important for transformation potential of BRAFV600E in human melanoma cells
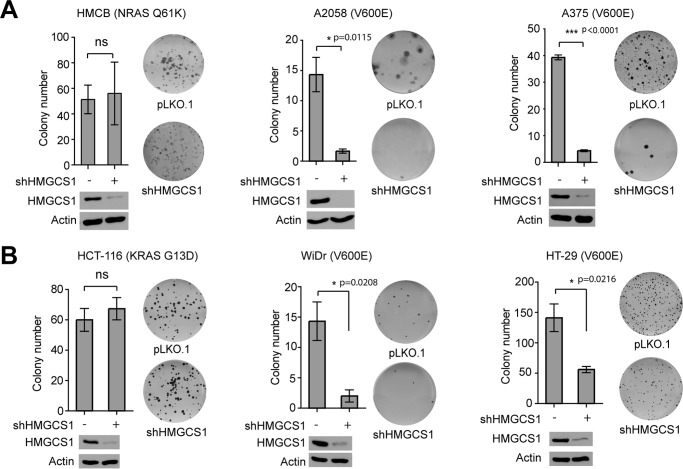
We next examined whether HMGCS1 is required for BRAFV600E-induced transformation. We found that stable knockdown of HMGCS1 resulted in attenuated colony-formation potential of BRAFV600E-expressing A2058 and A375 cells but not control HMCB cells expressing NRAS Q61K mutant (Fig. 6A). Similar results were obtained using KRAS G13D-expressing HCT-116 and BRAFV600E-expressing WiDr and HT-29 colon cancer cells (Fig. 6B).
Figure 6.
HMGCS1 knockdown decreases colony formation potential in BRAFV600E-expressing melanoma cells. A and B, effect of HMGCS1 knockdown on monolayer colony formation of diverse melanoma (A) and colon cancer (B) cells. Knockdown efficiency of HMGCS1 in corresponding cells are shown by Western blot. Error bars indicate ± S.D.; n = 3 each. p values were obtained by two-tailed Student's t test.
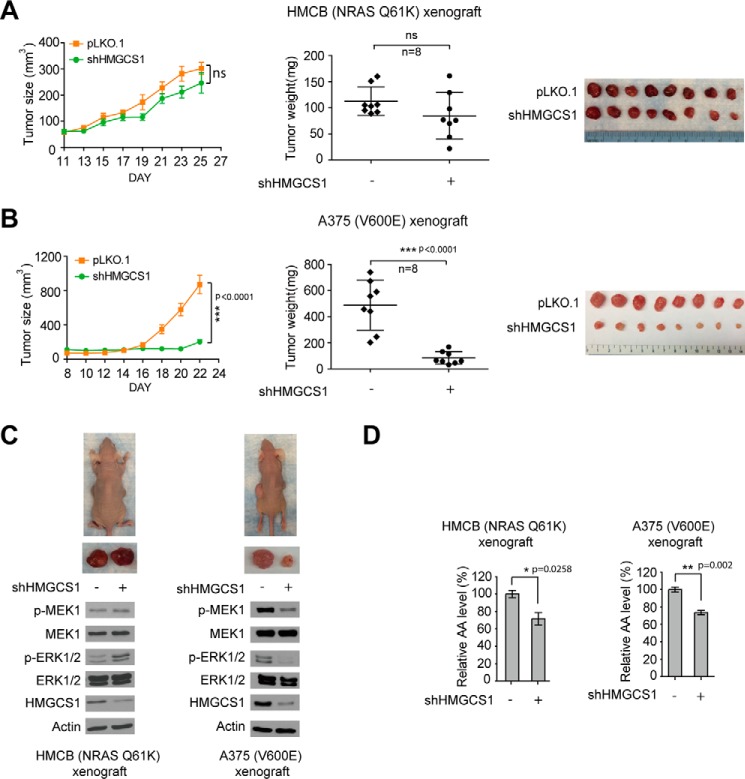
Moreover, in a xenograft nude mouse model, silencing HMGCS1 did not affect tumor growth rates, masses, or sizes of tumors harvested from xenograft nude mice injected with HMCB cells or cells with stable knockdown of HMGCS1 (Fig. 7A). In contrast, tumors derived from A375 cells with stable knockdown of HMGCS1 demonstrated decreased growth rates and masses with reduced tumor sizes compared with those derived from control A375 cells in xenograft nude mice (Fig. 7B). Mechanistic studies revealed that HMGCS1 knockdown resulted in decreased phosphorylation levels of MEK1 and ERK1/2 only in tumors derived from A375 cells but not HMCB cells in xenograft nude mice (Fig. 7C), despite the fact that reduced intracellular acetoacetate levels were observed in both A375 and HMCB xenograft tumors with stable knockdown of HMGCS1 compared with xenograft tumors derived from corresponding parental cells (Fig. 7D). These data together suggest a synthetic lethal interaction between BRAFV600E and HMGCS1 in human cancer cells.
Figure 7.
HMGCS1 is specifically important for tumor growth potential of BRAFV600E-expressing melanoma cells. A and B, growth rates, masses, and sizes of tumors harvested from xenograft mice injected with NRAS Q61K-expressing HMCB cells (A) or BRAFV600E-positive A375 cells (B) harboring an empty vector or HMGCS1 shRNA. Error bars indicate ± S.D. p values of tumor size and weight were obtained by two-way ANOVA. C and D, phosphorylation levels of MEK1 and ERK1/2 assessed by Western blot (C) and relative acetoacetate levels (D) in tumors obtained from xenograft mice injected with control or HMGCS1 knockdown HMCB or A375 cells. p values of results detecting acetoacetate concentrations were obtained by two-tailed Student's t test. Error bars indicate ± S.D.; n = 3 each.
Discussion
Our results suggest that a key ketogenic enzyme, HMGCS1, is specifically important for BRAFV600E-dependent transformation signaling in human melanoma cells. HMGCS1 regulates the intracellular levels of acetoacetate, the product of its downstream enzyme HMGCL in the ketogenesis pathway, to selectively promote BRAFV600E-MEK1 binding and consequent activation of MEK-ERK pathway. This is consistent with our previous findings that support a novel “feed-forward” model, where BRAFV600E activates a ketogenesis pathway by up-regulating HMGCL and subsequent intracellular levels of acetoacetate, which specifically binds to BRAFV600E mutant but not BRAF WT, leading to enhanced MEK1 and ERK1/2 activation in human melanoma cells (3). Interestingly, although mitochondrial HMGCS2 has been suggested to be the main regulator of ketone body production, it was not identified as one of the top candidates for BRAFV600E synthetic lethal partner (3), and consistently our results herein suggest that HMGCS2 is not selectively important for BRAFV600E cancer cell proliferation. Our results suggest that both HMGCS1 and HMGCL are localized in cytosol with BRAF proteins in human melanoma cells expressing either BRAF WT or V600E mutant, suggesting that ketogenesis and acetoacetate production in cytosol are more critical than those in mitochondria for BRAFV600E-dependent transformation in human melanoma cells.
Our results demonstrate that although oncogenic BRAFV600Emutant “rewires” metabolic and cell signaling networks in human cancer cells by up-regulating HMGCL subsequent intracellular acetoacetate levels through activation of transcription factor Oct-1, the expression levels of HMGCS1 do not correlate with BRAFV600E mutational status in human melanoma cells, suggesting that HMGCS1 is not directly involved in the process by which oncogenic BRAFV600E rewires the ketogenesis pathway to MEK-ERK signaling. However, physiological activity of HMGCS1 plays an important role in such a rewiring and is sufficient to cooperate with up-regulated HMGCL by BRAFV600E to achieve elevated production of acetoacetate that enhances BRAFV600E transformation potential in human melanoma cells. Interestingly, knockdown of HMGCS1 appears to cause a compensating increase in downstream HMGCL protein level because of attenuated protein degradation through an unknown mechanism, which, however, could not reverse the reduced ketogenesis in cells with HMGCS1 knockdown. Future studies are warranted to explore the molecular and signaling basis underlying HMGCS1-dependent regulation of protein stability in cells.
Our findings also suggest that the ketogenic HMGCS1-HMGCL-acetoacetate axis represents a promising therapeutic target in treatment of BRAFV600E-positive human cancers. Indeed, in our recent report, DHAA functions as an inhibitory derivative of acetoacetate by binding to BRAFV600E with a higher affinity that enables DHAA to compete with acetoacetate for BRAFV600E binding, which selectively inhibits cell proliferation and tumor growth potential of melanoma cells expressing BRAFV600E but not melanoma cells expressing wild type BRAF (15). Thus, future anti-ketogenic drugs such as small molecule inhibitors of HMGCS and HMGCL, as well as DHAA and its next generation derivatives, may represent alternative clinical treatments for patients with BRAFV600E-positive cancer. Furthermore, these findings provide insights into the development of a precision diet (15), which should be designed based on individual genetic background to lower cancer risk and provide cancer prevention.
Experimental procedures
Reagents
Acetoacetate was purchased from Sigma-Aldrich. Stable knockdown of endogenous human HMGCS1 was achieved by using lentiviral vector-harboring shRNA construct (Open Biosystems) (5′-CCCATCATTTGGTCAACTATA-3′). Antibodies against HMGCS1 and HMGCL were purchased from Abcam. Antibodies against p-MEK1, MEK1, p-ERK1/2, ERK1/2, and PARP were from Cell Signaling Technology. Antibody against β-actin was from Sigma-Aldrich. Antibodies against BRAF and TOM40 were from Santa Cruz Biotechnology.
Cell culture
PMWK, A2058, and A375 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). HMCB, WM-266-4, and SK-MEL-2 cells were cultured in Eagle's minimal essential medium (EMEM) with 10% FBS. AA treatment was performed by incubating 10 mm AA for about 12 h. For the cell proliferation assay, 2 mm AA were added in the cell culture medium.
Acetoacetate extraction and measurement
To determine the cellular concentration of AA, 100 μl of packed cell pellets were homogenized in 500 μl of lysis buffer (10 mm HEPES, pH 7.0, 150 mm NaCl, 1% Nonidet P-40, 5 mm NaF, 5 mm sodium pyrophosphate and protease inhibitor mixture). The cell lysates were filtered with 10-kDa Amicon® Ultra-4 Centrifugal Filters (Millipore). The flow through containing the metabolites was used for AA measurement according to the instructions for the Acetoacetate Assay Kit (BioVision).
Cell proliferation
Cell proliferation assays were performed by seeding 5 × 104 cells in a 6-well plate and culturing the cells at 37 °C in 5% CO2. Cell proliferation was determined by cell numbers recorded at 1 to 6 days after being seeded and normalized to that of each of the cell lines at the starting time (T = 0 h) by trypan blue exclusion using a TC10 Automated Cell Counter (Bio-Rad).
Quantitative RT-PCR
Total RNA was isolated from human melanoma tissues using TRIzol Reagent (Invitrogen). Total cDNA was prepared by using a two-step reverse transcription kit (TaKaRa). Primers for real-time PCR were obtained from Integrated DNA Technologies. Reactions were run on an Applied Biosystems Prism machine using Fast SYBR® Green Master Mix (Applied Biosystems). An amount of GAPDH was used to normalize results from gene-specific reactions. Primer sequences for gene-specific amplicons are 5′-TATTCCAAGCCCTGCCAAGA-3′ (HMGCS1/forward); 5′-TCCAACTGTTCCCATACCCC-3′ (HMGCS1/reverse); 5′-CTGGGCTACACTGAGCACC-3′ (GAPDH/forward); 5′-AAGTGGTCGTTGAGGGCAATG-3′ (GAPDH/reverse).
Colony-forming assay
Melanoma cell lines transduced with lentivirus harboring pLKO.1 or shHMGCS1 (300 cells) were seeded in 6-well plates for monolayer colony formation. After 2 to 3 weeks, colonies were stained with 0.5% crystal violet, counted microscopically, and photographed.
Co-immunoprecipitation assays
For co-immunoprecipitation, cells were harvested in 1 ml Nonidet P-40 buffer as described previously. 1-mg cell lysates were incubated with anti-BRAF antibody overnight at 4 °C. After incubation, protein G-Sepharose beads were used for precipitation. The beads were washed and eluted with SDS sample buffer for immunoblotting analysis.
Cell fractionation
Nuclear and cytoplasmic extraction was performed according to NE-PER Nuclear and Cytoplasmic Extraction Kit instructions (Pierce). Mitochondrial subfractionation was performed according to Mitochondria Isolation Kit instructions (Thermo Scientific).
Xenograft studies
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Nude mice (nu/nu, female 4–6 weeks old) (Harlan Laboratories) were subcutaneously injected with 2 × 106 melanoma A375 or HMCB cells harboring empty vector on the left flank, and cells with stable knockdown of endogenous HMGCS1 on the right flank. Tumor sizes were measured every 2 days by using Vernier calipers. Tumor growth was recorded by measurement of two perpendicular diameters using the formula 4π/3 × (width/2)2 × (length/2). The tumors were harvested and weighed at the experimental end point, and the masses of tumors (mg) derived from cells with or without stable knockdown HMGCS1 were compared. Statistical analyses were optioned by two-way analysis of variance (ANOVA). No blinding was done for these animal studies.
Study approval
Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. All clinical samples were obtained with informed consent with approval by the Emory University Institutional Review Board. Clinical information for the patients was obtained from the pathology files at Emory University Hospital under the guidelines and with approval from the Institutional Review Board of Emory University School of Medicine and according to the Health Insurance Portability and Accountability Act. Tissues were harvested for total RNA extraction.
Statistics
In studies in which statistical analyses were performed, a 2-tailed Student's t test was used to generate p values. p values less than or equal to 0.05 were considered significant. A two-way ANOVA was used for tumor volume and tumor weight.
Reproducibility of experiments
The results of one representative experiment from at least two independent experiments are shown except data obtained from primary patient samples and animal experiments shown in Figs. 1D and 7, respectively. There is no estimate of variation in each group of data and the variance is similar between the groups. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All data are expected to have normal distribution.
Author contributions
L. Z., R. L., F. C., and J. C. designed the study and wrote the manuscript. L. Z., R. L., J. F., S. X., Y. P., Z. Q., S. Liu, G. Q., and H.-B. K. performed experiments, analysis, and interpretation of the data. J. L. A., B. P. P., R. R. K., D. H. L., M. R., L. H. B., O. A.-W., T. M., H. J. K., F. R. K., and S. Lonial provided critical reagents and contributed to data analyses and interpretation.
Acknowledgment
We thank Anthea Hammond for critical reading and editing of the manuscript.
This work was supported in part by National Institutes of Health Grants CA140515, CA183594, CA174786 (J.C.), and AR47901 (J.L.A.); Joel A. Katz Music Medicine Fund supported by the T.J. Martell Foundation/Winship Cancer Institute (J.C. and R.L.); the Jamie Rabinowitch-Davis Foundation for Melanoma Research (J.L.A.); the Charles Harris Run For Leukemia, Inc. (H.J.K.); and the Melanoma Research Foundation and the Winship Cancer Institute Melanoma & Skin Cancer Fund (B.P.P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HMGCL
- HMG-CoA lyase
- HMGCS1
- HMG-CoA synthase 1
- AA
- acetoacetate
- 3HB
- d-β-hydroxybutyrate
- DHAA
- dehydroacetic acid
- ANOVA
- analysis of variance
- WT
- wild type.
References
- 1. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 3. Kang H. B., Fan J., Lin R., Elf S., Ji Q., Zhao L., Jin L., Seo J. H., Shan C., Arbiser J. L., Cohen C., Brat D., Miziorko H. M., Kim E., Abdel-Wahab O., Merghoub T., et al. (2015) Metabolic rewiring by oncogenic BRAF V600E links ketogenesis pathway to BRAF-MEK1 signaling. Mol. Cell 59, 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bollag G., Tsai J., Zhang J., Zhang C., Ibrahim P., Nolop K., and Hirth P. (2012) Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discov. 11, 873–886 [DOI] [PubMed] [Google Scholar]
- 5. Johnson D. B., and Sosman J. A. (2013) Update on the targeted therapy of melanoma. Curr. Treat. Options Oncol. 14, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibney G. T., Messina J. L., Fedorenko I. V., Sondak V. K., and Smalley K. S. (2013) Paradoxical oncogenesis–the long-term effects of BRAF inhibition in melanoma. Nat. Rev. Clin. Oncol. 10, 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sebolt-Leopold J. S., and Herrera R. (2004) Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4, 937–947 [DOI] [PubMed] [Google Scholar]
- 8. Benlloch S., Payá A., Alenda C., Bessa X., Andreu M., Jover R., Castells A., Llor X., Aranda F. I., and Massutí B. (2006) Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J. Mol. Diagn. 8, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arcaini L., Zibellini S., Boveri E., Riboni R., Rattotti S., Varettoni M., Guerrera M. L., Lucioni M., Tenore A., Merli M., Rizzi S., Morello L., Cavalloni C., Da Vià M. C., Paulli M., and Cazzola M. (2012) The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood 119, 188–191 [DOI] [PubMed] [Google Scholar]
- 10. Chapman M. A., Lawrence M. S., Keats J. J., Cibulskis K., Sougnez C., Schinzel A. C., Harview C. L., Brunet J. P., Ahmann G. J., Adli M., Anderson K. C., Ardlie K. G., Auclair D., Baker A., Bergsagel P. L., et al. (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balasse E. O., and Féry F. (1989) Ketone body production and disposal: effects of fasting, diabetes, and exercise. Diabetes Metab. Rev. 5, 247–270 [DOI] [PubMed] [Google Scholar]
- 12. McPherson P. A., and McEneny J. (2012) The biochemistry of ketogenesis and its role in weight management, neurological disease and oxidative stress. J. Physiol. Biochem. 68, 141–151 [DOI] [PubMed] [Google Scholar]
- 13. Morris A. A. (2005) Cerebral ketone body metabolism. J. Inherit. Metab. Dis. 28, 109–121 [DOI] [PubMed] [Google Scholar]
- 14. Cotter D. G., Schugar R. C., and Crawford P. A. (2013) Ketone body metabolism and cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 304, H1060–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia S., Lin R., Jin L., Zhao L., Kang H. B., Pan Y., Liu S., Qian G., Qian Z., Konstantakou E., Zhang B., Dong J. T., Chung Y. R., Abdel-Wahab O., Merghoub T., et al. (2017) Prevention of dietary-fat-fueled ketogenesis attenuates BRAF V600E tumor growth. Cell Metabol. 25, 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hegardt F. G. (1999) Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem. J. 338, 569–582 [PMC free article] [PubMed] [Google Scholar]