Abstract
Small ubiquitin-like modifier (SUMO) conjugation is a reversible post-translational modification process implicated in the regulation of gene transcription, DNA repair, and cell cycle. SUMOylation depends on the sequential activities of E1 activating, E2 conjugating, and E3 ligating enzymes. SUMO E3 ligases enhance transfer of SUMO from the charged E2 enzyme to the substrate. We have previously identified PIASy, a member of the Siz/protein inhibitor of activated STAT (PIAS) RING family of SUMO E3 ligases, as essential for mitotic chromosomal SUMOylation in frog egg extracts and demonstrated that it can mediate effective SUMOylation. To address how PIASy catalyzes SUMOylation, we examined various truncations of PIASy for their ability to mediate SUMOylation. Using NMR chemical shift mapping and mutagenesis, we identified a new SUMO-interacting motif (SIM) in PIASy. The new SIM and the currently known SIM are both located at the C terminus of PIASy, and both are required for the full ligase activity of PIASy. Our results provide novel insights into the mechanism of PIASy-mediated SUMOylation. PIASy adds to the growing list of SUMO E3 ligases containing multiple SIMs that play important roles in the E3 ligase activity.
Keywords: nuclear magnetic resonance (NMR), small ubiquitin-like modifier (SUMO), SUMO-interacting motif (SIM), sumoylation, UBC9, PARP1, PIASy, SIM, SUMO, TopoIIa
Introduction
Covalent attachment of small ubiquitin-like modifier (SUMO)3 to target proteins is an important post-translational modification that regulates multiple cellular processes including transcription, nuclear transport, DNA repair, and chromosome segregation (1, 2). Vertebrates express at least three functional SUMO isoforms, namely SUMO-1, SUMO-2, and SUMO-3, that can conjugate to other proteins. SUMO-1 shares less than 50% sequence identity to other isoforms, whereas SUMO-2 and SUMO-3 are virtually equivalent with 97% sequence identity and are commonly referred to as SUMO-2/3 (1–3). Similar to ubiquitylation, SUMO proteins are typically conjugated to the target lysine residue of the substrate by the sequential action of three enzymes: an E1 activating enzyme (Uba2/Aos1), an E2 conjugating enzyme (Ubc9), and an E3 ligase. A SUMO E3 ligase stabilizes and optimally positions the thioester-charged E2-SUMO complex to catalyze the transfer of SUMO from charged E2 to the substrate (2–5).
SUMOylation utilize the same E1 and E2 enzymes; however, different classes of SUMO-specific E3 ligases have been identified (2, 3). Among these are the protein inhibitor of activated STAT (PIAS) protein family and its yeast homolog Siz (4), RanBP2 (5, 6), Pc2 polycomb protein (7), and ZNF451 (8, 9). We have previously shown that PIASy, a member of the PIAS protein family, is essential for SUMO-2/3 conjugation of mitotic chromosomal proteins DNA topoisomerase IIα (TopoIIα) and poly(ADP-ribose) polymerase 1 (PARP1) and demonstrated that PIASy can efficiently catalyze SUMOylation of these substrates in reconstituted reactions as well (10–12).
Most known SUMO E3 ligases possess one or more SUMO-interacting motifs (SIMs) that mediate non-covalent interactions with SUMO (2, 3, 5–9, 13). SIMs are required for the E3 ligase activity and play an important role in positioning the donor SUMO from the E2-SUMO complex for efficient SUMO discharge and catalysis. SIMs consist of a core sequence of three or four hydrophobic aliphatic residues (isoleucine, leucine, and valine) with an acidic or polar residue at position 2 or 3, forming a triad or tetrad core sequence of (V/I)X(V/I)(V/I) or (V/I)(V/I)X(V/I/L) (where X is usually Asp, Glu, or Ser) (6, 14–16). The hydrophobic core of SIM is often flanked by a stretch of acidic amino acids that promote electrostatic SUMO-SIM interactions (14, 17). Structural studies of SUMO-SIM complexes have demonstrated that SIM peptides adopt a parallel or antiparallel β-strand conformation upon binding to SUMO in a hydrophobic groove formed by the β2-strand and α1-helix (5, 14, 17, 18).
The PIAS and Siz families of SUMO E3 ligases (2, 4, 10, 13) contain several conserved regions (see Fig. 1A) such as an N-terminal SAP (scaffold-associating region/Acinus/PIAS) domain involved in binding AT-rich DNA, a PINIT domain important for substrate recognition and subcellular localization, an SP-RING needed for interaction with the E2 Ubc9, and a C-terminal SIM needed for binding SUMO (2, 3, 13). Currently, PIASy is known to contain one SIM at the C-terminal region (4, 13). We report here the identification of a second SIM in PIASy, positioned between the SP-RING and the previously known SIM. NMR spectroscopy and mutational analysis revealed the significance of the new SIM in PIASy-mediated SUMO conjugation.
Figure 1.
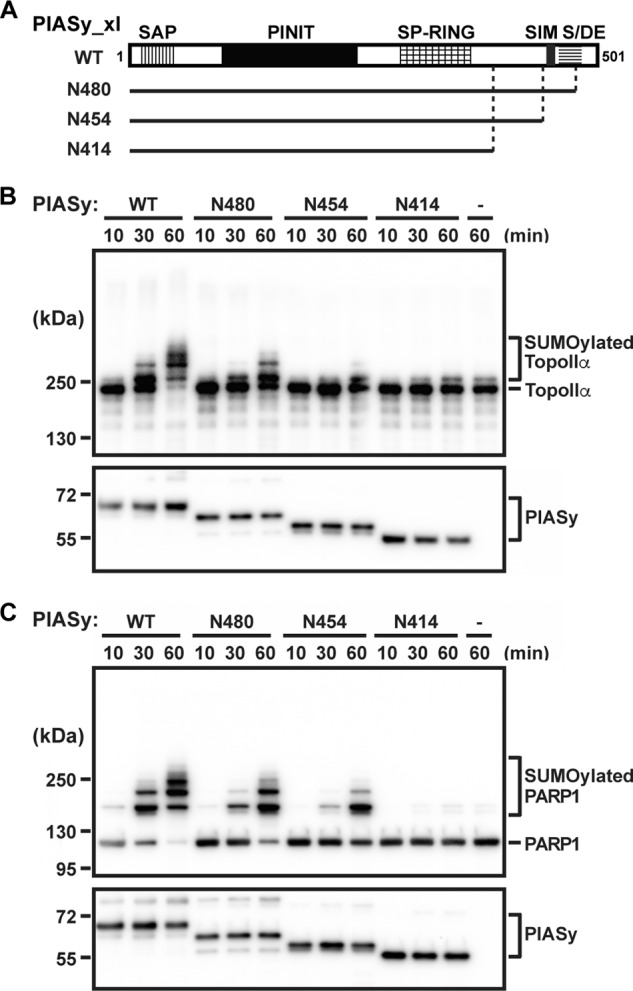
In vitro SUMO E3 ligase activity of C-terminal deletions of PIASy. A, schematic diagram showing the conserved domains of the Siz/PIAS family of E3 ligases. Solid lines indicate the PIASy truncation constructs tested in the SUMOylation assay. The time courses for in vitro SUMOylation of TopoIIα (B) and PARP1 (C) using WT PIASy and various truncation constructs of PIASy (N480, N454, and N414) as well as in the absence of PIASy (−) are shown. SUMOylated and unmodified forms of TopoIIα and PARP1 are indicated as are the various PIASy constructs used.
Results
SUMO ligase activity of C-terminal truncations of PIASy
We have previously demonstrated the crucial role of PIASy in SUMO-2/3 modification of mitotic substrates TopoIIα and PARP1 (10–12). Our initial deletion analysis of PIASy has shown that both N- and C-terminal regions of PIASy are essential for its activity (10). Although we demonstrated that the N terminus is required for localization of PIASy at the mitotic centromeres (19), the role of the C terminus of PIASy is unclear. To evaluate the role of the C-terminal domain of PIASy in SUMOylation, we tested C-terminal deletions of PIASy (Fig. 1A) in the reconstituted in vitro SUMOylation assay (11). At physiological concentration of the SUMOylation enzymes, PIASy N414 (a fragment lacking the 87 C-terminal residues) was inefficient in SUMOylating both TopoIIα and PARP1 (Fig. 1, B and C). The extent of SUMO conjugation using N414 was similar to the SUMOylation observed in the absence of PIASy (depicted by comparable higher shifted bands with N414 and no PIASy for both TopoIIα and PARP1; Fig. 1, B and C, last two lanes). Next, we examined a longer PIASy construct, N454, that lacked the currently known SIM (here after referred as original SIM) but retained all the components previously reported as sufficient for the ligase activity of Siz1, the PIAS homolog in yeast (4, 20). Surprisingly, N454 lacking the original SIM was capable of SUMOylating both the mitotic substrates (Fig. 1, B and C) albeit at a lower efficiency compared with full-length PIASy (WT). Consistent with the previous observation in Siz1 (4), inclusion of the original SIM and the flanking acidic domain (PIASy N480) further enhanced the SUMO conjugation (Fig. 1, B and C).
PIASy lacking the original SIM interacts with SUMO
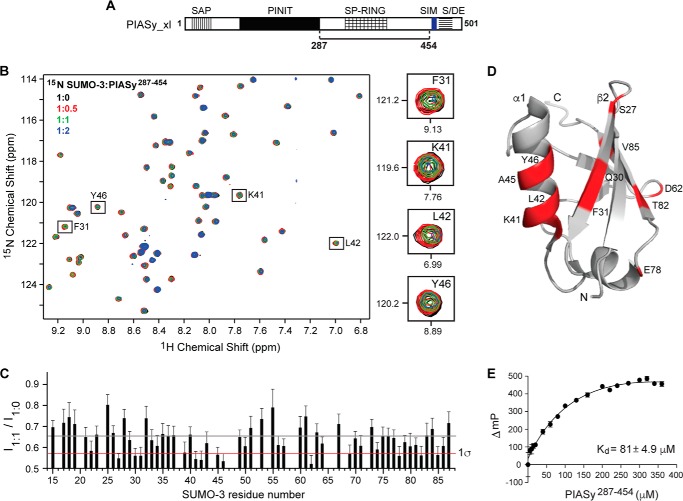
Both PIASy N414 and N454 could recruit E2 Ubc9 through the SP-RING domain; however, only N454 stimulated in vitro SUMO conjugation as an active ligase (Fig. 1). Previous studies have revealed that efficient E3-mediated SUMO conjugation requires optimal positioning of both E2 and SUMO from the E2-SUMO thioester complex (4, 5, 8, 9, 21). To determine whether the original SIM-less N454 orients the E2-SUMO complex for productive catalysis by additional stabilization of SUMO, we tested the direct binding between SUMO and the original SIM-less PIASy using NMR spectroscopy. A shorter construct, PIASy(287–454) (Fig. 2A), was more amenable for NMR studies due to better expression compared with N454 and was thus used in the NMR titrations. 15N-Labeled SUMO-3 was titrated with increasing concentrations of unlabeled PIASy(287–454) at 1:0, 1:0.5, 1:1, and 1:2 molar ratios, and the titration was monitored by acquiring 2D 1H-15N HSQC spectra (Fig. 2B). The stepwise addition of PIASy(287–454) resulted in peak broadening of specific resonances of SUMO-3 (Fig. 2B), indicating interaction in the intermediate exchange NMR time scale. To identify the SUMO-3 residues that were perturbed the most by PIASy(287–454), we calculated the peak intensity ratio (I1:1/I1:0) for each non-overlapped SUMO-3 peak at a SUMO-3:PIASy(287–454) molar ratio of 1:1 (Fig. 2C). Residues displaying significant peak intensity reduction (average intensity minus the standard deviation) included Ser-27, Gln-30, Phe-31, Lys-41, Leu-42, Ala-45, Tyr-46, Asp-62, Glu-78, and Thr-82. The affected residues were mapped onto the structure of SUMO-3 (Fig. 2D). Surprisingly, even though PIASy(287–454) lacks the original SIM, the majority of the affected residues clustered together spatially in the β2-strand and α1-helix of SUMO-3, which have been well characterized as the binding surface for the SIM on SUMO (6, 14, 17, 18). Our NMR results suggested the existence of another SIM in PIASy within residues 287–454.
Figure 2.
Analysis of the interaction of SUMO-3 with PIASy lacking the original SIM. A, PIASy schematic as in Fig. 1A with the solid line indicating the fragment, residues 287–454, used in the NMR and FP experiments. B, 2D 1H-15N HSQC spectra of 15N-labeled SUMO-3 titrated with increasing molar ratios of PIASy(287–454). Expanded sections of representative SUMO-3 residues affected upon binding of PIASy(287–454) are shown on the right. C, plot of relative peak intensity for all assigned, non-overlapping SUMO-3 resonances in the ligand-bound versus -free form (I1:1/I1:0). Gray and red lines depict mean and one standard deviation from the mean (1σ), respectively. D, SUMO-3 residues displaying significant peak intensity reduction upon complex formation with PIASy(287–454) are highlighted red. E, fluorescence polarization binding assay of fluorescein maleimide-labeled SUMO-3 titrated with increasing concentrations of PIASy(287–454). ΔmP, change in millipolarization. Error bars represent S.D.
Furthermore, results of a fluorescence polarization binding assay using SUMO-3 labeled with fluorescein maleimide and PIASy(287–454) showed an apparent Kd of 81 μm (Fig. 2E). The value obtained is consistent with the observed intermediate exchange NMR behavior and previously reported dissociation constants for other SUMO-SIM interactions, which are in the 1–120 μm range (6, 9, 14, 18, 22).
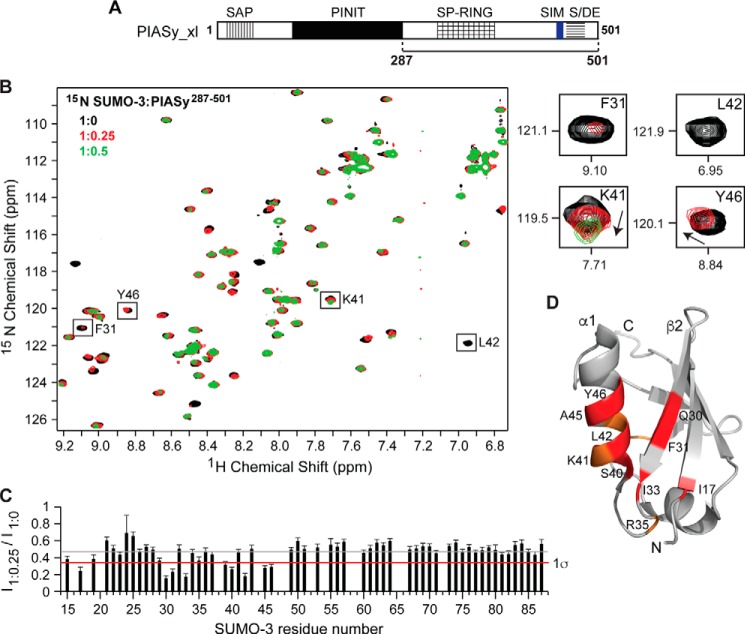
We also titrated a longer construct containing the original SIM, PIASy(287–501), into 15N-labeled SUMO-3 (Fig. 3A). As expected, incorporation of the original SIM and the acidic domain resulted in more pronounced chemical shift perturbations when compared with results of the NMR titrations using the shorter PIASy(287–454) (Fig. 3B). Stepwise addition of PIASy(287–501) caused a concentration-dependent decrease in intensities of SUMO-3 peaks. Some SUMO-3 residues showed changes in peak positions in addition to reduced peak intensities (e.g. Arg-35, Lys-41, Tyr-46, and Asn-67), and several peaks broadened beyond detection (Fig. 3B). Although most SUMO-3 residues showed some degree of line broadening from the beginning of the titration, which is not uncommon with the formation of a high molecular mass complex (SUMO-3, 13 kDa; PIASy(287–501), 28 kDa) slowing down the tumbling rate (23), specific residues were more affected than the others as illustrated by a peak intensity ratio graph (Fig. 3C). The SUMO-3 residues strongly affected by PIASy(287–501) were identical to the residues affected by PIASy(287–454) and mapped to the β2-strand and α1-helix, which form the SIM-binding surface on the SUMO (Fig. 3D).
Figure 3.
NMR titrations of SUMO-3 and PIASy(287–501) containing both original and new SIMs. A, schematic of PIASy with the solid bar indicating the fragment 287–501 used in the experiment. B, 2D 1H-15N HSQC spectra of 15N-labeled SUMO-3 titrated with increasing amounts of PIASy(287–501). Expanded sections of selected SUMO-3 residues affected by PIASy(287–501) are shown on the right. C, relative peak intensity graph for SUMO-3 resonances in the ligand-bound versus -free state (I1:0.25/I1:0) where gray and red lines correspond to the mean and one standard deviation from the mean (1σ), respectively. D, the SUMO-3 residues affected upon binding of PIASy(287–501) are highlighted (red, significant peak intensity reduction; orange, significant change in the peak position). Error bars represent S.D.
Identification of a new SIM in PIASy
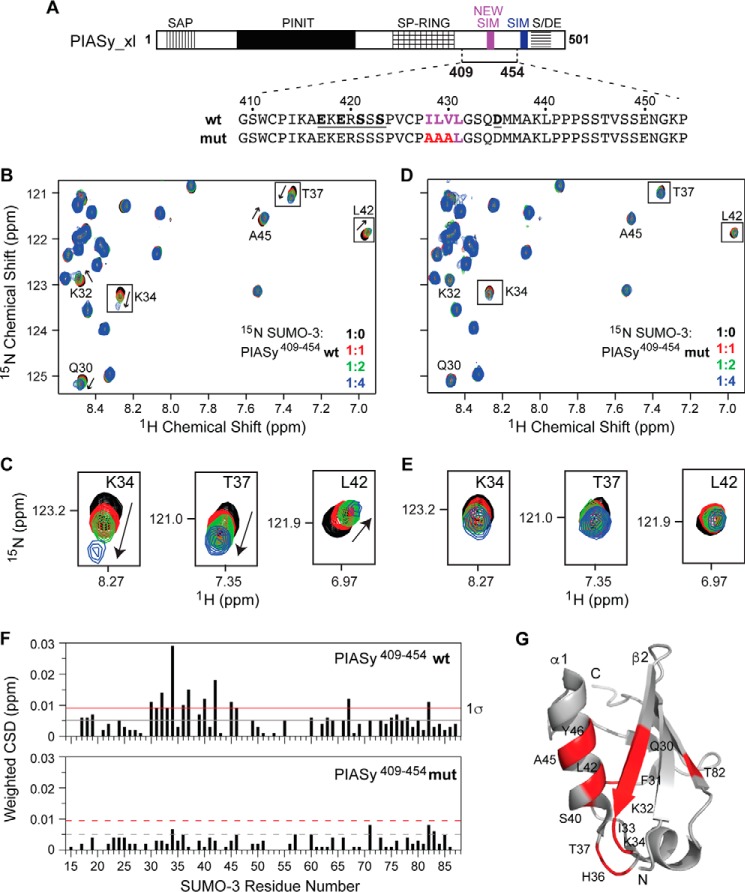
NMR titrations of 15N-labeled SUMO-3 with PIASy(287–454), which lacked the original SIM (Fig. 2), also perturbed the same SUMO-3 residues that a SIM peptide would, suggesting that an additional SIM may be present within the PIASy(287–454) fragment. Furthermore, a SIM prediction web server (24) suggested that there is a high probability that the PIASy(287–454) fragment contains a SIM (herein termed as new SIM) within residues 417–435 (Fig. 4A). The new SIM harbors a conserved hydrophobic core with the sequence “ILVL” from amphibian to mammalian PIASy and “VVDI” in fish PIASy and is preceded by a few acidic and serine residues, which are potential sites for phosphorylation (Fig. 4A). These features of the new SIM are similar to the characteristic known SIM sequences that bind SUMO in antiparallel orientation (9, 14, 15, 22). However, unlike known SIMs, the new SIM lacks a long stretch of acidic residues. (The acidic stretch in PIAS proteins varies from 5 to 7 residues, but the acidic stretch in PIASy is exceptionally long with 17 residues following the known SIM.)
Figure 4.
Characterization of the binding of SUMO-3 and the new SIM of PIASy. A, PIASy schematic as in Fig. 1A with new SIM shown in magenta. The solid bar corresponds to the PIASy fragment 409–454 (which contains the new SIM but lacks the original SIM) used in the experiment. The sequences of the WT and mut PIASy construct are shown. B, a section of the 2D 1H-15N HSQC spectra of 15N-labeled SUMO-3 titrated with increasing molar ratios of WT PIASy(409–454) demonstrates specific SUMO-SIM binding. C, expanded sections of selected SUMO-3 residues affected upon binding of WT PIASy(409–454). D and E, similar to B and C for SUMO-3 titrations with mut PIASy(409–454). F, plots of weighted CSD from titrations of SUMO-3 with WT PIASy(409–454) (top) and mut PIASy(409–454) (bottom). Gray and red lines correspond to the mean and one standard deviation (1σ) from the mean, respectively. G, SUMO-3 residues that are strongly affected by the binding of WT PIASy(409–454) are colored red.
To confirm the role of the new SIM in SUMO binding, we subcloned and expressed a 45-residue peptide encompassing the PIASy region containing the new SIM fragment (PIASy(409–454) WT; Fig. 4A). We also mutated the three critical ILV hydrophobic residues of the new SIM into triple alanines (PIASy(409–454) mut; Fig. 4A). Both WT and mut PIASy(409–454) showed greater stability and solubility in comparison with the longer PIASy fragments used for NMR that contained the SP-RING and were thus ideal for NMR studies. NMR titrations using 15N-labeled SUMO-3 with either WT or mut PIASy(409–454) were used to test the role of the ILV hydrophobic residues in the new SIM in binding SUMO-3. The WT fragment resulted in concentration-dependent changes in peak positions of SUMO-3 resonances (Fig. 4, B and C), indicating PIASy(409–454) WT interaction with SUMO-3 at the fast exchange NMR time scale. In contrast, NMR titrations with the mut fragment barely induced any discernible chemical shift perturbations on SUMO-3 (Fig. 4, D and E), indicating that mutating the ILV residues of the new SIM into triple alanines interfered with SUMO-3 interaction. Analysis of the weighted chemical shift deviations (CSDs; Fig. 4F) identified that the strongly affected SUMO-3 residues upon binding of PIASy(409–454) WT are clustered near the known SIM-binding groove of SUMO-3 (Fig. 4G). These results highlight the importance of the ILV hydrophobic residues of the new SIM in PIASy-SUMO recognition.
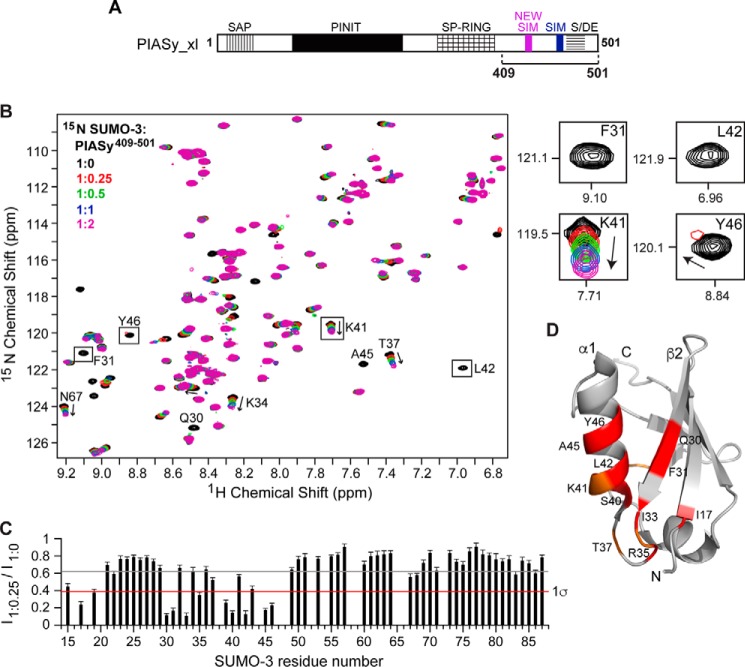
We also titrated 15N-labeled SUMO-3 with a longer construct, PIASy(409–501), which contains both the new SIM and the original SIM plus the acidic domain (Fig. 5A). Similar to the earlier titrations, identical SIM-binding residues of SUMO-3 were most strongly perturbed upon the addition of PIASy(409–501) (Fig. 5, B, C, and D). Comparison of the results of NMR titrations of SUMO-3 with PIASy(409–501) (where both SIMs are present) and PIASy(409–454) (where only the new SIM is present) indicated that the presence of both SIMs conferred a higher magnitude of change in the chemical shift perturbations of specific SUMO-3 resonances even at a lower ligand concentration. Important SUMO-3 SIM-binding residues (such as Gln-30, Phe-31, Leu-42, Ala-45, and Tyr-46) showed severe peak intensity reduction starting at 1:0.25 molar ratios and complete peak broadening at a 1:0.5 molar ratio (Fig. 5, B and C), indicating binding in the intermediate exchange NMR time scale.
Figure 5.
NMR titrations of SUMO-3 and PIASy(409–501) containing both original and new SIMs. A, schematic of PIASy indicating fragment 409–501 containing both the original SIM and the new SIM used in the experiment. B, 2D 1H-15N HSQC spectra of 15N-labeled SUMO-3 titrated with increasing amounts of PIASy(409–501). Expanded sections of representative SUMO-3 residues affected by the binding of PIASy(409–501) are on the right. C, plot of relative peak intensity for SUMO-3 resonances in the ligand-bound versus -free form (I1:0.25/I1:0). Gray and red lines depict mean and one standard deviation from the mean (1σ), respectively. D, the SUMO-3 residues affected upon complex formation with PIASy(409–501) are highlighted (red, residues displaying significant peak intensity reduction; orange, residues with significant change in the peak position). Error bars represent S.D.
The new SIM is crucial in PIASy-mediated SUMOylation
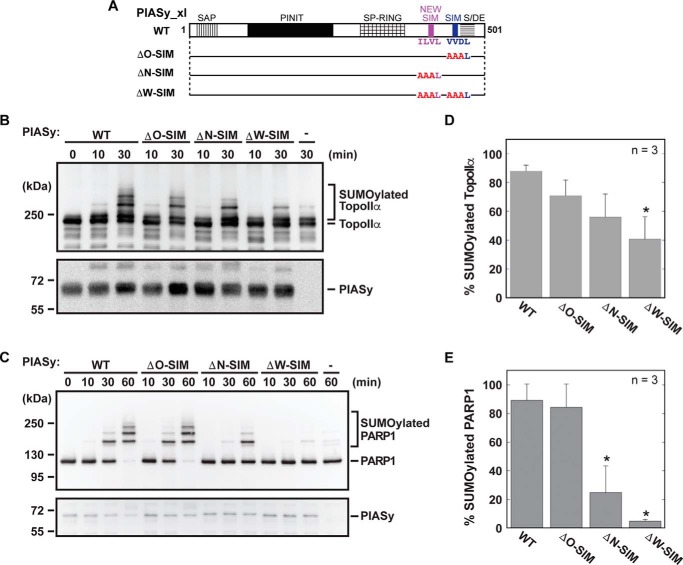
To evaluate the significance of the new and original SIMs for the E3 ligase activity of PIASy, we mutated the three essential core residues of the new and original SIMs into alanines (Fig. 6A). Three full-length PIASy constructs with multiple alanine mutations were introduced in the original SIM (construct ΔO-SIM; Fig. 6A), the new SIM (construct ΔN-SIM), and both new and original SIMs (construct ΔW-SIM). The mutants were examined for their ability to facilitate SUMOylation of TopoIIα and PARP1 in the in vitro SUMOylation assay (Fig. 6, B and C). Mutation of the hydrophobic core of original SIM (ΔO-SIM) did not interfere with the E3 ligase role of PIASy and exhibited high SUMOylation levels close to the wild-type PIASy for both TopoIIα and PARP1 (Fig. 6, B, C, D, and E). However, mutation of the new SIM (ΔN-SIM) resulted in less efficient SUMOylation of TopoIIα (Fig. 6, B and D) and significantly low SUMOylation of PARP1 compared with the WT PIASy (Fig. 6, C and E). Furthermore, mutation of both the original and new SIMs (ΔW-SIM) significantly reduced SUMOylation of TopoIIα and nearly abolished the E3 ligase activity of PIASy on PARP1 (Fig. 6, D and E). Overall, both TopoIIα and PARP1 showed a similar trend in SUMOylation, displaying a reproducible reduction in SUMOylation with the ΔN-SIM and the ΔW-SIM PIASy mutants in comparison with the WT PIASy. However, the observed effects were more distinct for PARP1 compared with TopoIIα (largely due to the ability of TopoIIα to undergo SUMOylation even in the absence of PIASy). Our results suggest that the new SIM plays an important role in PIASy-mediated SUMOylation, and both SIMs together are needed for the full ligase activity of PIASy.
Figure 6.
Functional role of the new SIM of PIASy. A, schematic of PIASy with solid bars illustrating the various SIM site-directed mutations examined in the SUMOylation assay. The time courses for SUMOylation of TopoIIα (B) and PARP1 (C) using the indicated PIASy SIM mutant in the in vitro SUMO conjugation assay are shown. SUMOylated and unmodified forms of TopoIIα and PARP1 along with the input PIASy are indicated. Quantification of the SUMOylation of TopoIIα (D) and PARP1 (E) at the final time point of the assay is shown. Assays were performed in triplicate. Error bars represent one standard deviation, and an asterisk indicates statistically significant differences from the WT activity.
Discussion
Results of our NMR titrations and in vitro SUMOylation assays revealed a new SIM in PIASy that was found to be essential for efficient PIASy-dependent SUMO-2/3 conjugation. In contrast to the original SIM of PIASy (or the known C-terminal SIM of other PIAS family members), the new SIM contains an uninterrupted core of four hydrophobic residues and lacks the conventional extended acidic residue stretch (Fig. 4). Previous studies of SUMO-SIM interaction have identified the importance of the hydrophobic core of SIM in SUMO binding (6, 14, 16). Consistent with these findings, our NMR results showed that hydrophobic core residues of the new SIM are essential in binding to SUMO-3, and mutation of these critical residues abolishes binding (Fig. 4). Others have shown the role of the acidic stretch surrounding the hydrophobic core of SIM in SUMO paralog binding specificity (14). Although presence of the negative charges in SIM has been shown to contribute more toward SUMO-1 binding, SIMs lacking the acidic stretch preferentially bind SUMO-2 (7, 14). The absence of the acidic stretch in the new SIM, together with the essential role of the new SIM in PIASy-mediated SUMOylation (Fig. 6), provides a likely explanation for the preference of PIASy in conjugation of SUMO-2/3 over SUMO-1 to substrates TopoIIα and PARP1 (10–12).
To address the question of why there are two SIMs in PIASy, the following observations are instructive. First, in addition to the covalent interaction between the E2 conjugating enzyme Ubc9 and SUMO to form the E2-SUMO thioester complex, the backside of Ubc9 also recruits a second molecule of SUMO through non-covalent interactions to form the E2-SUMO complex (25, 26). Second, the E3 ligase ZNF451 exploits these interactions to preferentially interact with two molecules of SUMO, the donor SUMO from E2-SUMOD and the backside E2-SUMO, via its dual SIMs (8, 9). In ZNF451, the first SIM plus the inter-SIM region positions the E2-SUMOD thioester complex for productive catalysis, whereas the second SIM acts as an anchor to bind to the backside SUMO to facilitate poly-SUMOylation (8, 9). Finally, although PIAS proteins use the SP-RING domain to directly bind to E2 Ubc9, others have shown that the (original) SIM of PIAS proteins also interacts with the backside E2-SUMO to form a non-covalent ternary complex (27). Combining our discovery of the new SIM in PIASy with the above observations (8, 9, 25–27), we propose a working hypothesis that, analogously to ZNF451, the dual SIMs of PIASy may also interact simultaneously with the donor SUMO (E2-SUMOD) and the backside SUMO (E2-SUMO) to mediate efficient SUMOylation.
Our results can be interpreted in line with this working hypothesis. Our NMR titrations with a PIASy construct containing both the new and original SIMs showed improved SUMO-binding affinity (Fig. 5) compared with a shorter construct consisting only of the new SIM (Fig. 4). Even with small amounts of PIASy(409–501) (containing dual SIMs), specific SUMO-3 SIM-interacting residues including Gln-30, Phe-31, Leu-42, Ala-45, and Tyr-46, were in intermediate exchange (where peaks were broadened) (Fig. 5); however, with PIASy(409–454) (containing the new SIM alone), these residues were in the fast exchange NMR time scale (Fig. 4), suggesting relatively weaker binding compared with PIASy(409–501). Because both SIMs individually bind to the same surface on SUMO-3 through classical SUMO-SIM interactions (27), the stronger NMR shift perturbations observed with PIASy(409–501) are probably reflective of a synergistic effect occurring upon the binding of different SUMO-3 molecules to the tandem SIM.
Additionally, our in vitro SUMOylation assays confirmed that PIASy stimulated SUMO conjugation in a SIM-dependent manner and that both SIMs are important for effective ligase activity. Although mutation of the new SIM (ΔN-SIM) rendered SUMOylation slow and inefficient, probably implying its role in the donor SUMO (E2-SUMOD) positioning and chain initiation, mutation of both SIMs together (ΔW-SIM; Fig. 6) or deletion of the dual SIM region (N414; Fig. 1) nearly eliminated the PIASy E3 ligase activity. In concordance, the presence of both SIMs together resulted in the robust ligase activity of PIASy (N480; Fig. 1). Our results suggest a hypothesis where the SP-RING domain and the new SIM of PIASy may configure the E2-SUMOD thioester complex, and the original SIM may engage the backside SUMO for efficient catalysis and SUMO conjugation by PIASy. To summarize, we have identified a new SIM in PIASy that is critical for the efficient ligase activity of PIASy and distinct from other members of the PIAS family that contain a single SIM at their C terminus.
Experimental procedures
Molecular cloning
Xenopus laevis PIASy sharing 85% sequence similarity to the human homolog (10) was used in these studies. The expression plasmids for N-terminal His6-tagged human SUMO-2-GG, SUMO-3-GG, and Xenopus full-length PIASy have been described previously (10, 28). C-terminal truncations of PIASy (N480, N454, and N414; Fig. 1) and PIASy NMR constructs (PIASy(287–454) and PIASy(287–501); Figs. 2 and 3) were made by PCR amplification from the full-length construct and subcloned into pET28a using the EcoRI and XhoI restriction sites. PIASy fragments 409–454 and 409–501 (Figs. 4 and 5) were obtained by digesting PIASy(287–454) and PIASy(287–501) in pET28a with BamHI/XhoI, respectively, and ligated into pET28a vector. PIASy constructs with mutations in the SIM region (PIASy(409–454) mut, ΔO-SIM, ΔN-SIM, and ΔW-SIM; Figs. 4 and 6) were made using a QuikChange mutagenesis kit (Stratagene). All mutations were verified by DNA sequencing.
Protein expression and purification
For protein expression, plasmids were freshly transformed in Escherichia coli BL21(DE3) cells and grown in 1 liter of culture medium containing kanamycin at 30 μg/ml. 15N-Labeled SUMO-3 was expressed in M9 minimal medium supplemented with 1 g of [15N]ammonium chloride. Unlabeled SUMO-3 (for fluorescence polarization studies) and PIASy fragments (for NMR studies) were obtained by cell growth in LB medium. PIASy constructs containing the RING domain were also supplied 0.1 mm ZnSO4 before and after induction. Cells were grown at 37 °C until A600 was ∼0.7–0.8 and induced with 0.5 mm isopropyl β-d-thiogalactopyranoside, and cell growth was continued overnight in a 15 °C shaker incubator to a final A600 ∼2.5 (∼2.1 for PIASy constructs). Cells were harvested by centrifugation (4000 rpm for 10 min), resuspended in ∼30 ml of binding buffer (500 mm NaCl, 20 mm Tris-HCl, pH 8.0, 5 mm imidazole), and lysed by sonication. Cellular debris was removed by centrifugation (13,000 rpm for 10 min), and 600 μl of 5% (v/v) polyethyleneimine was added to the supernatant to precipitate the nucleic acids. After another centrifugation (13,000 rpm for 10 min), the supernatant was loaded to a precharged Ni2+ affinity column (Gold Biotechnology). The column was washed with 100 ml of binding buffer and eluted with 50 ml of elution buffer (500 mm NaCl, 20 mm Tris-HCl, pH 8.0, 250 mm imidazole). Elution fractions containing purified protein were pooled and dialyzed in NMR buffer (100 mm NaCl, 10 mm NaPO4, pH 6.8, 5 mm DTT). Proteins were concentrated using Amicon Ultra 3000 centrifugal filter (Millipore), and concentrations were determined by absorbance at A280. Recombinant proteins used in the SUMOylation assays, the E1 complex (Uba2/Aos1), E2 Ubc9, PIASy (full length and truncation and mutant constructs), PARP1, TopoIIα, and SUMO-2 were expressed and purified as described previously (10–12, 28).
In vitro SUMOylation assay
SUMOylation reactions were performed as described previously (11, 12). Briefly, 5 μm SUMO-2, 2 mm ATP, 20 nm E1, 30 nm E2, 20 nm PIASy, and 500 nm T7-tagged PARP1 or TopoIIα in reaction buffer (20 mm HEPES, pH 7.8, 100 mm NaCl, 5 mm MgCl2, 0.05% Tween 20, 5% glycerol, 0.5 mm DTT) were incubated at 25 °C for 60 min. Although at a lower efficiency, TopoIIα can be SUMOylated in the absence of PIASy, and this effect becomes more prominent with longer reaction times (12, 19). Therefore, to better understand the contribution of SIMs of PIASy in TopoIIα SUMOylation, TopoIIα reactions with the SIM mutants were incubated for up to 30 min (Fig. 6B). Aliquots were taken from the reaction at various time points, mixed with SDS-PAGE loading buffer, and boiled. Samples were resolved by SDS-PAGE and analyzed by Western blotting using HRP-conjugated anti-T7 monoclonal antibody.
NMR spectroscopy
NMR data were acquired at 25 °C on a Bruker Avance III 600-MHz spectrometer equipped with a TXI-RT probe. Data processing and analysis were carried out using NMRPipe (29) and NMRView (30), respectively. For NMR chemical shift mapping, 2D 1H-15N HSQC spectra were acquired using 0.2 mm 15N-labeled SUMO-3 titrated with increasing molar ratios of unlabeled PIASy. All titration samples contained 10% (v/v) D2O. The published backbone amide assignment of SUMO-2/3 was used in the analysis (14, 31). The weighted CSD was calculated using 0.5[(ΔH)2 + (ΔN/5)2]1/2 (32). Residues perturbed during the titrations were mapped onto the NMR structure of SUMO-3 (Protein Data Bank code 1U4A) (31).
Fluorescence polarization (FP)
Purified SUMO-3 and PIASy(287–454) were dialyzed into 1× PBS buffer (phosphate-buffered saline, pH 7.4) overnight. The single native cysteine of SUMO-3 (residue 47) was utilized to covalently attach the fluorophore, fluorescein maleimide (FM) following the manufacturer's protocol (Invitrogen). Briefly, a 20-fold molar excess of FM prepared in dimethyl sulfoxide was added to the tris(2-carboxyethyl)phosphine-reduced protein and allowed to react in the dark at room temperature for ∼3 h. The unbound excess dye was removed by extensive dialysis in PBS followed by passage through an Amicon Ultra 3000 centrifugal filter. Increasing concentrations of PIASy (0–360 μm) were titrated into 25 nm FM-labeled SUMO-3. Samples were incubated at room temperature for 30 min, and FP was measured with excitation and emission wavelengths of 492 and 515 nm, respectively, using a Varian Cary Eclipse fluorescence spectrophotometer. G-factor-corrected polarization values were obtained using the following formula.
| (Eq. 1) |
Background fluorescence was subtracted from the average of five data points, and the change in millipolarization (ΔmP) was plotted against increasing concentrations of PIASy. The dissociation constant (Kd) was estimated by non-linear hyperbolic fitting of the data using GraphPad Prism 5.0.
Author contributions
K. K. and Y. A. designed, performed, and interpreted the majority of the experiments. H. P. and N. P. assisted in some of the SUMOylation experiments. R. N. D. G. helped in designing and interpreting NMR experiments. K. K. drafted the manuscript, and everyone contributed in revising the manuscript.
Acknowledgments
We thank Asokan Anbanandam for assistance with NMR spectroscopy and Mark Richter for the use of the fluorescence spectrophotometer. The University of Kansas Biomolecular NMR Core Facility was supported by Grant P30-GM110761 from the National Institutes of Health.
This work was supported by National Institutes of Health Grants RO1GM080278-S1 (to Y. A.) and AI074856 (to R. N. D. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- PIAS
- protein inhibitor of activated STAT
- TopoIIα
- DNA topoisomerase IIα
- PARP1
- poly(ADP-ribose) polymerase 1
- HSQC
- heteronuclear single quantum correlation
- mut
- mutant
- CSD
- chemical shift deviation
- FP
- fluorescence polarization
- FM
- fluorescein maleimide
- PINIT
- proline isoleucine asparagine isoleucine threonine
- SAP
- scaffold-associating region/Acinus/PIAS
- SP
- Siz/PIAS.
References
- 1. Meulmeester E., and Melchior F. (2008) Cell biology: SUMO. Nature 452, 709–711 [DOI] [PubMed] [Google Scholar]
- 2. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geiss-Friedlander R., and Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 4. Yunus A. A., and Lima C. D. (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reverter D., and Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., and Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merrill J. C., Melhuish T. A., Kagey M. H., Yang S.-H., Sharrocks A. D., and Wotton D. (2010) A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One 5, e8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eisenhardt N., Chaugule V. K., Koidl S., Droescher M., Dogan E., Rettich J., Sutinen P., Imanishi S. Y., Hofmann K., Palvimo J. J., and Pichler A. (2015) A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 22, 959–967 [DOI] [PubMed] [Google Scholar]
- 9. Cappadocia L., Pichler A., and Lima C. D. (2015) Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 22, 968–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Azuma Y., Arnaoutov A., Anan T., and Dasso M. (2005) PIASy mediates SUMO-2 conjugation of topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S. P., Dasso M., and Azuma Y. (2010) PIASy Mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 285, 14415–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryu H., Furuta M., Kirkpatrick D., Gygi S. P., and Azuma Y. (2010) PIASy-dependent SUMOylation regulates DNA topoisomerase II activity. J. Cell Biol. 191, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., and Palvimo J. J. (2009) PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66, 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hecker C. M., Rabiller M., Haglund K., Bayer P., and Dikic I. (2006) Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 15. Jardin C., Horn A. H., and Sticht H. (2015) Binding properties of SUMO-interacting motifs (SIMs) in yeast. J. Mol. Model. 21, 50. [DOI] [PubMed] [Google Scholar]
- 16. Song J., Zhang Z., Hu W., and Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 17. Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., and Shirakawa M. (2008) Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 18. Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H.-Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J., Naik N. M., Kung C. C., Lin S. Y., Chen R. H., Chang K. S., et al. (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 [DOI] [PubMed] [Google Scholar]
- 19. Ryu H., and Azuma Y. (2010) Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J. Biol. Chem. 285, 32576–32585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi Y., and Kikuchi Y. (2005) Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J. Biol. Chem. 280, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 21. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., and Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Namanja A. T., Li Y.-J., Su Y., Wong S., Lu J., Colson L. T., Wu C., Li S. S., and Chen Y. (2012) Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J. Biol. Chem. 287, 3231–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williamson M. P. (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 [DOI] [PubMed] [Google Scholar]
- 24. Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., and Ren J. (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knipscheer P., van Dijk W. J., Olsen J. V., Mann M., and Sixma T. K. (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26, 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Capili A. D., and Lima C. D. (2007) Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 369, 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mascle X. H., Lussier-Price M., Cappadocia L., Estephan P., Raiola L., Omichinski J. G., and Aubry M. (2013) Identification of a non-covalent ternary complex formed by PIAS1, SUMO1, and UBC9 proteins involved in transcriptional regulation. J. Biol. Chem. 288, 36312–36327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azuma Y., Arnaoutov A., and Dasso M. (2003) SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 30. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 31. Ding H., Xu Y., Chen Q., Dai H., Tang Y., Wu J., and Shi Y. (2005) Solution structure of human SUMO-3 C47S and its binding surface for Ubc9. Biochemistry 44, 2790–2799 [DOI] [PubMed] [Google Scholar]
- 32. Grzesiek S., Bax A., Clore G. M., Gronenborn A. M., Hu J. S., Kaufman J., Palmer I., Stahl S. J., and Wingfield P. T. (1996) The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 3, 340–345 [DOI] [PubMed] [Google Scholar]