Abstract
Sirtuin 2 (SIRT2) is a sirtuin family deacetylase, which maintains genome integrity and prevents tumorigenesis. Although Sirt2 deficiency in mice leads to tumorigenesis, the functional significance of somatic SIRT2 mutations in human tumors is unclear. Using structural insight combined with bioinformatics and functional analyses, we show that naturally occurring cancer-associated SIRT2 mutations at evolutionarily conserved sites disrupt its deacetylation of DNA-damage response proteins by impairing SIRT2 catalytic activity or protein levels but not its localization or binding with substrate. We observed that these SIRT2 mutant proteins fail to restore the replication stress sensitivity, impairment in recovery from replication stress, and impairment in ATR-interacting protein (ATRIP) focus accumulation of SIRT2 deficiency. Moreover, the SIRT2 mutant proteins failed to rescue the spontaneous induction of DNA damage and micronuclei of SIRT2 deficiency in cancer cells. Our findings support a model for SIRT2's tumor-suppressive function in which somatic mutations in SIRT2 contribute to genomic instability by impairing its deacetylase activity or diminishing its protein levels in the DNA-damage response. In conclusion, our work provides a mechanistic basis for understanding the biological and clinical significance of SIRT2 mutations in genome maintenance and tumor suppression.
Keywords: acetylation, cancer, DNA, DNA damage, DNA damage response, genomic instability, sirtuin, tumor suppressor gene
Introduction
Sirtuin 2 (SIRT2)2 is a class III NAD+-dependent deacetylase, which regulates a broad range of biological functions, including aging, metabolism, differentiation, genome maintenance, and tumor suppression (1–6). SIRT2 overexpression prolongs longevity in mice hypomorphic for BubR1 (7), and mice deficient in Sirt2 develop breast, liver, and other cancers (8–10), suggesting that SIRT2 functions in both aging and tumor suppression. Indeed SIRT2 expression is decreased in a number of cancers, including human breast, liver, glioma, renal, prostate, lung, uterine, and basal cell carcinomas (8, 11–16), and moreover SIRT2 is mutated or deleted in ovarian, adenoid cystic, cervical, uterine, lung, pancreatic, gastric, esophageal, colorectal, liver, melanoma, testicular germ cell, thyroid, and breast cancers (17, 18) Genetic loss of Sirt2 in mice results in genomic instability (8, 9), suggesting that SIRT2 prevents tumorigenesis at least in part by protecting cells against DNA damage and thereby promoting the DNA-damage response (DDR). SIRT2 depletion in cells results in hypersensitivity to replication stress, spontaneous induction of DNA damage, delayed S phase progression following replication stress, and an impaired G2/M checkpoint (9, 18–20). SIRT2 deacetylates ATR-interacting protein (ATRIP) at lysine (Lys) 32 to drive the ATR checkpoint by facilitating binding to replication protein A-coated single-stranded DNA (18) and deacetylates cyclin-dependent kinase 9 (CDK9) at Lys-48 to promote its activity in facilitating recovery from replication stress (18). SIRT2 also promotes the mitotic checkpoint through deacetylation of α-tubulin at Lys-40 (21, 22), anaphase-promoting complex/cyclosome co-activators CDH1 and CDC20 (23), histone H4 at Lys-16 and PR-Set7 at Lys-90 (9), and BubR1 at Lys-250 (24). Thus, SIRT2 is a critical regulator of the DDR in maintaining genome integrity. Although Sirt2 deficiency in mice leads to genomic instability and tumorigenesis and SIRT2 is often dysregulated in human cancers, the functional significance of somatic SIRT2 mutations in cancer is not known. Here, we show that naturally occurring cancer-associated SIRT2 mutations contribute to genomic instability of cancer cells by impairing its deacetylase activity or protein level in the DDR.
Results
Cancer-associated SIRT2 mutations are evolutionarily conserved and predicted to be functionally significant
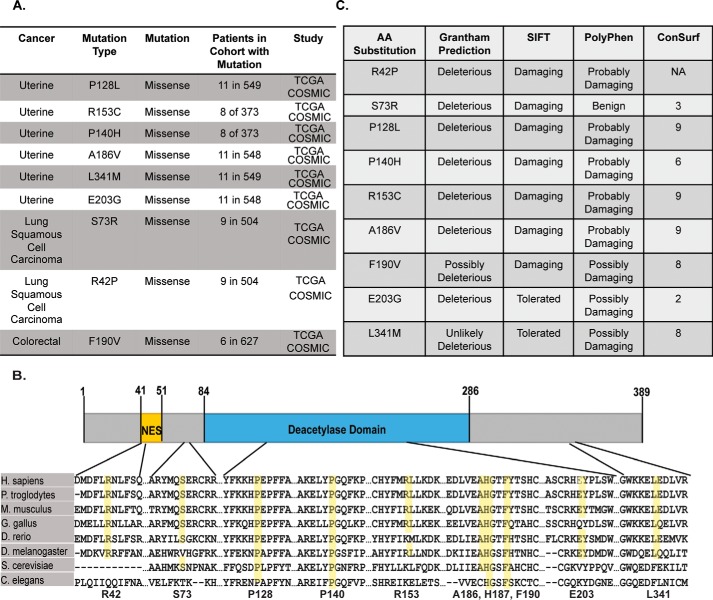
To determine the functional significance of somatic SIRT2 mutations in human cancers, we analyzed sequencing data from patient-derived tumor samples reported in The Cancer Genome Atlas through the cBioPortal for Cancer Genomics (16, 25). The Cancer Genome Atlas currently contains 70 SIRT2 missense mutations and five SIRT2 truncation mutations across multiple cancer types, including breast, cervical, colorectal, head and neck, lung, stomach, testicular germ cell, thyroid, and uterine cancer, most of which overlap with the 66 somatic SIRT2 mutations found in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (17). We focused our investigation on nine SIRT2 mutations present in uterine, lung, and colorectal cancer, which were found in both data sets, predicted to be high impact mutations by cBioPortal, and occurred at appreciable frequencies within their study cohort (Fig. 1A). All mutations selected were missense mutations, occurred throughout the length of the protein, and were highly conserved across distantly related species (Fig. 1B). To validate the potential importance of these mutations, we performed in silico bioinformatics analyses for functional significance using several conservation and impact prediction platforms, including Grantham, Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping (PolyPhen), and ConSurf (Fig. 1C) (26–34). Grantham scores predict the evolutionary distance between substituted amino acids and thus the potential for missense mutations to be deleterious or neutral. This analysis was completed using the Align-GVGD program through the Huntsman Cancer Institute at the University of Utah. SIFT and ConSurf predictions are based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences collected through Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) (35–37). ConSurf determines the conservation of an amino acid and rates it on a scale of 1–9 with 9 indicating highly conserved. PolyPhen predicts the possible impact of missense mutations on the structure and function of a given human protein by comparing physical and alignment-based comparisons. Consistently, mutations P128L, P140H, R153C, A186V, and F190V were found to be highly conserved and deleterious if mutated, and notably all are located within the SIRT2 deacetylase domain. R42P was also well conserved and predicted to have a high impact on SIRT2 function with its localization within the nuclear export sequence (NES) of SIRT2 (38, 39). Collectively, these data indicate that cancer-associated SIRT2 mutations are evolutionarily conserved and are predicted to be functionally significant, suggesting that they are more likely to be driver mutations that benefit cancer progression rather than random carrier mutations.
Figure 1.
Cancer-associated SIRT2 mutations are evolutionarily conserved and predicted to be functionally significant. A, table listing cBioPortal data for nine naturally occurring SIRT2 mutations in human cancer samples along with occurrence in patient cohort, cancer type, and study type. B, conservation of cancer-associated SIRT2 mutation amino acids among diverse species, including chimpanzee, mouse, chicken, zebrafish, fruit fly, yeast, and nematode, and their location in the protein structure. The NES is in yellow, and the deacetylase domain is highlighted in blue. C, impact prediction of cancer mutations by Grantham, SIFT, and PolyPhen, and their conservation scores as calculated by ConSurf. TCGA, The Cancer Genome Atlas; COSMIC, Catalogue of Somatic Mutations in Cancer.
Cancer-associated mutations impair SIRT2 in vitro deacetylase activity and protein level but not localization
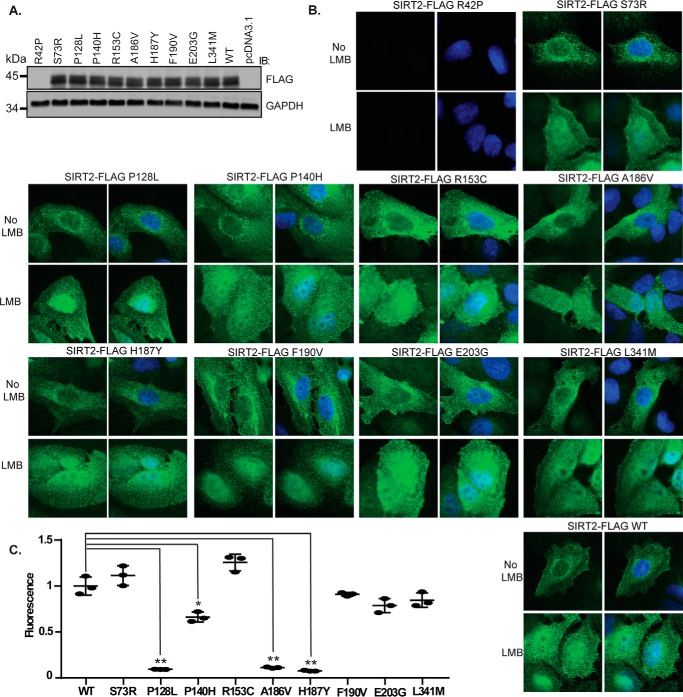
Each of the cancer-associated SIRT2 mutations was created by site-directed mutagenesis in a SIRT2-FLAG construct and expressed in 293T cells along with SIRT2 wild type (WT) and H187Y deacetylase-inactive mutant controls. All of the mutants were expressed at comparable levels to SIRT2 WT except for R42P, which showed lack of protein expression similar to an empty vector (Fig. 2A), suggesting that R42P affects either protein stability or mRNA expression. SIRT2 shuttles between the nucleus and cytoplasm through active nuclear export in a CRM1-dependent manner (38). To determine whether cancer-associated mutations are important for SIRT2 localization, we examined localization of SIRT2 WT and mutants expressed in U2OS cells before and after treatment with leptomycin B (LMB), which inhibits CRM1-mediated nuclear export, by indirect immunofluorescence. All of the mutants expressed localized predominantly in the cytoplasm and demonstrated nuclear retention following treatment of cells with LMB (Fig. 2B), suggesting that SIRT2 cancer mutants are capable of proper cytoplasmic and nuclear localization. To determine whether cancer-associated mutations are important for SIRT2 deacetylase activity, we performed in vitro measurements using the Fluor De Lys in vitro deacetylase assay by Enzo Life Sciences, which uses an acetylated p53 Lys-320 peptide as a substrate. Three of the mutants, SIRT2 P128L, P140H, and A186V, demonstrated significantly decreased deacetylase activity compared with SIRT2 WT with SIRT2 P128L and A186V impairing deacetylase activity similarly to catalytically inactive SIRT2 H187Y (Fig. 2C). All three of these mutations are located within the deacetylase domain of SIRT2 and likely affect NAD+ cofactor and/or substrate binding or catalytic activity.
Figure 2.
Cancer-associated mutations impair SIRT2 deacetylase activity and protein level but not localization. A, Western blot analysis demonstrating protein levels of SIRT2-FLAG WT and mutants expressed in U2OS cells. B, U2OS cells were transfected with SIRT2-FLAG WT or mutants. 72 h after transfection, cells were incubated with or without 5 nm LMB for 4 h to inhibit SIRT2 nuclear export, fixed, and processed for indirect immunofluorescence using anti-FLAG antibodies (green) and DAPI staining. Representative images are shown. C, in vitro measurements of SIRT2 deacetylase activity using the Fleur de Lys deacetylase assay are shown. SIRT2-FLAG WT and mutants were purified from 293T cells and eluted from FLAG M2 beads. Protein concentration was measured by Coomassie staining with a BSA standard control. 120 ng of SIRT2-FLAG WT and mutants was incubated in a reaction with an acetylated p53 Lys-320 peptide as a substrate. The mean of the relative fluorescence measured from three replicas is shown. Error bars represent S.D. Deacetylase activities of mutants were compared with that of SIRT2-FLAG WT. *, p < 0.05; **, p < 0.01. IB, immunoblotting.
Structural analysis of SIRT2 mutations yields insights into their functional significance
To gain further insight into how cancer-associated mutations might affect SIRT2 biological function, we examined the crystal structure of SIRT2 obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank, specifically structure 4RMH (40). We first focused on four mutants of interest based on their predicted impact scores, deacetylase activity, and structural location: P128L, P140H, A186V, and F190V. All four were predicted to negatively impact protein function, and three of these demonstrated decreased deacetylase activity in vitro. All four were also located in the active site of SIRT2 (Fig. 3A). Pro-128 and Pro-140 make up the ends of an important α helix responsible for composing part of the active site for NAD+ binding. Mutation of these prolines could therefore alter the orientation of the helix and result in decreased binding of NAD+ and thus lower deacetylase activity. Pro-140 is also important for the formation of the binding pocket for NAD+ along with Phe-190; these two amino acids stack on either side of NAD+ (Fig. 3A, blue molecule). Mutating either of these amino acids therefore would also likely result in decreased binding of NAD+ and lower deacetylase activity. F190V, however, does not yield a deacetylase-defective phenotype (Fig. 2C), suggesting that this mutation alone is not sufficient to inhibit cofactor binding. Ala-186 lies adjacent to the active site amino acid His-187 and thus could affect its neighboring amino acid's ability to deacetylate substrate (Fig. 3, B and C). In fact, Ala-186 lies in a tight pocket with little space for amino acid structure variation as better illustrated in the space-filling model in Fig. 3C. Even the small addition of two methyl groups is likely enough to blow out this pocket and upset the orientation of His-187, thereby inhibiting deacetylation of substrate. Overall, these four amino acids are heavily involved in proper deacetylase activity and/or cofactor binding. The other five mutations for this study lie outside of the active site and binding pocket of SIRT2. Only four of these five amino acids (Glu-203, Arg-153, Leu-341, and Ser-73) have been verified for orientation and structure as the current crystal structures of SIRT2 lack a portion of the N terminus where Arg-42 lies (Fig. 3D). Of these amino acids, three are on the surface of the protein where they potentially interact with other proteins (Glu-203 and Arg-153) and/or perhaps undergo post-translational modification (Ser-73). Therefore, mutation of these amino acids to non-compatible substitutes could upset other aspects of SIRT2 function regulated by protein-protein interaction but that are not necessary for deacetylase activity. The fourth amino acid, Leu-341, is hydrophobic and forms a portion of an α helix. Mutation of this amino acid therefore could lead to a change in structure of the helix and upset the surface structure without affecting SIRT2 deacetylase activity. Fig. 3E shows a close-up of the two types of amino acids discussed, one surface amino acid likely involved in protein-protein interaction (Arg-153) and the other a hydrophobic amino acid serving a more structural purpose (Leu-341). Fig. 3F shows a hypothetical structure and orientation of Arg-42 produced using the PyMOL Molecular Graphics System, version 1.8 (Schrödinger, LLC) that may be involved in protein-protein interaction because it lies on the protein surface and is in a more open portion of the protein. Arg-42 also lies within the NES and may be necessary for proper nuclear export (38, 39).
Figure 3.
Structural analysis of SIRT2 mutations yields insights into their functional significance. A, of the nine mutations selected from cBioPortal, four are found in either the NAD+-binding pocket or the catalytic domain. Mutations P128L, P140H, and F190V create the NAD+-binding pocket. NAD+ can be seen in blue, and the amino acids of interest are in red. Ala-186 lies next to the catalytic amino acid His-187. The protein structure was obtained from the Structural Bioinformatics Protein Data Bank, specifically structure 4RMH (40). B, a close-up of Ala-186 in a non-space-filling model. C, a close-up of Ala-186 in a space-filling model. D, of the nine mutations selected from cBioPortal, four are located in non-catalytic domains of the SIRT2 protein structure. These mutations are highlighted in purple. E, a close-up of two of the four mutations from D reveals they are mainly surface or hydrophobic amino acids. F, hypothetical crystal model of amino acid Arg-42.
Cancer-associated mutations impair SIRT2 deacetylation of DDR substrates in vitro and in cells but not interaction with substrate
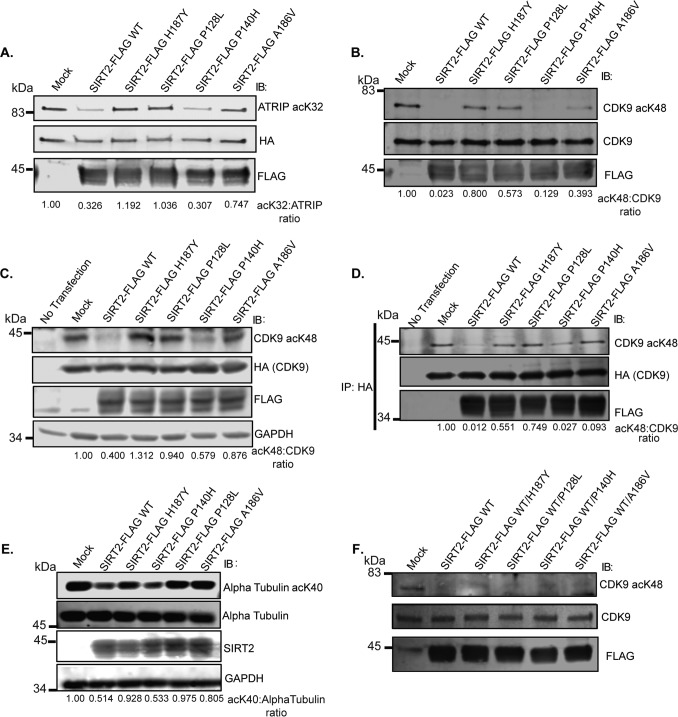
To determine whether the impairment in deacetylase activity of SIRT2 P128L, P140H, and A186V observed with an acetylated p53 Lys-320 peptide used in the Fluor De Lys assay is substrate-specific, we tested for their ability to deacetylate additional SIRT2 substrates involved in the DDR in vitro. Consistent with findings using an acetylated p53 Lys-320 peptide, SIRT2-FLAG P128L showed the greatest impairment in in vitro deacetylase activity compared with SIRT2-FLAG WT when using acetylated ATRIP Lys-32 and CDK9 Lys-48 as substrates (Fig. 4, A and B). SIRT2-FLAG P140H and A186V failed to show the same degree of impairment in deacetylation of ATRIP Lys-32 and CDK9 Lys-48 (Fig. 4, A and B) compared with p53 Lys-320 (Fig. 2C), suggesting that these mutants are less impactful on SIRT2 deacetylase activity and/or impair deacetylation of specific substrates. To determine whether cancer-associated mutations impair the deacetylation of DDR substrates in cells, we expressed SIRT2-FLAG WT and mutants in 293T cells and determined their ability to deacetylate CDK9-HA and endogenous α-tubulin. Again, SIRT2-FLAG P128L but not P140H demonstrated significant impairment in deacetylation of CDK9 Lys-48 and α-tubulin Lys-40 in cells (Fig. 4, C and E). Interestingly, SIRT2 A186V also demonstrated decreased deacetylase activity against these substrates in cells (Fig. 4, C and E), implying that A186V impairs SIRT2 deacetylase activity for these substrates to a greater degree in cells potentially due to factors present in cells but not in vitro conditions. To determine whether the impairment in deacetylation of CDK9 Lys-48 in cells by SIRT2 P128L and A186V may be due to differences in binding with substrate, we analyzed their interaction by co-IP. No differences in co-IP of CDK9-HA with SIRT2-FLAG WT and mutants was observed despite impairment in deacetylation of co-immunoprecipitated CDK9-HA by SIRT2-FLAG P128L and A186V (Fig. 4D), providing strong evidence that the impairment in deacetylation of CDK9 Lys-48 by SIRT2-FLAG P128L and A186V mutants is not due to an impairment in binding with substrate. As the majority of cancer-associated SIRT2 mutations are in a heterozygous state in patients, to determine whether they exert dominant negative effects on SIRT2 WT deacetylase activity, we performed an in vitro deacetylase assay in which acetylated CDK9-GFP was incubated with SIRT2-FLAG WT alone or with equal amounts of both SIRT2-FLAG WT and one of four SIRT2-FLAG mutants. We observed no significant difference in deacetylation of CDK9 Lys-48 by SIRT2-FLAG WT and mutants compared with SIRT2-FLAG WT alone in this in vitro deacetylase assay (Fig. 4F), suggesting that SIRT2 mutants do not exert a dominant negative effect on SIRT2 WT activity in this context.
Figure 4.
Cancer-associated mutations impair SIRT2 deacetylation of DDR substrates in vitro and in cells but not interaction with substrate. Shown are representative Western blots of several replicas. A, acetylated ATRIP was isolated from 293T cells transfected with HA-ATRIP and histone acetyltransferases (HATs) pCAF, p300, and CBP and incubated in an in vitro deacetylation assay with SIRT2-FLAG WT or mutants isolated from 293T cells and in the presence of TSA with or without NAD and nicotinamide. The reaction mixtures were separated by SDS-PAGE and immunoblotted (IB) with site-specific anti-acetyl ATRIP Lys-32, HA, and FLAG antibodies. B, acetylated CDK9 was isolated from 293T cells transfected with CDK9-GFP and HATs and incubated in an in vitro deacetylation assay with SIRT2-FLAG WT or mutants isolated from 293T cells and in the presence of TSA with or without NAD and nicotinamide. The reaction mixtures were separated by SDS-PAGE and immunoblotted with site-specific anti-acetyl CDK9 Lys-48, HA, and FLAG antibodies. C, 293T cells were transfected with CDK9-HA and HATs or were left untransfected. After 24 h, cells were subsequently transfected with SIRT2-FLAG WT or mutants and incubated in the presence of TSA for 24 h. Deacetylation of CDK9-HA was assessed by Western blot analysis using site-specific anti-acetyl CDK9 Lys-48, HA, FLAG, and GAPDH antibodies. D, immunoprecipitation of samples from Fig. 2C. 293T cells were transfected with CDK9-HA and HATs or were left untransfected. After 24 h, cells were subsequently transfected with SIRT2-FLAG WT or mutants and incubated in the presence of TSA for 24 h. Lysate were immunoprecipitated with an anti-HA antibody, separated by SDS-PAGE, and immunoblotted with site-specific anti-acetyl CDK9 Lys-48, HA, and FLAG antibodies. E, 293T cells were transfected with HATs. After 24 h, cells were subsequently transfected with SIRT2-FLAG WT or mutants and incubated in the presence of TSA for 24 h. Deacetylation of endogenous α-tubulin was assessed by Western blot analysis using site-specific anti-acetyl-α-tubulin Lys-40, α-tubulin, SIRT2, and GAPDH antibodies. F, acetylated CDK9 was isolated from 293T cells transfected with CDK9-GFP and HATs and incubated in an in vitro deacetylation assay without SIRT2 (Mock), with SIRT2-FLAG WT only, or with equal amounts of both SIRT2-FLAG WT and one of four SIRT2-FLAG mutants isolated from 293T cells (SIRT2-FLAG H187Y, P128L, P140H, or A186V) in the presence of TSA with NAD+. The reaction mixtures were separated by SDS-PAGE and immunoblotted with site-specific anti-acetyl CDK9 Lys-48, CDK9, and FLAG antibodies.
Cancer-associated SIRT2 mutations fail to rescue replication stress response (RSR) defects of SIRT2 deficiency
To determine whether cancer-associated mutations are important for the RSR activities of SIRT2, we analyzed recovery from replication arrest in HCT-116 SIRT2-depleted cells complemented with SIRT2-FLAG WT and mutants (Fig. 5A). Ten hours following release from HU-induced replication arrest, SIRT2-depleted cells expressing SIRT2-FLAG P128L, P140H, and A186V showed a similar impairment in recovery from replication arrest to SIRT2-depleted cells expressing deacetylase-inactive H187Y compared with WT (Fig. 5, B and C). SIRT2-depleted cells expressing SIRT2-FLAG P128L, P140H, and A186V also showed comparable HU hypersensitivity to H187Y compared with WT (Fig. 5D), suggesting that P128L, P140H, and A186V impair SIRT2's activity in responding to replication stress. Moreover, SIRT2-FLAG P128L and A186V but not P140H failed to alleviate the impairment in HU-induced GFP-ATRIP foci of SIRT2 deficiency to a similar level as SIRT2-FLAG WT (Fig. 5, E and F), suggesting that these mutants impair the recruitment of ATRIP to stalled replication forks.
Figure 5.
Cancer-associated SIRT2 mutations fail to rescue RSR defects of SIRT2 deficiency. A, Western blot analysis demonstrating efficiency of SIRT2 and ATR knockdown and expression of SIRT2-FLAG WT and mutants in HCT-116 cells. B, quantitation of cells with 4N DNA content following 10-h release from HU treatment in cells transfected with NS, ATR, or SIRT2-10 UTR siRNA with or without complementation with SIRT2-FLAG WT or mutants. The mean from two replicas is shown, and error bars represent S.D. NS indicates p ≥ 0.05; *, p < 0.05; **, p < 0.01. C, HCT116 cells were transfected with NS, ATR, or SIRT2-10 UTR siRNA, treated with 3 mm HU for 24 h (arrested), and released into nocodazole for 10 h (released). SIRT2–10 UTR knockdown was complemented with SIRT2-FLAG WT and mutants. DNA content was analyzed by flow cytometry. Representative cell cycle profiles are shown. D, HU sensitivity as measured by alamarBlue cell viability staining of HCT116 cells transfected with NS, siATR, or SIRT2-10 UTR siRNA with or without complementation with SIRT2-FLAG WT or mutants and treated with 1.6 mm HU for 24 h followed by a 24-h release. The mean of HU-treated to untreated viability relative to NS siRNA in triplicate is shown, and error bars represent S.D. E, quantitation of cells with >5 GFP-ATRIP foci following 24-h 3 mm HU treatment in a GFP-ATRIP stable U2OS cell line transfected with NS or SIRT2-5 siRNA with or without complementation with EV (pcDNA3.1) or SIRT2-FLAG WT or mutants (SIRT2-FLAG WT constructs contained wobble mutations to protect against knockdown). The mean from three replicas of 100 cells counted in each is shown, and error bars represent S.D. NS indicates p ≥ 0.05; *, p < 0.05; **, p < 0.01. F, representative images of conditions quantified and outlined in E with GFP-ATRIP in green, FLAG in red, and DAPI. IB, immunoblotting.
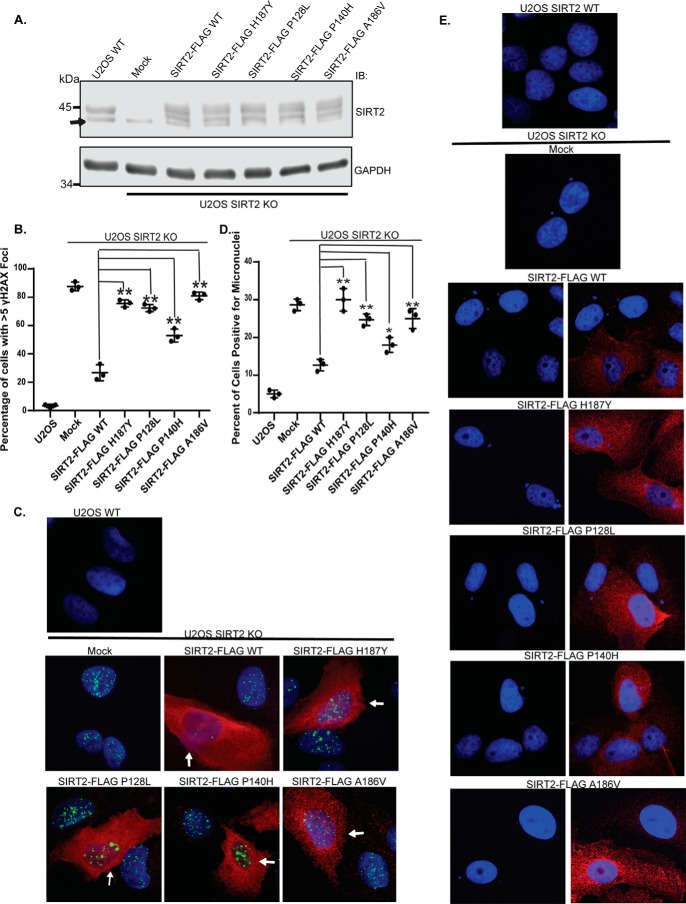
Cancer-associated SIRT2 mutations fail to rescue genomic instability of SIRT2 deficiency
To determine whether cancer-associated mutations contribute to impairment in genome stability, we generated U2OS SIRT2 knock-out (KO) cells utilizing CRISPR/Cas9 and performed complementation experiments with expression of SIRT2-FLAG WT and mutants (Fig. 6A). U2OS SIRT2 KO cells demonstrated a significant increase in spontaneous γH2AX foci, a marker for DNA double-strand breaks, which can form from the collapse of stalled replication forks, compared with U2OS SIRT2 WT cells, and this was reduced by expression of exogenous SIRT2-FLAG WT (Fig. 6, B and C). Expression of SIRT2-FLAG P128L, P140H, and A186V all showed impairment in rescue of the spontaneous induction of spontaneous γH2AX foci observed with SIRT2 deficiency, suggesting that P128L, P140H, and A186V impair SIRT2's ability to maintain genome integrity. As a more direct measure for genomic instability, we analyzed induction of spontaneous micronuclei, a marker for chromosomal breaks, in U2OS SIRT2 KO cells complemented with SIRT2-FLAG WT and mutants. Expression of SIRT2-FLAG P128L, P140H, and A186V also failed to reduce the spontaneous micronuclei of SIRT2 deficiency to a similar extent as expression of SIRT2-FLAG WT (Fig. 6, D and E). Collectively, these data suggest that P128L, P140H, and A186V impair SIRT2's activity in the DDR, which leads to genomic instability.
Figure 6.
Cancer-associated SIRT2 mutations fail to rescue genomic instability of SIRT2 deficiency. A, U2OS SIRT2 KO cells generated by CRISPR/Cas9 were complemented with or without SIRT2-FLAG WT or mutants. Western blot analysis demonstrating efficiency of expression of SIRT2-FLAG WT and mutants in U2OS SIRT2 KO cells or endogenous SIRT2 in U2OS WT cells is shown. The arrow indicates a nonspecific band beneath the SIRT2 protein band. B, U2OS SIRT2 KO cells demonstrate increased spontaneous γH2AX foci and were complemented with or without SIRT2-FLAG WT or mutants. The degree of alleviation of spontaneous γH2AX foci observed was quantified. Quantitation of the percentage of cells with >5 spontaneous γH2AX foci is shown. The mean was calculated from three replicas of 100 cells for each condition, and error bars represent S.D. *, p < 0.05; **, p < 0.01. C, representative images of U2OS SIRT2 KO cells complemented with or without SIRT2-FLAG WT or mutants and stained for γH2AX foci (green), FLAG (red), and DAPI. U2OS SIRT2 KO cells with SIRT2 construct expression stain positive for FLAG (red), and examples of this are highlighted by white arrows, whereas cells in the same population that did not express SIRT2 construct do not exhibit red staining in the cytoplasm. D, U2OS SIRT2 KO cells were transfected with or without SIRT2-FLAG WT or mutants and stained for FLAG in red and DAPI. Induced micronuclei were counted. Quantitation of micronuclei is shown. The mean was calculated from three replicas of 100 cells for each condition, and error bars represent S.D. *, p < 0.05; **, p < 0.01. E, representative images of micronuclei conditions from D. IB, immunoblotting.
Discussion
Using structural insight combined with bioinformatics and functional analyses, we found that naturally occurring cancer-associated SIRT2 mutations at evolutionarily conserved sites disrupt its deacetylation of DDR proteins by impairing its catalytic activity or protein level but not localization or binding with substrate, demonstrating that cancer-associated SIRT2 mutations are functionally significant. We further found that SIRT2 mutant proteins fail to restore the replication stress sensitivity, impairment in recovery from replication stress, and spontaneous induction of DNA damage of SIRT2 deficiency in cancer cells, providing evidence that cancer-associated SIRT2 mutations impair genomic integrity of cancer cells. Thus, somatic SIRT2 mutations in human tumors contribute to genomic instability by impairing its deacetylase activity or protein level in the DDR, providing a model for the loss of the tumor suppressor function of SIRT2 in human tumors.
Of the original nine SIRT2 mutations selected, only four demonstrated functional significance by impacting either deacetylase activity or protein level. These four mutations are located within the deacetylase active site, NES, or NAD+ cofactor-binding site. R42P resulted in decreased protein levels and likely affects either protein stability or expression. P128L, P140H, and A186V resulted in decreased SIRT2 deacetylase activity but had no significant effect on SIRT2 protein levels or localization.
P128L showed the greatest deficit in SIRT2 deacetylase activity against all substrates tested, including ATRIP Lys-32, CDK9 Lys-48, α-tubulin Lys-40, and p53 Lys-320. In addition, P128L was unable to rescue the replication stress sensitivity, impairment in recovery from replication arrest, impairment in HU-induced ATRIP focus accumulation, and spontaneous induction of γH2AX foci and micronuclei of SIRT2 deficiency. Structural analyses indicate that this lack of activity is due to decreased binding of the SIRT2 cofactor NAD+. The P128L proline is essential to the proper orientation of the α helix involved in forming the binding pocket for NAD+ and for the formation of the stacking interaction of Pro-140 with NAD+. Consequently, its mutation would result in improper orientation/folding and therefore proper interaction with the essential cofactor for deacetylation reactions.
P140H appears to be partially substrate-specific as it was unable to deacetylate p53 Lys-320 in vitro to the same extent as SIRT2 WT but was able to deacetylate other known substrates CDK9 Lys-48, ATRIP Lys-32, and α-tubulin Lys-40 both in vitro and in cells. Consistently, P140H rescued the impairment in HU-induced ATRIP focus accumulation of SIRT2 deficiency to a comparable level as SIRT2 WT. However, P140H was unable to rescue the genome maintenance defects of SIRT2 deficiency, suggesting that its effect on the DDR activities of SIRT2 may be mediated through p53 or an untested substrate. Interestingly, even in the absence of exogenous damage, SIRT2-depleted cells complemented with SIRT2 P140H showed a significant increase in the percentage of cells in S and G2/M phases even compared with SIRT2 depletion alone, suggesting a gain of function in impairing cell cycle progression. Structural analyses indicate that this mutation impairs the ability of SIRT2 to bind NAD+ as it affects the orientation of the same α helix as P128L and is involved in a stacking interaction between Phe-190 and NAD+ in the cofactor-binding pocket. It is likely that this mutation is not as impactful as P128L in disrupting the binding of NAD+ to impair deacetylase activity for the majority of known SIRT2 substrates but may impart a gain of function in impairing cell cycle progression, which leads to increased genomic instability.
A186V also exhibited decreased deacetylase activity for p53 in vitro but only displayed decreased deacetylase activity on CDK9 Lys-48 and α-tubulin Lys-40 in cells but not on CDK9 Lys-48 or ATRIP Lys-32 in vitro. A186V was unable to rescue the deficiency in ATRIP focus formation in response to replication stress after SIRT2 knockdown to the same extent as SIRT2 WT and P140H likely due to its reduced deacetylase activity of RSR substrates in cells. Additionally, there was an increase in the percentage of cells in S and G2/M phases as seen with the P140H mutation as well as impairment in SIRT2 DDR phenotypes. Structural analyses reveal that Ala-186 lies adjacent to H187Y, which is an essential catalytic amino acid for deacetylation reactions. Specifically, Ala-186 lies in a narrow pocket that is size-restricting. The addition of methyl groups found in the valine amino acid structure would interfere with the structure of the deacetylation pocket and orientation of His-187, leading to decreased deacetylation of substrate. It is possible that this mutation may be more impactful on certain substrates in cells due to the presence of an unknown regulatory partner, for example CDK9 complexes with several regulatory cyclins in cells.
The other five mutations, S73R, R153C, F190V, E203G, and L341M, did not significantly affect SIRT2 protein levels, localization, or deacetylase activity. S73R and L341M lie outside of well defined functional SIRT2 protein domains, and although it is possible that S73R may affect protein function as a site of post-translational modification by phosphorylation, it was predicted to be a benign mutation by PolyPhen and ConSurf based on conservation and structural analyses. Leu-341 is a hydrophobic amino acid likely involved in maintenance of a C-terminal α helical structure and, although highly conserved, was predicted to be a tolerated mutation by SIFT and Grantham prediction. Its higher score on the ConSurf scale is likely due to its functionality in an α helix structure that seems to lack an essential role in SIRT2 deacetylase activity. R153C, F190V, and E203G lie in the SIRT2 deacetylase domain, but only F190V lies in the active site (specifically in the NAD+-binding pocket). Despite its seemingly essential placement in the stacking interaction with NAD+ and Pro-140, the mutation of Phe-190 alone was not sufficient to induce a deacetylase-deficient phenotype. R153C and E203G lie on the surface of the protein structure as opposed to inside the active site and are likely involved in protein-protein interactions that are not critical for SIRT2's ability to bind and deacetylate substrate.
Interestingly, we found no evidence that cancer-associated SIRT2 mutants exert a dominant negative effect on the deacetylase activity of SIRT2 WT using an in vitro deacetylase assay. However, it is possible that this could differ in a heterozygous state in human cancers in vivo.
In summary, our findings support a model for the tumor-suppressive function of SIRT2 in which somatic mutations in SIRT2 contribute to genomic instability by impairing its deacetylase activity or protein level in the DDR and provide a mechanistic basis for understanding the biological and clinical significance of SIRT2 mutations in genome maintenance and tumor suppression. In addition, as SIRT2 depletion in cancer cells results in hypersensitivity to replication stress caused by many types of cancer therapies (18–20), our findings provide insight into the rationale-driven design of targeted therapies against SIRT2 activity and potential resistance mechanisms in tumors that may be utilized for individualized cancer therapy.
Experimental procedures
Transfections
siRNAs were obtained from Thermo Scientific or Qiagen. Transfections were performed using Lipofectamine 2000 (Invitrogen) or RNAiMAX (Invitrogen) following the manufacturer's instructions. Individual siRNAs sequences include: 1) siSIRT2-10 UTR, TGGGCAGAAGACATTGCTTAT; 2) siSIRT2-5, GGAGAAAGCTGGCCAGTCG; 3) siATR, CCTCCGTGATGTTGCTTGA; and 4) nontargeting siRNA, ATGAACGTGAATTGCTCAATT.
Cell cycle recovery
HCT116 cells were transfected with ATR, NS, or SIRT2 siRNA and 24 h later with SIRT2-FLAG WT (Addgene 13813) (21) or mutants, treated with or without 3 mm HU 72 h postknockdown for 20 h (arrested) (41), and released into 0.5 μg/ml nocodazole (Fisher Scientific) for 10 h. Cells were harvested and fixed in ice-cold 70% ethanol, and DNA was stained with 25 μg/ml propidium iodide (Sigma-Aldrich) with 100 μg/ml RNase A (Qiagen). DNA content was measured by flow cytometry using a BD FACSCanto II flow cytometer and then analyzed by the FlowJo software gating analysis tool (Tree Star).
Immunofluorescence
For SIRT2 localization experiments, U2OS cells were transfected with SIRT2-FLAG WT or mutants, treated 72 h posttransfection with 5 nm leptomycin B (Sigma) for 4 h, fixed in 4% paraformaldehyde for 10 min, and permeabilized in 0.5% Triton X-100 for 10 min. Cells were blocked in 5% BSA and immunostained with anti-FLAG (Cell Signaling Technology, 2368P) and Alexa Fluor 555 anti-rabbit secondary antibody (Invitrogen) followed by DAPI stain (Southern Biotech). For GFP-ATRIP experiments, a stably transfected GFP-ATRIP U2OS cell line was used. Cells were transfected with nontargeting siRNA or siSIRT2-5 followed by transfection with pcDNA3.1 empty vector (EV) or SIRT2-FLAG WT or mutants. SIRT2-FLAG constructs contained wobble mutations to protect exogenous expression from knockdown. Cells were treated with 3 mm HU for 24 h prior to fixation, fixed, and blocked as described above. Cells were immunostained with anti-FLAG (Cell Signaling Technology, 2368P) followed by Alexa Fluor 555 secondary antibody (red) and DAPI Stain. GFP-ATRIP did not require antibody (green). SIRT2-FLAG-positive cells were used for quantitation of these experiments with the exception of EV-transfected conditions. For spontaneous γH2AX focus experiments, we utilized SIRT2 WT or KO U2OS cells transfected with or without SIRT2-FLAG WT or mutants (42). Cells were fixed and blocked as described above, immunostained with anti-FLAG (Cell Signaling Technology, 2368P) and anti-γH2AX (Millipore, 05-636) followed by Alexa Fluor 488/555 secondary antibodies and DAPI stain. SIRT2-FLAG-positive cells were used for quantitation of these experiments with the exception of non-SIRT2-transfected conditions. The percentage of cells positive for GFP-ATRIP or γH2AX foci was counted from three replicas of 100 cells each. Micronuclei experiments were fixed and processed as described above. Only SIRT2-FLAG-positive cells were used in quantitation analysis. The percentage of cells positive for micronuclei was counted from three replicas with 100 cells each. All images were captured on a Zeiss Observer Z1 microscope equipped with AxioVision Rel 4.8 software.
Immunoblotting
Harvested cells were lysed for 30 min on ice in Nonidet P-40 buffer (200 mm NaCl, 1% Nonidet P-40,50 mm Tris·HCl, pH 8.0) freshly supplemented with protease inhibitors. Protein samples were resolved by SDS-PAGE and probed with the indicated antibodies: SIRT2 (Santa Cruz Biotechnology, sc-20966), GAPDH (Santa Cruz Biotechnology, sc-25778 or sc-47724), FLAG (Sigma, F4042), CDK9 (Santa Cruz Biotechnology, sc-13130), GFP (Abcam, Ab6556), HA (Sigma, H9658), Ac-CDK9 Lys-48 (custom generated through Epitomics), Ac-tubulin (Sigma, T7451), tubulin (Sigma, T6074), Ac-ATRIP Lys-32 (custom generated through Epitomics), and ATR (Santa Cruz Biotechnology, sc-1887). Detection was performed with the Odyssey system. All Western blot analyses were performed at least four times.
Immunoprecipitation
Harvested cells were lysed for 40 min on ice in IP lysis buffer (0.75% CHAPS, 10% (v/v) glycerol, 150 mm NaCl, 50 mm Tris, pH 7.5) freshly supplemented with protease inhibitors. Supernatants were diluted to adjust the CHAPS concentration to 0.375%. Target proteins from 3 mg of lysate were captured with anti-GFP antibody (Abcam, Ab6556) and protein A-agarose beads (Invitrogen) or with HA-preconjugated beads (Sigma, A2095). Complexes were washed four times with IP washing buffer (0.375% CHAPS, 10% glycerol, 150 mm NaCl, 50 mm Tris, pH 7.5) supplemented with protease inhibitors. Immunoprecipitation experiments were performed at least four times.
Generation of CRISPR/Cas9 SIRT2 knock-out cells
U2OS/HCT116cells were transfected with plasmid containing guide RNAs targeting an early SIRT2 exon or a nontargeting control and Cas9-GFP construct (Sigma). 72 h after transfection, cells were harvested and sorted by FACS into 96-well plates with one cell per well based on a high level of GFP expression. Single cells that survived and gave rise to cell lines were tested for SIRT2 knock-out by Western blot analysis.
In vitro or cellular deacetylation experiments
For in vitro deacetylation analysis of CDK9 and ATRIP, 293T cells were transiently transfected with CDK9-GFP or HA-ATRIP and histone acetyltransferase (p300/CBP/pCAF) or SIRT2-FLAG WT or mutants and treated with 0.5 μm TSA and 20 mm nicotinamide for 12 h. Cells were lysed with IP buffer (20 mm Hepes, pH 7.4, 180 mm KCl, 0.2 mm EGTA, 1.5 mm MgCl2, 20% glycerol, 1.0% Nonidet P-40) supplemented with 1 μm TSA and fresh protease inhibitors. Acetyl-CD Lys-9 or acetyl-ATRIP were immunoprecipitated using anti-GFP (Abcam, Ab6556) and protein A-agarose beads (Invitrogen) or anti-HA-preconjugated beads (Sigma), washed with IP buffer containing 1 μm TSA to remove nicotinamide, and washed an additional two times with deacetylation buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm MgCl2). SIRT2-FLAG constructs were immunoprecipitated using anti-FLAG M2-agarose beads (Sigma), washed with IP buffer and TBS (50 mm Tris, pH 7.5, 150 mm NaCl), and eluted with 0.15 mg/ml 3× FLAG peptide (Sigma) for 30 min at 4 °C. Protein concentration was determined by Coomassie staining. Substrate on beads (HA-ATRIP or CDK9-GFP) was incubated with or without 1 μg of SIRT2-FLAG WT or mutants in 24 μl of deacetylation reaction buffer (1 μm TSA, 50 mm Tris·HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 1 mm NAD+) at 30 °C for 3 h. For the dominant negative assay, CDK9-GFP was incubated with 1 μg of SIRT2 WT or with 0.5 μg of SIRT2-WT and 0.5 μg of one of the four mutants tested. The reaction was stopped by the addition of 5× SDS loading buffer, and samples were analyzed for acetylation by Western blotting with anti-Ac-ATRIP Lys-32 (19–21) and Ac-CDK9 Lys-48 (18). For cellular deacetylation analysis, 293T cells were transiently cotransfected with CDK9-HA, histone acetyltransferases (p300/CBP/pCAF), and SIRT2-FLAG WT or mutants. Cells were cultured with 0.5 μm TSA for 12 h prior to being lysed with IP buffer containing 1 μm TSA, and protein lysates were immunoprecipitated using HA-preconjugated beads. The immunocaptured protein were analyzed for deacetylation by immunoblotting with anti-AcCDK9 K48, anti-HA, α-tubulin, and Ac-tubulin Lys-40. For the Flour de Lys in vitro experiments, SIRT2-FLAG WT and mutants were purified from 293T cells and eluted from FLAG M2 beads as described above. 120 ng of SIRT2-FLAG was used following the manufacturer's instructions (Enzo Life Sciences). All deacetylation experiments, both in vitro and in cells, were performed at least four times.
Cell viability assay
HCT116 cells were transfected with 40 nm siRNA using RNAiMAX reagent and split followed by transfection with SIRT2-FLAG WT or mutants, treatment with or without 1.6 mm HU 72 h postknockdown for 24 h, and recovery for 24 h. Viability was measured using alamarBlue (Thermo Fisher) on a Spectra Plus plate reader. A ratio of treated/untreated viability was calculated and normalized to that of nontargeting siRNA (43).
Sequence alignment
Protein BLAST (blastp) was used through the NCBI website to compare sequence homology among closely and distantly related organisms with the human SIRT2 protein sequence obtained from UniProt and to align protein sequences.
Author contributions
P. E. H. and D. S. Y. conceived and coordinated the study, designed experiments, and wrote the paper. P. E. H. performed and analyzed the experiments shown in Figs. 2–6. H. Z. assisted in study conception, experimental design, and experiments for figures regarding GFP-ATRIP foci. A. J. B. designed, performed, and analyzed the experiments shown in Fig. 1. A. E. K. also contributed to the design of Fig. 6, C and D. A. E. W. contributed to the design of Fig. 2A. W. B. D. provided assistance in generation of the SIRT2 knock-out lines by CRISPR/Cas9. X. C. assisted in conception and production of Fig. 3. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Yu laboratory for helpful discussion. We kindly thank Eric Verdin for the SIRT2-FLAG plasmid.
This work was supported by NCI, National Institutes of Health Grants R01CA178999 and R01CA178999S1, Basser Center for BRCA Innovation Award 32356, and Winship Cancer Institute/Brenda Nease Breast Cancer Research Fund Pilot Award 53237 (all to D. S. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SIRT2
- sirtuin 2
- ATR
- ATM and Rad3-related protein
- ATM
- ataxia telangiectasia-mutated
- ATRIP
- ATR-interacting protein
- DDR
- DNA-damage response
- CDK9
- cyclin-dependent kinase 9
- SIFT
- Sorting Intolerant from Tolerant
- PolyPhen
- Polymorphism Phenotyping
- NES
- nuclear export sequence
- LMB
- leptomycin B
- IP
- immunoprecipitation
- RSR
- replication stress response
- HU
- hydroxyurea
- CRISPR
- clustered regularly interspaced short palindromic repeats
- NS
- nonspecific
- EV
- empty vector
- CBP
- cAMP-response element-binding protein (CREB)-binding protein
- pCAF
- p300/CBP-associated factor
- TSA
- trichostatin A
- HAT
- histone acetyltransferase.
References
- 1. Guarente L. (2011) Franklin H. Epstein Lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 364, 2235–2244 [DOI] [PubMed] [Google Scholar]
- 2. Finkel T., Deng C. X., and Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saunders L. R., and Verdin E. (2007) Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 26, 5489–5504 [DOI] [PubMed] [Google Scholar]
- 4. Donmez G., and Outeiro T. F. (2013) SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 5, 344–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang H., Head P. E., and Yu D. S. (2016) SIRT2 orchestrates the DNA damage response. Cell Cycle 15, 2089–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi J. E., and Mostoslavsky R. (2014) Sirtuins, metabolism, and DNA repair. Curr. Opin. Genet. Dev. 26, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. North B. J., Rosenberg M. A., Jeganathan K. B., Hafner A. V., Michan S., Dai J., Baker D. J., Cen Y., Wu L. E., Sauve A. A., van Deursen J. M., Rosenzweig A., and Sinclair D. A. (2014) SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 33, 1438–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H. S., Vassilopoulos A., Wang R. H., Lahusen T., Xiao Z., Xu X., Li C., Veenstra T. D., Li B., Yu H., Ji J., Wang X. W., Park S. H., Cha Y. I., Gius D., et al. (2011) SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serrano L., Martínez-Redondo P., Marazuela-Duque A., Vazquez B. N., Dooley S. J., Voigt P., Beck D. B., Kane-Goldsmith N., Tong Q., Rabanal R. M., Fondevila D., Muñoz P., Krüger M., Tischfield J. A., and Vaquero A. (2013) The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 27, 639–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song H. Y., Biancucci M., Kang H. J., O'Callaghan C., Park S. H., Principe D. R., Jiang H., Yan Y., Satchell K. F., Raparia K., Gius D., and Vassilopoulos A. (2016) SIRT2 deletion enhances KRAS-induced tumorigenesis in vivo by regulating K147 acetylation status. Oncotarget 7, 80336–80349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiratsuka M., Inoue T., Toda T., Kimura N., Shirayoshi Y., Kamitani H., Watanabe T., Ohama E., Tahimic C. G., Kurimasa A., and Oshimura M. (2003) Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 309, 558–566 [DOI] [PubMed] [Google Scholar]
- 12. Temel M., Koç M. N., Ulutaş S., and Göğebakan B. (2016) The expression levels of the sirtuins in patients with BCC. Tumour. Biol. 37, 6429–6435 [DOI] [PubMed] [Google Scholar]
- 13. Li Z., Huang J., Yuan H., Chen Z., Luo Q., and Lu S. (2016) SIRT2 inhibits non-small cell lung cancer cell growth through impairing Skp2-mediated p27 degradation. Oncotarget 7, 18927–18939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartosch C., Monteiro-Reis S., Almeida-Rios D., Vieira R., Castro A., Moutinho M., Rodrigues M., Graça I., Lopes J. M., and Jerónimo C. (2016) Assessing sirtuin expression in endometrial carcinoma and non-neoplastic endometrium. Oncotarget 7, 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Xie Q. R., Chen Z., Lu S., and Xia W. (2013) Regulation of SIRT2 levels for human non-small cell lung cancer therapy. Lung Cancer 82, 9–15 [DOI] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., and Stuart J. M. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Forbes S. A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C. Y., Jia M., De T., Teague J. W., Stratton M. R., et al. (2015) COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Park S. H., Pantazides B. G., Karpiuk O., Warren M. D., Hardy C. W., Duong D. M., Park S. J., Kim H. S., Vassilopoulos A., Seyfried N. T., Johnsen S. A., Gius D., and Yu D. S. (2013) SIRT2 directs the replication stress response through CDK9 deacetylation. Proc. Natl. Acad. Sci. U.S.A. 110, 13546–13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang H., Head P. E., Daddacha W., Park S. H., Li X., Pan Y., Madden M. Z., Duong D. M., Xie M., Yu B., Warren M. D., Liu E. A., Dhere V. R., Li C., Pradilla I., et al. (2016) ATRIP deacetylation by SIRT2 drives ATR checkpoint activation by promoting binding to RPA-ssDNA. Cell Rep. 14, 1435–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsushita N., Takami Y., Kimura M., Tachiiri S., Ishiai M., Nakayama T., and Takata M. (2005) Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells 10, 321–332 [DOI] [PubMed] [Google Scholar]
- 21. North B. J., Marshall B. L., Borra M. T., Denu J. M., and Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 22. Inoue T., Hiratsuka M., Osaki M., Yamada H., Kishimoto I., Yamaguchi S., Nakano S., Katoh M., Ito H., and Oshimura M. (2007) SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 26, 945–957 [DOI] [PubMed] [Google Scholar]
- 23. Suematsu T., Li Y., Kojima H., Nakajima K., Oshimura M., and Inoue T. (2014) Deacetylation of the mitotic checkpoint protein BubR1 at lysine 250 by SIRT2 and subsequent effects on BubR1 degradation during the prometaphase/anaphase transition. Biochem. Biophys. Res. Commun. 453, 588–594 [DOI] [PubMed] [Google Scholar]
- 24. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tavtigian S. V., Deffenbaugh A. M., Yin L., Judkins T., Scholl T., Samollow P. B., de Silva D., Zharkikh A., and Thomas A. (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 43, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar P., Henikoff S., and Ng P. C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 28. Ng P. C., and Henikoff S. (2006) Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7, 61–80 [DOI] [PubMed] [Google Scholar]
- 29. Ng P. C., and Henikoff S. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng P. C., and Henikoff S. (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12, 436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng P. C., and Henikoff S. (2001) Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., and Sunyaev S. R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., and Ben-Tal N. (2013) ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53, 199–206 [Google Scholar]
- 34. Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 36. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gish W., and States D. J. (1993) Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272 [DOI] [PubMed] [Google Scholar]
- 38. North B. J., and Verdin E. (2007) Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2, e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rumpf T., Schiedel M., Karaman B., Roessler C., North B. J., Lehotzky A., Oláh J., Ladwein K. I., Schmidtkunz K., Gajer M., Pannek M., Steegborn C., Sinclair D. A., Gerhardt S., Ovádi J., et al. (2015) Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun 6, 6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith S. C., Petrova A. V., Madden M. Z., Wang H., Pan Y., Warren M. D., Hardy C. W., Liang D., Liu E. A., Robinson M. H., Rudra S., Wang J., Ehdaivand S., Torres M. A., Wang Y., et al. (2014) A gemcitabine sensitivity screen identifies a role for NEK9 in the replication stress response. Nucleic Acids Res. 42, 11517–11527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hall W. A., Petrova A. V., Colbert L. E., Hardy C. W., Fisher S. B., Saka B., Shelton J. W., Warren M. D., Pantazides B. G., Gandhi K., Kowalski J., Kooby D. A., El-Rayes B. F., Staley C. A. 3rd, Volkan Adsay N., et al. (2014) Low CHD5 expression activates the DNA damage response and predicts poor outcome in patients undergoing adjuvant therapy for resected pancreatic cancer. Oncogene 33, 5450–5456 [DOI] [PubMed] [Google Scholar]
- 43. Colbert L. E., Petrova A. V., Fisher S. B., Pantazides B. G., Madden M. Z., Hardy C. W., Warren M. D., Pan Y., Nagaraju G. P., Liu E. A., Saka B., Hall W. A., Shelton J. W., Gandhi K., Pauly R., et al. (2014) CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 74, 2677–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]