Abstract
In plasmodia, the dihydrofolate reductase (DHFR) enzyme is the target of the pyrimethamine component of sulfadoxine-pyrimethamine (S/P). Plasmodium vivax infections are not treated intentionally with antifolates. However, outside Africa, coinfections with Plasmodium falciparum and P. vivax are common, and P. vivax infections are often exposed to S/P. Cloning of the P. vivax dhfr gene has allowed molecular comparisons of dhfr alleles from different regions. Examination of the dhfr locus from a few locations has identified a very diverse set of alleles and showed that mutant alleles of the vivax dhfr gene are prevalent in Southeast Asia where S/P has been used extensively. We have surveyed patient isolates from six locations in Indonesia and two locations in Papua New Guinea. We sequenced P. vivax dhfr alleles from 114 patient samples and identified 24 different alleles that differed from the wild type by synonymous and nonsynonymous point mutations, insertions, or deletions. Most importantly, five alleles that carried four or more nonsynonymous mutations were identified. Only one of these highly mutant alleles had been previously observed, and all carried the 57L and 117T mutations. P. vivax cannot be cultured continuously, so we used a yeast assay system to determine in vitro sensitivity to pyrimethamine for a subset of the alleles. Alleles with four nonsynonymous mutations conferred very high levels of resistance to pyrimethamine. This study expands significantly the total number of novel dhfr alleles now identified from P. vivax and provides a foundation for understanding how antifolate resistance arises and spreads in natural P. vivax populations.
Plasmodium vivax causes a severe and debilitating febrile illness. This mosquito-borne parasite infects an estimated 80 million people each year and is a prevalent cause of malaria outside sub-Saharan Africa (39). Unfortunately, P. vivax cannot be maintained in continuous culture, so despite its prevalence, the study of vivax has lagged markedly behind that of Plasmodium falciparum. This difference is particularly true with respect to the genetics and epidemiology of drug resistance in P. vivax. Chloroquine has been the first-line antimalarial treatment in most areas of endemicity, but resistance to this drug has virtually eliminated its usefulness against P. falciparum (61, 63, 65) and chloroquine resistance has now been observed with P. vivax as well (1, 21, 34, 45, 52, 55, 58, 59). Fansidar, a fixed combination of sulfadoxine and pyrimethamine (S/P), is most often the alternative chosen when chloroquine-resistant P. falciparum parasites render chloroquine ineffective (4). However, S/P has not been recommended for primary therapy of vivax malaria due to the poor clinical efficacy reported when the drug was introduced in the 1950s (13, 14, 26, 49, 64, 68).
In both P. falciparum and P. vivax, the dihydrofolate reductase (DHFR, E.C. 1.5.1.3) enzyme is the therapeutic target of the pyrimethamine component of S/P (12, 18, 23, 26, 27, 43, 44, 60). A combination of in vitro analysis of cultured parasites and molecular analysis of field isolates has allowed a clear definition of the genetic basis for pyrimethamine resistance in P. falciparum (reviewed in references 25 and 53). A point mutation from serine to asparagine at codon 108 of the falciparum dhfr gene increases resistance slightly, and the subsequent addition of changes at combinations of 6 codons (A16V, C50R, N51I, C59R, S108N, and I164L) increases the resistance markedly (8, 47, 48). The simplicity of this picture is remarkable; the mutations that contribute to pyrimethamine resistance appear to be the same worldwide, synonymous substitutions are very rarely observed, and only one insertion has ever been reported (7). These patterns support the idea that the present P. falciparum populations have evolved recently from a few founders and spread virtually worldwide from these foci, probably influenced by the strong selection pressure of drug treatment (6, 42, 51, 67).
Limited studies to date have demonstrated that there is a far higher level of overall genetic diversity in the P. vivax population than one observes in the P. falciparum population (9, 16, 29, 41). This difference is likely to be important to the evolution of drug resistance. Vivax infections are not often treated intentionally with antifolates like pyrimethamine, yet coinfections with P. falciparum and P. vivax are common in southern and Southeast Asia and South America (36, 38, 57). As a result, P. vivax organisms are often exposed to S/P because fevers are presumptively treated or mixed infections are misdiagnosed. Because P. vivax cannot be continuously cultured, in vitro determination of drug sensitivity is extremely difficult. However, cloning of the dhfr gene from P. vivax (11) has allowed molecular comparisons of alleles from different regions. Examination of the dhfr locus from only a few locations has already yielded more unique dhfr alleles in vivax populations than the far more extensive examination of P. falciparum has demonstrated (12, 23, 24, 26, 27, 60). In particular, these studies showed that mutant alleles of the vivax dhfr gene are prevalent in Southeast Asia in areas where there is a long history of extensive S/P use. In contrast, wild-type vivax dhfr has been found more commonly in regions with limited or no historical use of S/P (12, 26, 27, 60).
So far, these studies have not been sufficient to define the overall patterns of the genetic diversity in P. vivax, but two previous reports have identified different characteristic subsets of vivax dhfr alleles in Central Java and Papua (23, 60), and another distinct subset has been reported in Thailand (27). In this study, we surveyed vivax dhfr alleles at six locations in the Indonesian archipelago and two locations in Papua New Guinea (PNG). We sequenced alleles from 114 patient samples and used a Saccharomyces cerevisiae assay system to determine the relative in vitro sensitivity to pyrimethamine for a subset of the alleles.
This study adds significantly to the number of unique dhfr alleles identified in previous data sets. The identification of novel pyrimethamine-resistant vivax dhfr alleles and characterization of their distribution in the Indonesian archipelago is the first step toward understanding how antifolate resistance arises and spreads in natural P. vivax populations.
MATERIALS AND METHODS
Patient samples.
The Indonesian isolates evaluated in this study are comprised of subsets of samples from clinical studies conducted in the Armopa region of northeastern Papua (1996 to 1999) (2, 31), Purworejo in southern Central Java (2000) (3, 32), Legundi Island in South Lampung, Sumatra (July, 2000) (33), Gag Island, Papua Province (April 1997) (19), Ketapang, West Kalimantan (July 1996) (20), and Maumere, East Flores (April 1996). In addition, a single sample was available from Laut Island, South Kalimantan (June 2002). The isolates from PNG evaluated in this study are a subset of samples from a clinical study conducted in the Wosera (1998 to 1999) and Liksul (2000) areas. This study has also been described in detail elsewhere (37). All subjects had slide- and PCR-proven infection by P. vivax. None of the subjects received S/P therapy; infections were treated with chloroquine or another antimalarial drug or combination, as per the protocol of each study. The work with Indonesian subjects in this study was reviewed (Department of Defense protocol no. 30820, 30839, and 30833) and approved by U.S. Navy and Indonesian institutional review boards. All Indonesian subjects provided written informed consent to participate in the study in accordance with U.S. Navy regulations governing the use of human subjects of medical research (SECNAVINST 3900.39C and BUMEDINST 3900.6B.). All Papuan subjects provided informed consent, and the samples from PNG were analyzed with the permission of the PNG Medical Research Advisory Committee (MRAC no. 02.14).
Cloning and sequencing of dhfr.
Genomic P. vivax DNA from the Indonesian samples was extracted from dried blood blots on filter paper by using the blood blot DNA extraction protocol from a commercial kit (QIAamp DNA mini kit; QIAGEN). The samples from PNG were prepared at the time of collection from whole blood by using QIAamp 96 spin blood kits (37). The dhfr gene was then amplified by PCR in a 100-μl reaction mixture, with 2 μl of Taq polymerase (Promega), 10 μl of 10× PCR buffer, 8 μl of 25 mM MgCl2, 1 μl of each 10 μM primer, 1 μl of 10 mM deoxynucleoside triphosphates, 10 μl of template DNA, and 67 μl of PCR H2O. The primers used have been described previously (24). The 5′ end of each primer is complementary to the sequence of the shuttle plasmid at the desired insertion position to facilitate homologous recombination in yeast (7, 54). The cycling parameters were as follows: (i) initial denaturation for 3 min at 94°C, followed by 5 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min; (ii) 5 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min; and (iii) 20 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min.
The S. cerevisiae yeast strain used lacks endogenous DHFR activity and has been described previously (66). Yeast cells were transformed by using a high-efficiency lithium acetate protocol (28) and were then plated onto medium lacking tryptophan, to select for the plasmid, and dTMP, to select for functional DHFR activity. Escherichia coli strain DH5α was used for propagation and preparation of the shuttle plasmid from the transformed yeast. The plasmid was isolated and purified from the bacteria by using a commercial kit (QIAprep spin miniprep kit; QIAGEN). Sequencing was conducted via fluorescent dye chemistry (MegaBACE; Amersham Pharmacia Biotech) and analyzed with Sequencher software (Gene Codes Corporation) and the ClustalW multiple-sequence alignment program in SDSC Biology Workbench, version 3.2. At least two isolates were sequenced from each patient sample to check for mixed genotypes; if the two were not in agreement, additional isolates were sequenced to verify the mixed genotype. If a particular allele was not observed again after 10 isolates from that sample were sequenced, it was assumed to be a PCR artifact and discounted from subsequent analysis.
In vitro susceptibility classification.
The 50% inhibitory concentration (IC50) assays were performed as previously described to obtain quantitative measures of drug sensitivity (23). The growth of the yeast in this assay depends upon the antifolate resistance of the dhfr allele expressed. An IC50 is defined as the concentration of drug at which the growth of the yeast culture is inhibited by 50% relative to the untreated control. Yeast cells transformed with dhfr from P. vivax were grown overnight in a 96-well culture dish in complete medium (yeast extract-peptone-dextrose broth) lacking supplemental dTMP. The growth of the yeast cells in each well was assessed by spectrophotometry after approximately 24 h of incubation at 30°C, the amount of time required for the untreated control yeast cells to reach mid-log phase. The average reading of six wells for each allele at each drug concentration was then used to plot the percent growth relative to the control. The average reading of 12 wells was used for the control yeast cells. The numerical IC50 was calculated from the slope and intercept of the line defined by the two averaged data points that bracket 50% relative growth. Comparisons of the IC50s of the mutant alleles to the wild-type allele were used to assess the relative drug resistance level of each allele. IC50 assays were performed at least twice for each allele to determine the standard deviation and to ensure reproducibility.
Nucleotide sequence accession number.
The complete sequence of the alleles identified has been submitted to GenBank and assigned accession numbers AY772063 to AY772087.
RESULTS
P. vivax dhfr alleles identified in Indonesia and PNG.
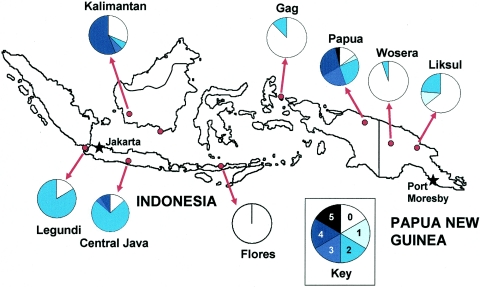
In this study, we examined samples from eight geographic locations, including six sites in Indonesia and two sites in PNG, and identified 137 dhfr alleles from 114 patients. Figure 1 shows the approximate position of each sampling location; the pie charts indicate the proportion of the alleles from each location that carried particular numbers of mutations. The genotypes of all alleles that we identified are listed in Table 1. The reference sequence was the wild-type Burma-6 sequence provided in GenBank (accession no. AJ222633), and we considered insertions, deletions, and synonymous or nonsynonymous point mutations. Each unique nucleotide sequence was counted as a separate allele. The exact sequence of each allele has been submitted to GenBank (accession numbers AY772063 to AY772087).
FIG. 1.
Map of locations from which samples were derived. All locations had numerous samples, except the southernmost Kalimantan site which had only a single patient sample. That location is indicated by a dot only, and the sample was included with the West Kalimantan samples for analysis. The pie charts show the proportions of alleles that showed a particular number of point mutations. The darkest color indicates 4 or more nonsynonymous mutations, and the progressively lighter shades show 3, 2, or 1 mutation; white indicates the wild type.
TABLE 1.
Unique dhfr allele sequences observed
| No. of isolates | % of total | Change(s) in sequence from wild type
|
No. of locations | ||
|---|---|---|---|---|---|
| Mutation(s)
|
Indel | ||||
| Nonsynonymous | Synonymous | ||||
| 51 | 37.2 | None | None | None | 7 |
| 18 | 13.1 | 58R/117N | None | None | 4 |
| 16 | 11.7 | 58R/117N | None | Deletion A | 2 |
| 10 | 7.3 | 57L/58R/61M/117T | None | None | 2 |
| 7 | 5.1 | 57L/111L/117T/173F | None | None | 1 |
| 6 | 4.4 | 57L/58R | 38GGC | None | 1 |
| 5 | 3.6 | 58R/117N/199V | None | None | 2 |
| 2 | 1.5 | 58R | None | None | 1 |
| 2 | 1.5 | None | 215CGA | None | 1 |
| 2 | 1.5 | None | None | Insertion A | 2 |
| 2 | 1.5 | None | None | Insertion B | 1 |
| 2 | 1.5 | None | None | Deletion B | 2 |
| 2 | 1.5 | None | None | Deletion A | 1 |
| 1 | 0.7 | None | None | Insertion C | 1 |
| 1 | 0.7 | None | None | Insertion D | 1 |
| 1 | 0.7 | 199V | None | None | 1 |
| 1 | 0.7 | 117N | None | Insertion C | 1 |
| 1 | 0.7 | 57L/58R | None | Insertion A | 1 |
| 1 | 0.7 | 58R/117N | 196ATA | None | 1 |
| 1 | 0.7 | 58R/117N/99S | None | None | 1 |
| 1 | 0.7 | 58R/61M/117T | None | None | 1 |
| 1 | 0.7 | 57L/58R/117T | None | None | 1 |
| 1 | 0.7 | 15S/57L/117T/173F | None | None | 1 |
| 1 | 0.7 | 57L/111L*/117T/173Fa | None | None | 1 |
| 1 | 0.7 | 49R/57L/58R/61M/117T | None | None | 1 |
The asterisk denotes CTG at codon 111 instead of TTG.
Our goal was to identify a broad range of dhfr alleles. We amplified and cloned the dhfr allele(s) from each patient sample and sequenced at least two independent clones to identify mixed infections and eliminate any mutations that may have been introduced by polymerase error. If the first two sequences were not in agreement, additional isolates were sequenced to verify each allele. If a particular allele was not observed again after 10 isolates from that sample were sequenced, it was assumed to be a PCR artifact and discounted from subsequent analysis. Polyclonal P. vivax infections were common in this sample set, and we have not exhaustively identified all alleles in each patient sample. As a result, our survey does not measure either the population frequency of any given allele or the proportion of each allele in a polyclonal infection. Our data are indicative only of general trends and patterns in Indonesia and PNG, and the allele frequencies reported are specific to the set of samples analyzed in our study.
Commonly observed alleles.
The most common allele in this data set matched the wild-type dhfr sequence (37.2%), with no point mutations, insertions, or deletions. Most of the wild-type alleles came from three study sites: Gag, Indonesia; Flores, Indonesia; and the Wosera, PNG. Two alleles with point mutations resulting in an arginine at position 58 and an asparagine at position 117 (58R/117N) accounted for another quarter of the isolates (24.8%). One of these double-mutant alleles has a deletion of codons 98 to 103 and was found in Central Java, Indonesia, and Legundi, Indonesia. The other 58R/117N allele has no insertions or deletions and was found in four of the six Indonesian study sites. Two alleles with four point mutations, 57L/58R/61M/117T and 57L/111L/117T/173F, were also common, accounting for 7.3 and 5.1% of the alleles, respectively. The 57L/58R/61M/117T allele was primarily found in Papua, Indonesia, but was also isolated from a single sample from South Kalimantan and a polyclonal sample from West Kalimantan. The 57L/111L/117T/173F allele was the most common allele found in Ketapang, West Kalimantan, but also was observed in a single sample from Java. A 57L/58R allele with a silent mutation at position 38 was common in the Liksul, PNG study site and accounted for 4.4% of all alleles identified in this study. The 58R/117N/199V allele, found primarily in the Papua study site, accounted for 3.6% of alleles. Thus, this commonly observed set of 7 alleles accounted for approximately 80% of the alleles identified.
Less commonly observed alleles.
An additional 18 alleles were found only once or twice, for a grand total of 25 unique alleles. Fifteen of these alleles have not been previously identified in the published literature; only the deletion B, 58R, and 57L/58R/117T alleles have been previously identified (11, 23). Two of the rare alleles were defined by the presence of a synonymous point mutation: wild type with CGA at codon 215 and 58R/117N with ATA at codon 196. There were three rare alleles with three nonsynonymous point mutations (58R/117N/99S, 58R/61M/117T, and 57L/58R/117T), two alleles with four nonsynonymous point mutations (15S/57L/117T/173F and 57L/111L*/117T/173F), and one allele with five nonsynonymous point mutations (49R/57L/58R/61M/117T).
Insertions and deletions.
We identified several alleles defined by the presence of an insertion or deletion. Details of the insertions and deletions (indels) are given in Table 2. All of the indels were localized to a central GGDN repeat region of the gene that is thought to be nonessential for substrate binding and which is missing entirely from the P. falciparum dhfr coding sequence (50). Furthermore, all of the indels involved 18 bp of sequence, except for one 36-bp insertion found in an allele from the Wosera. The exact nucleotide coordinates of the indels could not always be determined due to the repetitive nature of the GGDN repeat region but was estimated based on previous reports in the literature.
TABLE 2.
Nucleotide sequences and coordinates of insertions and deletions observeda
| Insertion or deletion | Codons | No. of bp | Sequence |
|---|---|---|---|
| Insertion A | 103, 104 | 18 | ACACACGGTGGTGACAAC |
| Insertion B | 103, 104 | 18 | ACAAGCGGTGGTGACAAC |
| Insertion C | 98, 99 | 18 | AGCGGTGGTGACAACACA |
| Insertion D | 98, 99 | 36 | AGCGGTGGTGACAACACAC ACGGTGGTGACGACACA |
| Deletion A | 98-103 | 18 | ACACACGGTGGTGACAAC |
| Deletion B | 92-97 | 18 | AACACAAGCGGTGGTGAC |
No examples of alleles with both an insertion and a deletion were identified.
Codon usage.
Some alleles that encoded identical polypeptides differed in their nucleotide sequences. For example, there were two 57L/111L/117T/173F alleles, both found in Kalimantan. The leucine at position 111 was encoded by CTG in one allele and by TTG in the other, more common allele. Similarly, the leucine at position 57 was encoded by CTC in the 57L/58R/61M/117T allele and by TTG in the 57L/111L/117T/173F and 57L/58R alleles. The arginine at position 58 was encoded by AGA in the 57L/58R allele in Liksul and in the 58R and 58R/117N/199V alleles and by AGG in all other alleles. The CGC codon for arginine at position 58 was not found in this sample set nor was the TTA codon for leucine at position 57.
Geographic trends.
Despite the fact that all of the study sites are located in the same archipelago, most of the dhfr alleles were found in only one or two of the study locations (Table 3). Only the wild-type allele was found in a majority (7 of 8) of the sites. The 58R/117N allele was found in 4 study sites, all in Indonesia. Papua and Kalimantan were the only locations in which the majority of the alleles had more than two nonsynonymous point mutations (55.5 and 62.6%, respectively). The Wosera, Gag, and Flores samples had primarily wild-type alleles.
TABLE 3.
Geographic distribution of allelesa
| Location | No. (%) of alleles | Change(s) in sequence from wild type
|
||
|---|---|---|---|---|
| Mutation(s)
|
Indel | |||
| Nonsynonymous | Synonymous | |||
| Wosera | 17 (89.5) | None | None | None |
| 1 (5.3) | None | None | Insertion D | |
| 1 (5.3) | 57L/58R | None | Insertion A | |
| Liksul | 12 (48.0) | None | None | None |
| 6 (24.0) | 57L/58R | 38GGC | None | |
| 2 (8.0) | 58R | None | None | |
| 2 (8.0) | None | 215CGA | None | |
| 1 (4.0) | None | None | Insertion A | |
| 1 (4.0) | None | None | Deletion B | |
| 1 (4.0) | 199V | None | None | |
| Papua | 8 (29.6) | 57L/58R/61M/117T | None | None |
| 7 (25.9) | 58R/117N | None | None | |
| 4 (14.8) | None | None | None | |
| 4 (14.8) | 58R/117N/199V | None | None | |
| 1 (3.7) | 117N | None | Insertion C | |
| 1 (3.7) | 58R/61M/117T | None | None | |
| 1 (3.7) | 57L/58R/117T | None | None | |
| 1 (3.7) | 49R/57L/58R/61M/117T | None | None | |
| Gag | 5 (62.5) | None | None | None |
| 2 (25.0) | None | None | Insertion B | |
| 1 (12.5) | 58R/117N | 196ATA | None | |
| Legundi | 5 (41.7) | 58R/117N | None | None |
| 5 (41.7) | 58R/117N | None | Deletion A | |
| 1 (8.3) | None | None | Insertion C | |
| 1 (8.3) | None | None | Deletion B | |
| Central Java | 11 (50.0) | 58R/117N | None | Deletion A |
| 5 (22.7) | 58R/117N | None | None | |
| 2 (9.1) | None | None | None | |
| 1 (4.5) | None | None | Insertion A | |
| 1 (4.5) | 58R/117N/99S | None | None | |
| 1 (4.5) | 15S/57L/117T/173F | None | None | |
| 1 (4.5) | 57L/111L/117T/173F | None | None | |
| Flores Kali- mantan | 8 (100) | None | None | None |
| 7 (43.8) | 57L/111L/117T/173F | None | None | |
| 3 (18.8) | None | None | None | |
| 2 (12.5) | None | None | Deletion A | |
| 2 (12.5) | 57L/58R/61M/117T* | None | None | |
| 1 (6.3) | 58R/117N | None | None | |
| 1 (6.3) | 58R/117N/199V | None | None | |
The starred allele derived from a single sample from South Kalimantan and a sample from Ketapang, West Kalimantan.
Polyallelic samples.
Multiple vivax dhfr alleles were identified in 18 (15.8%) of the 114 patient samples. This is a conservative estimate of the number of polyclonal infections, as it is based on only one locus and the patient samples were not exhaustively evaluated to identify all dhfr alleles present. The majority of the patient samples from Legundi were of mixed genotype; the Papua, Kalimantan, and Liksul study sites also had more than one polyallelic sample. The largest number of dhfr alleles found in any one sample was four, from a patient in the West Kalimantan study site. Three alleles were found in one patient sample from the Papua study site; only two different alleles were found in the other polyallelic samples.
In vitro resistance to pyrimethamine.
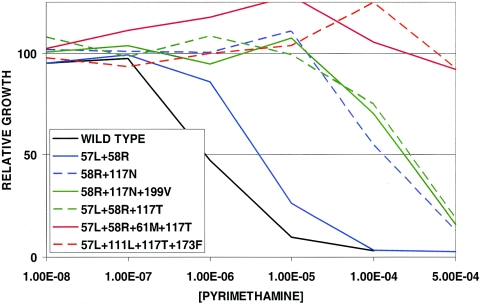
We have established an expression system for the P. vivax dhfr gene in yeast cells that lack endogenous DHFR activity. For alleles of P. falciparum, the in vitro sensitivity of these engineered yeast lines to antifolate drugs reflects the sensitivity of the parasite from which the dhfr allele was isolated (17, 22, 66). To define the range of pyrimethamine sensitivity represented in these P. vivax alleles, we created yeast lines dependent upon each of 7 alleles: the wild type and two double-, two triple-, and two quadruple-mutant alleles. These were grown in 0 to 500 μM pyrimethamine, and the concentration of drug required to inhibit yeast growth by 50% (the IC50) was measured. Figure 2 shows representative data. The IC50 value of the yeast dependent upon the wild-type allele was approximately 1 μM. The 57L/58R double mutant was only about sevenfold more resistant than the wild type. The 58R/117N double-mutant allele and the two triple-mutant alleles all showed similar, higher levels of resistance, about 200- to 300-fold higher than that of the wild-type allele. Finally, both of the quadruple-mutant alleles tested were highly pyrimethamine resistant; the growth of these strains of yeast was unaffected by up to 500 μM, the maximum concentration of pyrimethamine that can be tested in the assay. Thus, an IC50 could not be measured for these alleles.
FIG. 2.
Pyrimethamine sensitivity of yeast strains dependent upon P. vivax dhfr alleles. Yeast strains were grown in liquid culture with the indicated concentrations of pyrimethamine, and growth relative to the same strain without pyrimethamine was measured after 24 h as described in Materials and Methods.
DISCUSSION
The enormous diversity of the P. vivax dhfr alleles identified in this relatively small sample set is in striking contrast to the very limited degree of polymorphism that is observed in dhfr alleles from P. falciparum. This is particularly interesting considering that the antifolate drug treatments that drive the selection of mutated alleles are presumably the same in this region, where P. falciparum and P. vivax are sympatric and coinfections and simultaneous treatment are common. The reasons for the contrast cannot be clearly defined, but there are numerous biological, demographic, and epidemiologic differences between the two species. For example, the repeated relapses of hypnozoites from P. vivax in patients may be one element that contributes to its genetic diversity. It has also been proposed that P. vivax has been a primate parasite for much longer than P. falciparum and thus has had far longer to accumulate polymorphisms (16). Certainly, differences in both biological patterns and evolutionary history must play a role, and whatever the origin of the diversity, it will complicate development of both drug treatments and vaccines against P. vivax.
The samples in our set were taken from a variety of locations, and the patterns of allelic diversity varied enormously. Several populations, like Gag Island and the Wosera region of PNG, contained wild-type dhfr almost exclusively, whereas samples from the Indonesian regions of Papua and Kalimantan carried numerous different alleles, some with four or five mutations that confer high levels of pyrimethamine resistance. The small sample size does not allow one to draw definitive conclusions about the patterns of gene flow, but the extent of mixing of human populations and the history of antifolate drug use have varied enormously among these sites. Both factors are likely to have a profound effect on the diversity of each local P. vivax population. Moreover, the biogeographically fragmented populations of plants, animals, and humans in this island archipelago have been noted as a fantastic source of genetic diversity for more than a century (62), and the P. vivax populations certainly follow this pattern.
It is clear that areas where antifolate use has been intensive, like Thailand and Indonesian Papua, show a higher prevalence of alleles that carry more than two mutations and encode DHFR enzymes that are more resistant to pyrimethamine (5, 24, 26, 27, 60). Treatment of mixed-species infections, the use of pyrimethamine or S/P in suppressive treatment, and use of S/P in presumptive treatment have all likely exerted selective pressure on P. vivax dhfr in Indonesia. A massive effort at suppressive treatment with pyrimethamine was attempted in the 1950s in Papua (40) and was more recently undertaken with S/P in the transmigrant camps in Papua and Kalimantan (2, 20, 31). This is almost surely the reason for the high frequency of quadruple-mutant alleles in these study sites. In other areas, such as Java and Legundi, presumptive treatment likely plays a dominant role. Inadvertent treatment of P. vivax in mixed infections, where the P. vivax component is cryptic, is also a well-documented phenomenon in the literature; it is quite common for a P. vivax infection to follow a treated P. falciparum infection by a few weeks or vice versa (35, 36, 49, 56, 57). Unfortunately, teasing apart the various mechanisms behind the selection of the resistant alleles in each study site was beyond the scope of this paper, as there is not enough baseline information for these study sites. The presence of highly resistant P. vivax dhfr alleles in areas where pyrimethamine and S/P use have been particularly intensive provides evidence that drug measures against P. falciparum malaria exert correspondingly significant pressures against P. vivax, whether or not this is intentional
Perusal of the larger data set also demonstrates that there are two main branches of the selection process, based on dhfr sequence. Those alleles that carry the 117N mutation are common but do not confer exceedingly high levels of pyrimethamine resistance. However, the alternate 117T mutation is always found among the highly pyrimethamine-resistant alleles with four or five nonsynonymous point mutations. This apparent dichotomy has some practical consequences. For example, the common 58R/117N allele apparently cannot accommodate an additional mutation at position 57. This particular triple-mutant allele has not been observed in field isolates, and it failed to complement the DHFR-deficient yeast line when it was created in the lab. Thus, the 58R/117N allele, while moderately resistant to pyrimethamine, may represent a dead end with respect to the evolution of high levels of drug resistance. Interestingly, alleles with the 117N mutation do not demonstrate significantly increased resistance to the experimental DHFR inhibitor WR99210 (24). In contrast, all five highly pyrimethamine-resistant alleles identified in this study carried both the 117T mutation and the 57L mutation. This pattern is consistent with three highly pyrimethamine-resistant alleles recently identified in Thailand (26). Thus, the presence of threonine at position 117 rather than arginine seems to allow more key mutations to accumulate without significant loss of enzyme function, resulting in higher levels of resistance to pyrimethamine. These alleles also show a moderate increase in resistance to WR99210 (data not shown).
The combined data sets also allow correlations to be made between the molecular analysis of the dhfr alleles and the clinical effectiveness of S/P against P. vivax infections. A recent study showed that Indonesian patients infected with P. vivax that carried up to three mutations in dhfr were still responsive to S/P treatment (23). In contrast, patients whose parasites carried the 57L/61M/117T/173F allele were far more likely to fail S/P treatment (23, 60). The presence of parasites with four or five mutations in dhfr has also been associated with reduced parasite clearance rates in Thailand (26). The novel quadruple and quintuple mutant alleles identified in this study have not been tested for their clinical relevance, but in the yeast assay, the quadruple-mutant alleles all conferred comparably high levels of pyrimethamine resistance (data not shown). Thus, the evidence indicates that clinical resistance to S/P is not inherent, as it has been previously assumed, but rather is associated with point mutations in vivax dhfr.
A limited number of mutations in dhfr are commonly associated with antifolate resistance in P. falciparum. This regularity has allowed the development of relatively simple allele-specific oligonucleotide PCR and hybridization methods, and these have been productively employed in molecular surveillance (10, 15). The extremely high level of diversity in P. vivax dhfr alleles argues strongly against such focused approaches. There are too many mutations that need to be identified, requiring complete sequencing of the gene. The two-branch structure of the mutation families suggests that a simple PCR-based method that would identify the 117T mutation may be helpful as a first step, since all of the alleles identified to date that encode highly pyrimethamine-resistant enzymes contain this change. Only alleles that carry the 117T mutation would then need to be followed up with direct sequencing to confirm the presence of other mutations associated with high levels of S/P resistance.
For decades, it has been assumed that S/P is not effective against P. vivax. However, our findings suggest that in areas where the prevalence of dhfr quadruple- and quintuple-mutants is low, a DHFR inhibitor could provide one component of an effective combination treatment against the erythrocytic stages of vivax malaria. One possible partner would be a sulfa or sulfone, although based on the information currently available, one may need to search for a partner other than sulfadoxine or dapsone (30). The molecular studies of dhfr mutations so far are not a useful guide for individual clinical decisions; the relationship between parasites' genotype and outcome of chemotherapy is too complex (46). The molecular data can, however, provide important information for decisions on population-based drug use in a region. The paucity of information on P. vivax underlines the need to extend molecular analyses to other areas where P. vivax is common, e.g., the Indian subcontinent, Central Asia, the Middle East, Ethiopia, and South and Central America (39). Based on the small amount of information gathered so far, highly mutated alleles of dhfr may not yet be common in these regions and S/P may still provide effective treatment for the asexual erythrocytic stages of vivax in areas where chloroquine treatment failure has been reported. However, history suggests that selection of high-level resistance will be rapid. Therefore, any drug should be introduced in combination with a partner having a different pharmacokinetic mechanism to retard the selection or spread of resistant alleles. Our data contribute to the small but growing body of evidence suggesting that antifolates can provide one component in the design of such a combination (23, 60).
Acknowledgments
We thank the patients and health care workers who participated in the clinical studies from which these samples were derived and the Indonesian Ministry of Health, National Institute of Health Research and Development, for assistance in collecting samples in Indonesia.
This work was supported by grants from the National Institutes of Health, AI 55604 to C.H.S. and AI 46919 to P.A.Z., and by an NSF graduate fellowship to M.D.H. Collection of the Indonesian samples was supported by funding from the U.S. Naval Medical Research Command, Silver Spring, Md., and the Department of Defense Global Emerging Infections Surveillance (GEIS) Program.
REFERENCES
- 1.Baird, J. K., M. F. Sustriayu Nalim, H. Basri, S. Masbar, B. Leksana, E. Tjitra, R. M. Dewi, M. Khairani, and F. S. Wignall. 1996. Survey of resistance to chloroquine by Plasmodium vivax in Indonesia. Trans. R. Soc. Trop. Med. Hyg. 90:409-411. [DOI] [PubMed] [Google Scholar]
- 2.Barcus, M. J., Krisin, I. R. Elyazar, H. Marwoto, T. L. Richie, H. Basri, I. Wiady, D. J. Fryauff, J. D. Maguire, M. J. Bangs, and J. K. Baird. 2003. Primary infection by Plasmodium falciparum or P. vivax in a cohort of Javanese migrants to Indonesian Papua. Ann. Trop. Med. Parasitol. 97:565-574. [DOI] [PubMed] [Google Scholar]
- 3.Barcus, M. J., F. Laihad, M. Sururi, P. Sismadi, H. Marwoto, M. J. Bangs, and J. K. Baird. 2002. Epidemic malaria in the Menoreh Hills of Central Java. Am. J. Trop. Med. Hyg. 66:287-292. [DOI] [PubMed] [Google Scholar]
- 4.Bloland, P., S. Kachur, and H. Williams. 2003. Trends in antimalarial drug deployment in sub-Saharan Africa. J. Exp. Biol. 206:3761-3769. [DOI] [PubMed] [Google Scholar]
- 5.Brega, S., F. de Monbrison, C. Severini, R. Udomsangpetch, I. Sutanto, P. Ruckert, F. Peyron, and S. Picot. 2004. Real-time PCR for dihydrofolate reductase gene single-nucleotide polymorphisms in Plasmodium vivax isolates. Antimicrob. Agents Chemother. 48:2581-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortese, J. F., A. Caraballo, C. E. Contreras, and C. V. Plowe. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999-1006. [DOI] [PubMed] [Google Scholar]
- 7.Cortese, J. F., and C. V. Plowe. 1998. Antifolate resistance due to new and known Plasmodium falciparum dihydrofolate reductase mutations expressed in yeast. Mol. Biochem. Parasitol. 94:205-214. [DOI] [PubMed] [Google Scholar]
- 8.Cowman, A. F. 1997. The mechanisms of drug action and resistance in malaria. Mol. Gen. Drug Resist. 3:221-246. [Google Scholar]
- 9.Cui, L., C. N. Mascorro, Q. Fan, K. A. Rzomp, B. Khuntirat, G. Zhou, H. Chen, G. Yan, and J. Sattabongkot. 2003. Genetic diversity and multiple infections of Plasmodium vivax malaria in Western Thailand. Am. J. Trop. Med. Hyg. 68:613-619. [DOI] [PubMed] [Google Scholar]
- 10.de Pecoulas, P. E., B. Abdallah, M. K. Dje, L. K. Basco, J. leBras, and A. Mazzabraud. 1995. Use of a semi-nested PCR diagnosis test to evaluate antifolate resistance of Plasmodium falciparum isolates. Mol. Cell. Probes 9:391-397. [DOI] [PubMed] [Google Scholar]
- 11.de Pecoulas, P. E., L. K. Basco, R. Tahar, T. Ouatas, and A. Mazabraud. 1998. Analysis of the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene sequence. Gene 211:177-185. [DOI] [PubMed] [Google Scholar]
- 12.de Pecoulas, P. E., R. Tahar, T. Ouatas, A. Mazabraud, and L. K. Basco. 1998. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol. Biochem. Parasitol. 92:265-273. [DOI] [PubMed] [Google Scholar]
- 13.Doberstyn, E. B., C. Teerakiartkamjorn, R. G. Andre, P. Phintuyothin, and S. Noeypatimanondh. 1979. Treatment of vivax malaria with sulfadoxine-pyrimethamine and with pyrimethamine alone. Trans. R. Soc. Trop. Med. Hyg. 73:15-17. [DOI] [PubMed] [Google Scholar]
- 14.Doi, H., A. Kaneko, W. Panjaitan, and A. Ishii. 1989. Chemotherapeutic malaria control operation by single dose of Fansidar plus primaquine in North Sumatra, Indonesia. Southeast Asian J. Trop. Med. Public Health 20:341-349. [PubMed] [Google Scholar]
- 15.Duraisingh, M. T., J. Curtis, and D. C. Warhurst. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 89:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Feng, X., J. M. Carlton, D. A. Joy, J. Mu, T. Furuya, B. B. Suh, Y. Wang, J. W. Barnwell, and X. Z. Su. 2003. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc. Natl. Acad. Sci. USA 100:8502-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferlan, J. T., S. Mookherjee, I. N. Okezie, L. Fulgence, and C. H. Sibley. 2001. Mutagenesis of dihydrofolate reductase from Plasmodium falciparum: analysis in Saccharomyces cerevisiae of triple mutant alleles resistant to pyrimethamine or WR99210. Mol. Biochem. Parasitol. 113:139-150. [DOI] [PubMed] [Google Scholar]
- 18.Foote, S. J., D. Galatis, and A. F. Cowman. 1990. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc. Natl. Acad. Sci. USA 87:3014-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fryauff, D. J., I. Sumawinata, Purnomo, T. L. Richie, E. Tjitra, M. J. Bangs, A. Kadir, and G. Ingkokusumo. 1999. In vivo responses to antimalarials by Plasmodium falciparum and Plasmodium vivax from isolated Gag Island off northwest Irian Jaya, Indonesia. Am. J. Trop. Med. Hyg. 60:542-546. [DOI] [PubMed] [Google Scholar]
- 20.Fryauff, D. J., S. Tuti, A. Mardi, S. Masbar, R. Patipelohi, B. Leksana, K. C. Kain, M. J. Bangs, T. L. Richie, and J. K. Baird. 1998. Chloroquine-resistant Plasmodium vivax in transmigration settlements of West Kalimantan, Indonesia. Am. J. Trop. Med. Hyg. 59:513-518. [DOI] [PubMed] [Google Scholar]
- 21.Garg, M., N. Gopinathan, P. Bodhe, and N. A. Kshirsagar. 1995. Vivax malaria resistant to chloroquine: case reports from Bombay. Trans. R. Soc. Trop. Med. Hyg. 89:656-657. [DOI] [PubMed] [Google Scholar]
- 22.Hankins, E. G., D. C. Warhurst, and C. H. Sibley. 2001. Novel alleles of the Plasmodium falciparum dhfr highly resistant to pyrimethamine and chlorcycloguanil, but not WR99210. Mol. Biochem. Parasitol. 117:91-102. [DOI] [PubMed] [Google Scholar]
- 23.Hastings, M. D., K. M. Porter, J. D. Maguire, I. Susanti, W. Kania, M. J. Bangs, C. H. Sibley, and J. K. Baird. 2004. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J. Infect. Dis. 189:744-750. [DOI] [PubMed] [Google Scholar]
- 24.Hastings, M. D., and C. H. Sibley. 2002. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc. Natl. Acad. Sci. USA 99:13137-13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyde, J. E. 2002. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 4:165-174. [DOI] [PubMed] [Google Scholar]
- 26.Imwong, M., S. Pukrittakayamee, S. Looareesuwan, G. Pasvol, J. Poirreiz, N. J. White, and G. Snounou. 2001. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 45:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imwong, M., S. Pukrittayakamee, L. Renia, F. Letourneur, J. P. Charlieu, U. Leartsakulpanich, S. Looareesuwan, N. J. White, and G. Snounou. 2003. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 47:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joy, D. A., X. Feng, J. Mu, T. Furuya, K. Chotivanich, A. U. Krettli, M. Ho, A. Wang, N. J. White, E. Suh, P. Beerli, and X. Z. Su. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 300:318-321. [DOI] [PubMed] [Google Scholar]
- 30.Korsinczky, M., K. Fischer, N. Chen, J. Baker, K. Rieckmann, and Q. Cheng. 2004. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother. 48:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krisin, H. Basri, D. J. Fryauff, M. J. Barcus, M. J. Bangs, e. Ayomi, H. Marwoto, I. R. F. Elyazar, T. L. Richie, and J. K. Baird. 2003. Malaria in a cohort of Javanese migrants in Indonesian Papua. Ann. Trop. Med. Parasitol. 97:543-556. [DOI] [PubMed] [Google Scholar]
- 32.Maguire, J. D., M. D. Lacy, Sururi, P. Sismadi, Krisin, I. Wiady, B. Laksana, M. J. Bangs, S. Masbar, I. Susanti, W. Basuki, M. J. Barcus, H. Marwoto, M. D. Edstein, S. Tjokrosonto, and J. K. Baird. 2002. Chloroquine or sulfadoxine-pyrimethamine for the treatment of uncomplicated, Plasmodium falciparum malaria during an epidemic in Central Java, Indonesia. Ann. Trop. Med. Parasitol. 96:655-668. [DOI] [PubMed] [Google Scholar]
- 33.Maguire, J. D., I. W. Sumawinata, S. Masbar, B. Laksana, P. Prodjodipuro, I. Susanti, P. Sismadi, N. Mahmud, M. J. Bangs, and J. K. Baird. 2002. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet 360:58-60. [DOI] [PubMed] [Google Scholar]
- 34.Marlar-Than, Myat-Phone-Kyaw, Aye-Yu-Soe, Khaing-Khaing-Gyi, Ma-Sabai, and Myint-Oo. 1995. Development of resistance to chloroquine by Plasmodium vivax in Myanmar. Trans. R. Soc. Trop. Med. Hyg. 89:307-308. [DOI] [PubMed] [Google Scholar]
- 35.Mayxay, M., S. Pukritrayakamee, K. Chotivanich, M. Imwong, S. Looareesuwan, and N. J. White. 2001. Identification of cryptic coinfection with Plasmodium falciparum in patients presenting with vivax malaria. Am. J. Trop. Med. Hyg. 65:588-592. [DOI] [PubMed] [Google Scholar]
- 36.Mayxay, M., S. Pukrittayakamee, P. N. Newton, and N. J. White. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20:233-240. [DOI] [PubMed] [Google Scholar]
- 37.Mehlotra, R. K., L. J. Kasehagen, M. Baisor, K. Lorry, J. W. Kazura, M. J. Bockarie, and P. A. Zimmerman. 2002. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am. J. Trop. Med. Hyg. 67:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehlotra, R. K., K. Lorry, W. Kastens, S. M. Miller, M. P. Alpers, M. Bockarie, J. W. Kazura, and P. A. Zimmerman. 2000. Random distribution of mixed species malaria infections in Papua New Guinea. Am. J. Trop. Med. Hyg. 62:225-231. [DOI] [PubMed] [Google Scholar]
- 39.Mendis, K., B. J. Sina, P. Marchesini, and R. Carter. 2001. The neglected burden of Plasmodium vivax malaria. Am. J. Trop. Med. Hyg. 64:97-106. [DOI] [PubMed] [Google Scholar]
- 40.Meuwissen, T. 1964. The use of a medicated salt in an antimalaria campaign in West New Guinea. Trop. Geogr. Med. 16:245-255. [PubMed] [Google Scholar]
- 41.Mueller, I., J. Kaiok, J. C. Reeder, and A. Cortes. 2002. The population structure of Plasmodium falciparum and Plasmodium vivax during an epidemic of malaria in the Eastern Highlands of Papua New Guinea. Am. J. Trop. Med. Hyg. 67:459-464. [DOI] [PubMed] [Google Scholar]
- 42.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, D. S., W. K. Milhous, and T. E. Wellems. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. USA 87:3018-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson, D. S., D. Walliker, and T. E. Wellems. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. USA 85:9114-9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips, E. J., J. S. Keystone, and K. C. Kain. 1996. Failure of combined chloroquine and high-dose primaquine therapy for Plasmodium vivax malaria acquired in Guyana, South America. Clin. Infect. Dis. 23:1171-1173. [DOI] [PubMed] [Google Scholar]
- 46.Plowe, C. V. 2003. Monitoring antimalarial drug resistance: making the most of the tools at hand. J. Exp. Biol. 206:3745-3752. [DOI] [PubMed] [Google Scholar]
- 47.Plowe, C. V., A. Djimde, M. Bouare, O. Doumbo, and T. E. Wellems. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 52:565-568. [DOI] [PubMed] [Google Scholar]
- 48.Plowe, C. V., J. G. Kublin, and O. K. Doumbo. 1998. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. Updat. 1:389-396. [DOI] [PubMed] [Google Scholar]
- 49.Pukrittayakamee, S., M. Imwong, S. Looareesuwan, and N. J. White. 2004. Therapeutic responses to antimalarial and antibacterial drugs in vivax malaria. Acta Trop. 89:351-356. [DOI] [PubMed] [Google Scholar]
- 50.Rastelli, G., S. Pacchioni, and M. D. Parenti. 2003. Structure of Plasmodium vivax dihydrofolate reductase determined by homology modeling and molecular dynamics refinement. Bioorg. Med. Chem. Lett. 13:3257-3260. [DOI] [PubMed] [Google Scholar]
- 51.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174-1181. [DOI] [PubMed] [Google Scholar]
- 52.Ruebush, T. K., II, J. Zegarra, J. Cairo, E. M. Andersen, M. Green, D. R. Pillai, W. Marquino, M. Huilca, E. Arevalo, C. Garcia, L. Solary, and K. C. Kain. 2003. Chloroquine-resistant Plasmodium vivax malaria in Peru. Am. J. Trop. Med. Hyg. 69:548-552. [PubMed] [Google Scholar]
- 53.Sibley, C. H., J. E. Hyde, P. F. Sims, C. V. Plowe, J. G. Kublin, E. K. Mberu, A. F. Cowman, P. A. Winstanley, W. M. Watkins, and A. M. Nzila. 2001. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 17:582-588. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh, R. K. 2000. Emergence of chloroquine-resistant vivax malaria in south Bihar (India). Trans. R. Soc. Trop. Med. Hyg. 94:327. [PubMed] [Google Scholar]
- 56.Siripoon, N., G. Snounou, P. Yamogkul, K. Na-Bangchang, and S. Thaithong. 2002. Cryptic Plasmodium falciparum parasites in clinical P. vivax blood samples from Thailand. Trans. R. Soc. Trop. Med. Hyg. 96:70-71. [DOI] [PubMed] [Google Scholar]
- 57.Snounou, G., and N. J. White. 2004. The co-existence of Plasmodium: sidelights from falciparum and vivax malaria in Thailand. Trends Parasitol. 20:333-339. [DOI] [PubMed] [Google Scholar]
- 58.Soto, J., J. Toledo, P. Gutierrez, M. Luzz, N. Llinas, N. Cedeno, M. Dunne, and J. Berman. 2001. Plasmodium vivax clinically resistant to chloroquine in Colombia. Am. J. Trop. Med. Hyg. 65:90-93. [DOI] [PubMed] [Google Scholar]
- 59.Sumawinata, I. W., Bernadeta, B. Leksana, A. Sutamihardja, Purnomo, B. Subianto, Sekartuti, D. J. Fryauff, and J. K. Baird. 2003. Very high risk of therapeutic failure with chloroquine for uncomplicated Plasmodium falciparum and P. vivax malaria in Indonesian Papua. Am. J. Trop. Med. Hyg. 68:416-420. [PubMed] [Google Scholar]
- 60.Tjitra, E., J. Baker, S. Suprianto, Q. Cheng, and N. M. Anstey. 2002. Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob. Agents Chemother. 46:3947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trape, J. F. 2001. The public health impact of chloroquine resistance in Africa. Am. J. Trop. Med. Hyg. 64:12-17. [DOI] [PubMed] [Google Scholar]
- 62.Wallace, A. R. 1860. On the zoological geography of the Malay Archipelago (communicated by Charles Darwin to the LSL meeting of 3 November 1859). J. Proc. Linn. Soc. Zool. 4:172-184. [Google Scholar]
- 63.Wellems, T. E., and C. V. Plowe. 2001. Chloroquine-resistant malaria. J. Infect. Dis. 184:770-776. [DOI] [PubMed] [Google Scholar]
- 64.Wilairatana, P., U. Silachamroon, S. Krudsood, P. Singhasivanon, S. Treeprasertsuk, V. Bussaratid, W. Phumratanaprapin, S. Srivilirit, and S. Looareesuwan. 1999. Efficacy of primaquine regimens for primaquine-resistant Plasmodium vivax malaria in Thailand. Am. J. Trop. Med. Hyg. 61:973-977. [DOI] [PubMed] [Google Scholar]
- 65.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 66.Wooden, J. M., L. H. Hartwell, B. Vasquez, and C. H. Sibley. 1997. Analysis in yeast of antimalaria drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol. Biochem. Parasitol. 85:25-40. [DOI] [PubMed] [Google Scholar]
- 67.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]
- 68.Young, M. D., and R. W. Burgess. 1959. Pyrimethamine resistance in Plasmodium vivax malaria. Bull. W. H. O. 20:27-36. [PMC free article] [PubMed] [Google Scholar]