Abstract
Purpose
To illustrate the role of the exposome in child health while highlighting unique aspects of this research pertinent to children, such as the time dependency of environmental exposures on fetal programming, as well as the time dependent nature of child behavior, diet, and motor function, which alter the probability of exposure to different compounds. Future environmental health research will be more hypothesis generating but will also need to heed lessons learned from other “omic” sciences. The NIH Child Health Environmental Analysis Resource (CHEAR) is a major step towards providing the infrastructure needed to study the exposome and child health.
Recent Findings
Environmental exposures have overlapping mechanisms such as endocrine disruption and oxidative stress among others. The nature of the long term health impact of an exposure is dependent not only on dose, but also on the timing of exposure. Advances in exposure science, toxicology and biostatistics will create new opportunities to identify and better define windows of susceptibility to environmental exposures.
Summary
As exposure science matures, we will better understand the role of environment on health. Linking the exposome with genomics will unlock the root origins of multiple complex diseases.
Keywords: Exposome, windows of susceptibility, environmental health
Introduction: The Exposome-a new science
The “exposome” is defined as the totality of environmental exposure from conception to death1,2. Although characterizing the exposome is a monumental goal, many of the tools to measure environment on an “omic” scale already exist, and recent advances in analytical chemistry, geospatial statistics, and the same scientific and cultural developments that made smart phones ubiquitous now make the goal of estimating the exposome possible. In the next 10 years, more and more research will be conducted in “exposomics”. To build toward this larger goal, environmental health researchers must integrate a vast array of tools from multiple, seemingly disparate, scientific disciplines to successfully measure the exposome. In many ways, environmental health is at a similar development stage as genomics was 15 years ago. We can therefore learn several lessons from genomics, which already transitioned from a hypothesis driven science to a hypothesis generating “omic” science. The growth of genomics was propelled by embracing the best of multiple fields and building teams, and did not emanate from laboratories that were focused on a single aspect of genomics: a combination of biologists, physicians, bioinformaticists, computer scientists, statisticians, and an appreciation of “big data” drove the growth of genomics. By merging multiple fields, geneticists forced a need for the development of new technology, the enrollment of large study populations, and the creation of bioinformatic tools that could manage larger and larger amounts of data efficiently. This led to consortiums and further collaboration among epidemiologists, molecular biologists, bioinformaticians, statisticians and clinicians. The key to the rapid development of exposomics will be to follow this blueprint of success, and the NIH CHEAR (Child Health Exposure Analysis Resource) program provides a strong foundation. CHEAR will provide researchers with the tools to measure the exposome. Similar to genomics, the initial measures will be targeted (just as single nucleotide polymorphisms in microarrays were the mainstay assay 5–10 years ago) but as exposomic technology improves, environment will be measured on larger and larger scales.
Environment is key to understanding the Genome
In the last 2 decades, genomics profoundly changed the fundamental approach to science by moving away from hypothesis-driven research to agnostic, hypothesis-generating research followed by hypothesis testing replication. While genomics has rapidly advanced our understanding of the underlying biology of disease, actual disease-causing genetic variants are rare and account for no more than ~20% of the variance that explains the cause of complex diseases. Even in diseases such as autism, in which the genetic contribution was once predicted to be 80–100% of the heritability, the concept that measuring genetics alone could explain the root cause of the disease has fallen out of favor. Much debate has occurred over the source of this “missing heritability”..The underlying assumption of heritability estimates is that genetics and environment act independently to cause disease, and by doing, we typically overestimate the genetic contributions. In reality, genes and environment cannot work independently; they must interact, as even mutated genes require substrates to generate gene products. Research in the last 20 years has focused on unraveling the genetic component of disease risk, while the effect of environmental stimuli received limited attention. In large part, the lack of focus on environmental causes was driven by the absence of tools to measure the environment on an “omic” scale. Until recently, while millions of SNPs could be measured in a population, interactions with environmental risk factors were limited to a handful of measurements. This discordance tended to minimize the importance of environment. Further, these environmental risks were chosen not because they were known to interact with given genetic variants but primarily because they were easiest to measure. Environmental exposure assessment technology has exploded in the last few years and continues to develop. In recent years, many have concluded that Gene x Environment interactions likely explain the largest portion of the missing heritability3,4 The time is now ripe to leverage these gains and conduct gene-environment interaction investigations on a scale that can discover and replicate findings via both genomic and exposomic approaches.
Linking Exposomics with Genomics
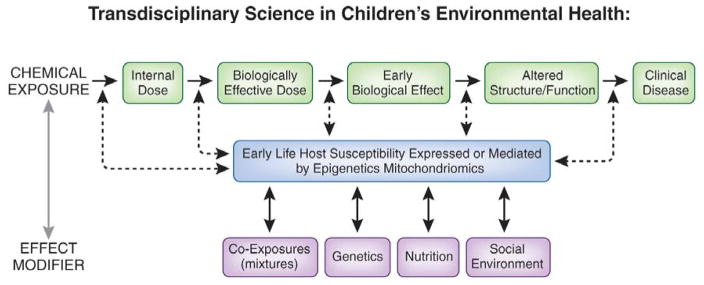
Although an emerging consensus suggests that prevalent complex diseases in humans develop as a result of multiple biologically unique gene–environment and environment by environment interaction (Figure 1), this conceptual framework remains limited. In fact, the development of disease in humans is far more complex and is not even a 3-dimensional issue (i.e., involving multiple interactions) but a 4-dimensional issue (i.e., changes in interaction-related risk over time). Environmental exposures affect those who are vulnerable temporally (age at time of exposure) and by unique circumstance (co-morbid disease, nutritional status, economic status, culture, genetics). Even this paradigm fails to address the complex interaction of endogenous and exogenous chemicals that ultimately interact to cause disease. Human genetics provided an unparalleled insight to understanding how genes and genetic variants interact with environmental exposures to either preserve health or cause disease. These advances have led to the concept of systems biology research and the study of genetic variants as networks instead of individual predictors. However, as currently practiced, Systems Biology is an incomplete science. Without accounting for the temporal, spatial, and other unique components of an individual’s microenvironment, our understanding of genetics and gene networks remains incomplete. Even the downstream products of genes (transcriptomics, proteomics and metabolomics etc) are only partial pieces of a much larger puzzle. Understanding the complex relationships among our genetic code, our metabolic machinery, and our temporally changing environment in populations and within affected individuals is precisely the opportunity and challenge that scientists involved in both environmental health and genomics face.
Figure 1. Transdisciplinary Science in Children’s Environmental Health.
Schematic of relationship between environmental exposures and clinical disease. These effects are exposure timing dependent as well.
The Uniqueness of Children’s Health Research: Opportunities and Pitfalls
Over the last 2 decades a growing appreciation for the role of fetal/childhood environments on child and adult health has come to the forefront. The field of “developmental origins of health and disease (DOHaD) has even developed to the point where it now hosts an international society with an annual conference(https://dohadsoc.org/). Concurrent to these developments has been the rise of “omic” science (genomics, epigenomics, proteomics and metabolomics among others). These technology driven fields have led to an explosion of measures that can be incorporated into population based research. Combining the lessons learned from developmental origins with these new technologies will require thoughtful, interdisciplinary approaches that can integrate chemistry, pediatrics, developmental biology, epidemiology and biostatistics/bioinformatics. For example, factoring in the role of child development in metabolomic assays will be considerably more complex in children than in adults. Adult metabolism can be grouped by years (i.e. the average 35 year old is not particularly different then a 40 years old) while in child health, developmental metabolism changes occur in time sets of months or even weeks. Overly simplistic approaches of adjusting for age cannot capture this complexity and will lead to erroneous interpretations of data. In addition, all “omic” analyses are hypothesis generating and must be replicated, otherwise they will inevitably produce primarily false positive results5–7. Relative to genomics, time varying measures, such as the exposome, metabolome, proteome and epigenome, must factor age of the child, as the risk of exposure (behaviors, motor development, diet) and response to exposure(metabolism, gene expression variation, cell differentiation etc) operate at varying degrees of intensity over childhood life stages. Replication must also be careful to factor in these issues to avoid type 2 error as well as type 1 error. While the pitfalls are many, so are the potential rewards. The rapid time varying life stages of childhood are an opportunity to build upon the current state of science towards a better understanding of disease etiology, prevention and treatment. With a growing understanding that the childhood environment is often a root cause for adult health and disease, a focused effort to conduct exposomic research during childhood will touch almost every field of biology.
Susceptibility Windows and Exposure Assessment
Fetal life is a state of sequential physiological shifts, driven by cell specific gene expression changes in which there are dynamic changes in growth rates as well as the establishment of hormonal and metabolic circadian rhythms. This timed cascade of biological events means that the fetus is highly vulnerable to even subtle environmental insults, as cell and tissue differentiation is most active at this life stage, yet defense mechanisms against toxic environmental factors are underdeveloped.8 These events occur over varying time scales of years, months, weeks or even days. To produce programmed effects in the absence of overt cellular toxicity, environmental exposures must coincide with these timed developmental processes inducing changes in the developmental trajectory. Some of the earliest reports on DOHaD, came from David Barker’s research team, which conducted a series of seminal studies on fetal nutritional environment and subsequent adult cardiometabolic health9–11. Increasingly, the observation that early life nutritional famine was a strong predictor of later life hypertension, obesity and even behavioral disorders such as schizophrenia12,13 has become an accepted biological tenet. Research on the fetal chemical and social environment has found many parallel effects to the Barker hypothesis, which have been termed “windows of susceptibility” or “critical windows”. To advance the field of developmental origins of disease, exposure assessment must occur at time scales that can assess multiple life stages, sometimes as brief as a few months. Prospective, longitudinally assessed data on environment in multiple life stages is the key to this endeavor. CHEAR can measure multiple toxicants on extant samples to begin the process of systematically searching for critical windows of susceptibility.
Example of Environmental Programming: Oxidative Stress
While many mechanisms may contribute to nutritional or toxicant-elicited disruption of fetal development, a growing body of evidence underscores a central role for oxidative stress (OS) 14,15. Oxidative stress may be a source of toxicity but it is also a signal for normal development16,17. Reactive oxygen species are needed to induce the timed transcription of genes critical to cell differentiation and proliferation16. During pregnancy, increased placentation and oxygenated blood flow coincide with rapid growth and increased oxygen needs, creating a switch in the fetal redox state from reduced to oxidized. This switch is critical to timed gene expression changes and drives cell and tissue differentiation. The placenta regulates the delicate balance between normal and excess ROS generation, preventing toxicity and promoting cell differentiation. Hypoxemia and poor nutrition cause oxidative stress 18,19,20 and can be driven by maternal health/environment, nutrient/waste exchange and placental vascular reactivity. Chemical oxidants can be directly fetal toxic if transferred across the placenta, but may also induce cytokines and other inflammatory mediators that constrict placental blood flow, mimicking famine like effects 21,22. In either case, oxidants alter normal gene expression signals, producing a stressed fetal environment. The key concept is that the fetus may react to a hostile in utero environment by programming gene expression in preparation for a similarly hostile ex utero life. However, if ex utero resources are plentiful, these programmed gene expression changes can become detrimental.
Mechanisms of Oxidative Stress and Fetal Programming
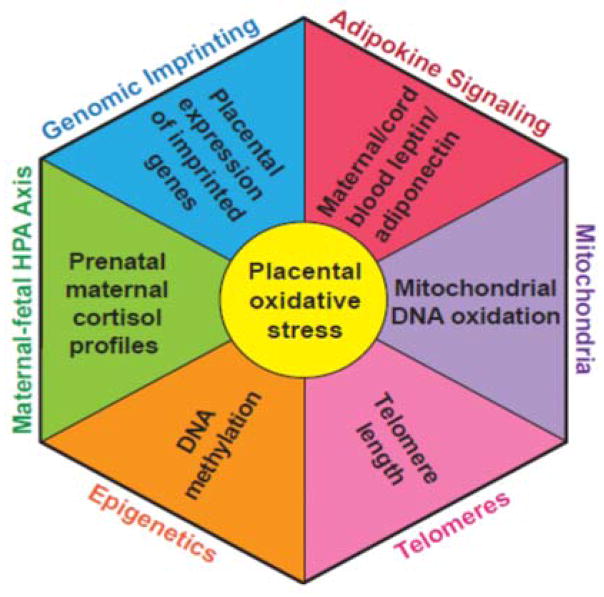
How might oxidative stress cause programming? Oxidative stress impacts multiple heritable cellular processes including DNA methylation/imprinting, mitochondrial DNA (mtDNA) function, and telomere biology, all of which influence programming 23–27. The placenta, which is the gateway to the fetus, is highly sensitive to these pro-oxidant stressors. All have the potential to alter biochemical, molecular, epigenetic and gene expression processes (see Figure 2). Thus, the placenta provides a readily available source of integrated molecular information on the cumulative past oxidative stress that occurred over pregnancy. Moreover, which pathways are disrupted, and/or which disorder originates, may depend as much on timing as on differences in exposures26,28. Key challenges for future research include whether we can identify biomarkers at the maternal-fetal interface that reflect an integrated measure of mixed oxidant exposures over pregnancy and how to better estimate the influence of exposure timing.
Figure 2. Central Role of Oxidative Stress in Fetal Programming.
Figure illustrates the role of oxidative stress experienced in the placenta as a mediator of programmed health effects experienced in later life.
Untargeted Chemical Profiling
The vast majority of children’s environmental health studies have focused on one or, at most, a few candidate chemicals or metabolites which may cause disease or disorders in children. While there are strengths to such an approach, including biologic plausibility and clear a priori hypotheses, there are also limitations to selecting only a few environmental exposures in a single study. Human population studies take years to conduct and are expensive, and this pace of research is very limiting with regards to informing health care providers and public policy makers. In addition, given the multifaceted nature of biologic interactions, it is difficult to conceive that one environmental exposure can ever primarily account for the rise of complex diseases such as asthma, autism, obesity, etc. Finally, there are certainly chemicals/metabolites important to disease pathogenesis or child development that are not yet known, and keeping in mind that more than 80,000 chemicals are registered with the EPA for commercial use in the U.S,29 a candidate exposure approach may never be able to identify the chemical and nonchemical mixtures driving disease processes. The selection of environmental exposures in candidate studies is always open to bias. That is, we tend to study what we already believe or suspect is toxic. If we are to keep pace, we must rethink our approach to environmental health research.2,30 Similar to the field of genetics that capitalized on the genome wide scan to advance new genetic “discoveries”, environmental health must begin to utilize untargeted scans allowing for screening of the exposome to “discover” novel environmental risk factors and their effects.1,31 Advances in analytical chemistry methods now allow assessment of exposure to hundreds to thousands of untargeted classes of chemicals by performing global screening of small molecules using methods such as liquid chromatography coupled with quadrupole time-of-flight mass spectrometry metabolomics. Our team has even applied this technology to novel matrices such as teeth that can reconstruct exposures from fetal life and infancy32. We found (a) more than 12,000 unique chemical signatures in trimester-specific dentine layers, (b) high inter- and intra-child variability in chemical profiles, and (c) ‘known unknown’ and ‘suspected unknown’ compounds. Because we used teeth, reconstruction of exposures was done 7 to 10 years after prenatal and early childhood exposure. A similar approach could be applied to ongoing studies, even those that did not collect prenatal biospecimens enabling exposomic research on prenatal exposure even with prospective sample collection in pregnancy.
Statistical methods must integrate with exposure and biology
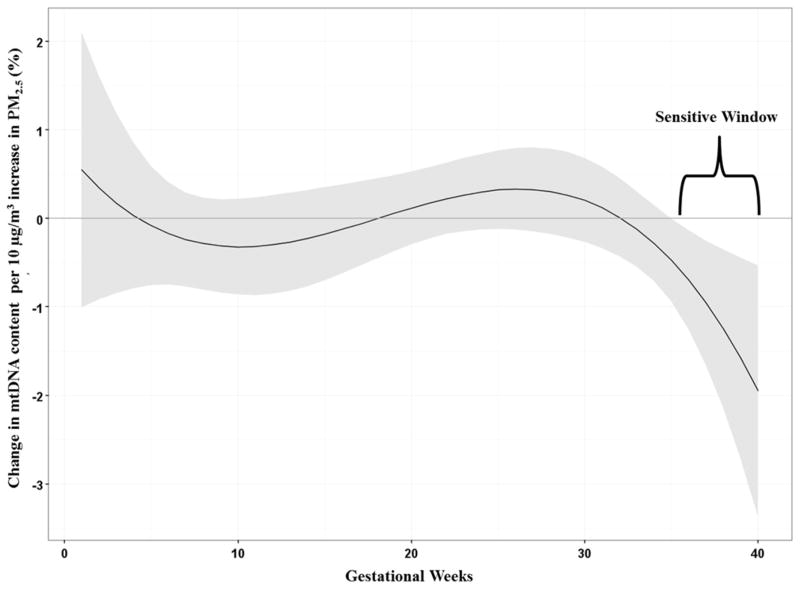
Given its complexity, exposome research will require a team-based approach to assess the relationship between environment and health, integrating exposure science, with pediatrics, developmental biology, and statistics. To illustrate, our team recently developed a new approach to identifying developmental windows of susceptibility using air pollution exposure data collected daily. The concept of “windows of susceptibility” implies that exposure timing determines subsequent health effects28,33,34. Due to the rapidity of development, windows of susceptibility can be missed if exposure measures are made outside the appropriate window, even if the correct chemical is assessed; however, the time boundaries of windows are rarely known and researchers effectively are left to guess when the windows occur. If we could measure exposure repeatedly over short, frequent time periods rather than collecting serial exposure measures months or years apart, then we could use data-driven methods to objectively identify windows of susceptibility. To facilitate such an approach, we have proposed novel methods that provide highly temporally resolved exposure data thereby coupling our novel exposure methods with data-driven statistics to determine the windows of susceptibility in which air pollution (PM2.5 –particulate matter<2.5 microns in diameter) exposure predict health outcomes. Fig 3 is a previously published result illustrating this concept using prenatal PM2.5 measured weekly across pregnancy to estimate mitochondrial DNA copy number, a biomarker of cumulative oxidative stress. The Distributed Lag Regression Model reveals that the critical window for this mixture is found in the late 3rd trimester. These data driven methods to identify critical windows may well be a “rosetta stone” in biological science35,36, as critical susceptibility windows are in nearly all cases, ill defined.
Figure 3.
Associations between weekly prenatal PM2.5 and mtDNA content in cord blood adjusted for sex, maternal age at delivery, year of birth, maternal education, prenatal exposure to environmental tobacco smoke and batch. The y-axis represents the change in mtDNA content associated with a 10μg/m3 increase in PM2.5; the x-axis is gestational age in weeks. Solid lines show the predicted change in mtDNA content. Gray areas indicate 95% CIs. A sensitive window is identified for the weeks where the estimated pointwise 95% CI (shaded area) does not include zero (i.e. the length of time the regression is statistically significant.)
Conclusions
These are exciting times for Environmental Health and Exposure Science. The field has made tremendous strides toward developing an exposomics approach to its research in recent years37–39. CHEAR represents the type of infrastructure achievements that create the foundation for the development of new methods and new consortia that can address the big picture questions of how environment impacts health and development. Future research will no doubt generate new methods for exposure assessment, data harmonization and statistical approaches but with respect to children’s health will need to factor in the time varying biological processes that define childhood. The CHEAR program is well positioned to deliver the research tools that will bring the nascent field of exposomics to the forefront of medical science.
The Exposome is a new science that assesses the totality of human exposure from conception to death.
Measuring environment rigorously will improve our understanding of genomics and developmental biology.
The next decade will see enormous advances in our ability to measure human environment across all life stages.
Windows of susceptibility are the key to understanding how genes and environment interact.
Multidisciplinary teams are needed to avoid pitfalls that arise from focus on technology rather than biology and rigorous study design and statistics.
Acknowledgments
I would like to thank Ms Rozalyn Paupaw for her assistance with the manuscript
Financial support and sponsorship
This work was supported in part by grants from the National Institutes of Health, U2CES026561; R01ES026033; UG3 OD023337; P30 ES023515 and R01 ES013744
Footnotes
Conflicts of interest
None
References
- 1**.Rappaport SM. Implications of the exposome for exposure science. Journal of exposure science & environmental epidemiology. 2011;21(1):5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- 2**.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. References 1 and 2 are landmark papers illustrating the potential of the exposome to identify the underlying root causes of complex diseases. [DOI] [PubMed] [Google Scholar]
- 3.Rappaport SM. Genetic Factors Are Not the Major Causes of Chronic Diseases. PloS one. 2016;11(4):e0154387. doi: 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon PH, Sylvestre MP, Tremblay J, Hamet P. Key Considerations and Methods in the Study of Gene-Environment Interactions. American journal of hypertension. 2016;29(8):891–899. doi: 10.1093/ajh/hpw021. [DOI] [PubMed] [Google Scholar]
- 5.Begley CG, Ioannidis JP. Reproducibility in science: improving the standard for basic and preclinical research. Circulation research. 2015;116(1):116–126. doi: 10.1161/CIRCRESAHA.114.303819. [DOI] [PubMed] [Google Scholar]
- 6.Goodman SN, Fanelli D, Ioannidis JP. What does research reproducibility mean? Science translational medicine. 2016;8(341):341ps312. doi: 10.1126/scitranslmed.aaf5027. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology (Cambridge, Mass) 2008;19(5):640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 8.Hogg K, Price EM, Hanna CW, Robinson WP. Prenatal and perinatal environmental influences on the human fetal and placental epigenome. Clinical pharmacology and therapeutics. 2012;92(6):716–726. doi: 10.1038/clpt.2012.141. [DOI] [PubMed] [Google Scholar]
- 9.Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341(8850):938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- 10.Osmond C, Barker DJ, Slattery JM. Risk of death from cardiovascular disease and chronic bronchitis determined by place of birth in England and Wales. Journal of epidemiology and community health. 1990;44(2):139–141. doi: 10.1136/jech.44.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravelli AC, van Der Meulen JH, Osmond C, Barker DJ, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. The American journal of clinical nutrition. 1999;70(5):811–816. doi: 10.1093/ajcn/70.5.811. [DOI] [PubMed] [Google Scholar]
- 12*.Barker DJ, Lampl M, Roseboom T, Winder N. Resource allocation in utero and health in later life. Placenta. 2012;33(Suppl 2):e30–34. doi: 10.1016/j.placenta.2012.06.009. References 9–12 are among the first to illustrate the role of the fetal environment in adult onset disease. [DOI] [PubMed] [Google Scholar]
- 13.Neugebauer R. Accumulating evidence for prenatal nutritional origins of mental disorders. Jama. 2005;294(5):621–623. doi: 10.1001/jama.294.5.621. [DOI] [PubMed] [Google Scholar]
- 14.Myatt L, Cui X. Oxidative stress in the placenta. Histochemistry and cell biology. 2004;122(4):369–382. doi: 10.1007/s00418-004-0677-x. [DOI] [PubMed] [Google Scholar]
- 15.Herrera EA, Krause B, Ebensperger G, et al. The placental pursuit for an adequate oxidant balance between the mother and the fetus. Fronteirs in Pharmacology. 2014;5:49. doi: 10.3389/fphar.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennery PA. Oxidative stress in development: nature or nurture? Free radical biology & medicine. 2010;49(7):1147–1151. doi: 10.1016/j.freeradbiomed.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Hitchler MJ, Domann FE. An epigenetic perspective on the free radical theory of development. Free radical biology & medicine. 2007;43(7):1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolton JL, Bilbo SD. Developmental programming of brain and behavior by perinatal diet: focus on inflammatory mechanisms. Dialogues in clinical neuroscience. 2014;16(3):307–320. doi: 10.31887/DCNS.2014.16.3/jbolton. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Patot MC, Ebensperger G, Gassmann M, Llanos AJ. The hypoxic placenta. High altitude medicine & biology. 2012;13(3):176–184. doi: 10.1089/ham.2012.1046. [DOI] [PubMed] [Google Scholar]
- 20.Jansson T, Powell TL. Role of the placenta in fetal programming: underlying mechanisms and potential interventional approaches. Clin Sci (Lond) 2007;113(1):1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- 21.Chertok RJ, Kullgren B, Burbank D. The effects of CdCl2 on the maternal-to-fetal clearance of 67Cu and placental blood flow. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine (New York, N.Y.) 1984;176(2):138–142. doi: 10.3181/00379727-176-41853. [DOI] [PubMed] [Google Scholar]
- 22.Lasuncion MA, Lorenzo J, Palacin M, Herrera E. Maternal factors modulating nutrient transfer to fetus. Biology of the neonate. 1987;51(2):86–93. doi: 10.1159/000242637. [DOI] [PubMed] [Google Scholar]
- 23.Byun HM, Baccarelli A. Environmental expousre and mitochondrial epigenetics: study design and analytical challenges. Human Genetics. 2014;133:247–257. doi: 10.1007/s00439-013-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron N, Demerath EW. Critical periods in human growth and their relationship to diseases of aging. American Journal Physical Anthropology. 2002;35:159–184. doi: 10.1002/ajpa.10183. [DOI] [PubMed] [Google Scholar]
- 25.Entinger S, Buss C, Wadhwa PD. Prenatal stress, telomere biology, and fetal programming of health and disease risk. Science Signaling. 2012;5:112. doi: 10.1126/scisignal.2003580. [DOI] [PubMed] [Google Scholar]
- 26.Janssen BG, Byun HM, Cox B, et al. Variation of DNA methylation in candidate age-related targets on the mitochondrial-telomere axis in cord blood and placenta. Placenta. 2014;35:665–672. doi: 10.1016/j.placenta.2014.06.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaughnessy DT, McAllister K, Worth L, et al. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environmental health perspectives. 2014;122(12):1271–1278. doi: 10.1289/ehp.1408418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells JC. Adaptive variability in the duration of critical windows of plasticity: Implications for the programming of obesity. Evolution, medicine, and public health. 2014;2014(1):109–121. doi: 10.1093/emph/eou019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandjean P, Landrigan PJ. Developmental neurotoxicity of industrial chemicals. Lancet. 2006;368(9553):2167–2178. doi: 10.1016/S0140-6736(06)69665-7. [DOI] [PubMed] [Google Scholar]
- 30.Wild CP. The exposome: from concept to utility. International journal of epidemiology. 2012;41(1):24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 31.Kortenkamp A, Faust M, Scholze M, Backhaus T. Low-level exposure to multiple chemicals: reason for human health concerns? Environmental health perspectives. 2007;115(Suppl 1):106–114. doi: 10.1289/ehp.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andra SS, Austin C, Wright RO, Arora M. Reconstructing pre-natal and early childhood exposure to multi-class organic chemicals using teeth: Towards a retrospective temporal exposome. Environment international. 2015;83:137–145. doi: 10.1016/j.envint.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fudvoye J, Bourguignon JP, Parent AS. Endocrine-disrupting chemicals and human growth and maturation: a focus on early critical windows of exposure. Vitamins and hormones. 2014;94:1–25. doi: 10.1016/B978-0-12-800095-3.00001-8. [DOI] [PubMed] [Google Scholar]
- 34.Luo ZC, Fraser WD, Julien P, et al. Tracing the origins of “fetal origins” of adult diseases: programming by oxidative stress? Medical hypotheses. 2006;66(1):38–44. doi: 10.1016/j.mehy.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 35*.Chiu YH, Hsu HH, Coull BA, et al. Prenatal particulate air pollution and neurodevelopment in urban children: Examining sensitive windows and sex-specific associations. Environment international. 2016;87:56–65. doi: 10.1016/j.envint.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Rosa MJ, Just AC, Guerra MS, et al. Identifying sensitive windows for prenatal particulate air pollution exposure and mitochondrial DNA content in cord blood. Environment international. 2016 doi: 10.1016/j.envint.2016.11.007. References 35 and 36 are the first to illustrate how time dependent exposure assessed in short time intervals can be analyzed to identify windows of susceptibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Y, Balshaw DM, Kwok RK, Thompson CL, Collman GW, Birnbaum LS. The Exposome: Embracing the Complexity for Discovery in Environmental Health. Environmental health perspectives. 2016;124(8):A137–140. doi: 10.1289/EHP412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeBord DG, Carreon T, Lentz TJ, Middendorf PJ, Hoover MD, Schulte PA. Use of the “Exposome” in the Practice of Epidemiology: A Primer on -Omic Technologies. American journal of epidemiology. 2016;184(4):302–314. doi: 10.1093/aje/kwv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland N. Future of environmental research in the age of epigenomics and exposomics. Reviews on environmental health. 2016 doi: 10.1515/reveh-2016-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]