Abstract
Background.
The nuclear factor I-A (NFIA) transcription factor promotes glioma growth and inhibits apoptosis in glioblastoma (GBM) cells. Here we report that the NFIA pro-survival effect in GBM is mediated in part via a novel NFIA–nuclear factor-kappaB (NFκB) p65 feed-forward loop.
Methods.
We examined effects of gain- and loss-of-function manipulations of NFIA and NFκB p65 on each other’s transcription, cell growth, apoptosis and sensitivity to chemotherapy in patient-derived GBM cells and established GBM cell lines.
Results.
NFIA enhanced apoptosis evasion by activating NFκB p65 and its downstream anti-apoptotic factors tumor necrosis factor receptor-associated factor 1 (TRAF1) and cellular inhibitor of apoptosis proteins (cIAPs). Induction of NFκB by NFIA was required to protect cells from apoptosis, and inhibition of NFκB effectively reversed the NFIA anti-apoptotic effect. Conversely, NFIA knockdown decreased expression of NFκB and anti-apoptotic genes TRAF1 and cIAPs, and increased baseline apoptosis. NFIA positively regulated NFκB transcription and NFκB protein level. Interestingly, NFκB also activated the NFIA promoter and increased NFIA level, and knockdown of NFIA was sufficient to attenuate the NFκB pro-survival effect, suggesting a reciprocal regulation between NFIA and NFκB in governing GBM cell survival. Supporting this, NFIA and NFκB expression levels were highly correlated in human GBM and patient-derived GBM cells.
Conclusions.
These data define a previously unknown NFIA-NFκB feed-forward regulation that may contribute to GBM cell survival.
Keywords: apoptosis, chemoresistance, glioblastoma (GBM), NFκB, nuclear factor I-A (NFIA)
Importance of this study
In this study we identify a previously unknown feed-forward loop between two pro-GBM transcription factors, NFIA and NFκB. We show that NFIA and NFκB promote each other’s expression and function. This feed-forward loop contributes to enhanced GBM cell survival and suppression of apoptosis. The novel NFIA-NFκB positive feedback loop that we have discovered in GBM may prove helpful in development of improved therapeutic approaches against glioblastomas.
Gliomas are the most common primary tumors in the CNS, representing nearly half of all primary intracranial neoplasms. Malignant gliomas constitute 80% of malignant tumors in the CNS. Glioblastoma (GBM) is the most common malignant glioma and is mostly incurable.1–3 Despite advances in treatment using surgery, chemotherapy, and radiation, median survival remains ~15 months.4–6 GBM often exhibits aberrant differentiation, suggesting a deregulated neurodevelopmental program. Increasing evidence by our group and others suggests that nuclear factor I-A (NFIA), a glial fate determinant, plays pivotal roles in both embryonic development and tumorigenesis of the nervous system.
The NFI family (NFIA, NFIB, NFIC, and NFIX) of site-specific DNA-binding proteins was first described as genes required for viral replication and regulation of gene expression.7–9 NFI genes comprise a family of vertebrate nuclear proteins that recognize and bind the palindromic DNA sequence 5'-TTGGC(N)5GCCAA-3' and are capable of activating or repressing transcription and DNA replication.10 In the CNS, the NFIA transcription factor is a principal player in glial development as it mediates glial lineage specification, maintains glial progenitors, and regulates astrocyte terminal differentiation.11 In addition to its role in glial development, NFIA has been implicated in human glial tumors.12–14 We and others reported that NFIA is highly expressed in astrocytomas of all grades compared with nonneoplastic brains and that high NFIA confers growth advantage in gliomas. We recently reported that NFIA is necessary and sufficient to promote glioma growth, proliferation, and inhibition of apoptosis in a manner mediated in part through negative regulation of tumor suppressors, such as p53 and p21, identifying NFIA as a critical component of the oncogenic network in glioma.13 The molecular mechanisms by which NFIA induces glioma cell survival and drug resistance are not fully understood.
The NFκB family of transcription factors mediates a variety of cellular responses and disease processes, including apoptosis resistance, immune response, and cancer development.15–18 NFκB activation in cancer may be the result of either exposure to pro-inflammatory stimuli, such as tumor necrosis factor (TNF), or upregulation of signaling from upstream regulators.19–22 NFκB is constitutively active in many cancers, including gliomas,23–25 and aberrant regulation of NFκB signaling is involved in apoptosis evasion and tumor promotion.26 In GBM, elevated NFκB signaling is associated with enhanced chemo- and radiation resistance in the mesenchymal subtype.27 NFκB plays a fundamental role in inhibiting apoptosis by inducing anti-apoptotic factors, such as TRAF1 and cellular inhibitors of apoptosis proteins 1 and 2 (cIAP1/2).28 Conversely, inhibition of the NFκB p65 pathway induces caspase-mediated apoptosis. We now have identified putative NFI-binding sites in the human NFκB p65 promoter, and therefore set out to determine if NFκB mediated the pro-survival and apoptosis-resisting effects of NFIA in GBM. Here we demonstrate a novel and previously unknown reciprocal feed-forward loop involving NFIA and NFκB, which contributes to the anti-apoptotic effect in GBM.
Materials and Methods
Materials
Primary antibodies used for immunoblotting, immunofluorescent staining, and chromatin immunoprecipitation (ChIP) were: anti-NFIA, rabbit polyclonal (Active Motif, and V. Dawson, Johns Hopkins University); anti-NFκB p65, anti-TRAF1, anti-cIAP1, anti-cIAP2, anti–caspase-8, anti–signal transducer and activator of transcription 3 (STAT3), rabbit polyclonal (Cell Signaling Technology); anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and mouse monoclonal (Meridian Life Science). Temozolomide and etoposide were purchased from Sigma, NFκB inhibitor from Millipore (cat# 481407).
Brain Tumors, Cell Culture, Growth, and Tumorsphere Assay
GBM tumors and nonneoplastic brain tissues from epilepsy surgery were obtained from the New York University Human Brain Tumor Bank after institutional review board (7658) approval. U251MG and U87MG cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and L-glutamine (2mM), and maintained in 5% CO2 at 37°C. Human patient derived GBM cells (glioma sphere cultures [GSC]) were previously described13,29 and were cultured in serum-free neurobasal media supplemented with N2, B27, and L-glutamine (Invitrogen), along with epidermal growth factor and basic fibroblast growth factor (20 ng/mL each). Cells were used according to the needs of the experimental design (ie, transduction efficacy, growth as monolayer, etc). For cell growth analysis, glioma cells were enumerated by direct microscopic cell counting using trypan blue exclusion at day 3 posttransduction by 2 independent observers in a blinded fashion. GBM tumorsphere assay was performed using human patient derived GSC expressing NFIA constructs (NFIA, vector, short hairpin NFIA [shNFIA], and control shRNA [shCont]), and tumorspheres larger than 0.1mm were quantified as described before.13
Apoptosis Assays, Caspase-3 Activity
Apoptosis was evaluated by labeling DNA breaks with fluorescein isothiocyanate–deoxyuridine triphosphate (FITC-dUTP) followed by flow cytometry, using the Apo-Direct kit (BD Biosciences Pharmingen), according to the manufacturer’s instructions. Caspase activity was measured using the ApoTarget Caspase-3 Colorimetric Protease Assay (BioSource), and was determined in 200 μg of lysate proteins according to the manufacturer’s instructions. Absorbance at 400/405 nm was determined after 16 h of incubation (37°C) with the substrate.
Plasmids
To express NFIA, we used a lentiviral vector that contains human influenza hemagglutinin (HA)–NFIA cDNA.13 To achieve efficient knockdown of endogenous NFIA, we used a lenti-shRNA specific to human NFIA (shNFIA) and compared it with a control shRNA (shCont) as previously described.13 The human NFκB p65 (−507 to 67) and NFIA (−1400 to −450) promoter plasmids were generated using gene synthesis (Genewiz). The inserts were ligated into pGL3-basic vector (Promega) using KpnI/XhoI sites. Mutagenesis was performed using the QuikchangeII site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The plasmid containing green fluorescent protein (GFP)–p65 was purchased from Addgene (cat# 23255).30 The 3x κB-luc-reporter was previously described.31
Lentiviral Transduction, Transient Transfection, and Promoter Reporter Assays
GBM cells (30000 cells) were incubated at 37°C for 8–12 hours with lentivirus (FUGW-NFIA, vector, shNFIA, or shCont) in a 100 µL volume. Transduction efficiency was analyzed 3 days after transduction with either flow cytometry or immunofluorescence microscopy. Transient transfection was performed using calcium phosphate. One day after transfection with expression constructs encoding NFIA (NFIA or shNFIA), p65 (GFP-p65 or small interfering [si]NFκB p65) or controls (vector or shRNA or siRNA control), cells were co-transfected with Renilla luciferase reporter plasmid (20ng) and pGL3 Firefly luciferase plasmid (50ng). After 2 days of transfection, luciferase activities were measured using the Dual Luciferase Reporter assay system (Promega) according to the manufacturer’s instructions.
RNA Extraction and Real-Time PCR
RNA was extracted using TRIzol, and first-strand cDNA was synthesized using Super Script II Reverse Transcriptase (Invitrogen). Real-time (RT)-PCR analysis was done using the one-step RT-PCR system (Invitrogen). The primers used are the following: NFIA 5'-TAATCCAGGGCTCTGTGTCC-3' and 5'-CCTGCAGCTATTGGTGTCTG-3'; p65 5'-CCGCACCTC CACTCCATCC-3' and 5'-ACATCAGCACCCAAGGACACC-3'; GAPDH 5'-GAGTCAACGGATTTGGTCGT-3' and 5'-GACAA GCTTCCCGTTCTCAG-3'.
Chromatin Immunoprecipitation Assays
GBM cells were fixed with 1% formaldehyde for 10 minutes. Cross-linked chromatin was then sheared by sonication. The samples were precleared using magnetic beads. The antibody was coupled to the magnetic beads, the complex was added to the precleared chromatin, and the reaction mix was incubated for 12–16 hours. Immunoprecipitated complexes were isolated, the cross-links reversed, and proteins digested with proteinase K. The DNA was purified, and quantitative (q)PCR was performed using region-specific primers: NFIA 5'-ATGGCGTCTGTGTTCAAACC-3' and 5'-GTGAGGAATCAGTGCCCACA-3'; p65 5'-AAACAAAGTGA GTAATCGGCGG-3' and 5'-GACGAAGAGGCTGGCGTG-3'.
Immunoblotting
Whole cell lysates were fractionated on 10% or 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Milipore). Blots were incubated overnight (4°C) with primary antibodies, probed with horseradish peroxidase–conjugated secondary antibodies, and visualized by enhanced chemiluminescence (Amersham). Immunodensitometry was measured using ImageJ (National Institutes of Health).
Immunofluorescence Staining
Cells were fixed with 4% paraformaldehyde and incubated at 4°C overnight with primary antibodies, followed by incubation at room temperature with AlexaFluor-conjugated secondary antibodies (Invitrogen). After labeling nuclei (Hoechst), cells were analyzed using an EclipseE800 microscope (Nikon Instruments), a Nikon FDX-35 camera (Axiovision software, Carl Zeiss), and an EVOS Auto FL Cell Imaging System (Thermo Fisher Scientific).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0c for MacIntosh software. All experiments were performed in triplicates and repeated at least 3 times unless indicated otherwise. Data are expressed as mean ± SD. P-values were calculated by unpaired Student’s t-test for comparison between 2 groups and an ANOVA test for multiple groups. Significance level was set at P < .05.
Results
NFIA Enhances Glioma Cell Survival and Resistance to Chemotherapy
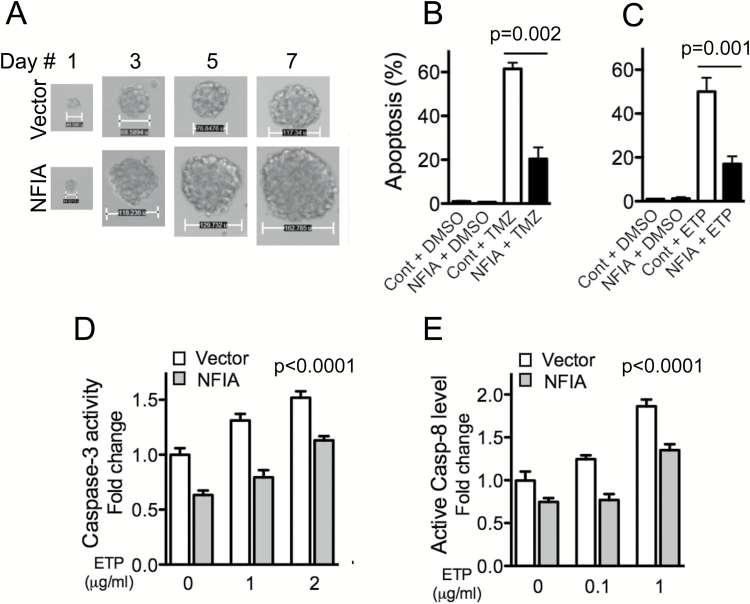
We previously reported that NFIA is abundantly expressed in malignant gliomas compared with nonneoplastic brains and that high NFIA confers growth advantage on GBM cells.13 To evaluate whether NFIA contributes to resistance to chemotherapy-induced cell death, we compared patient-derived GBM cell tumorsphere formation using NFIA gain-of-function in the presence or absence of temozolomide or etoposide in culture. Overexpression of NFIA increased the tumorsphere-forming capacity compared with vector control (Fig. 1A, Fig. S1). Furthermore, ectopic NFIA diminished temozolomide- and etoposide-induced apoptosis by 50%–70% (Fig. 1B–C, Fig. S2). Overexpression of NFIA also diminished 2 other measures associated with apoptosis: activity of caspase-3 and cleavage of caspase-8 (Fig. 1D–E). Together, these data suggest that NFIA contributes to survival and chemoresistance of GBM cells. However, analysis using the dataset of The Cancer Genome Atlas (TCGA) (Cell 2013, cBioPortal) 32,33 showed no significant correlation between NFIA mRNA expression and O6-methylguanine DNA methyltransferase (MGMT) methylation status, a predictive marker for temozolomide sensitivity,34 suggesting that the anti-apoptotic effect of NFIA may be independent of MGMT methylation status.
Fig. 1.
NFIA enhances GBM cell survival. (A) Growth of GBM1 primary tumorspheres stably expressing NFIA or vector (also see Fig. S1). (B, C) GBM1 cells stably expressing NFIA or vector control (Cont) were treated with temozolomide (TMZ 25 μg/mL, B) or etoposide (ETP 1 μg/mL, C; Fig. S2) or vehicle (dimethyl sulfoxide [DMSO]) for 24 h. Apoptosis (%) was assessed using the Apo-Direct kit; means ± SD from 3 experiments. (D) NFIA inhibits caspase-mediated apoptosis induced by ETP; caspase-3 activity measured in GBM cells expressing NFIA or vector following treatment with ETP. (E) Caspase-8 cleavage measured in GBM cells expressing NFIA or vector following treatment with the increasing amount of etoposide (ETP; also see Fig. S3).
NFκB p65 Is a Transcriptional Target of NFIA in GBM Cells
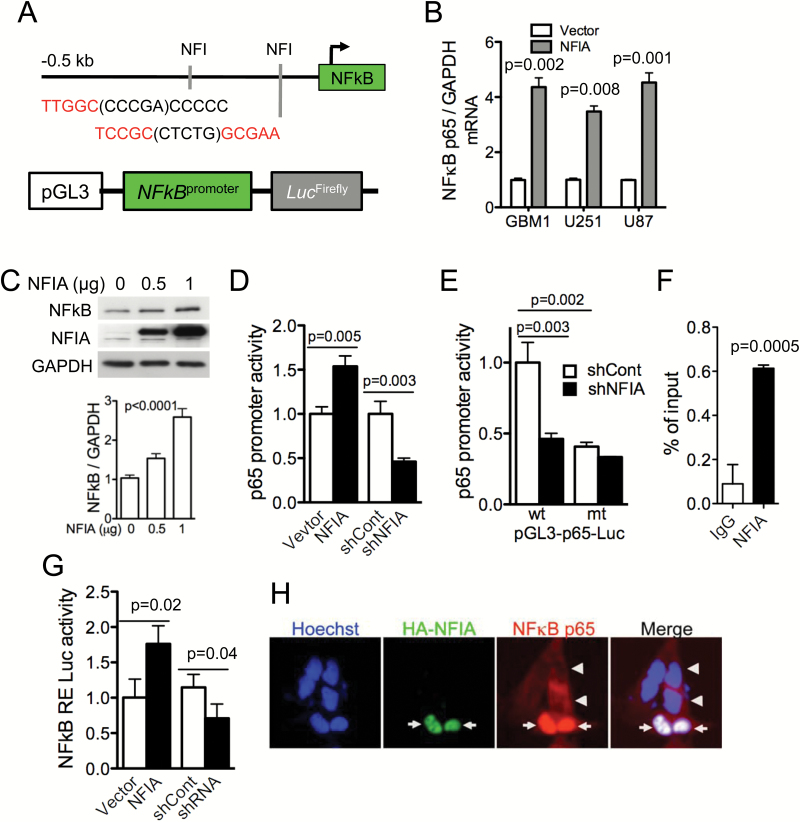
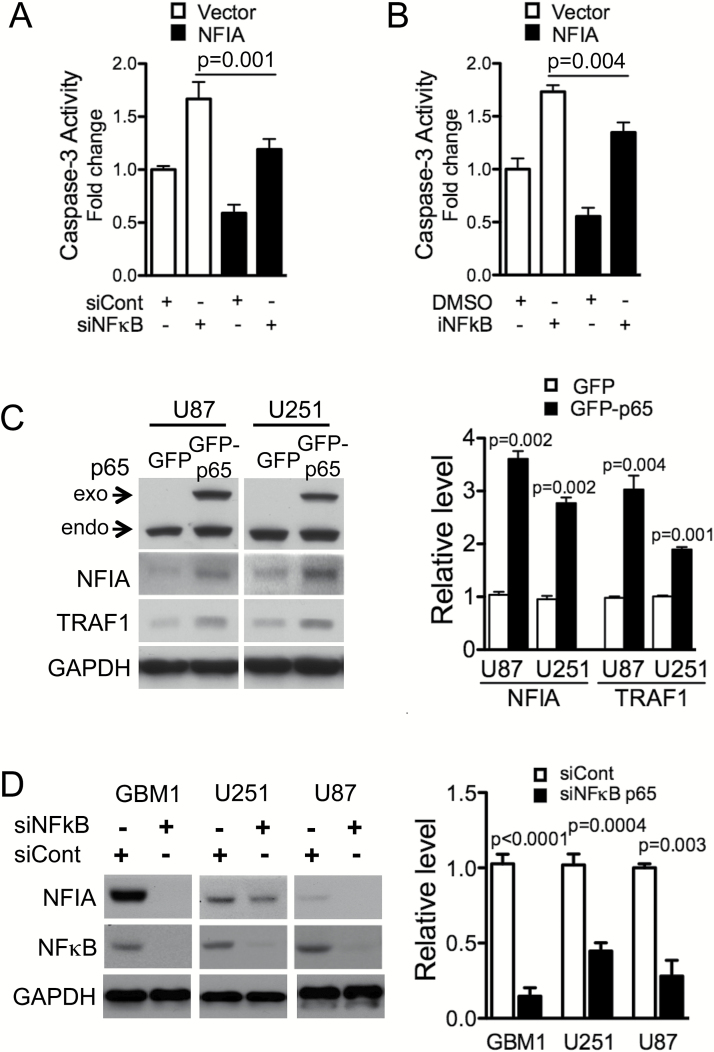
The NFκB p65 canonical pathway contributes to apoptosis evasion in many cancers, including gliomas, by suppressing caspase-8 and activating anti-apoptotic signaling.15 The NFIA transcription factor binds to the consensus sequence TTGGC(N)5GCCAA to regulate gene expression (Fig. 2A). We now find putative NFI-binding sites in the human NFκB p65 promoter, which contains the NFIA binding motifs -237 TTGGC(CCCGA)CCCCC -223 and -75 TCCGC(CTCTG)GCGAA -61. Therefore, we reasoned that regulation of apoptosis evasion by NFIA may be mediated through the NFκB pathway. Indeed, in assessing mRNA expression of NFκB p65 in GBM cells transduced with NFIA or vector control, we found that p65 mRNA was increased by 3- to 4-fold in GBM cells overexpressing NFIA (Fig. 2B). In addition, transient expression of NFIA enhanced the NFκB p65 protein level in a dose-dependent manner (Fig. 2C). To investigate further whether NFIA-dependent NFκB p65 expression is through transcriptional regulation, we generated an NFκB p65 luciferase reporter construct that contains the putative wild-type NFIA-binding sites (p65-Luc-wt) and measured p65 promoter activity in the presence or absence of NFIA (Fig. 2D). NFIA overexpression increased wild-type p65 promoter activity in GBM cells and shRNA knockdown of NFIA decreased it (Fig. 2D). Mutation of the NFIA binding sites in the p65 promoter reporter (p65-Luc-mt; TTGGC to TTAAA and GCGAA to AAAAA) abolished the endogenous NFIA-induced p65 promoter activity to a level similar to NFIA-depleted GBM cells (Fig. 2E). Consistent with these findings, NFIA associates with the p65 promoter, suggesting that NFIA directly regulates p65 transcription (ChIP assays, Fig. 2F). Furthermore, ectopic NFIA expression resulted in activation of NFκB (Fig. 2G) and nuclear translocation of NFκB p65 in GBM cells (Fig. 2H). These data suggest that NFIA transcriptionally and posttranscriptionally regulates NFκB p65.
Fig. 2.
NFIA increases NFκB p65 transcription and induces NFκB activity and nuclear translocation. (A) Putative NFIA binding sites in the human NFκB p65 promoter. (B) Relative mRNA expression of NFκB p65 to GAPDH in GBM cells expressing NFIA or vector was determined by RT-PCR and densitometric analysis. (C) Immunoblots of whole cell lysates from GBM1 cells transiently transfected with the increasing amount of a lentivector encoding NFIA (bottom; densitometry). (D) Luciferase activities were measured 48 h after transfection of NFκB p65 luciferase reporter into U87 GBM cells expressing NFIA, shNFIA, or controls (vector or shCont); n = 3. (E) Relative luciferase activity of pGL3 wild-type (wt) or mutant (mt) NFκB p65 promoter transfected into U87 GBM cells expressing shNFIA or shCont similar to the left panel. (F) ChIP-qPCR assays on GBM cells using anti-NFIA or control immunoglobulin G. (G) Luciferase assay with the 3x κB-luc-reporter in U87 cells transfected with NFIA constructs (NFIA, vector only, shNFIA, or shCont). (H) NFIA promotes nuclear translocation of NFκB p65. Immunofluorescence staining of GBM1 cells expressing HA-NFIA or vector control with Hoechst (nuclei; blue), anti-HA (green), anti-p65 NFκB (red): arrows: nuclear p65 in HA-positive cells, arrowheads: cytoplasmic p65 in HA-negative control cells.
NFIA Diminishes Apoptosis Partly Through Positive Regulation of NFκB p65 in GBM Cells
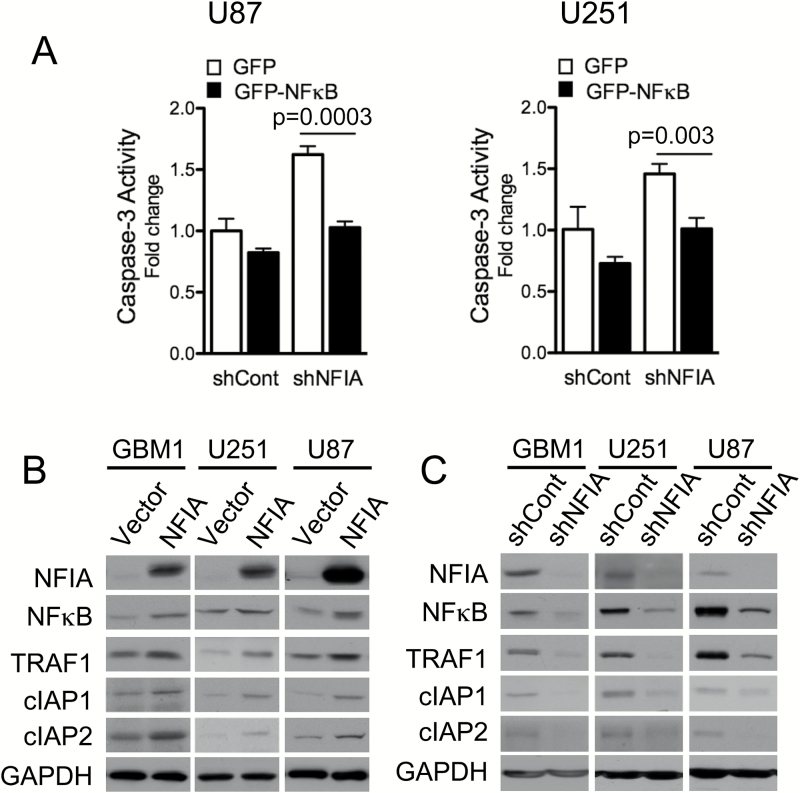
We previously observed that shNFIA increased activity of caspase-3, consistent with a pro-apoptotic effect for loss of NFIA (Fig. 3A).13 Ectopic NFκB p65 effectively diminished this shNFIA-induced caspase-3 activity, further solidifying a role for NFκB p65 as a functional downstream mediator of the pro-survival effect of NFIA (P < .005 compared with shCont, Fig. 3A). Overexpression of NFκB p65 was also sufficient to increase TRAF1 in NFIA-deficient shNFIA GBM cells (Fig. S4). These results indicate that NFκB mediates at least part of the NFIA-induced suppression of apoptosis. NFIA overexpression in patient-derived GBM cells (GBM1) and GBM cell lines (U251, U87), which enhanced the level of NFκB protein, also accompanied concurrent 2- to 3-fold increase in levels of NFκB downstream anti-apoptotic factors, TRAF1 and cIAP1/2 proteins (Fig. 3B). Conversely, NFIA knockdown (shNFIA) effectively reduced levels of NFκB p65, TRAF1, and cIAPs (Fig. 3C). These results show that NFIA positively regulates NFκB p65 expression and function and affects downstream inhibitors of apoptotic signaling in GBM cells.
Fig. 3.
NFIA pro-survival effect is mediated through NFκB p65 (A) Caspase-3 activity measured in NFIA-deleted GBM cells (transduced with shNFIA or shCont) treated with GFP-NFκB p65 or GFP vector control. NFκB p65 and TRAF1 levels in GBM cells used in (A) were verified by immunoblotting (Fig. S4). (B, C) Immunoblots of GBM1, U251, and U87 GBM cells transduced with NFIA, vector control, shRNA (shNFIA), or shRNA control (shCont).
NFκB p65 Regulates NFIA Transcription
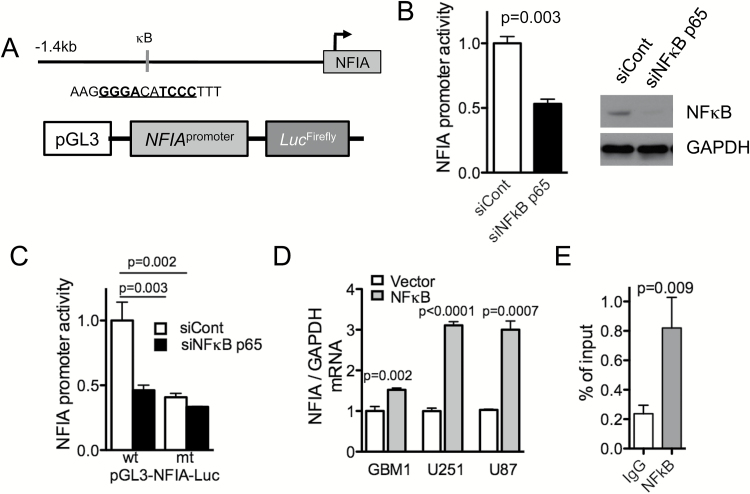
Considering that NFκB p65 is a transcription factor that binds to the consensus motif GGGRNNYYCC (R: A or G, Y: T or C), we asked whether NFκB also regulated NFIA, thus potentially forming a feed-forward loop NFIA → NFκB → NFIA. Interestingly, we found that the human NFIA promoter harbors a putative NFκB p65 binding sequence (-1267 GGGACATCCC -1257), suggesting that NFIA may indeed be a downstream transcriptional target gene of NFκB p65. In testing NFIA promoter activity in GBM cells expressing NFIA luciferase reporter constructs containing wild-type or mutant NFκB-binding sites (pGL3-NFIA-Luc; GGGACATCAA; Fig. 4A), we found that silencing NFκB (siNFκB p65) indeed decreased NFIA promoter activity (Fig. 4B). Consistent with this, mutating the NFκB-binding site in the NFIA promoter decreased NFIA promoter activity to the levels comparable to NFκB-deficient cells (Fig. 4C). Along with this, NFκB p65 overexpression increased NFIA mRNA levels (Fig. 4D). Furthermore, ChIP assays demonstrated that p65 associates with the NFIA promoter, suggesting that transcriptional regulation of NFIA by p65 is direct (Fig. 4E). These results indicate that NFIA is a novel transcriptional target of NFκB p65, and its transcription and expression are positively regulated by NFκB p65. Together with the findings above that NFIA activates NFκB p65 transcription, this suggests the presence of a tumor-promoting feed-forward loop between NFIA and NFκB in GBM.
Fig. 4.
NFκB p65 regulates NFIA transcription activity. (A) A putative NFκB p65-binding site in the human NFIA promoter luciferase reporter. (B) Luciferase activity was measured 48 h after transfection of NFIA luciferase reporter into U87 GBM cells expressing NFκB p65 siRNA (siNFκB p65) or control (siCont); n = 3; western blot of whole cell lysate of GBM cells used in the experiment B. (C) Relative luciferase activity of pGL3 wild-type (wt) or mutant (mt) NFIA promoter transfected into U87 cells expressing siNFκB p65 or siCont. (D) NFIA mRNA expression measured by RT-PCR in GBM cells transfected with NFκB p65 or vector control, n = 3. (E) ChIP-qPCR assays on GBM cells using anti-NFκB p65 or immunoglobulin G control.
NFIA Restores Cell Survival in NFκB p65–Depleted GBM Cells
The results above showed that NFIA expression is regulated by NFκB p65, suggesting that NFIA may be a novel functional target of NFκB in cell survival. To test this, we overexpressed NFIA in GBM cells depleted of NFκB-p65 by siRNA (siNFκB) or a pharmacologic inhibitor (iNFκB) and assessed caspase-3 activity as a surrogate for apoptosis. In NFκB-intact cells, NFIA overexpression was sufficient to attenuate caspase-3 activity (Fig. 5A–B), consistent with our previous finding.13 Knockdown or inhibition of endogenous NFκB increased caspase-3 activity in control cells (Fig. 5A–B), as expected. NFIA overexpression in U87 cells with NFκB knockdown (siNFκB p65) or NFκB inhibition (iNFκB) partially reduced the increased caspase-3 activity compared with vector control (Fig. 5A–B). However, in 2 other cell lines NFIA was not able to reduce caspase-3 activity induced by decreased NFκB (Fig. S5), indicating that additional mechanisms may contribute to apoptosis induced by loss of NFκB signaling. Moreover, overexpression of NFκB p65 increased NFIA protein level with a concurrent increase in TRAF1 level (Fig. 5C), and siRNA knockdown of NFκB p65 (siNFκB) decreased the level of NFIA protein (Fig. 5D). These results suggest that NFκB-mediated GBM cell survival is in part via activation of the NFIA pathway.
Fig. 5.
NFκB p65-induced inhibition of apoptosis is mediated in part by NFIA. (A, B). Apoptosis induced by knockdown (siNFκB) or pharmacologic inhibition (iNFκB) of NFκB is reversed by introduction of NFIA in U87 GBM cell (also see Fig. S5). Caspase-3 activity in NFκB p65-depleted GBM cells (siNFκB or siCont, or NFκB inhibitor or DMSO) transduced with NFIA or vector control (also see Fig. S6). (C) Immunoblots of whole cell lysates from GBM cells expressing GFP-p65 or GFP only. Right-densitometric analysis relative to GAPDH. (D) Knockdown of NFκB p65 decreases NFIA level. NFIA levels in GBM cells transfected with NFκB p65 siRNA (siNFκB p65) or siRNA control (siCont).
Downregulation of NFIA Sensitizes GBM Cells to Chemotherapy-Induced Cell Death
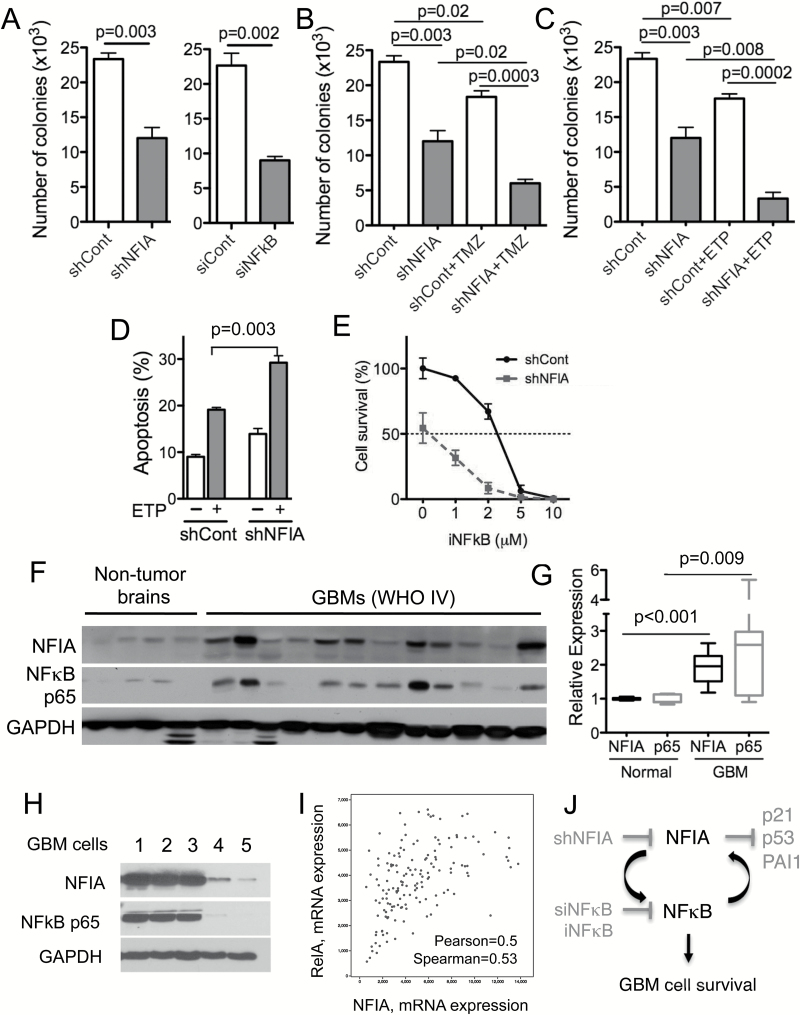
Our data above demonstrated that high NFIA or NFκB p65 levels enhance GBM cell survival and contribute to its resistance to apoptosis in culture (Fig. 1) and that NFIA and NFκB positively regulate each other’s transcription and protein levels (Figs. 2–5). We therefore asked whether downregulation of both NFIA and NFκB p65 may sensitize GBM cells to cell death more efficiently than each alone. Knockdown of NFIA (shNFIA) or NFκB p65 (siNFκB p65) each decreased GBM cell tumorsphere formation (Fig. 6A). Downregulation of NFIA augmented chemotherapy-induced GBM cell death (ie, temozolomide or etoposide) (Fig. 6B–D). Growth inhibition was greater when both NFIA was decreased and NFκB p65 was inhibited, compared with blocking each individually (Fig. 6A, E). Furthermore, NFIA overexpression attenuated NFκB inhibitor-induced apoptosis (Figs. 5B, S6). This suggests that the NFIA-NFκB p65 loop may contribute to inhibition of apoptosis in GBM.
Fig. 6.
Downregulation of NFIA sensitizes GBM cells to chemotherapy-induced cell death. (A) Knockdown of NFIA by shNFIA or NFκB p65 by siNFκB p65 decreased GBM tumorsphere formation compared with control (shCont or siCont). (B, C) Greater reduction in GBM tumorsphere formation in NFIA-depleted GBM cells treated with ETP or TMZ. GBM1 cells transduced with shNFIA or shCont, treated with TMZ or ETP for 24 h. (D) Downregulation of NFIA sensitized GBM cells to ETP-induced apoptosis. Apoptosis was measured using Apo-Direct kit followed by cell sorting analysis. (E) Greater reduction in GSC survival in NFIA-depleted GSCs treated with NFκB inhibitor (iNFκB). (F) Immunoblot of whole cell lysates from human GBM (n = 12) and nonneoplastic brains (n = 4). (G) Densitometric analysis of NFIA and NFκB p65 levels relative to GAPDH. (H) Immunoblot of whole cell lysates from patient derived GBM cells (GBM 1–5) and normal astrocytes (Fig. S8). (I) Correlation between NFIA and NFκB mRNA expression in GBM (580 GBM samples in total, 291 samples with completed information; n = 291; TCGA, Cell 2013; cbioportal.org). (J) Schematic model of an NFIA-NFκB feed-forward mechanism in GBM cell survival.
NFIA and NFκB p65 Levels Are Highly Correlated in Human GBM and Patient-Derived GBM Cells
The NFκB p65 pathway is constitutively active in GBM. We previously reported that NFIA mRNA and protein level are higher in GBM compared with normal brains.12,13 Our data here—that NFIA positively regulates NFκB p65 and that NFκB p65 positively regulates NFIA in GBM cells—raised the question of whether NFIA and NFκB p65 are correlated in human GBM. In a cohort of human GBM and patient-derived human GSC, we found that protein levels of NFIA and NFκB p65 were significantly higher in human GBM tumors (World Health Organization grade IV) compared with nonneoplastic brains: by 2-fold for NFIA and 2.5-fold for NFκB (Fig. 6F–G). Additionally, levels of NFIA and NFκB proteins were highly correlated in the human GBM and human patient derived GBM cell samples: GBM with high NFIA expression also showed high NFκB and vice versa (Fig. F–H). Furthermore, in the patient-derived GBM cells, NFIA levels were highly correlated with STAT3, another transcription factor induced by NFκB-mediated drug resistance and mesenchymal differentiation27 (Fig. S9). Consistent with this, ectopic NFIA increased STAT3 phosphorylation and protein level in GBM cells (Fig. S9). Moreover, the glioblastoma TCGA dataset (Cell 2013, cBioPortal, cbioportal.org) reveals that NFIA and NFκB p65 (also known as RelA) are highly correlated at the mRNA level (Pearson’s correlation = 0.50, Spearman’s correlation = 0.53; Fig. 6I), whereas other NFκB family members, such as NFκB1 (p50), NFκB2 (p52), RelB, and c-Rel, show no correlation with NFIA mRNA level, further supporting the presence of a feed-forward loop between NFIA and NFκB p65. Interestingly, the coexpression analysis reveals high correlation between NFIA and other master transcription factors (ie, STAT3, transcriptional coactivator with PDZ-binding motif [TAZ]) induced by NFκB-mediated mesenchymal differentiation27,35,36 (Fig. S10). Our results therefore suggest that NFIA and NFκB expression may be coupled in GBM and that the feed-forward link we identified between these 2 pathways contributes to GBM cell survival and drug resistance (Fig. 6J).
Discussion
Our findings here demonstrate that the 2 tumor-promoting transcription factors, NFIA and NFκB, function together to form a feed-forward loop that increases their expression and contributes to GBM cell survival and resistance to etoposide and temozolomide.
Our prior work13 and the experiments here demonstrate that NFIA promotes GBM through suppression of apoptosis, promotion of proliferation and cell survival, and resistance to chemotherapy. NFIA shares significant sequence homology in the DNA-binding domain with other NFI transcription factors (NFIB, NFIC, and NFIX).8 To date, neither NFIA nor any of the other NFI transcription factors have been reported to affect NFκB or be affected by NFκB, making the NFIA-NFκB regulation we uncovered the first published report of a functional link between an NFI transcription factor and NFκB.
NFκB p65 is a well-recognized anti-apoptotic transcription factor15,37 that is implicated in many cancers, including gliomas.18,31,38,39 NFκB activity is primarily regulated by interaction with inhibitor of kappaB (IkB) proteins40 with contributions from additional positive and negative upstream regulators.41 Our findings that NFIA increased NFκB transcription activity, NFκB mRNA, and NFκB protein and that NFκB attenuated shNFIA-induced apoptosis suggest that the pro-GBM effect of NFIA is mediated in part via positive transcriptional regulation of NFκB p65.
Constitutive activation of NFκB is found in many cancers, including GBM.23,24,42 Our study showed that NFIA increases expression of NFκB p65, which at least in part contributes to NFIA-induced GBM survival and drug resistance in culture. Interestingly, we found that a small increase in NFκB p65 expression by NFIA (Fig. 2C) was accompanied by effective nuclear translocation, and therefore the NFIA effect on NFκB is likely a combination of increased expression as well as activation. The mechanism of such NFκB activation by NFIA is currently unknown. Possible scenarios include NFIA modulation of the upstream inhibitory IkB proteins and/or NFIA regulation of secreted molecules that lead to activation of NFκB signaling.
NFκB was shown to promote mesenchymal differentiation via induction of master transcription factors, such as STAT3 and TAZ in GBM.27,35,36 NFIA is highly correlated with NFκB, and NFκB-induced mesenchymal signature (ie, STAT3 and TAZ) and ectopic NFIA increased STAT3 level. This further suggests that the NFIA-NFκB feed-forward pathways may be active in GBM and may contribute to GBM cell survival. It would be important in the future to determine the relationship between NFIA and these genes and pathways in promoting mesenchymal differentiation. Additionally, it remains to be seen whether the current findings in cell culture extend to the more complex in vivo situations.
Since mutation of the consensus recognition sites for NFIA and NFκB in each other’s promoters abolishes their effects on each other’s promoter reporter activation, and ChIP assays demonstrate interaction of NFIA and NFκB with the other’s promoter, this indicates that the transcriptional regulation in this feed-forward loop is direct. In this respect, it is interesting that of the 3 GBM cells we tested, we only found NFIA-induced mitigation of the NFκB-blockage dependent caspase-3 activity in the p53 wild-type cells (U87), but not in the p53 mutant cells (U251, GBM1; Fig. 5 and Fig. S5. Previously we found that NFIA effects are mediated in both p53-dependent and p53-independent manners.14 It is not yet known if differences in p53 are the cause of the differential effect of NFIA in attenuating caspase-3 activity induced by NFκB depletion/inhibition that we observed. Other possible explanations for the NFIA-mediated drug resistance effect include via its other downstream effectors, such as p21 and PAI1.13
Our studies here provide a novel mechanism for upregulation of NFIA in GBM cells by NFκB p65-induced transcriptional activation of NFIA. Other possible mechanisms for high NFIA in GBM include regulation by other upstream transcriptional regulators, such as Sox943; epigenetic regulators, such as miR-22314,44 and methylation; and posttranslational modifications. It is unknown whether these NFIA upstream regulators have a role in NFIA-mediated GBM cell survival. Through mining of TCGA data (Cell 2013, cBioportal), we also found that NFIA shows copy number variation in some cases (approximately 8%, Fig. S7). Compared with normal copy number status, NFIA copy loss or gain shows corresponding down- and upregulation of gene expression levels, suggesting minor dosage effect on its transcription in the dataset examined. An analysis of a dataset of a larger cohort may strengthen this finding.
In summary, in this study we report a previously unknown feed-forward cycle between NFIA and NFκB p65 that may contribute to GBM cell survival and protect GBM cells from chemotherapy-induced apoptosis. Our report provides new insights into the role of such an NFIA-NFκB loop in glioma, and may prove important in understanding of the mechanisms of drug resistance.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the National Institutes of Health (K08NS064297), the Child Neurology Foundation Shields Award, the Concern Foundation, the St. Baldrick’s Research Grant, the Perlmutter Cancer Center Support Grant (P30CA016087), and support from the Department of Neurosurgery (H.R.S.).
Conflict of interest statement
None.
Supplementary Material
Acknowledgments
We thank Valina Dawson for the NFIA antibody and Jinhua Wang, Anat Erdreich-Epstein, and Andrew Chi for helpful discussions.
References
- 1. Central Brain Tumor Registry of the United States. CBTRUS Statistical Facts Primary brain and other CNS tumor diagnosed in the United States in 2009–2013. www.cbtrus.org. [Google Scholar]
- 2. Dolecek TA, Propp JM, Stroup NE, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stupp R, Hegi ME, Mason WP, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 4. Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62(4):753–764; discussion 264–266. [DOI] [PubMed] [Google Scholar]
- 5. Keles GE, Chang EF, Lamborn KR, et al. Volumetric extent of resection and residual contrast enhancement on initial surgery as predictors of outcome in adult patients with hemispheric anaplastic astrocytoma. J Neurosurg. 2006;105(1):34–40. [DOI] [PubMed] [Google Scholar]
- 6. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 7. Gronostajski RM, Adhya S, Nagata K, et al. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol Cell Biol. 1985;5(5):964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249(1-2):31–45. [DOI] [PubMed] [Google Scholar]
- 9. Santoro C, Mermod N, Andrews PC, et al. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334(6179):218–224. [DOI] [PubMed] [Google Scholar]
- 10. de Jong RN, van der Vliet PC. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236(1):1–12. [DOI] [PubMed] [Google Scholar]
- 11. Deneen B, Ho R, Lukaszewicz A, et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52(6):953–968. [DOI] [PubMed] [Google Scholar]
- 12. Song HR, Gonzalez-Gomez I, Suh GS, et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro Oncol. 2010;12(2):122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee JS, Xiao J, Patel P, et al. A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro Oncol. 2014;16(2):191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glasgow SM, Laug D, Brawley VS, et al. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci. 2013;33(33):13560–13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–227. [DOI] [PubMed] [Google Scholar]
- 16. Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. [DOI] [PubMed] [Google Scholar]
- 17. Bassères DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–6830. [DOI] [PubMed] [Google Scholar]
- 18. Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol. 2011;12(8):715–723. [DOI] [PubMed] [Google Scholar]
- 19. Nogueira L, Ruiz-Ontañon P, Vazquez-Barquero A, et al. The NFκB pathway: a therapeutic target in glioblastoma. Oncotarget. 2011;2(8):646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puliyappadamba VT, Chakraborty S, Chauncey SS, et al. Opposing effect of EGFRWT on EGFRvIII-mediated NF-κB activation with RIP1 as a cell death switch. Cell Rep. 2013;4(4):764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang W, Xia Y, Cao Y, et al. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48(5):771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanaka K, Babic I, Nathanson D, et al. Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1(6):524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364(7):627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nozell S, Laver T, Moseley D, et al. The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol Cell Biol. 2008;28(21):6632–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Wang H, Zhang W, et al. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84(8):941–951. [DOI] [PubMed] [Google Scholar]
- 26. Tran NL, McDonough WS, Savitch BA, et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res. 2006;66(19):9535–9542. [DOI] [PubMed] [Google Scholar]
- 27. Bhat KP, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang CY, Mayo MW, Korneluk RG, et al. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–1683. [DOI] [PubMed] [Google Scholar]
- 29. Son MJ, Woolard K, Nam DH, et al. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Lf, Fischle W, Verdin E, et al. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. [DOI] [PubMed] [Google Scholar]
- 31. Espinosa L, Cathelin S, D’Altri T, et al. The Notch/Hes1 pathway sustains NF-κB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18(3):268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dutta J, Fan Y, Gupta N, et al. Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene. 2006;25(51):6800–6816. [DOI] [PubMed] [Google Scholar]
- 38. Vilimas T, Mascarenhas J, Palomero T, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13(1):70–77. [DOI] [PubMed] [Google Scholar]
- 39. Baker RG, Hayden MS, Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011;13(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012;12(2):121–132. [DOI] [PubMed] [Google Scholar]
- 41. Gray GK, McFarland BC, Nozell SE, et al. NF-κB and STAT3 in glioblastoma: therapeutic targets coming of age. Expert Rev Neurother. 2014;14(11):1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang H, Wang H, Zhang W, et al. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84(8):941–951. [DOI] [PubMed] [Google Scholar]
- 43. Kang P, Lee HK, Glasgow SM, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74(1):79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.