Abstract
Sulfonamide resistance in meningococci is associated with mutations in the chromosomal gene folP, which encodes dihydropteroate synthase. Several mutations associated with resistance have been previously described, including amino acid substitutions at codons 31 and 194, a glycine-serine insertion at codons 195 and 196, and, recently, an additional mutation at nucleotide 682 (C682A). In this study, sulfisoxazole MICs were determined for 424 geographically diverse clinical isolates of Neisseria meningitidis, including all major subtypes. A subset of 134 isolates with MICs ranging from 0.5 to >64 μg/ml were assayed for the C682A mutation by real-time PCR, and 25 isolates were selected for folP gene sequencing. All isolates for which the sulfisoxazole MIC was ≥8 possessed the C682A mutation by real-time PCR or folP sequencing, and 34 of 35 isolates with a MIC of ≤2 lacked this mutation. Of 16 sequenced isolates for which the sulfisoxazole MIC was ≥4, 15 possessed previously described mutations, including 10 at codon 31, 1 at codon 194, and 4 with the 2-amino-acid insertion codons 195 and 196; all 16 possessed the C682A mutation. The C682A mutation predicted elevated sulfonamides MICs for a large number of geographically diverse clinical isolates of meningococci. Detection of this mutation by real-time PCR or other methods may allow more wide-scale detection of meningococcal isolates with for which the sulfonamide MICs are elevated without resorting to multiple assays or folP gene sequencing, providing a simple, high-throughput screening method for use in public health and epidemiologic settings.
Sulfonamide drugs showed striking effectiveness in the therapy of invasive disease caused by Neisseria meningitidis in the early part of the 20th century, leading to reduced morbidity and mortality (8, 10). In 1942, implementation of sulfonamide prophylaxis on a U.S. Marine Corps base in San Diego, California, during a meningococcal outbreak was met with dramatic success (5), and in 1943, large-scale case control studies demonstrated the effectiveness of sulfonamide prophylaxis (16). Sulfonamides were mainstays in the treatment and prophylaxis of invasive meningococcal disease until 1963, when sulfonamide prophylaxis at the U.S. Marine Corps base in San Diego failed during multiple outbreaks, leading to a number of fatalities. In vitro studies revealed sulfonamide resistance; this resistant strain moved into the civilian population within the year (17). Sulfonamide resistance across several serogroups in both invasive meningococcal isolates and those from carriers was increasingly recognized over the next decade worldwide (11, 14, 24); a surveillance study by the U.S. Centers for Disease Control and Prevention (CDC) found that 27% meningococcal isolates in 1974 were sulfonamide resistant (1). This led to recommendations against the use of sulfonamides unless susceptibility data were available (4). Thus, a cheap, inexpensive therapeutic and prophylactic agent appeared to be lost.
Sulfonamide resistance remains common today despite the discontinuation of the use of these agents in meningococcal disease; a recent CDC survey found that only 46% of clinical isolates were susceptible to sulfonamide agents (23). Sulfonamide resistance has been associated with increased mortality, even when these agents were not used in therapy (2). Identification of sulfonamide-resistant strains may be used in the study of the epidemiology of meningococcal disease (24) and may provide information for the future development of newer generations of sulfonamides. In addition, identification of sulfonamide-susceptible strains may be important in resource-limited environments where these agents may be useful for prophylaxis or therapy.
Sulfonamide agents target bacterial dihydropteroate synthase (DHPS) enzymes, acting as competitive inhibitors. Resistance to these agents in meningococci is mediated by alterations in the chromosomal gene encoding this enzyme, folP (15). Investigators have described several mutations associated with resistance, including a 2-amino-acid insertion at codons 195 and 196 and substitutions at codons 31, 84, and 194 (22). Bennett and Cafferkey recently investigated a single mutation of adenine (A) to cytosine (C) at nucleotide position 682 in several isolates from the United Kingdom that was consistently associated with sulfonamide resistance, irrespective of the presence of other previously described mutations (7). This mutation leads to an amino acid substitution in codon 228 and was also identified in the sequences of previously described resistant isolates (7).
As part of a study to define susceptibility breakpoints for 14 drugs in meningococci, we determined sulfisoxazole MICs for 424 geographically diverse clinical isolates of Neisseria meningitidis. To evaluate the molecular mechanisms of sulfonamide resistance in these isolates, we designed a real-time PCR assay to detect the mutation at position 682 described by Bennett and Cafferkey in a subset of these isolates.
MATERIALS AND METHODS
Isolates.
Sulfisoxazole MICs were determined for 424 geographically diverse clinical isolates of N. meningitidis, including serotypes A, B, C, W135, X, Y, and Z. From these, a subset of 134 isolates from seven countries and five continents were selected for molecular testing. One-hundred six isolates originated in the United States (representing 13 states), and 22 isolates were from other countries, including Australia (13), Saudi Arabia (1), Burkina Faso (1), Hong Kong (1), Bangladesh (2), and Spain (4). Geographic origin was unavailable for six isolates. Sulfisoxazole MICs ranged from 0.5 to >64 μg/ml. Serotypes represented included A (7), B (37), C (32), W135 (11), X (1), Y (39), and Z (1); 6 isolates were nontypeable.
Broth microdilution MIC susceptibility tests.
Sulfisoxazole MICs were determined using the broth microdilution procedure described in NCCLS document M7-A6 (18). It included use of cation-adjusted Mueller-Hinton broth supplemented with 3% lysed horse blood as the test medium. Test inocula were prepared from meningococcal colonies grown on chocolate agar plates (Becton-Dickinson) that had been incubated for 18 to 24 h in 5% CO2. Colonies were suspended in 0.9% saline to obtain a suspension equivalent to the turbidity of a 0.5 McFarland standard, and they were further diluted to provide a final inoculum density of 5 × 105 CFU/ml in the wells of the microdilution panels.
Quality control strains.
For quality control of the broth microdilution tests, Escherichia coli ATCC 25922 was employed, using approved NCCLS control ranges (19).
DNA extraction.
Genomic DNA was extracted as described previously (3). One square centimeter of confluent 20- to 22-h growth on chocolate agar (Becton-Dickinson) was suspended in 500 μl of distilled water. The bacterial suspension was subjected to one freeze-thaw cycle, boiled at 100°C for 3 min, and centrifuged at 10,000 × g for 5 min.
Real-time PCR.
A real-time PCR allelic discrimination assay for the C682A mutation was designed using Primer Express software, version 2.0 (Applied Biosystems, Inc., Foster City, Calif.). Primers and probes were synthesized by Applied Biosystems, Inc. The sequence of the forward primer (NMDHPS_A>C682FOR) was CCGAATTGATGGCGGAAAC, and the reverse primer (NMDHPS_A>C682REV) sequence was GCCACGCTGCCGTGT; these primers amplified a 115-bp portion of the folP gene. Two fluorescently labeled probes were designed to detect the wild-type folP sequence (C at nucleotide position 682) and mutated sequence (A at nucleotide position 682). The sequence of the wild-type probe (NMDHPS_C682) was 6FAM-TGCGTTTGCGCGACA-MGBNFQ, and the mutant probe (NMDHPS_A682) sequence was VIC-TGCTTTTGCGACAC-MGBNFQ; both probes were constructed with minor groove-binding nonfluorescent quenchers on the 3′ end (MGB probes).
The 50-μl PCR mixture included 25 μl of TaqMan 2× Universal Master Mix (Applied Biosystems), forward and reverse primers at a final concentration of 900 mM, and the wild-type probe and mutant probe at a final concentration of 200 mM each. One microliter of extracted template DNA was added to each reaction mixture. Real-time PCR was performed with the ABI 7000 Sequence Detection System, with the following thermocycling conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles consisting of 15 s at 95°C followed by 1 min at 60°C. A positive reaction for wild-type sequence was indicated by a FAM signal; the presence of the mutation was indicated by the VIC signal.
folP gene sequencing.
Isolates yielding no signal by real-time PCR with either probe were subjected to folP gene sequencing, as were selected representative isolates representing the full range of MICs. The folP gene was sequenced using primers NM1, NM2, and NM3, as described previously (22). Amplified product was purified using the QIAQuick PCR purification kit (QIAGEN, Inc., Valencia, Calif.). Sequencing was performed at the University of Texas Health Science Center Nucleic Acids Core Facility (San Antonio, Tex.) using Big Dye Terminator v3.1 chemistry and 3100 capillary sequencers (Applied Biosystems).
Nucleotide sequence accession numbers.
The GenBank and EMBL accession numbers for the sequences in this paper are as follows: NM006 (AY721983), NM031 (AY721984), NM086 (AY721985), NM103 (AY721986), NM107 (AY721987), NM108 (AY721988), NM113 (AY721989), NM145 (AY721990), NM146 (AY721991), NM185 (AY721992), NM194 (AY721993), NM217 (AY721994), NM231 (AY721995), NM245 (AY721996), NM252 (AY721997), NM255 (AY721998), NM271 (AY721999), NM309 (AY722000), NM310 (AY722001), NM333 (AY722002), NM400 (AY722003), NM403 (AY722004), NM417 (AY722005), NM419 (AY722006), and NM421 (AY722007).
RESULTS
Sulfisoxazole MICs for the 424 clinical isolates showed a bimodal distribution, with the MIC for a majority of isolates being either ≤2 or ≥8 μg/ml (Table 1). Of the 134 isolates tested for the C682A mutation by real-time PCR, 129 showed a signal with either wild-type or mutant probe; of these, all 34 isolates for which the MIC was ≤2 were identified as wild type at position 682, whereas all isolates for which the MIC was ≥8 possessed the C682A mutation.
TABLE 1.
Frequency of sulfisoxazole MICs and detection of the C682A mutation by real-time PCR
| Sulfisoxazole MIC (μg/ml) | No. of isolates | C682A mutation detection by real-time PCR
|
||
|---|---|---|---|---|
| No. of isolates tested | Wild typea | Mutanta | ||
| 0.25 | 6 | 0 | ||
| 0.5 | 24 | 3 | 3 | 0 |
| 1 | 78 | 15 | 15 | 0 |
| 2 | 67 | 17 | 16 | 1b |
| 4 | 24 | 18 | 6 | 18 |
| 8 | 48 | 28 | 0 | 28c |
| 16 | 25 | 7 | 0 | 7 |
| 32 | 28 | 16 | 0 | 16 |
| 64 | 69 | 20 | 0 | 20 |
| >64 | 55 | 10 | 0 | 10 |
Wild type, C at nucleotide position 682; mutant, A at nucleotide position 682.
One isolate negative by real-time PCR; C682A mutation present by folP gene sequencing (see the text).
Four isolates negative by real-time PCR; C682A mutation present by folP gene sequencing (see the text).
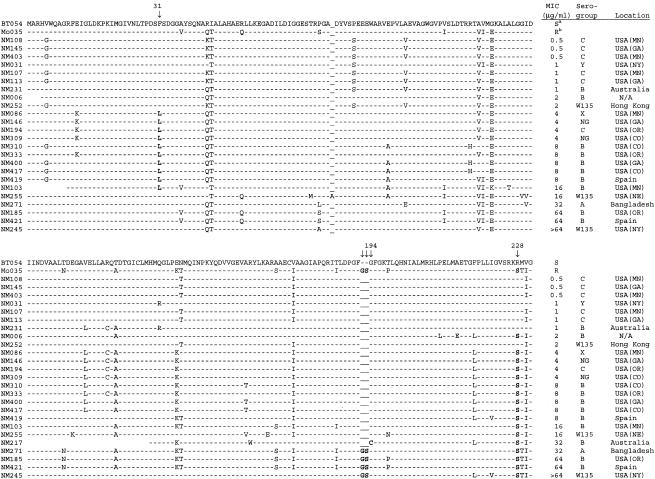
Aligned DHPS amino acid sequences predicted by folP gene sequence for 25 clinical isolates and two reference strains are shown in Fig. 1. Real-time PCR results for the presence or absence of the C682A mutation were confirmed in all 20 isolates for which real-time PCR data was available. The Phe31-Leu mutation was present in 10 isolates, with sulfisoxazole MICs ranging from 4 to 16 μg/ml. The 195 to 196 Gly-Ser insertion was present in four isolates lacking the Phe31-Leu mutation; MICs for these isolates ranged from 32 to >64 μg/ml. The MIC for one isolate with the Gly194-Cys mutation was 32 μg/ml. One isolate (NM255) for which the MIC was 16 μg/ml showed only the C682A mutation, predicting an Arg228-Ser substitution, and contained none of the other mutations previously described as associated with resistance.
FIG. 1.
Aligned DHPS amino acid sequence predicted by folP gene sequence in selected isolates. Footnote a indicates BT054, a characterized strain sensitive to sulfonamides (GenBank accession no. X68067) (22). Footnote b indicates MO035, a characterized strain resistant to sulfonamides (GenBank accession no. X68062) (22). Arrows indicate amino acid substitutions discussed in the text. NG, nongroupable. N/A, location not available.
Real-time PCR was negative for either wild-type or mutant probe in five of the clinical isolates (NM006, NM310, NM400, NM417, and NM419); the sulfisoxazole MIC for four of these isolates was 8 μg/ml. Upon sequencing, all five isolates showed the C682A mutation, predicting the substitution of serine for arginine at codon 228 (Fig. 1). These isolates also contained a silent A681G mutation (data not shown). The MIC for one isolate (NM006) was 2 μg/ml, and this isolate had no signal by real-time PCR; sequencing revealed the presence of the C682A mutation as well as the same silent A681G mutation identified in the other four isolates negative by real-time PCR. This isolate also lacks mutations in codons 31, 84, and 194; it does not possess the 195 to 196 Gly-Ser insertion. Repeated MIC determinations confirmed a sulfisoxazole MIC of 2 μg/ml for this isolate.
DISCUSSION
The folP genes of several sulfonamide-resistant meningococcal isolates have been extensively characterized, leading to the description of two types of folP genes present in these isolates (12, 20). One type is similar to the folP gene of susceptible isolates, but it possesses mutations leading to one or more single amino acid changes in conserved regions, including Phe31-Leu and Gly194-Cys; a third amino acid substitution, Pro84-Ser, is believed to improve fitness of isolates with these mutations. The second type shows less similarity to the susceptible folP strains and possesses an additional 6-bp sequence leading to a 2-amino-acid insertion (Gly-Ser) at codons 195 and 196. Site-directed mutagenesis creating the Phe31-Leu and Gly194-Cys substitutions has led to elevated sulfonamide MICs (12), which have been confirmed by in vitro enzyme kinetic studies (13). Deletion of the Gly-Ser insertion in a resistant strain eliminated sulfonamide resistance (12); however, introduction of this same insertion into the folP gene of a susceptible strain did not produce a resistant gene, but rather a defective enzyme (20). In their analysis of folP gene sequences in meningococci, these same investigators also noted an amino acid change immediately following a conserved SRK sequence, and they suggested that this was likely to be important in resistance to sulfonamides (25).
Bennett and Cafferkey analyzed nine published folP sequences in an attempt to identify mutations found in all sulfonamide-resistant N. meningitidis (7). Of four candidate mutations, only one, C682A, was consistently present in resistant isolates and absent in susceptible isolates among nine recent clinical isolates from Ireland; this mutation was identified in 10 additional Irish isolates with elevated sulfonamide MICs. This mutation predicts the substitution of serine for arginine at position 228 in a highly conserved region of the DHPS enzyme involved in substrate binding (6), the same amino acid change noted a decade earlier by Swedberg and colleagues (25). Those isolates tested by Bennett and Cafferkey were all clinical isolates from a restricted geographic area during a period of hyperendemicity (7). In this study, we were able to show that the C682A mutation predicts elevated sulfisoxazole MICs for a large number of isolates from around the world and representing several serogroups.
Of the 25 isolates in this study that were selected for folP sequencing, 10 possessed the Phe31-Leu mutation and 4 possessed the Gly-Ser insertion. One isolate for which the MIC was 32 μg/ml possessed the Gly194-Cys mutation; sequence information at position 31 was not available for this isolate. The four isolates for which the MICs were 32 or greater all possessed alterations at position 194 (Gly194-Cys or the Gly-Ser insertion at positions 195 and 196). These alterations may have a dosage effect, conferring a greater level of resistance than the Arg228-Ser substitution alone. One sequenced isolate (NM255) for which the MIC was 16 μg/ml showed the C682A mutation by both real-time PCR and folP gene sequencing, but it did not possess either the Phe31-Leu mutation or the Gly-Ser insertion. This supports the theory that the Arg228-Ser alteration may be related to the mechanism of sulfonamide resistance. A single isolate for which the sulfonamide MIC was 2 μg/ml (NM006) possessed the C682A mutation. This isolate did not possess the Phe31-Leu mutation. Two additional unique amino acid alterations were identified at positions 210 and 214, which may modify the phenotypic expression of resistance in this isolate.
Site-directed mutagenesis studies may clarify the role of the C682A mutation in sulfonamide-resistant meningococci. Our own review of several folP sequences in GenBank, informative at position 682 (21) in commensal nasopharyngeal isolates of Neisseria spp. defined as sulfonamide resistant (GenBank accession no. AJ457071) or susceptible (AJ457072 and AJ457076), showed that the C682A mutation is associated with sulfonamide resistance in these nonmeningococcal isolates as well. However, regardless of its functional effects, the C682A mutation identified by Bennett and Cafferkey (7) appears to be a single robust marker for elevated sulfonamide MICs for meningococci.
Due in part to previously administered antibiotics, cerebrospinal fluid cultures may be negative in cases of meningococcal meningitis, and definitive diagnosis is more frequently made by PCR; isolates are therefore not available for susceptibility testing (7). In the appropriate setting, detection of meningococcal DNA or RNA by PCR or other nucleic acid amplification methods may lead to a rapid diagnosis of invasive meningococcal disease, which is particularly important for infection control (9). Identification of molecular markers of antibiotic resistance in the absence of a culture isolate would be helpful not only for therapeutic and prophylaxis decisions but also in defining the evolving epidemiology of meningococcal disease and detection of emerging resistance. With increasing knowledge of the structure of DHPS and its interactions with competitive inhibitors (6), an understanding of the mutations associated with resistance to present sulfonamide agents is vital in the rational design of new antibiotics targeting DHPS.
Sulfonamide drugs are relatively cheap and have been proven effective for therapy and prophylaxis against sulfonamide-susceptible strains of N. meningitidis in the past. These agents are no longer recommended for meningococcal disease, because isolates are not predictably susceptible to these drugs and treatment and prophylaxis decisions are made before susceptibility results could be available. However, the ability to predict susceptibility with a simple, rapid molecular test may be useful in identifying cases and outbreaks due to sulfonamide-susceptible strains. This may offer additional options for treatment and prophylaxis, particularly in resource-limited environments, although additional studies on how quickly sulfonamide resistance develops in modern invasive meningococcal isolates in the presence of sulfonamides may be warranted prior to their use in these circumstances. This real-time PCR assay provides a simple, high-throughput screening method for the identification of sulfonamide-susceptible strains in the public health and epidemiologic setting. A second set of probes or modification of the probe sequence to include a neutral base at position 681 would allow the detection of this mutation in strains with the A681G polymorphism. This would allow a more wide-scale molecular detection of isolates for which the sulfonamide MIC is elevated without resorting to multiple assays or folP sequencing.
Acknowledgments
This study was supported by RS1 grant number CCR622402 from the Centers for Disease Control and Prevention.
Many of the isolates included in this study were generously provided by Fred Tenover, Tanja Popovic, and Nancy Rosenstein of the CDC. Additional United States isolates were kindly provided by the Minnesota Department of Health, the New York State Department of Health, and by the Oregon Department of Health Services. Non-United States isolates were generously provided by John Turnidge from Adelaide, Australia.
REFERENCES
- 1.Allen, J. R., and The Meningococcal Disease Surveillance Group. 1976. Meningococcal disease: secondary attack rate and chemoprophylaxis in the United States, 1974. JAMA 235:261-265. [PubMed] [Google Scholar]
- 2.Andersen, B. 1978. Mortality in meningococcal infections. Scand. J. Infect. Dis. 10:277-282. [DOI] [PubMed] [Google Scholar]
- 3.Antignac, A., P. Kriz, G. Tzanakaki, J.-M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 4.Artenstein, M. S. 1975. Prophylaxis for meningococcal disease. JAMA 231:1035-1036. [PubMed] [Google Scholar]
- 5.Awe, C. D., R. W. Babione, and J. N. DeLamater. 1943. Meningococcic meningitis in the San Diego area during 1942. U.S. Navy Med. Bull. 41:625-634. [Google Scholar]
- 6.Baca, A. M., R. Sirawaraporn, S. Turley, W. Sirawaraporn, and W. G. J. Hol. 2000. Crystal structure of Mycobacterium tuberculosis 6-hydroxymethyl-7,8-diydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J. Mol. Biol. 302:1193-1212. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, D. E., and M. T. Cafferkey. 2003. PCR and restriction endonuclease assay for detection of a novel mutation associated with sulfonamide resistance in Neisseria meningitidis. Antimicrob. Agents Chemother. 47:3336-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant, J., and H. D. Fairman. 1939. Chemotherapy of cerebrospinal fever in the field. Lancet i:923-926. [Google Scholar]
- 9.Bryant, P. A., H. Y. Li, A. Zaia, J. Griffith, G. Hogg, N. Curtis, and J. R. Carapetis. 2004. Prospective study of a real-time PCR that is highly sensitive, specific, and clinically useful for diagnosis of meningococcal disease in children. J. Clin. Microbiol. 42:2919-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle, J. H., L. Thomas, and A. R. Morton. 1941. Treatment of meningococcic meningitis and meningococcemia with sulfadiazine. JAMA 116:2666-2668. [Google Scholar]
- 11.Eickhoff, T. C., and M. Finland. 1965. Changing susceptibility of meningococci to antimicrobial agents. N. Engl. J. Med. 272:395-398. [DOI] [PubMed] [Google Scholar]
- 12.Fermér, C., B. E. Kristiansen, O. Sköld, and G. Swedberg. 1995. Sulfonamide resistance in Neisseria meningitidis as defined by site-directed mutagenesis could have its origin in other species. J. Bacteriol. 177:4669-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fermér, C., and G. Swedberg. 1997. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases for N. meningitidis expressed in a knockout mutant of Escherichia coli. J. Bacteriol. 179:831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, R. C., D. J. Clark, M. D. Yow, L. H. Taber, and F. F. Barrett. 1971. In vitro studies of 100 strains of meningococci: clinical implications. South. Med. J. 64:627-629. [DOI] [PubMed] [Google Scholar]
- 15.Kristiansen, B. E., P. Rådstrom, A. Jenkins, E. Ask, B. Facinelli, and O. Sköld. 1990. Cloning and characterization of a DNA fragment that confers sulfonamide resistance in a serogroup B, serotype 15 strain of Neisseria meningitidis. Antimicrob. Agents Chemother. 34:2277-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhns, D. M., C. T. Nelson, H. A. Feldman, and L. R. Kuhn. 1943. The prophylactic value of sulfadiazine. JAMA 123:335-339. [Google Scholar]
- 17.Millar, J. W., E. E. Siess, H. A. Feldman, C. Silverman, and P. Frank. 1963. In vivo and in vitro resistance to sulfadiazine in strains of Neisseria meningitidis. JAMA 186:139-141. [DOI] [PubMed] [Google Scholar]
- 18.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. NCCLS, Wayne, Pa.
- 19.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing. Supplement M100-S14. NCCLS, Wayne, Pa.
- 20.Qvarnstrom, Y., and G. Swedberg. 2000. Additive effects of a two-amino-acid insertion and a single-amino-acid substitution in dihydropteroate synthase for the development of sulphonamide-resistant Neisseria meningitidis. Microbiology 146:1151-1156. [DOI] [PubMed] [Google Scholar]
- 21.Qvarnstrom, Y., and G. Swedberg. 2002. Sulphonamide resistant commensal Neisseria with alterations in the dihydropteroate synthase can be isolated from carriers not exposed to sulphonamides. BMC Microbiol. 2:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rådström, P., C. Férmer, B. E. Kristiansen, A. Jenkins, O. Sköld, and G. Swedberg. 1992. Transformational exchanges in the dihydropteroate synthase gene of Neisseria meningitidis: a novel mechanism for acquisition of sulfonamide resistance. J. Bacteriol. 174:6386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenstein, N. E., S. A. Stocker, T. Popovic, F. C. Tenover, B. A. Perkins, and the Active Bacterial Core Surveillance Team. 2000. Antimicrobial resistance of Neisseria meningitidis in the United States, 1997. Clin. Infect. Dis. 30:212-213. [DOI] [PubMed] [Google Scholar]
- 24.Salmi, I., O. Pettay, I. Simula, A. K. Kallio, and O. Waltimo. 1976. An epidemic due to sulphonamide-resistant group A meningococci in the Helsinki area (Finland). Scand. J. Infect. Dis. 8:249-254. [DOI] [PubMed] [Google Scholar]
- 25.Swedberg, G., C. Fermer, and O. Skold. 1993. Point mutations in the dihydropteroate synthase gene causing sulfonamide resistance, p. 555-558. In J. E. Ayling (ed.), Chemistry and biology of pteridines and folates. Plenum Press, New York, N.Y. [DOI] [PubMed]