Abstract
Five azole-susceptible Candida glabrata isolates obtained before 1975 became resistant to fluconazole, itraconazole, and voriconazole within 4 days of in vitro fluconazole exposure. This cross-resistance was stable for at least 4 months after removal of fluconazole and was associated with increased CgCDR1 and CgCDR2 expression.
Candida glabrata has become one of the most common causes of Candida bloodstream infections (BSI) worldwide, now accounting for 7.5 to 18.3% of cases in different countries (20). The emergence of C. glabrata is of concern because of recent reports that up to 18% of incident bloodstream isolates are resistant to fluconazole (6, 8, 20, 21). Moreover, resistance to azole antifungal agents, including fluconazole and itraconazole, can emerge rapidly when patients with C. glabrata infection are treated with these drugs (10, 25). Resistance can also emerge when patients, such as hematopoietic stem cell transplant recipients, are given prolonged periods of azole prophylaxis (2). In these cases, both the acquisition of new strains and increased resistance in existing strains have been described (2, 10, 15). Multiple mechanisms of resistance have been reported, including increased expression of the ABC transporter genes CgCDR1 and CgCDR2 (PDH1), which result in decreased accumulation of intracellular fluconazole (2, 11, 17, 23), and increased expression of the CgERG11 gene encoding the fluconazole target, 14α-lanosterol demethylase (15, 22). Recent studies have described up-regulation of these genes in C. glabrata isolates upon fluconazole exposure (2, 9, 15, 22), including some for which the previous fluconazole exposure history was unknown.
We therefore sought to determine whether the rapid acquisition of fluconazole resistance by C. glabrata is an innate characteristic of this organism or whether multiple exposures to azoles over time are required. To ensure that the isolates tested had not been previously exposed to azole antifungal agents, we studied susceptible isolates of C. glabrata obtained between 1917 and 1975, a time period before the first introduction of oral or parenteral formulations of azole drugs. These C. glabrata isolates were then examined for their capacity to become resistant to azoles after in vitro exposure to fluconazole and for the length of time required for isolates to become resistant. In addition, we studied the molecular mechanisms associated with the acquired resistance and the stability over time of such resistance after the removal of fluconazole.
Isolates.
Three clinical C. glabrata isolates, CBS 138, CBS 860, and CBS 4692, which were isolated in 1917, 1935, and 1960, respectively, were obtained from the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands. Two other clinical C. glabrata isolates, 73/124 and 75/015, isolated in 1973 and 1975, were obtained from the University of Aberdeen, Aberdeen, United Kingdom. Species identification was confirmed by using species-specific Candida DNA probes in a PCR-enzyme immunoassay as described previously (7).
Antifungal susceptibility testing.
Prior to testing, isolates were subcultured onto Sabouraud dextrose agar plates for 24 h at 35°C. The MICs of fluconazole, itraconazole, and voriconazole were determined by the National Committee for Clinical Laboratory Standards (NCCLS) M27-A2 broth microdilution method (18). Standard powders of fluconazole and voriconazole were received as gifts from Pfizer Pharmaceuticals Group (Groton, Conn.), and itraconazole drug powder was purchased from Research Diagnostics, Inc. (Flanders, N.J.). The final concentrations of the antifungal agents ranged from 0.125 to 64 μg of fluconazole/ml and 0.015 to 8 μg of itraconazole and voriconazole/ml. The MIC endpoints were read visually following 24 and 48 h of incubation and were defined as the lowest drug concentration that produced a prominent reduction in growth (∼50%) compared with that of the drug-free growth control. For fluconazole and itraconazole, MIC interpretations were assigned according to the NCCLS criteria (18). No interpretive breakpoints have yet been established for voriconazole. NCCLS-recommended quality control and reference strains (Candida krusei ATCC 6258 and Candida parapsilosis ATCC 90018) were included in each test run, and MICs were within the recommended range for each test.
Induction of resistance.
To study the induction of azole resistance, a 10-μl loopful of yeast cells, grown on Sabouraud dextrose agar plates at 35°C for 24 h, was suspended in 1 ml of sterile water, and 100 μl of this suspension was used to inoculate 10 ml of RPMI 1640 medium (Sigma Chemical Co., St. Louis, Mo.) buffered to pH 7.0 with 0.165 mol of 3-(N-morpholino)propanesulfonic acid (MOPS; Sigma)/liter and containing 16 μg of fluconazole/ml. The cultures were incubated at 35°C with constant agitation. Every 48 h, 1 ml of each culture was diluted in 9 ml of fresh fluconazole-containing medium for a period of 14 days. At day 8, the fluconazole concentration in the medium was increased from 16 to 64 μg/ml for isolate CBS 4692. From day 14 onwards, all cultures were subcultured every 48 to 72 h in medium without fluconazole (days 14 to 76 in liquid medium and days 77 to 136 on solid medium). At each passage, an aliquot of the cultures was stored at −70°C in glycerol. To control for spontaneous changes in azole susceptibility, each isolate was also passaged in RPMI medium without fluconazole in the manner described above.
DNA extraction.
For each isolate, genomic DNA was extracted from approximately 107 CFU with the DNeasy tissue kit (QIAGEN, Valencia, Calif.) according to the manufacturer's specifications. DNA was eluted in 100 μl of elution buffer and stored at −20°C until used.
RAPD typing.
Random amplified polymorphic DNA (RAPD) typing was performed as follows: 25-μl reaction mixtures containing approximately 2 ng of template DNA, 2.5 μl of 10× PCR buffer, 1.75 mM MgCl2, 1.5 U of Taq DNA polymerase, 200 μM deoxynucleoside triphosphates, and 0.4 μM primer were amplified in 45 cycles of 1 min at 94°C, 2 min at 36°C, and 2 min at 73°C in a Perkin-Elmer model 9700 thermal cycler. Amplification products were separated in a 1.3% agarose gel and detected by ethidium bromide staining and UV illumination. The following primer sequences were used: OPA-01, CAGGCCCTTA; OPA-02, TGCCGAGCTG; OPA-04, AATCGGGCTG; OPA-10, GTGATCGCAG; OPA-16, AGCCAGCGAA; OPA-18, AGGTGACCGT; OPE-04, GTGACATGCC; OPE-18, GGACTGCAGA (14).
RNA extraction.
For each isolate, an overnight culture grown in 10 ml of Sabouraud dextrose broth was diluted 1:50 in fresh Sabouraud dextrose broth and grown to mid-logarithmic phase. Aliquots of 5 ml were then centrifuged to harvest the cells, and cells were washed once with sterile distilled water. Cell lysis was performed by resuspending cells in sterile distilled water plus lysis binding solution (Qbiogene, Carlsbad, Calif.), transferring them to lysis Matrix C tubes (Qbiogene), and homogenizing twice for 45 s at speed level 6 in a FastPrep instrument (Qbiogene). Following the directions of the manufacturer (Ambion, Austin, Tex.), total RNA was extracted from the C. glabrata cells by using an RNAqueous kit. RNA samples were stored at −70°C until analyzed.
Northern blotting.
For each isolate, 5 μl of total RNA (∼1.5 μg) was separated in a 1.2% agarose gel containing 6% formaldehyde in MESA buffer (43 g of MOPS, 6.8 g of sodium acetate, and 3.8 g of EDTA per liter; pH 7.0). RNA was transferred to a positively charged nylon membrane (Roche Diagnostics Corporation, Indianapolis, Ind.) and immobilized by UV cross-linking. Digoxigenin (DIG)-labeled CgCDR1, CgCDR2, and CgERG11 probes were synthesized using the PCR DIG probe synthesis kit (Roche) with approximately 0.1 μg of genomic DNA from C. glabrata strain ATCC 2001 as template. The following primer sequences were used for probe synthesis (5′ to 3′): CgCDR1 forward, CTCCGCTTACCTACGTCAACC, and reverse, CCCAAGTACTCACCACAAGTC; CgCDR2 forward, GGGATAACGCTAGGAGAGG, and reverse, GGTATGCAAGAGGGTTGATG; CgERG11 forward, CGTTGAACTATTGGAGTACG, and reverse, GAGGCAAGTTAGGGAAGACG (22). PCR conditions were as follows: 5 min at 90°C, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C in a Perkin-Elmer model 9700 thermal cycler. A subsequent elongation step was conducted for 7 min at 72°C. Hybridization and posthybridization washes were carried out using FastHyb hybridization solution and the wash-block-buffer system (Roche). Hybridization signals were detected with CDP-Star following the manufacturer's directions (DIG application manual for filter hybridization; Roche). Fluorescent signals were visualized and quantified using a Lumi-Imager F1 and accompanying software (Roche). To quantify the amount of RNA loaded in the gel used for Northern blotting, another 5-μl aliquot of RNA was loaded and run in a second agarose gel, detected with ethidium bromide, and quantified using the Lumi-Imager after UV illumination. Expression of CgCDR1, CgCDR2, and CgERG11 was normalized to the amount of RNA loaded in the gel for each isolate.
Screening for mitochondrial deficiency.
The mitochondrial function of the isolates was analyzed by examining their ability to grow on medium containing glycerol as the sole carbon source. The parental isolates, the first resistant mutants, and the last mutants collected at day 136 were cultured simultaneously on yeast extract-peptone-glucose agar (5 g of yeast extract/liter, 10 g of peptone/liter, 20 g of glucose/liter, 20 g of agar/liter) and on yeast extract-peptone agar containing 2% glycerol.
Results.
Prior to fluconazole exposure, all parental C. glabrata isolates examined were susceptible to fluconazole (MICs ≤ 4 μg/ml) and susceptible or susceptible dose dependent to itraconazole (MICs ≤ 0.25 μg/ml) (Table 1). In addition, all parental isolates exhibited low voriconazole MICs (0.125 μg/ml). Isolates CBS 138 and CBS 860 and isolates 73/124 and 75/015, respectively, became fully resistant (MICs ≥ 64 μg/ml) to fluconazole after 2 and 4 days of exposure to 16 μg of fluconazole/ml. Isolate CBS 4692 remained susceptible after 8 days of exposure to 16 μg of fluconazole/ml, but it became resistant within 4 days when the fluconazole concentration in the medium was increased to 64 μg/ml. None of the isolates developed resistance spontaneously during repeated subculture in the absence of fluconazole.
TABLE 1.
In vitro susceptibilities of C. glabrata parental isolates, first resistant mutants, and last isolates examined
| Isolate no. | Fluconazole MIC (μg/ml)
|
Itraconazole MIC (μg/ml)
|
Voriconazole MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Parent | Firsta | Lastb | Parent | Firsta | Lastb | Parent | Firsta | Lastb | |
| CBS 138 | 2 | 64 (day 2) | 32 | 0.125 | 2 (day 2) | 1 | 0.125 | 4 (day 2) | 1 |
| CBS 860 | 4 | 64 (day 2) | 64 | 0.25 | 1 (day 2) | 1 | 0.125 | 2 (day 2) | 1 |
| CBS 4692 | 1 | 64 (day 4)c | 64 | 0.25 | 2 (day 4)c | 1 | 0.125 | 4 (day 4)c | 1 |
| 73/124 | 4 | 64 (day 4) | 64 | 0.125 | 1 (day 4) | 1 | 0.125 | 2 (day 4) | 2 |
| 75/015 | 4 | 64 (day 4) | 64 | 0.125 | 0.5 (day 4) | 1 | 0.125 | 2 (day 4) | 2 |
The first resistant mutant to emerge during fluconazole exposure; day of culture when resistance was noted is shown in parentheses.
The last isolate examined (exposed to fluconazole for 14 days then cultivated in the absence of fluconazole for 122 days).
Isolate CBS 4692 remained susceptible after 8 days of exposure to 16 μg of fluconazole/ml but became resistant within 4 days when the fluconazole concentration in the medium was increased to 64 μg/ml.
Acquired fluconazole resistance remained stable after 122 days of cultivation in the absence of fluconazole (end point of study) for all but one isolate. The fluconazole MIC of this isolate (CBS 138) decreased from 64 μg/ml (resistant) to 32 μg/ml (susceptible dose dependent) after 73 days of cultivation in the absence of fluconazole, a time point that coincided with the change in subculturing conditions from liquid to solid medium.
Fluconazole-resistant mutants of each parental strain exhibited cross-resistance to itraconazole and increased voriconazole MICs (Table 1). Upon removal of drug pressure, isolates CBS 138 and CBS 4692, respectively, showed a twofold and a fourfold decrease in itraconazole and voriconazole MICs, although MICs remained high compared to those of the parental isolates. Isolate CBS 860 showed a twofold decrease in voriconazole MIC and no change in itraconazole MIC, whereas the remaining isolates showed no decrease in either the itraconazole or voriconazole MIC upon removal of drug (Table 1).
To verify that the azole-resistant mutants of each isolate were indeed descendants of their parental strain, RAPD typing was performed using genomic DNA from cells collected at the following time points: day 0 (parent), day 136 (122 days after fluconazole was removed), and day 136 (for parental isolates subcultured in the absence of fluconazole). RAPD patterns using the eight different primers showed no or minor changes among resistant mutants of a given strain (e.g., some differences in band intensity were observed). Representative results are shown in Fig. 1.
FIG. 1.
RAPD patterns for C. glabrata isolate 75/015. Lane 1, parental isolate; lane 2, no-fluconazole control isolate at day 136; lane 3, resistant mutant at day 136 (exposed to fluconazole for 14 days and then cultivated in the absence of fluconazole for 122 days). (A) Primer OPE-18; (B) primer OPE-04; (C) primer OPA-01; (D) primer OPA-02; (E) primer OPA-04; (F) primer OPA-10; (G) primer OPA-16; (H) primer OPA-18.
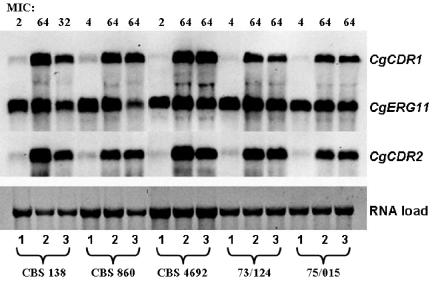
Mechanisms known to be associated with azole resistance in C. glabrata were examined in mutant isolates displaying resistance. Two independent Northern blotting experiments were performed using the same RNA preparation, and the average of the results is reported in Table 2. Expression of the drug efflux genes, CgCDR1 and CgCDR2, was minimal in all parental isolates but was considerably higher in the resistant mutants (Table 2; Fig. 2). In contrast, changes in expression of CgERG11, the gene encoding the azole target, 14α-lanosterol demethylase, were small. For isolate CBS 860, expression of both CgCDR1 and CgCDR2 continued to increase after fluconazole exposure was discontinued. For the remaining isolates, expression of the efflux pump genes decreased.
TABLE 2.
Expression of drug resistance genes in C. glabrata parental isolates, the first resistant mutants, and last isolates examined
| Isolate no. and type | Expression (relative to parent)a
|
||
|---|---|---|---|
| CgCDR1 | CgCDR2 | CgERG11 | |
| CBS 138 | |||
| Parent | 1 | 1 | 1 |
| Firstb | 32.1 | 27.9 | 1.7 |
| Lastc | 10.2 | 8.3 | 0.9 |
| CBS 860 | |||
| Parent | 1 | 1 | 1 |
| Firstb | 7.0 | 5.8 | 0.7 |
| Lastc | 11.7 | 8.7 | 0.4 |
| CBS 4692 | |||
| Parent | 1 | 1 | 1 |
| Firstb | 31.9 | 25.5 | 1.2 |
| Lastc | 27.6 | 14.6 | 0.5 |
| 73/124 | |||
| Parent | 1 | 1 | 1 |
| Firstb | 10.1 | 11.1 | 0.8 |
| Lastc | 6.8 | 8.7 | 0.6 |
| 75/015 | |||
| Parent | 1 | 1 | 1 |
| Firstb | 15.1 | 7.9 | 0.9 |
| Lastc | 11.6 | 8.9 | 0.6 |
Expression values were normalized against the amount of RNA loaded on the gel for each isolate. The ratio of gene expression after fluconazole exposure compared to gene expression by the corresponding parental isolate is shown. Values are the averages of two independent Northern hybridization experiments.
The first resistant mutant to emerge during fluconazole exposure.
The last isolate examined (exposed to fluconazole for 14 days and then cultivated in the absence of fluconazole for 122 days).
FIG. 2.
Quantification of CgERG11, CgCDR1, and CgCDR2 mRNA by Northern hybridization. Lane 1, parent; lane 2, first resistant mutant; lane 3, last isolate collected. MICs are the fluconazole MICs (in micrograms per milliliter).
Discussion.
Specific reasons for the recent emergence of C. glabrata as a significant cause of BSI are unknown. However, the azole antifungal drug fluconazole has been used extensively since the early 1990s for the prophylaxis and treatment of Candida infections, and it has been suggested that this practice has led to an increased number of infections caused by intrinsically less susceptible or resistant species, such as C. glabrata and C. krusei (1, 19, 24, 28). Therefore, to better understand the role of fluconazole exposure in the emergence of C. glabrata infections, we studied isolates that were obtained from patients at a time in history before fluconazole was in use; these isolates should never have been previously exposed to azole antifungal agents.
We found, despite no prior exposure to azole antifungal agents, only 2 to 4 days of in vitro exposure to fluconazole were necessary for the development of azole drug resistance in previously susceptible, naïve C. glabrata isolates. These data indicate that prior exposure to azole drugs is not a prerequisite for the rapid development of fluconazole resistance.
Of the three resistance mechanisms examined, expression of the drug efflux pump genes, CgCDR1 and CgCDR2, was most closely associated with acquired resistance. In contrast, changes in CgERG11 expression were small, and mRNA levels of this gene in resistant mutants on the last day of the study were lower than those of the corresponding parental isolates. Although overexpression of CgERG11 has been reported to occur in azole-resistant C. glabrata isolates (9, 22), our results clearly demonstrated that CgERG11 overexpression did not contribute significantly to the stable resistance observed in our isolates after fluconazole exposure. Point mutations in the CgERG11 gene which are associated with azole resistance have not yet been identified in C. glabrata and were, therefore, not examined in the present study.
Another reported mechanism of azole resistance found in C. glabrata is a loss of mitochondrial function (5). These so-called “petite” yeast cells have a respiratory deficiency inhibiting their ability to utilize ethanol or glycerol as a carbon source. Kaur et al. (13) recently demonstrated that petites might be able to switch between mitochondrially competent (fluconazole-susceptible) and incompetent (fluconazole-resistant) states at high frequency. All of our isolates, however, including the parents as well as the resistant mutants, grew equally well on medium containing glycerol as the sole carbon source, indicating that they had not lost their mitochondrial function. This, and the fact that the acquired resistance was stable, implies that isolates in this study did not use a mitochondrial resistance mechanism to obtain their resistant phenotype. Petite C. glabrata cells have been observed after growth on medium containing 2% glucose (5). The much lower concentration of glucose (0.2%) in the medium that was used for the induction of resistance in our isolates may have restricted the occurrence of such mitochondrially deficient cells.
Induction of azole resistance in Candida albicans has been reported to occur either gradually over a long period of time (4, 26, 27) or more rapidly, but in the latter case resistance is always transitory (3, 16). In contrast, all C. glabrata isolates in this study rapidly acquired resistance and all but one remained azole resistant after 4 months of subculture in the absence of fluconazole. Indeed, for isolate CBS 860, expression of CgCDR1 and CgCDR2 continued to increase even after the removal of drug. Although mRNA levels for these genes decreased somewhat for all other isolates upon removal of fluconazole, expression levels remained high compared to those of the respective parental isolates and never returned to preinduction levels. These data indicate that C. glabrata maintains an elevated level of CDR1 and CDR2 expression after only a single fluconazole exposure. It will be of interest, in future studies, to expose these resistant mutants to fluconazole once again to determine how the first fluconazole exposure affects gene expression upon secondary exposure. Only one isolate, CBS 138, demonstrated a significant reduction in expression of both efflux pump genes after fluconazole was removed, and this reduction corresponded to a decrease in the fluconazole MIC from 64 to 32 μg/ml.
It is interesting that expression of both CgCDR1 and CgCDR2 was up-regulated in all of our resistant mutants. The sequences for these genes contain upstream elements that are nearly identical to the Pdr1p and Pdr3p response elements of Saccharomyces cerevisiae (17, 23). In S. cerevisiae, these transcription factor binding sites are essential for expression of the ABC transporter gene, PDR5, and are also found in other members of this superfamily of genes (12). It is possible that expression of CgCDR1 and CgCDR2 in C. glabrata is regulated by the same transcription factor, leading to coexpression of both efflux pump genes.
In conclusion, we observed the rapid development of azole drug resistance following in vitro exposure to fluconazole among C. glabrata isolates that had never previously been exposed to azole antifungal agents. Resistance was associated with increased expression of the ABC transporters CgCDR1 and CgCDR2, but not CgERG11. Acquired resistance was stable in these isolates, and CgCDR1 and CgCDR2 gene expression remained up-regulated for at least 4 months after the removal of fluconazole pressure. Although fluconazole use most likely is involved in the emergence of C. glabrata infections in those regions where resistant C. glabrata isolates are frequently encountered, other factors are clearly involved in the emergence of C. glabrata as an important BSI pathogen (8). Studies focusing on risk factors and possible confounding factors are needed to better understand causes associated with the increased prevalence of this species.
Acknowledgments
We thank Frank Odds and Teun Boekhout for kindly providing the C. glabrata isolates that were used in this study.
A.B. was a recipient of an American Society for Microbiology/National Center for Infectious Diseases postdoctoral research fellowship.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, J. E., K. Izumikawa, and K. A. Marr. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvet, H. M., M. R. Yeaman, and S. G. Filler. 1997. Reversible fluconazole resistance in Candida albicans: a potential in vitro model. Antimicrob. Agents Chemother. 41:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Defontaine, A., J. P. Bouchara, P. Declerk, C. Planchenault, D. Chabasse, and J. N. Hallet. 1999. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48:663-670. [DOI] [PubMed] [Google Scholar]
- 6.Diekema, D. J., S. A. Messer, A. B. Brueggemann, S. L. Coffman, G. V. Doern, L. A. Herwaldt, and M. A. Pfaller. 2002. Epidemiology of candidemia: 3-year results from the Emerging Infections and the Epidemiology of Iowa Organisms study. J. Clin. Microbiol. 40:1298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajjeh, R. A., A. N. Sofair, L. H. Harrison, G. M. Lyon, B. A. Arthington-Skaggs, S. A. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. A. Ciblak, L. E. Benjamin, L. T. Sanza, S. Huie, S. F. Yeo, M. E. Brandt, and D. W. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchcock, C. A., G. W. Pye, P. F. Troke, E. M. Johnson, and D. W. Warnock. 1993. Fluconazole resistance in Candida glabrata. Antimicrob. Agents Chemother. 37:1962-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izumikawa, K., H. Kakeya, H. F. Tsai, B. Grimberg, and J. E. Bennett. 2003. Function of Candida glabrata ABC transporter gene, PDH1. Yeast 20:249-261. [DOI] [PubMed] [Google Scholar]
- 12.Katzmann, D. J., T. C. Hallstrom, Y. Mahe, and W. S. Moye-Rowley. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271:23049-23054. [DOI] [PubMed] [Google Scholar]
- 13.Kaur, R., I. Castano, and B. P. Cormack. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart, S. R., S. Joly, C. Pujol, J. D. Sobel, M. A. Pfaller, and D. R. Soll. 1997. Development and verification of fingerprinting probes for Candida glabrata. Microbiology 143:3733-3746. [DOI] [PubMed] [Google Scholar]
- 15.Marichal, P., H. Vanden Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. Van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marr, K. A., C. N. Lyons, T. R. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. J. Falconer, D. J. Ward, K. Marsden, and J. E. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Nguyen, M. H., J. E. Peacock, Jr., A. J. Morris, D. C. Tanner, M. L. Nguyen, D. R. Snydman, M. M. Wagener, M. G. Rinaldi, and V. L. Yu. 1996. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am. J. Med. 100:617-623. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., and D. J. Diekema. 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10:11-23. [DOI] [PubMed] [Google Scholar]
- 21.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Geographic variation in the susceptibilities of invasive isolates of Candida glabrata to seven systemically active antifungal agents: a global assessment from the ARTEMIS Antifungal Surveillance Program conducted in 2001 and 2002. J. Clin. Microbiol. 42:3142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redding, S. W., W. R. Kirkpatrick, S. Saville, B. J. Coco, W. White, A. Fothergill, M. Rinaldi, T. Eng, T. F. Patterson, and J. Lopez-Ribot. 2003. Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal candidiasis receiving head and neck radiation. J. Clin. Microbiol. 41:619-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 25.Warnock, D. W., J. Burke, N. J. Cope, E. M. Johnson, N. A. von Fraunhofer, and E. W. Williams. 1988. Fluconazole resistance in Candida glabrata. Lancet 2:1310. [DOI] [PubMed] [Google Scholar]
- 26.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14-α demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wingard, J. R., W. G. Merz, M. G. Rinaldi, C. B. Miller, J. E. Karp, and R. Saral. 1993. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob. Agents Chemother. 37:1847-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]