Abstract
A novel gene for quinolone resistance was cloned from a transferable plasmid carried by a clinical isolate of Shigella flexneri 2b that was resistant to fluoroquinolones. The plasmid conferred low-level resistance to quinolones on Escherichia coli HB101. The protein encoded by the gene showed 59% amino acid identity with Qnr.
Quinolone resistance is usually caused by mutations in the chromosomal genes for DNA gyrase and DNA topoisomerase IV, the targets of quinolones (1, 3). Plasmid-mediated resistance was first discovered in a clinical strain of Klebsiella pneumoniae isolated in Alabama (5). This plasmid, pMG252, was a multiresistance-encoding plasmid and contained the gene for quinolone resistance, qnr. The qnr gene has been found to encode a 218-amino-acid protein, and the purified Qnr protein has been shown to protect DNA gyrase activity in vitro (8). The qnr gene has since been detected in more than 20 clinical strains of K. pneumoniae and E. coli isolated in the United States and China (4, 7, 10, 11).
At the beginning of October 2003, an outbreak of food poisoning caused by Shigella flexneri 2b occurred in Aichi Prefecture, Japan. Eight clinical strains isolated from the patients were subjected to pulsed-field gel electrophoresis analysis for epidemiological investigation. They were judged to be clones from the same origin on the basis of the pulsed-field gel electrophoresis pattern obtained. We also determined the drug sensitivities of these isolates. Seven of the eight clones had the same pattern of drug resistance. However, one had additional resistance to fluoroquinolones and cephaloridine. In the course of investigating the mechanism of acquisition of the additional drug resistance, we found a plasmid carrying resistance genes and cloned a novel gene responsible for quinolone resistance. This is the first report of a gene for quinolone resistance on the conjugative plasmid in Shigella.
Strains and drug resistance.
Nine strains of S. flexneri 2b were isolated from eight patients. Strains 1 to 8 were isolated from different patients from 1 to 9 October 2003. Strains 8 and 8′ were from the same patient and were isolated on 8 and 20 October, respectively. This patient was treated with levofloxacin.
With antibiotic disks (KB disk; Eiken Chemical Co., Tokyo, Japan), patterns of resistance to 12 drugs were examined. Patterns of seven isolates of S. flexneri 2b were the same. They were resistant to ampicillin, streptomycin, kanamycin, chloramphenicol (CHL), tetracycline, and nalidixic acid (NAL). However, strain 8 (and strain 8′) was resistant to three additional drugs, norfloxacin, ciprofloxacin, and cephaloridine (CER). All isolates were sensitive to cefotaxime, fosfomycin, and trimethoprim-sulfamethoxazole.
The mutations in the quinolone resistance-determining region (12) of the gyrA and parC genes were examined. The quinolone resistance-determining region cloned by PCR (9) was sequenced by the IR dye terminator method with the LI-COR 4200 IR2 system (LI-COR, Lincoln, Nebr.). The following mutations were found in all of the isolates: Ser-83 to Leu in the gyrA gene and Ser-80 to Ile in the parC gene.
Plasmid profiles and conjugation experiments.
Plasmid profiles revealed that strain 8 (and strain 8′) had a unique plasmid of about 47 kb that was not present in the other seven isolates. The plasmid was designated pAH0376.
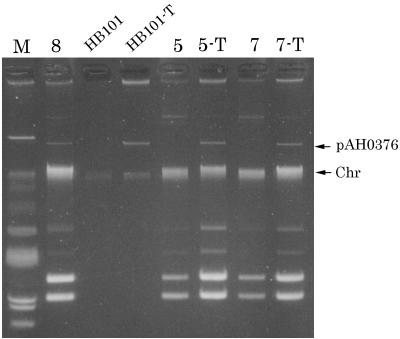
A conjugation experiment was carried out with E. coli HB101 (F− hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 [Strr] xyl-5 mtl-1 supE44 leuB6 thi-1) as the recipient, with selection for pAH0376-encoded cephaloridine resistance. Shigella strain 8 was the donor. Overnight cultures of Shigella cells (10 μl) and HB101 cells (1 ml) were added to fresh Luria-Bertani (LB) broth (5 ml) and incubated for 6 h. Transconjugants were selected on Sorbitol-MacConkey agar (Eiken Chemical Co.) plates containing cephaloridine at 5 μg/ml. HB101 cells generate red colonies on Sorbitol-MacConkey plates (Sorbitol fermented), whereas Shigella cells generate pinkish colonies (not Sorbitol fermented). Transconjugant HB101 appeared as red colonies on the plates. pAH0376 was transferred to HB101 (Fig. 1, lane HB101-T). Transconjugant HB101 showed low resistance to nalidixic acid (MIC, 16 μg/ml), ciprofloxacin (MIC, 0.25 μg/ml), and cephaloridine (MIC, 16 μg/ml) and in addition high resistance to ampicillin (MIC, 512 μg/ml). The transconjugant was sensitive to other drugs except streptomycin, to which HB101 is resistant.
FIG. 1.
Transmission of plasmid pAH0376 to E. coli HB101 and outbreak strains of S. flexneri 2b. Lanes: M, molecular size marker strain V517; 8, strain 8; HB101-T, transconjugant of HB101 with pAH0376 transmitted from strain 8; 5 and 7, strains 5 and 7; 5-T and 7-T, transconjugants of strains 5 and 7 with pAH0376 transmitted from HB101-T. The positions of plasmid pAH0376 and chromosomal DNA (Chr) are indicated on the right.
A second conjugation experiment was performed to transfer pAH0376 from transconjugant HB101 to the Shigella strains. Transconjugants were selected on Salmonella-Shigella agar (Eiken Chemical Co.) plates containing cephaloridine at 5 μg/ml. Growth of E. coli is suppressed on Salmonella-Shigella agar plates. Two S. flexneri 2b strains (5 and 7) were the recipients. The recipient strains were sensitive to ciprofloxacin (MIC, 1 μg/ml), cephaloridine (MIC, 4 μg/ml), and ampicillin (MIC, 128 μg/ml). Plasmid pAH0376 was transferred to these Shigella strains, and the transconjugants showed the same level of drug resistance as strain 8: the MIC of ciprofloxacin was 8 μg/ml, that of cephaloridine was 16 μg/ml, and that of ampicillin was 512 μg/ml (Fig. 1). The results indicated that the difference in the drug resistance patterns observed in the outbreak strains of S. flexneri 2b was caused by conjugative plasmid pAH0376.
Cloning of resistance-encoding genes.
pAH0376 was isolated from transconjugant HB101 and digested with EcoRI or HindIII. The resulting fragments were cloned into vector pBC SK (+) (Stratagene, La Jolla, Calif.), and recombinants were transformed into HB101. Transformants were selected on LB broth plates containing CHL (30 μg/ml; the selection drug for the pBC vector) and NAL (4 μg/ml) or CHL and cephaloridine (CER; 5 μg/ml). A clone carrying a plasmid with a 2.6-kb HindIII insert was isolated on the NAL-CHL plates. This clone (pBC-H2.6) showed the same level of resistance to quinolones as pAH0376. The nucleotide sequence of the 2,642-bp insert was determined. We identified an open reading frame (ORF) of 657 bp encoding a 218-amino-acid polypeptide. The PCR fragment of the 839 bp encompassing the entire transcription unit of the ORF (−96 from ATG to +86 from TAA) was amplified with pAH0376 as the template. The PCR product was cloned into a pGEM-T vector (Promega, Madison, Wis.) and transformed into JM109 (Takara Bio, Otsu, Japan). The resultant construct (pGEM-ORF) was isolated, its nucleotide sequence was verified, and it was transformed into HB101. The transformant was isolated on broth plates containing ampicillin at 50 μg/ml (the selection drug for the pGEM vector). pGEM-ORF conferred the same level of quinolone resistance as pAH0376, indicating that this ORF was responsible for quinolone resistance.
A BLAST analysis of deduced amino acid sequences revealed that the gene product exhibited significant homology to the Qnr protein, with 59% amino acid identity. Besides Qnr, two other proteins also revealed high similarity. Hypothetical proteins of Photobacterium profundum (CAG22829) and Vibrio vulnificus (AAO07889) showed 64 and 53% amino acid identity, respectively. An alignment of the ORF-encoded protein, Qnr, and the hypothetical proteins of P. profundum and V. vulnificus is shown in Fig. 2. The four proteins all consisted of 218 amino acids and had 43% of their amino acids in common. In view of the high similarity to the Qnr protein, it is conceivable that the product of the new gene is a homologue of Qnr. We designated the new gene qnrS and its product QnrS. The resistance-encoding activity of qnrS was similar to that of qnr. Both conferred a low level of quinolone resistance and had an additive effect on quinolone resistance due to defined DNA gyrase alterations (6).
FIG. 2.
Alignment of deduced amino acid sequences of the ORF-encoded protein examined in this study (QnrS), Qnr, and hypothetical proteins of P. profundum (CAG22829) and V. vulnificus (AAO07889). Identical amino acids are highlighted.
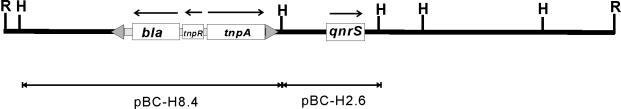
A recombinant clone carrying a plasmid with an ∼20-kb EcoRI insert (pBC-R20) was isolated on both NAL-CHL and CER-CHL plates, and a clone with an 8.4-kb HindIII insert (pBC-H8.4) was isolated on CER-CHL plates. A restriction map of pBC-R20 revealed that the 2.6- and 8.4-kb HindIII fragments were involved in the EcoRI insert (Fig. 3). The nucleotide sequence analysis identified a Tn3-like sequence containing a TEM-1 β-lactamase gene (2). There was no element for an integron in the region sequenced. Further investigations are required to resolve how the qnrS gene was acquired.
FIG. 3.
Map of the EcoRI fragment of pAH0376. The qnrS gene and the TEM-1 β-lactamase gene are indicated. The other two open squares are the genes for resolvase (tnpR) and transposase (tnpA), respectively. The triangles represent 38-bp inverted repeats on both sides of the Tn3-like transposon. H, HindIII; R, EcoRI.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the EMBL/GenBank/DDBJ databases and assigned accession number AB187515.
REFERENCES
- 1.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heffron, F., B. J. McCarthy, H. Ohtsubo, and E. Ohtsubo. 1979. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell 18:1153-1163. [DOI] [PubMed] [Google Scholar]
- 3.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Martínez, L., A. Pascual, I. García, J. Tran, and G. A. Jacoby. 2003. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51:1037-1039. [DOI] [PubMed] [Google Scholar]
- 7.Rodrígues-Martínez, J. M., A. Pascual, I. García, and L. Martínez-Martínez. 2003. Detection of the plasmid-mediated quinolone resistance determinant qnr among clinical isolates of Klebsiella pneumoniae producing AmpC-type β-lactamase. J. Antimicrob. Chemother. 52:703-706. [DOI] [PubMed] [Google Scholar]
- 8.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hopper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]