Abstract
Receptor-interacting protein 140 (RIP140) is a wide-spectrum coregulator for hormonal regulation of gene expression, but its activity in development/stem cell differentiation is unknown. Here, we identify RIP140 as an immediate retinoic acid (RA)-induced dual-function chaperone for LSD1 (lysine-specific demethylase 1). RIP140 protects LSD1’s catalytic domain and antagonizes its Jade-2-mediated ubiquitination and degradation. In RA-induced neuronal differentiation, the increased RIP140/LSD1 complex is recruited by RA-elevated Pit-1 to specifically reduce H3K4me2 modification on the Pax6 promoter, thereby repressing RA-induction of Pax6. This study reveals a new RA-induced gene repressive mechanism that modulates the abundance, enzyme quality, and recruitment of histone modifier LSD1 to neuronal regulator Pax6, which provides a homeostatic control for RA induction of neuronal differentiation.
Keywords: Embryonic stem cells, Neural differentiation, Retinoic acid, Transcription factors, RIP140
Introduction
Retinoic acid (RA), the principal active ingredient of vitamin A, exerts various biological activities particularly for embryonic and neural development [1]. Changes in RA homeostasis affect early stages of neural development such as in the posterior neuroectoderm (hindbrain and spinal cord) and eye (an outpocketing of the fore-brain) [2]. In vitro, embryonic stem cells (ESCs) aggregated embryoid bodies (EBs) treated with RA differentiate mainly into neurons [3]. Hence, RA is generally thought as a neural inducer. But how neuronal differentiation is controlled at a homeostatic state so that a proper neural stem cell population can be preserved is less clear.
Lysine-specific demethylase 1 (LSD1), the first identified histone demethylase, removes H3K4me1/2 in a flavin-adenine-dinucleotide-dependent oxidative reaction [4]. Mouse gene knockout study shows that LSD1 is essential for embryonic development beyond embryonic day 6.5 (e6.5), which also establishes LSD1 as a key regulator of neural stem cell proliferation [5]. Studies also suggest that LSD1 is crucial for the differentiation of ESCs. For instance, loss of LSD1 causes Pax6 gene upregulation and promotes ESC differentiation toward the neural lineage. PAX6 is an important transcription factor involved in the development of the central nervous system [6], as it controls the homeostasis of self-renewal and neurogenesis of neural stem cells. Specifically, increasing PAX6 levels drives the system toward neurogenesis; whereas decreasing PAX6 blunts neural stem cells’ self-renewal [7]. In the in vitro EB-neural differentiation model, before RA treatment, few cells express PAX6; but RA increases the number of PAX6-expressing, differentiating cells in a concentration-dependent manner [8]. Interestingly, the Pax6 gene promoter contains no functional RA response elements in spite of its regulation by RA [9]. The mechanism that fine-tunes PAX6 expression levels, particularly in RA-induced neural differentiation process, remains unclear.
It is suggested that Pax6 gene regulation involves Pituitary-specific positive transcription factor 1 (Pit-1). Pit-1 is an RA-induced transcription factor, and a member of the homeobox POU domain-containing transcription factor family that is involved in various developmental processes [10]. Mechanistically, Pit-1 can recruit opposing coregulatory proteins (such as CBP/p300 for activation, and NCoR for repression) to target gene promoters to provide a homeostatic control of the target gene [11]. But, there has been no study delineating whether and how Pit-1 modulates the response of Pax6 gene to RA. Deleting LSD1 causes Pax6 upregulation and enhances neural differentiation, which would indicate a critical role for certain LSD1-containing negative coregulatory complex in suppressing Pax6 gene. Whether and how the LSD-1 complex is involved in RA regulation of Pax6 is not known.
At the molecular level, LSD1 has been detected in CoREST and NcoR coregulator complexes and its activity can be regulated by associated factors [12]. To this end, studies have indicated that LSD1 is an unstable protein, subjected to extensive proteasome degradation. For instance, Jade-2, an E3 ubiquitin ligase, specifically targets LSD1 for degradation [13], and CoR-EST can protect LSD1 from ubiquitination to regulate certain neuronal genes such as human neuronal-specific sodium channel genes [5]. Apparently, LSD1 can be extensively and differentially regulated, most likely in a gene- and complex-dependent manner. The control over its enzymatic activity, quantity, and recruitment is critical for gene regulation.
Receptor-interacting protein 140 (RIP140, also known as NRIP1) is a wide spectrum transcription coregulator. It can be directly induced by RA [14] and recruited to RA receptors [15–17], which has been suggested to provide a negative feedback mechanism to limit RA activation of genes whose homeostasis (via positive and negative regulations) is crucial [18]. For most transcription factors, RIP140 acts as a corepressor by recruiting various repressive enzymes/factors such as HDACs, CtBP, and G9a, and so forth [19–21]. Animal studies have revealed its diverse functions in various biological/physiological processes including energy expenditure, behavior, immunity, lipid metabolism, insulin signaling, and neuron stress, and so forth [22–27]. In the RA-induced ESC differentiation process, RIP140 is required for the Brg/Brm switch on the Nanog and Oct4 gene promoters when they are silenced in differentiating cells [28]. In this study, we report new dual functions of RIP140 in suppressing RA-induced ESC differentiation. Specifically, RIP140 is directly induced by RA and acts to protect LSD1’s catalytic domain and to antagonize Jade-2-mediated ubiquitination/degradation of LSD1. The RIP140/LSD1 complex is recruited by Pit-1 to form a chromatin demethylating complex on Pax6 gene promoter to repress Pax6 expression, thereby dampening neuronal differentiation. This prevents overly activated neural differentiation following RA induction.
Materials and Methods
mESC Cultures
CJ7 ESCs were maintained in ESC medium (Dulbecco’s modified Eagle’s medium), supplemented with 17% ESC-qualified fetal bovine serum, 2 mM glutamine, 0.1 mM nonessential amino acids, 6 μM β-mercaptoethanol, 2 mM HEPES, and 1,000 U/ml recombinant leukemia inhibitory factor (LIF). Cells were grown on irradiated mouse embryonic feeder cells in 0.2% gelatin-coated plates. For differentiation, ESCs were dissociated and plated onto 0.2% gelatin-coated tissue culture plastic at a density of 0.5–1.5 × 104/cm2 in regular medium without LIF. Medium was renewed every day. Cells were treated with 1 μM all-trans RA (Cat. #R2625, Sigma, St. Louis, MO, http://www.sigmaaldrich.com) for indicated time periods.
In Vitro Differentiation
To induce EBs formation, cells were dissociated into a single-cell suspension with 0.05% Trypsin/EDTA and plated onto non-adherent bacterial culture dishes for 4 days at a density of 1–2 × 104 cells per square centimeter in ESC medium (without LIF). The EBs were then cultured for an additional 4 days in the presence of 5 μM RA. For EB cryosection, EBs were fixed in paraformaldehyde (4%) for 10 minutes at room temperature and embedded in OCT (Sigma); 15 mm cryosections were generated. For plating induced ESCs, the EBs were dissociated on the 9th day, grown on PORN/laminin-coated plates or coverslips and fed every 2 days with differentiation medium (N2/B27 medium) for up to 8 days in vitro.
Establishment of Stable Cell Lines
To obtain stable RIP140 shRNA ESCs, the lentiviral shRNA vector system was selected and RIP140 shRNA (gene target sequences: shRIP140 4, 5′-AAG CTT CTT TCT TTA ATC TAA-3′) was used. RIP140 shRNA was subcloned into the site of NotI and XbaI of pCDH (System Biosciences, Mountain View, CA, https://www.systembio.com/) to generate RIP140-silencing lentiviral vectors. Lentivirus was concentrated and titled as described [39]. Control cells received in a similar sized noneffective scrambled sequence. Additionally, for obtaining stable clones, cells were selected post-transfection using puromycin. Single-cell clones were expanded, passaged, and characterized before use.
Silencing
RIP140-siRNA was purchased from Qiagen. siRNAs were introduced into cells by HiPerFect transfection reagent (301704, Qiagen, Hilden, Germany, www1.qiagen.com). The cells were retransfected every 2 days and subcultured into a monolayer after the second transfection (i.e., on day 6). For knockdown of Pit-1, specific shRNA’s were purchased from University of Minnesota Biomedical Genomics Center.
Western Blotting and Immunoprecipitation
To prepare whole cell lysates, cells were washed twice with cold phosphate buffered saline (PBS) and harvested in RIPA buffer (50 mM Tris-HCl pH 7.4, 0.5% deoxycholic acid, 150 mM NaCl, 0.1% SDS, 4 Mm EDTA, and 1% Nonidet P40) with a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, https://lifescience.roche.com), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium fluoride, and 1 mM sodium orthovanadate. After centrifugation, supernatant was collected and protein concentrations were determined using Bradford method and subjected to SDS-PAGE. For immunoprecipitation, cells were lysed and collected with IP buffer (50 mM Tris-HCl pH 8.0, 10% glycerol, 100 mM NaCl, 1 Mm EDTA, and 0.1% Nonidet P40) containing protease inhibitor cocktail, 1 mM PMSF, 1 mM sodium fluoride, and 1 mM sodium orthovanadate for immunoprecipitation. One hundred micrograms of proteins was incubated with primary antibodies overnight, and supplemented with protein G beads for another 1 hour for immunoprecipitation. After five times washing with IP buffer, proteins were eluted by boiling at 100°C for 5 minutes in 2 × Laemmli loading buffer. Antibodies against LSD1, Pit-1, CoREST, HA, FLAG, and β-actin antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, http://www.scbt.com/). RIP140 and Pax6 antibody were purchased from Abcam (Cambridge, MA, www.abcam.com).
Reverse Transcription and Real-Time Polymerase Chain Reaction
Total RNA was extracted from ESCs using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). RNA was reverse transcribed using the Omniscript RT kit (205113, Qiagen). For real-time polymerase chain reaction (PCR) analysis, 2 × Brilliant II Master Mix (600804, Agilent Technologies, Santa Clara, CA, www.home.agilent.com) was used, and PCR was performed on an MX3000P Stratagene thermocycler. The relative values were normalized to β-actin and presented as ΔΔCt methods. All the primers (for Oct4, Ncam, Neurog1, Pax6, RIP140, and β-actin) were purchased from Qiagen.
IF Staining
Cells were fixed by 4% paraformaldehyde for 10 minutes and permeabilized using 0.2% Triton X-100 for 5 minutes at 4°C. Nonspecific binding was blocked by incubation of cells with 0.5% bovine serum albumin (BSA)-PBS 1 hour before incubation with primary antibodies. IF was performed using as primary antibody rabbit anti-PAX6 IgG (Abcam, ab5790), goat anti-MAP2 (Santa Cruz Biotechnology, sc-5359), Rabbit anti-RIP140 (Santa Cruz Biotechnology, sc-8997), or mouse anti-LSD1 (Santa Cruz Biotechnology, sc-271720) and as secondary antibody FICT-conjugated donkey anti-Rabbit IgG, Cy3-conjugated donkey anti-Rabbit IgG (Santa Cruz Biotechnology), Cy3-conjugated donkey anti-mouse IgG (Santa Cruz Biotechnology), and Cy3-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology). Nuclei were stained by DAPI (Sigma-Aldrich). Images were acquired with an Olympus FluoView 1000 IX2 upright confocal microscope (OLYMPUS, New York, NY, http://www.olympusamerica.com/).
Histone Extraction and Demethylation Assay
Cells were lysed for 10 minutes (4°C) in lysis buffer (PBS, 0.5% Triton X-100, 2 mM phenylmethanesulfonylfluoride, 5 mM Na Butyrate, protease inhibitor cocktail) at 107 cells per milliliter. Nuclei were pelleted by centrifugation at 2,000g for 10 minutes and washed in lysis buffer. Nuclei were extracted in 0.2 N HCl overnight at 4°C. Proteins were precipitated by 2,2,2-trichloroacetic acid (TCA) (100%) and pelleted at 16,000g for 10 minutes, acetone-washed, and air-dried, before dissolving in lysis buffer. LSD1 demethylation activity on free histones was carried out as previously described by Shi et al. [12]. Briefly, bulk histones were incubated with purified His-LSD1 with or without purified GST-RIP140 in the HDM assay buffer (50 mM Tris [pH 8.5], 50 mM KCl, 5 mM MgCl, 0.5% BSA, and 5% glycerol) from 10 or 30 minutes at 37°C. The demethylase activity of LSD1 was evaluated by Western blotting using K4-H3 dimethylation-specific antibodies.
Chromatin Immunoprecipitation Assay
ChIP assays were performed as described [28], using antibodies: H3K4me2 (07-030, Millipore, Billerica, MA, http://www.emdmillipore.com/), LSD1 (sc-49291, Santa Cruz Biotechnology), Pit-1 (sc-442, Santa Cruz Biotechnology), and receptor-interaction protein 140 (RIP140; ab42126, Abcam). Primers used for ChIP assay are shown in Supporting Information Table S1.
Luciferase Assay
To construct wild-type and deletion mutants, Pax6 promoter regions of various lengths with common 3′ end were prepared by polymerase chain reaction (PCR) amplification of mouse genomic DNA. The primers used for amplification are shown in Supporting Information Table S1. To facilitate directional cloning, two restriction sites, Mlu1 and Xho1, were incorporated into the primer at the 5′ and 3′ ends, respectively. Additionally, the introduction of a mismatched primer mutation into Pit 1 was used to generate pGL3-Pax6 ΔPit1 (ATTCA → AaaCA) mutant constructs. The primers used for amplification are shown in Supporting Information Table S1. The PCR products were verified by sequencing and cloned into promoterless luciferase reporter gene vector PGL3-Basic (Promega, Madison, WI, https://www.promega.com/). Luciferase assays were conducted using the dual luciferase reporter system (Promega), in which the relative luciferase activity was calculated by normalizing transfection efficiency according to the Renilla luciferase activities.
Pull-Down Assay
LSD1 and RIP140 (full-length [FL] and truncated fragments) were translated in vitro using TNT Quick Coupled Transcription/Translation Systems (Promega). Truncated RIP140 proteins were mixed with FL-LSD1 and immunoprecipitated with HA antibody. Reciprocally, truncated LSD1 fragments were mixed with FL-RIP140 and then immunoprecipitated with FLAG antibody. Following incubation with protein G beads for 1 hour, samples were washed with IP buffer, eluted in 2 × Laemmli loading buffer, subjected to SDS-PAGE, and detected by Western blotting using HA and FLAG antibodies.
Statistical Analyses
Statistical significance for multiple comparisons was determined by Student’s t test or analysis of variance as indicated in the figure legend using GraphPad Prism Program (GraphPad, San Diego, CA, http://www.graphpad.com/scientific-software/prism/) and summarized as the mean ± SEM of repeated measures. p < .05 was considered statistically significant at the 95% level.
Result
RIP140 Negatively Regulates RA Induction of PAX6- and MAP2-Positive Cells
The Rip140 gene is generally silenced in healthy, proliferating ESCs, and is rapidly and directly induced by RA in differentiating cultures [28, 29], suggesting a functional role for RIP140 in RA-induced differentiation of ESCs, particularly toward the neuronal lineage. We generated multiple ESC stable clones carrying constitutively expressed shRNAs against Rip140 (Supporting Information Fig. S1A). Two of three clones (#2 and #3) carrying Rip140 shRNA 4 showed >80% decrease in RA-induced RIP140 protein levels as compared to control clones carrying scramble shRNA. Under a standard ESC culture condition, cell cycle pattern and Nanog and Oct4 expression were similar between the RIP140shRNA clones (#2 and #3) and the control ESC (Supporting Information Fig. S1B, S1C). Thus, silencing RIP140 does not affect normal ESC proliferation.
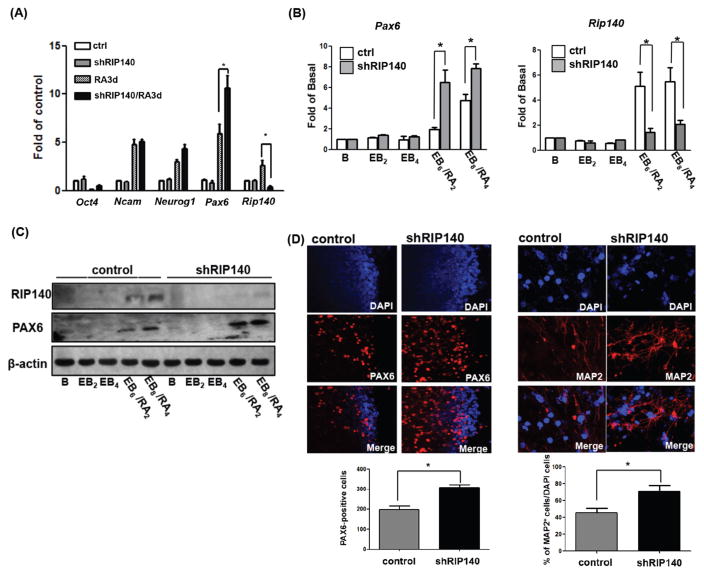
Using these RIP140-silenced ESC clones, we wanted to determine whether depleting RIP140 could affect RA-induced ESC differentiation into the neural lineage. Q-PCR analysis showed that RA effectively induced Rip140 expression, as well as the expected neuronal differentiation markers such as Ncam, Neurog1, and Pax6 (Fig. 1A, RA3d). Among these neuronal genes, Pax6 response was most interesting, because in the RA-treated, RIP140-silencing ESC clones (shRIP140/RA3d), Pax6 mRNA level was further increased (from approximately 6–12-folds) (Fig. 1A), suggesting RIP140 specifically modulates Pax6. We then examined whether the enhancing effect of silencing RIP140 on RA-induced Pax6 expression remained true in the EB/RA differentiation model. As shown (Fig. 1B), Pax6 mRNA was also upregulated by silencing RIP140 in RA-treated, differentiating EBs. Western blot analysis confirmed changes in RIP140 and PAX6 protein expression (Fig. 1C). Immunofluorescence (IF) staining also validated the effect of silencing RIP140 in elevating the expression of PAX6 in the nuclei (Fig. 1D, left) as well as neuronal marker MAP2 (Fig. 1D, right). These results demonstrate that silencing RIP140 enhances RA induction of PAX6- and MAP2-positive cells after differentiation, and suggest that RIP140 is a negative regulator of RA-induced differentiation, possibly by repressing the key early gene in neuronal differentiation, Pax6. Reducing RIP140 level specifically promotes RA-induction of Pax6 and increases the number of MAP2-positive cells.
Figure 1.
Silencing RIP140 increases MAP2- and PAX6-positive cells under retinoic acid (RA) induced differentiation. Expression during RA-induced (A) or embryoid body (EB)/RA-induced (B) differentiation of control embryonic stem cells (ESCs) and shRIP140 ESC clones, as detected by q-PCR. Embryoid body differentiation kinetics denoted as follows: B, unstimulated ESC; EB2, 2 days embryoid body; EB4, 4 days embryoid body, EB6/RA2 6 days embryoid body with 2 days RA treatment, and EB8/RA4 8 days embryoid body with 4 days RA treatment. (C): Western blot analyses of RIP140, PAX6, and β-actin in EB/RA-induced differentiation. (D): Immunofluorescence images of differentiated ESCs (EB8/RA4) stained with PAX6 (left panel). PAX6-positive cells were quantified from scoring seven random areas (left bottom). Neurons differentiated from control EBs or shRIP140 EBs were stained with neuronal marker MAP2 (right panel), and nuclei were visualized by DAPI staining. Total cells (DAPI+) and MAP2-positive neurons in five randomly selected areas were scored to determine the frequency of neuron production (n = 3 or more) (right bottom panel). Asterisks denote a statistically significant difference (p < .05).
RA Induces RIP140/LSD1/Pit-1 Complex Formation to Repress PAX6 Expression
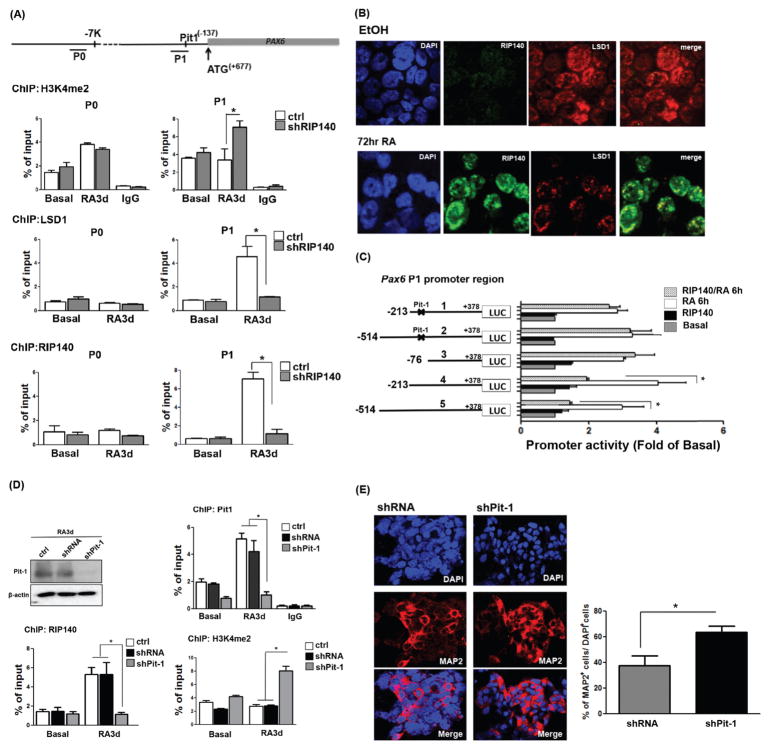
Since PAX6 was specifically affected by silencing RIP140 in RA-induced ESC neuronal differentiation, we wanted to determine the region responsible for RIP140 regulation. We first performed chromatin immunoprecipitation (ChIP) assays. As shown in Figure 2A, while RA enhanced active histone marks (H3K4me2) on the upstream region of the Pax6 promoter (P0, upper left panel), RIP140 silencing had no effect on chromatin modification on this region in the absence of RA. Importantly, silencing RIP140 very dramatically elevated active histone marks on the proximal promoter (P1, upper right panel) in RA-treated cultures. This reveals that RIP140 specifically targets P1 region to modulate RA-regulation of Pax6 gene expression. H3K4me2 is a well-characterized substrate of the lysine-specific demethylase, LSD1, which functions mainly as a corepressor [5, 12]. We then investigated whether LSD1 could be recruited to the Pax6 promoter. As shown in Figure 2A middle right panel, LSD1 was highly recruited to P1 of Pax6 gene in RA-treated cultures, which was completely abolished by silencing RIP140. This is consistent with the observation that, in RA-treated ESC cultures, RIP140 indeed also binds to P1 of Pax6 gene (Fig. 2A, bottom panels). With this observation, we speculated that RIP140 might recruit LSD1 to form a repressive complex to negatively modulate RA-induction of Pax6 expression. As shown in Figure 2B, IF staining demonstrated that, in RA-treated cultures where RIP140 began to be expressed, LSD1 was mainly found in RIP140-positive puncta (yellow in merged images).
Figure 2.
Retinoic acid (RA) treatment induces RIP140/LSD1 complex formation to repress Pax6 gene expression through Pit-1. (A): Top: Diagram of the 5′-flanking region of the Pax6 gene. Lines under the map indicate the two regions (P0 and P1) in chromatin immunoprecipitation (ChIP) analysis. Lower: ChIP assays for H3K4me2, LSD1, or RIP140 on the Pax6 promoter in control (ctrl) and RIP140-silenced (shRIP140) embryonic stem cells (ESCs) untreated or treated with RA for the indicated times. Experiments were performed in triplicate. (B): Immunofluorescence staining of RIP140 (green) and LSD1 (red) in ESCs treated with RA (3 days). (C): Left: Pax6 Luc P1 promoter constructs, with Pit-1 mutation indicated with x marks. Right: Reporter (wild-type, truncated or the Pit-1 mutated) activities assayed in ESCs in the absence or presence of RIP140, and treated with vehicle (basal) or RA for 6 hours. (D): ChIP assays for Pit-1, RIP140, or H3K4me2 on Pax6 promoter in control (ctrl), shRNA vector-transfected (shRNA), and Pit-1-silenced (shPit-1) ESCs untreated or treated with RA for the indicated times. Experiments were performed in triplicate. (E): Cultures differentiated from shRNA- and shPit-1-treated embryoid bodies were stained with MAP2 (left panel), and nuclei were visualized by DAPI staining. Total cells (DAPI+) and MAP2-positive neurons in five randomly selected areas were scored (n = 3 or more) (right panel). Asterisks denote a statistically significant difference (p < .05).
To determine the specific transcription factor that recruited RIP140/LSD1 repressive complex to modulate RA-regulation of Pax6 gene, we applied a promoter assay to identify the Pax6’s P1 promoter region specifically responsible for the effect of RIP140. As shown in Figure 2C, from a series of sequentially truncated Pax6-luciferase constructs, we identify the region −213 to −76 as the major target of RIP140, because only constructs 4 and 5 responded to the addition of RIP140 whereas constructs deleted in this sequence (construct 3) failed to respond. Interestingly, a Pit-1 binding site is located in this region. We then generated Pit-1 binding site mutated reporters (constructs 1 and 2) for the reporter assay. Consistently, both Pit-1 binding mutant reporters failed to respond to the addition of RIP140, supporting that RIP140 targets the specific Pit-1 binding region of the Pax6 gene promoter (Fig. 2C). The essential functional roles for Pit-1 in RA-induced RIP140 recruitment to the P1 of Pax6 promoters were validated using shRNA knockdown followed by ChIP experiments. Pit1 silencing resulted in a marked decrease in Pit1 and RIP140 recruitment, but increased the level of H3K4me2 after RA treatment (Fig. 2D). The efficiency of Pit-1 silencing was confirmed (upper panel). IF staining also validated the effect of silencing Pit-1 in RA-induced MAP2-postive cells (Fig. 2E, left panel). These results demonstrate that silencing Pit-1 enhances the H3K4me2 level of Pax6 p1 promoter and MAP2-positive cells after differentiation.
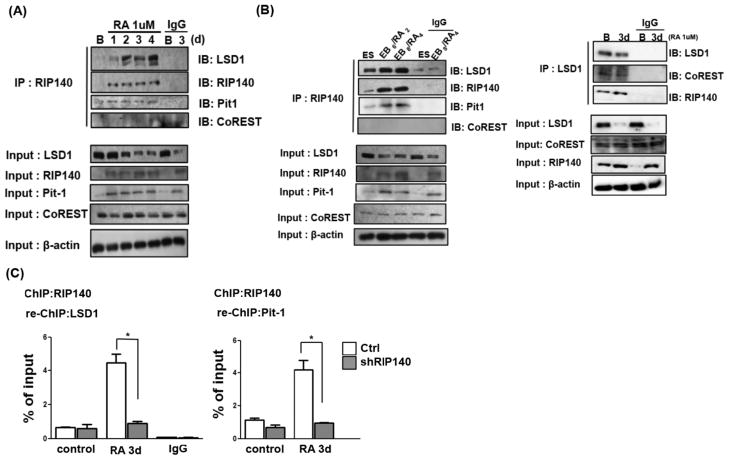
We then used a Co-IP assay to examine endogenous complex formation of RIP140 with LSD1 and Pit-1 in RA-treated ESC cultures. As shown in Figure 3A, the association of endogenous RIP140 with LSD1 and Pit-1 increased after RA treatment, even though the endogenous LSD1 protein level was decreasing (lower panel). This suggested that during the course of RA treatment, the majority of LSD1 is degraded but a small fraction is preserved and recruited by RIP140 to certain transcription factors like Pit-1 to form repressive complexes to dampen the expression of specific target genes such as Pax6. The RIP140/LSD-1/Pit-1 complex formation on the Pax6 gene is also validated in RA-treated EB differentiation (Fig. 3B, left panel). Interestingly, we did detect a small fraction of RIP140/LSD1 in undifferentiated ESC. A Re-ChIP assay further confirmed that RIP140/LSD1/Pit-1 repressive complex was recruited to P1 region after RA treatment (Fig. 3C). Together, these results show that RA induces repressive complex RIP140/LSD1 to be recruited by Pit-1 to the P1 region of Pax6 gene promoter.
Figure 3.
Co-immunoprecipitation (IP) assays to detect association of endogenous LSD1, CoREST and Pit-1 with RIP140 antibody in retinoic acid (RA)-treated embryonic stem cells (ESCs) (A) and RA-treated embryoid bodies (B left panel). (B right panel) Co-IP assays to detect association of endogenous CoREST, RIP140, and Pit-1 with LSD1 antibody in RA-treated ESCs. (C): Re-ChIP detection of RIP140-LSD1 and RIP140-Pit-1 complex on Pax6 P1 in ESC treated with RA (3 days).
RIP140 Directly Interacts with LSD1 to Regulate Its Activity
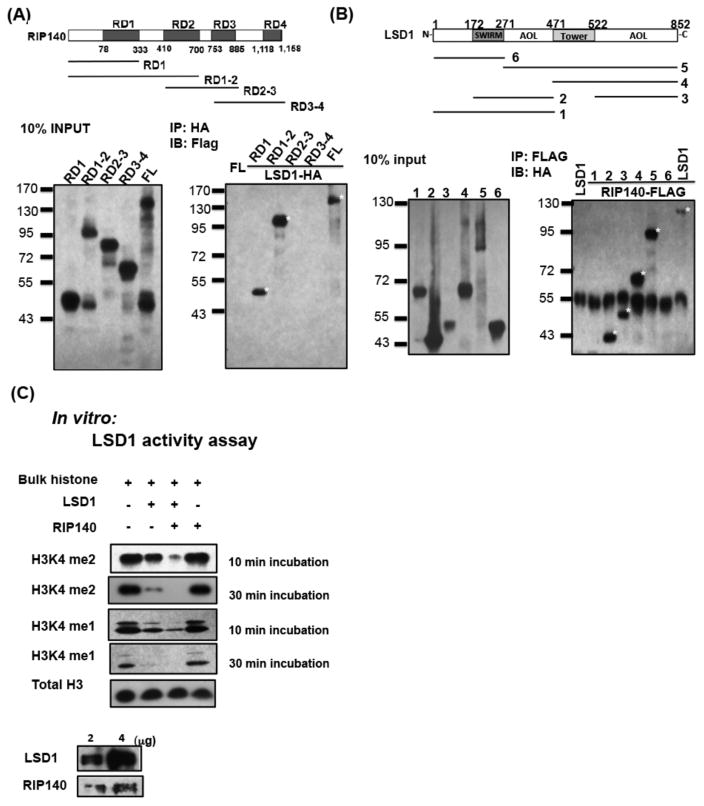
Given that RIP140 associated with LSD1 (Fig. 3), we speculated direction interaction of RIP140 with LSD1. A pull-down assay was carried out to confirm, and precisely define, the nature of RIP140 interaction with LSD1. RIP140 consists of four modular domains, RD1, RD2, RD3, and RD4. Thus, we generated four truncated RIP140 proteins for the pull down assay (Fig. 4A). These results revealed that the RD1 domain was responsible for interaction with LSD1 (Fig. 4A, lower right panel). Reciprocally, truncated LSD1 fragments were tested for interaction with RIP140 as shown in Figure 4B, which revealed that an amine oxidase-like (AOL) domain (construct 3) directly interacted with RIP140.
Figure 4.
RIP140 directly interacts with LSD1 to regulate its activity. (A): Pull-down assay for HA-LSD1 full-length (FL) interaction with Flag-RIP140 (FL), Flag-RD1, Flag-RD1-2, Flag-RD2-3, or Flag-RD3-4. Positive interaction denoted by *. (B): Pull-down assay for Flag-RIP140 (FL) interaction with HA-LSD1 (FL) or LSD1 truncated proteins. Positive interaction denoted by *. (C): In vitro LSD1 demethylase activity detected using free bulk histones as substrates and bacteria-expressed LSD1 and RIP140 (lower panel), analyzed by Western blotting using anti-H3k4me2 (upper) or anti-H3k4me1 (middle). Controls include antibody detected H3 (H3 panel).
Several studies have indicated that multiple factors can associate with LSD1 to regulate its histone demethylase activity (HDM), such as in CoREST and BHC80 [12]. We speculated that RIP140 might interact with LSD1 to regulate its enzymatic activity. We addressed this question using a direct in vitro enzyme essay on its natural substrate, H3K4me2. As shown in Figure 4C, the recombinant LSD1 itself could demethylate H3K4me2 on free histones, and the addition of recombinant RIP140 dramatically increased this activity. As a control, RIP140 itself had no effect on H3K4 demethylation. Furthermore, RIP140 could also enhance the ability of LSD1 to demethylate H3K4me1.
RIP140 Antagonizes Jade-2 to Stabilize LSD1
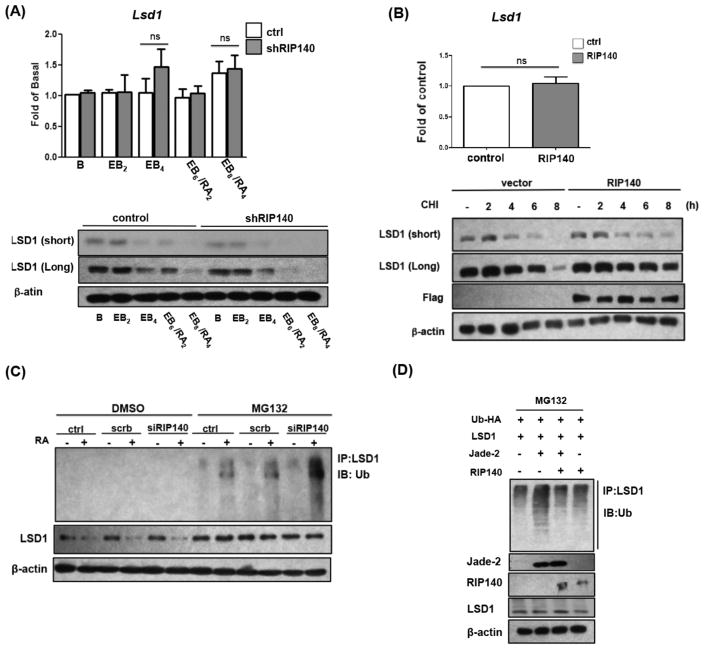
To further examine the molecular details of how RIP140 regulated LSD1, we first performed Western blot and q-PCR to monitor the effect of RIP140 on the expression levels of Lsd1 mRNA and protein. It appears that Lsd1 mRNA level was not altered by RIP140 silencing or RA treatment (Fig. 5A, upper), but LSD1 protein level was significantly decreased by silencing RIP140 (Fig. 5A, lower). This suggests that RIP140 could modulate LSD1 protein stability. To test this possibility, we compared protein stability of endogenous LSD1 with and without RIP140 over-expression in ESCs. As shown in Figure 5B bottom panel, LSD1 protein was more stable in RIP140-overexpressing cells, whereas Lsd1 mRNA level remained relatively constant (Fig. 5B, upper panel).
Figure 5.
RIP140 antagonizes Jade-2 to stabilize LSD1. (A): Western blot (lower) and q-PCR (upper) analyses of LSD1 and β-actin in embryoid body/retinoic acid-induced differentiation. Exposure time is indicated (short and long). (B): LSD1 protein stability is enhanced by RIP140. Embryonic stem cells (ESCs) were transfected with control or RIP140 plasmid. After cycloheximide (CHI) treatment, LSD1 and Flag-RIP140 were monitored on Western blots (bottom panel). Upper: The mRNA level of endogenous LSD1 determined by q-PCR (ns = nonsignificant). Exposure time is indicated (short and long). (C): RIP140 silencing enhances ubiquitination of LSD1. Western blot detection of endogenous LSD1 ubiquitination in control, scramble RNA, or RIP140 siRNA-treated ESCs, with or without 5 μM MG132 for 6 hours. (D): RIP140 antagonizes Jade-2-mediated ubiquitination of LSD1 in P19 cells with or without MG132 for 6 hours. Polyubiquitination of LSD1 was examined by Western blotting using ubiquitin antibody.
To address the issue of LSD1’s protein stability, we examined ubiquitin-mediated protein degradation using the proteasome inhibitor MG-132 to assess the amount of ubiquitinated LSD1 in RIP140-silenced ESCs and control ESCs. It appears that the amount of ubiquitinated LSD1 was significantly higher in RIP140 depleted ESCs than in control ESCs (Fig. 5C), supporting that RIP140 stabilizes LSD1 protein by blocking/inhibiting its ubiquitin-mediated protein degradation. To this end, it is known that Jade-2 is an E3 ubiquitin ligase known to target LSD1 [13]. Therefore, we asked whether RIP140 could antagonize Jade-2-induced polyubiquitination on LSD1. As shown in Figure 5D, a robust increase in LSD1 polyubiquitination is detected in the presence of Jade-2, which is dramatically reduced by expressing RIP140. Furthermore, full length RIP140 could decrease Jade-2 elicited polyubiquitination of LSD1 (Fig. 5D), revealing that intact molecular complex formation is required for RIP140 to block Jade-2 accessibility to the LSD-1 molecule for its ubiquitination and subsequent degradation.
Discussion
In this study, we uncover a new mechanism for ESC to modulate RA induction of neuronal differentiation, mediated by new dual functions of RIP140. The study identifies LSD1 as the target of these newly found activities of RIP140. Specifically, RIP140 directly interacts with LSD1 to protect its catalytic domain (the AOL domain), and competes with Jade-2 to prevent LSD1 from Jade-2-mediated ubiquitination and subsequent degradation (Figs. (4 and 5)). The RIP140/LSD1 complex is recruited to transcription factor Pit-1 to form a Pit-1/RIP140/LSD1 chromatin demethylating complex that binds to Pax6 proximal promoter to repress Pax6 expression (Figs. (2 and 3)). In RA-exposed cells, RIP140 and Pit-1 are both directly induced by RA (Fig. 3A, lower panel). While Lsd1 mRNA is maintained constant, LSD1 is a labile protein (Fig. 5). Thus, the RA-elevated RIP140 and Pit-1 produce increasing amounts of transcription complex Pit-1/RIP140 that protects and recruits LSD1 to provide a negative regulatory mechanism to specifically dampen Pax6 expression (Figs. 1–3). This would prevent overtly activated neural differentiation from stem/progenitor cells following RA induction.
The most remarkable changes are noticed at P1 of Pax6 promoter under silencing of RIP140. However, we did detect increased H3K4me2 induced by RA at P0, but this was not altered by loss of RIP140 expression. With regard to changes detected in H3K4me2 occupancy, it is important to note that H3K4me2 is more abundant on the P1 promoter than on the P0 promoter in undifferentiated ESCs. Since gene regulation involves multiple events such as histone modifications, binding of cis-regulatory elements by transcription factors, chromatin remodeling, DNA methylation, and so forth. It would be interesting to better understand the interplay between these two promoters in regulating Pax6 expression. But our analyses of promoters (Fig. 2C) indicated that P1 alone is able to robustly upregulate Pax6 expression under RA treatment.
Previous studies have shown that increasing PAX6 levels pushes neural progenitor differentiation into neurons [7, 30]. Negative regulation of PAX6 by RA-induced factors like RIP140/LSD1/Pit-1 complex is likely to provide a fine tuning mechanism for proper neuronal differentiation such that progenitor cells can be preserved. Interestingly, this negative activity of RIP140 is exerted at the level of protein quality/quantity control of its target—LSD1. Our data show that RIP140 ensures LSD1’s enzyme activity by directly targeting its catalytic domain, the AOL domain. Only the full length RIP140 protein is capable of protecting LSD1 from Jade-2-mediated ubiquitination (Fig. 5D), indicating RIP140 most likely acts as a chaperone protein for LSD1 to antagonize the action of this specific E3 ligase Jade-2. This is supported by the in vitro competition assay. These newly identified activities of RIP140, as well as its mode of action, are different from all previously demonstrated biological activities of RIP140, which responds to hormonal signals by associating with nuclear receptors that recruit histone modifying enzymes to repress the activation of hormone targeted gene. In the case of RA regulation of Pax6 promoter that contains no RA response element, the key transcription factor, Pit-1, is not a nuclear receptor. Nevertheless, the ultimate function of RIP140 in this context remains the same—to inhibit, or counteract, potential over-induction by hormones (in this case RA) in neuronal differentiation. Therefore, RIP140 seems to be able to adopt various mechanisms to provide a feedback control for multiple critical physiological processes. Gene knockout/silencing studies in animals reveal numerous abnormal phenotypes in the mouse [23–26, 28]. Most interestingly, behavior phenotypes are commonly observed in these studies. This further substantiates the notion that RIP140 is critically involved in regulating neural development, thus deleting RIP140 will cause abnormal brain function in animals.
PAX6 is a paired box family member and plays an essential role in brain development. It is required for preventing precocious cell-cycle exit and loss of PAX6 leads to a depletion of the neuron progenitor pools [7, 30]. Abnormal expression of PAX6 is known to have phenotypic consequences for eye and neuronal development [6]. DNA sequence suggests the proximal region P1 contains multiple transcription factor-binding sites in addition to Pit-1, such as AP1, AP2, Sp1, and c-Myb, but no RA response element [31]. With regard to RA-response, it is executed through Pit-1/RIP140, which are directly induced by RA.
RIP140, through its RD1 domain, binds directly to LSD1’s AOL domain (Fig. 4A, 4B), which is the catalytic center for substrate binding [4]. Interestingly, only full length RIP140 can protect LSD1 from Jade-2-mediated ubiquitination and degradation. Presumably, upon binding to LSD1, the RIP140/LSD1 complex undergoes a conformation change to hinder Jade-2’s attack, thereby preserving LSD1. This model will require rigorous biochemical studies in the future to validate conformational changes. Interestingly, LSD1 can also act on non-histone substrates such as p53 [32], DNMT1 [33], and MYPT1 [34], and RIP140 is also a lysine-methylated protein [35]. We tested whether LSD1 could remove RIP140’s methyl groups inside the cells, which turned out to not be the case.
In ESCs, LSD1 has been found to occupy the promoters of a subset of developmental genes to maintain their ground state, and it is progressively downregulated in ESC differentiation [36], which is consistent with our data (Figs. 3A, 3B, 5A). We also observed that after RA treatment, while the amount of LSD1 is dramatically reduced, the remaining immunologically positive LSD1 signals are mostly colocalized with those RIP140-positive foci in the nuclei (Fig. 2B). Co-IP data also show that the association of RIP140 with LSD1 is greatly increased after RA treatment, in spite of a dramatic reduction in the total LSD1 protein level. These data indicate that RA-elevated RIP140 recruits/protects LSD1 to form repressive complexes only on selected gene loci including Pax6. How extensively this RA-preserved RIP140/LSD1 repressive complex acts and whether it may affect global genome organization in RA-exposed cells are interesting questions to pursue in the future.
Another LSD1-interacting corepressor complex, CoREST, is a master regulator of key neural cell fate decision. We observed that the protein level of CoREST and its association with LSD1 are not affected by RA treatment (Fig. 3B, left panel), which is consistent with previous publications [37, 38]. Therefore, the RA-induced RIP140/LSD1 complex is different from the constitutive CoREST/LSD1 complex. It is tempting to speculate that the RIP140/LSD1 complex (induced by RA) and CoREST/LSD1 complex represent two distinguished families of corepressor complexes that differentially contribute to homeostatic control of neurogenesis and neuronal differentiation.
Conclusion
In conclusion, our data clearly indicate a new mechanism that the RIP140/LSD1 complex is recruited by Pit-1 to form a chromatin demethylating complex on Pax6 gene promoter to fine-tune Pax6 expression and neuronal differentiation.
Significance Statement.
This manuscript describes a novel retinoic acid-induced, ubiquitination-antagonizing co-repressor complex, RIP140/LSD1, which represents a novel gene-suppressive pathway to modulate the homeostasis of embryonic stem cell-neuronal differentiation. Mechanistically, this is mediated by new dual functions of transcription coregulator RIP140, which acts as a chaperone for LSD1, an important H3K4me2 demethylase, to ensure its enzyme quality and quantity to carry out critical gene repressive function. This study delineates a new developmentally relevant pathway of retinoic acid’s action in controlling timely neuronal differentiation and maintaining the homeostasis of neurogenesis.
Acknowledgments
We thank technical assistance from Yi-Wei Lin and Bomi Lee. This work was supported by DK54733, DK60521, DK54733-11S, Dean’s Commitment, and the Distinguished McKnight Professorship of University of Minnesota to L.N.W.
Author contributions: L.-N.W.: designed the study, provided reagents, technical/financial support, and conceptual advice; C.-Y.W.: designed the study, performed experiments, collected and analyzed data, and wrote the manuscript; S.D.P.: performed experiments and collected and analyzed data.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no competing financial interests.
See www.StemCells.com for supporting information available online.
References
- 1.Duester G. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin Cell Dev Biol. 2013;24:694–700. doi: 10.1016/j.semcdb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16:110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wichterle H, Lieberam I, Porter JA, et al. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Yang Y, Wang F, et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc Natl Acad Sci USA. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster CT, Dovey OM, Lezina L, et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol. 2010;30:4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osumi N, Shinohara H, Numayama-Tsuruta K, et al. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 7.Sansom SN, Griffiths DS, Faedo A, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada Y, Shimazaki T, Sobue G, et al. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 9.Oosterveen T, Kurdija S, Alekseenko Z, et al. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Dev Cell. 2012;23:1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 10.de Zegher F, Pernasetti F, Vanhole C, et al. The prenatal role of thyroid hormone evidenced by fetomaternal Pit-1 deficiency. J Clin Endocrinol Metab. 1995;80:3127–3130. doi: 10.1210/jcem.80.11.7593413. [DOI] [PubMed] [Google Scholar]
- 11.Voss TC, Demarco IA, Booker CF, et al. Functional interactions with Pit-1 reorganize co-repressor complexes in the living cell nucleus. J Cell Sci. 2005;118:3277–3288. doi: 10.1242/jcs.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi YJ, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Gui B, Xiong C, et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: An antibraking system in neural development. Mol Cell. 2014;55:482–494. doi: 10.1016/j.molcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Hu X, Chen Y, Farooqui M, et al. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J Biol Chem. 2004;279:319–325. doi: 10.1074/jbc.M307621200. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Kerimo A, Khan S, et al. Real-time analysis of molecular interaction of retinoid receptors and receptor-interacting protein 140 (RIP140) Mol Endocrinol. 2002;16:2528–2537. doi: 10.1210/me.2002-0124. [DOI] [PubMed] [Google Scholar]
- 16.Farooqui M, Franco PJ, Thompson J, et al. Effects of retinoid ligands on RIP140: Molecular interaction with retinoid receptors and biological activity. Biochemistry. 2003;42:971–979. doi: 10.1021/bi020497k. [DOI] [PubMed] [Google Scholar]
- 17.Lee CH, Wei LN. Characterization of receptor-interacting protein 140 in retinoid receptor activities. J Biol Chem. 1999;274:31320–31326. doi: 10.1074/jbc.274.44.31320. [DOI] [PubMed] [Google Scholar]
- 18.White KA, Yore MM, Warburton SL, et al. Negative feedback at the level of nuclear receptor coregulation. Self-limitation of retinoid signaling by RIP140. J Biol Chem. 2003;278:43889–43892. doi: 10.1074/jbc.C300374200. [DOI] [PubMed] [Google Scholar]
- 19.Persaud SD, Huang WH, Park SW, et al. Gene repressive activity of RIP140 through direct interaction with CDK8. Mol Endocrinol. 2011;25:1689–1698. doi: 10.1210/me.2011-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei LN, Hu X, Chandra D, et al. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 22.Chung HT. RIP140, a Janus metabolic switch involved in defense functions. Cell Mol Immunol. 2013;10:7–9. doi: 10.1038/cmi.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duclot F, Lapierre M, Fritsch S, et al. Cognitive impairments in adult mice with constitutive inactivation of RIP140 gene expression. Genes Brain Behav. 2012;11:69–78. doi: 10.1111/j.1601-183X.2011.00731.x. [DOI] [PubMed] [Google Scholar]
- 24.Feng X, Krogh KA, Wu CY, et al. Receptor-interacting protein 140 attenuates endoplasmic reticulum stress in neurons and protects against cell death. Nat Commun. 2014;5:4487. doi: 10.1038/ncomms5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flaisher-Grinberg S, Tsai HC, Feng X, et al. Emotional regulatory function of receptor interacting protein 140 revealed in the ventromedial hypothalamus. Brain Behav Immunity. 2014;40:226–234. doi: 10.1016/j.bbi.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho PC, Tsui YC, Feng X, et al. NF-kappaB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol. 2012;13:379–386. doi: 10.1038/ni.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steel JH, White R, Parker MG. Role of the RIP140 corepressor in ovulation and adipose biology. J Endocrinol. 2005;185:1–9. doi: 10.1677/joe.1.05896. [DOI] [PubMed] [Google Scholar]
- 28.Wu CY, Feng X, Wei LN. Coordinated repressive chromatin-remodeling of Oct4 and Nanog genes in RA-induced differentiation of embryonic stem cells involves RIP140. Nucleic Acids Res. 2014;42:4306–4317. doi: 10.1093/nar/gku092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim KC, White KA, Deng D, et al. Selective repression of retinoic acid target genes by RIP140 during induced tumor cell differentiation of pluripotent human embryonal carcinoma cells. Mol Cancer. 2007;6:57. doi: 10.1186/1476-4598-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bel-Vialar S, Medevielle F, Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Zheng JB, Zhou YH, Maity T, et al. Activation of the human PAX6 gene through the exon 1 enhancer by transcription factors SEF and Sp1. Nucleic Acids Res. 2001;29:4070–4078. doi: 10.1093/nar/29.19.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J, Sengupta R, Espejo AB, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 33.Jin L, Hanigan CL, Wu Y, et al. Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53- and DNMT1(DNA methyltransferase 1)-independent manner. Biochem J. 2013;449:459–468. doi: 10.1042/BJ20121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho HS, Suzuki T, Dohmae N, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 35.Huq MD, Ha SG, Barcelona H, et al. Lysine methylation of nuclear co-repressor receptor interacting protein 140. J Proteome Res. 2009;8:1156–1167. doi: 10.1021/pr800569c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamo A, Sese B, Boue S, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 37.Ballas N, Grunseich C, Lu DD, et al. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Gao Z, Ure K, Ding P, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]