Abstract
[NiFe]-hydrogenase, which catalyzes the reversible conversion between hydrogen gas and protons, is a vital component of the metabolism of many pathogens. Maturation of [NiFe]-hydrogenase requires selective nickel insertion that is completed, in part, by the metallochaperones SlyD and HypB. Escherichia coli HypB binds nickel with sub-picomolar affinity, and the formation of the HypB-SlyD complex activates nickel release from the high-affinity site (HAS) of HypB. In this study, the metal selectivity of this process was investigated. Biochemical experiments revealed that the HAS of full length HypB can bind stoichiometric zinc. Moreover, in contrast to the acceleration of metal release observed with nickel-loaded HypB, SlyD blocks the release of zinc from the HypB HAS. X-ray absorption spectroscopy (XAS) demonstrated that SlyD does not impact the primary coordination sphere of nickel or zinc bound to the HAS of HypB. Instead, computational modeling and X-ray absorption spectroscopy of HypB loaded with nickel or zinc indicated that zinc binds to HypB with a different coordination sphere than nickel. The data suggested that Glu9, which is not a nickel ligand, directly coordinates zinc. These results were confirmed through the characterization of E9A-HypB, which afforded weakened zinc affinity compared to wild-type HypB but similar nickel affinity. This mutant HypB fully supports the production of [NiFe]-hydrogenase in E. coli. Altogether, these results are consistent with the model that the HAS of HypB functions as a nickel site during [NiFe]-hydrogenase enzyme maturation and that the metal selectivity is controlled by activation of metal release by SlyD.
Graphical abstract
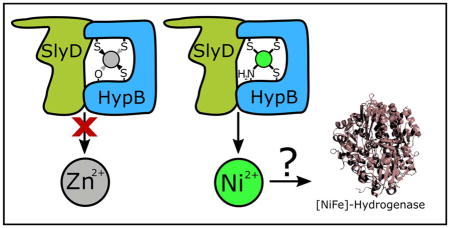
SlyD activates the release of nickel and blocks the release of zinc from the high-affinity metal site of HypB.
Introduction
Metalloenzymes rely on essential, yet toxic, metal cofactors to catalyze chemical transformations that are required for life.1–5 The availability and distribution of these hazardous nutrients are maintained through the efforts of metal homeostasis networks, which include metal ion importers, exporters, genetic regulators, storage proteins, and metallochaperones.1, 6–9 An example of a consequential metalloenzyme is [NiFe]-hydrogenase, which catalyzes the reversible conversion of hydrogen gas into protons and electrons in a wide variety of bacteria and archaea.10, 11 There is significant interest in understanding [NiFe]-hydrogenase, from maturation to mechanism, because it is a virulence factor in some pathogenic bacteria,12–14 and it could be utilized in a green energy economy as a source of biologically-derived hydrogen.10, 11 Furthermore, [NiFe]-hydrogenase is one of the few enzymes that employs nickel as a cofactor,9 so the maturation pathway for this enzyme can serve as a model system to study the intricate details of nickel ion delivery.
Nickel is located at the active site of [NiFe]-hydrogenase as part of a NiFe(CN)2(CO) bimetallic cofactor.10, 11 The biosynthesis of this complex cofactor in Escherichia coli proceeds through several discrete stages and requires at least seven accessory proteins.15, 16 The proteins that are assigned a specific role in the nickel insertion stage are HypA, HypB, and SlyD,15, 17 and extensive characterization of these nickel metallochaperones has defined how each individual protein binds nickel. However, one outstanding issue is how these metallochaperones selectively deliver nickel over other divalent transition metals in the face of the relative metal affinities dictated by the Irving-Williams series18 and the expected cytosolic concentrations of metals.19 One possible strategy, as outlined in several recent reports, is that partner protein interactions modulate selective metal binding and transfer.20–22
Escherichia coli HypB binds nickel ions at two distinct sites: a high-affinity nickel-binding site (HAS) located at the N-terminus, and a weaker, highly conserved site contained within the GTPase domain (G-domain) of the protein.23, 24 The G-domain metal site of HypB uses different sets of ligands to coordinate nickel versus zinc,25–27 and occupancy of this site modulates GTP hydrolysis, a property that is essential for hydrogenase maturation.27–29 Furthermore, the G-domain site rapidly and selectively transfers nickel, but not zinc, to HypA,21, 22 the accessory protein that is thought to serve as an adapter for nickel delivery to the large hydrogenase subunit.30, 31 In contrast to the G-domain site, the HAS is not as well conserved, binds nickel with much tighter affinity, and has an unexplored function. Nevertheless, in vivo studies confirmed that the HAS is necessary for [NiFe]-hydrogenase maturation in E. coli.25 A combination of mutagenesis, chemical modification, and spectroscopic studies revealed that nickel is ligated with sub-picomolar affinity in a square planar geometry by a CTTCGCG motif through three cysteine thiolate ligands and the N-terminal amine.23–25 Furthermore, experiments with a short peptide maquette of the HypB HAS found that other metal ions besides nickel, such as zinc, also bind with tight affinity to this sequence.32
SlyD, which interacts with HypB, is a member of the FK506 binding protein (FKBP) family of peptidyl-prolyl isomerases (PPIases),33, 34 augmented with additional domains that contribute to protein folding and metal binding.35–37 The C-terminal 50-residue metal-binding domain (MBD) contains 15 histidines, 6 cysteines, and 7 aspartate/glutamate amino acids. This MBD binds multiple metal ions, stores nickel in E. coli, and regulates PPIase activity.36–40 An E. coli strain with slyD knocked-out exhibited reduced nickel accumulation as well as stunted hydrogenase activity that can be fully restored by the addition of excess nickel to the growth media,41 indicating that SlyD contributes to the delivery of nickel to the hydrogenase precursor protein. Additional analysis of the SlyD-HypB complex suggested that the role of SlyD during hydrogenase biosynthesis is to stimulate nickel release from the HypB HAS in a process that depends on complex formation between the two proteins as well as a full-length MBD on SlyD.40, 42, 43 However, it is not understood how this activity is achieved or if SlyD stimulates the release of metals other than nickel from HypB.
In this study, zinc binding to the HAS of E. coli HypB was investigated, and the impact of SlyD on HypB loaded with nickel or zinc was studied. It was found that zinc is readily loaded into the HAS, but SlyD prevents zinc from leaving HypB, in contrast to the stimulation of dissociation observed for nickel. Spectroscopic studies reveal that nickel and zinc discrimination by SlyD does not occur through modification of the primary coordination environment of the metal loaded at the HypB HAS. Instead, the results indicate that zinc binds the HAS of HypB with a coordination different from that of nickel, including ligation by a distinct residue - Glu9. HypB with an E9A mutation was fully operational for hydrogenase biosynthesis in E. coli, providing additional evidence that the HAS of HypB functions as a nickel site in vivo, and that this selectivity is due to the mobilization of nickel from this site by SlyD. These results afford insight into the mechanisms of nickel selective delivery during [NiFe]-hydrogenase biosynthesis.
Experimental
Materials
NiSO4, ZnSO4, EDTA (ethylene diaminetetraacetic acid), EGTA (ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid), PAR (4-(2-pyridylazo)-resorcinol), PMB (p-hydroxymercuribenzoic acid), DTNB (5,5′-dithiobis(2-nitrobenzoic acid)), β-mercaptoethanol, and GDP (guanosine diphosphate) were purchased from Sigma-Aldrich at a minimum of 99% purity. Kanamycin, IPTG (isopropyl β-D-1-thiogalactopyranoside), TCEP (tris(2-carboxyethyl) phosphine), Tris (tris(hydroxymethyl)aminomethane), PMSF (phenylmethanesulfonylfluoride), and DTT (dithiothreitol) were purchased from BioShop (Toronto, ON), and all chromatography media were purchased from GE Healthcare. Buffers for all metal assays were prepared with Milli-Q water and treated with Chelex-100 (Bio-Rad) to minimize trace metal contamination in an anaerobic glovebox (Coy Lab Products, Michigan, USA) with an atmosphere of 95% N2 and 5% H2 for at least 24 hr prior to use. All assays were conducted in at least triplicate (n=3) to ensure reproducibility.
Plasmid construction
Plasmids bearing hypB and slyD genes were described in previous publications.23, 25, 41 The E9A mutation was introduced into the HypB-pET24b23 parent vector for protein production and the pBAD24-hypB25 parent vector for in vivo studies using QuikChange PCR mutagenesis. The plasmids were amplified using the forward primer 5′-CATGCGGTTGCGGTGCAGGCAACCTGTATATC-3′ and the reverse primer 5′-GATATACAGGTTGCCTGCACCGCAACCGCATG-3′ (Integrated DNA Technologies, the mutated bases are underlined). PCR products were transformed into NEB Turbo E. coli (New England Biolabs) and the plasmids were isolated by using the GeneJET plasmid mini-prep kit (ThermoFisher Scientific). All plasmids were sequenced in the forward and reverse directions (ACGT, Toronto, ON) to verify the fidelity of the mutagenesis.
Protein expression, purification, and preparation
SlyD was purified using nickel-nitrilotriacetic acid (Ni-NTA) chromatography followed by anion exchange chromatography on a MonoQ column and gel filtration on a Superdex-200 as described previously,41 and stored at −80°C in storage buffer (25 mM HEPES, pH 7.6, 200 mM NaCl, 1 mM TCEP). SlyD146, a construct comprised of the first 146 amino acids of SlyD that lacks the metal-binding domain, was purified with a DEAE column in lieu of the initial Ni-NTA column.37, 41 Only fully reduced SlyD, determined via an N-ethyl maleimide modification assay,37 was used for subsequent experiments.
Wild-type (WT) HypB and E9A-HypB were expressed and purified as previously described.23 The amount of metal bound to purified HypB was determined by treating 5 μM HypB with 100 μM p-hydroxymecuribenzoic (PMB) in the presence of 150 μM 4-(2-pyridylazo)resorcinol (PAR) for 90 min at room temperature followed by measuring the absorption at 495 nm. The resulting metal-PAR2 signal was compared to a metal standard curve (0 – 15 μM NiSO4 or ZnSO4) prepared under the same conditions to determine the fraction of metal bound. The protein was routinely found to contain > 0.95 equivalents of metal. Circular dichroism spectroscopy of E9A-HypB was unchanged from that of WT-HypB, confirming that this mutation does not affect the secondary structure of HypB (data not shown).
Protein concentrations were estimated by using the calculated ε280 coefficients (16500 M−1cm−1 for HypB and E9A, and 5960 M−1cm−1for SlyD and SlyD146) for the fully reduced proteins.44 Protein molecular weights were measured using electrospray-ionization mass spectrometry (ESI-MS) and compared with expected values.
Apo-HypB samples were prepared by incubating purified HypB in 25 mM HEPES, pH 7.5, 200 mM NaCl, 20mM EDTA and 5 mM TCEP, in an anaerobic glovebox for 72 hr at 4°C. HypB was desalted through two PD-10 columns equilibrated with protein buffer (25 mM HEPES, pH 7.5, 200 mM NaCl). All apo-proteins were confirmed to be >94% reduced using a DTNB assay. To perform the DTNB assay, protein (5 μM) was treated with 0.27 mM DTNB after dilution into 6 M guanidinium hydrochloride and 1.0 mM EDTA and incubated for 1 hr at room temperature, and the absorbance at 412 nm was compared to a β-mercaptoethanol standard curve (0 – 60 μM) prepared under the same conditions.
Zn(II)-HypB was prepared by incubating apo-HypB (100 μM) with ZnSO4 (100μM) in an anaerobic glovebox at 4°C overnight. The protein was desalted through two PD-10 columns equilibrated with protein buffer. The amount of zinc bound to HypB was determined by PAR assay, as described above (vide supra).
Zinc competition assay
A competition experiment was performed to calculate the apparent binding affinities of the HAS of WT-HypB or E9A-HypB for zinc. Apo-protein (0 – 50 μM) was titrated into 100 μM PAR and 10 μM ZnSO4 in protein buffer and incubated overnight in an anaerobic glovebox at 4 °C to reach equilibrium. Samples were analyzed by electronic absorption spectroscopy and the fraction of zinc bound to PAR was determined by monitoring the ratio of Abs500 nm : Abs500 nm + Abs412 nm. The absorption data were fit by using a custom DynaFit45 script and a dissociation constant for Zn(II)-PAR2 of 7.7 ×10−11 M.46
Metal release monitored by electronic absorption spectroscopy
Release of metal from HypB (5 μM) loaded with either nickel or zinc in the HAS to PAR (100 μM) was examined following the addition of 50 μM SlyD or SlyD146 where appropriate. Samples were allowed to equilibrate for ~1 min following transfer to a cuvette and data collection. The release of metal was monitored every 5 min for 2 hr by an increase in the absorbance at 495 nm, due to the formation of the metal-PAR2 complex.42, 47 The total metal in the samples (Amax) was calculated by treating an aliquot of the same sample with PMB (100 μM). Data were converted to % metal bound [100 ×(A−Amin)/(Amax−Amin)] where Amin is the absorbance of the first collected data point. The half-lives of the decay curves were calculated by fitting to a first-order exponential decay.
Metal release monitored by ESI-MS
All proteins were buffer exchanged using PD-10 columns into MS buffer (10 mM ammonium acetate, pH 7.5). Zn(II)-HypB or Ni(II)-HypB (30 μM) was incubated with 90 μM SlyD or SlyD146 and 1 mM EGTA for the indicated amount of time at room temperature in ananaerobic atmosphere (95% N2 and 5% H2) and diluted 10-fold in MS buffer prior to immediate analysis via ESI-MS. Control experiments with Zn(II)-HypB, SlyD, and chelator alone were carried out under identical experimental conditions.
The mass spectra were acquired on an AB/Sciex QStar XL mass spectrometer equipped with an ion spray source in the positive ion mode and a hot source-induced desolvation (HSID) interface (Ionics Mass Spectrometry Group Inc.). Ions were scanned from 800–3000 m/z with accumulation of 1 s per spectrum with no inter scan time delay and averaged over a 2–4 min period. The instrument parameters were as follows: ion source temperature 200 °C; ion source gas 50.0 psi; curtain gas 50.0 psi; ion spray voltage 5000.0 V; declustering potential 60.0 V; focusing potential 60 V; collision gas 3.0; MCP (detector) 2200.0V. The mass spectra were deconvoluted without further manipulation using the Bayesian protein reconstruction program over an appropriate mass range using a step mass of 1 Da, signal/noise ratio of 5, and the minimum intensity detected set to 1%. The amounts of holo-protein or apo-protein were determined based on the intensity of the signal for each species in the reconstructed spectra.
X-ray absorption spectroscopy (XAS) sample preparation
Purified protein solutions were pretreated with 1 mM TCEP and 20 mM EDTA overnight in an anaerobic glovebox at 4 °C to remove bound metal. Protein was buffer exchanged into 20 mM HEPES, pH 7.5, 200 mM NaCl, and 2 mM TCEP, and concentrated by using an AmiconUltra Centrifugal Filter Device (Millipore, MWCO = 3000 Da) to an approximate concentration of ~1.2 mM. Protein concentrations were determined from absorbance at 280 nm and placed on ice. NiSO4 or ZnSO4 (up to 0.7 equivalents) was added in multiple small aliquots to avoid protein aggregation. Glycerol, used as a cryoprotectant, was added to the protein solutions to final concentrations of 20% (v/v). Final protein concentrations were approximately 0.7–1 mM. Samples were transferred to 2 mm path length (2 mm × 4 mm × 20 mm) cuvettes of either Lucite (University of Saskatoon, Saskatoon, Canada) or polyoxymethylene copolymer (Vantec, Saskatoon, Canada), flash frozen with liquid nitrogen, and stored at −80°C until analysis.
XAS data collection and analysis
XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) in Menlo Park, California using data collection software, XAS Collect.48 Ni(II) and Zn(II) K-edge spectra were collected on the structural biology beam line 7-3 with the SPEAR storage ring containing 500 mA at 3 GeV. A Si(220) double crystal monochromator was used. Samples Ni(II)-HypB, Ni(II)-(HypB + SlyD), Ni(II)-(HypB + SlyD146), Zn(II)-HypB, Zn(II)-(HypB + SlyD), and Zn(II)-(HypB + SlyD146) were analyzed using a rhodium coated vertically collimating mirror to achieve harmonic rejection by adjusting the cut-off angle. Samples Ni(II)-E9A-HypB, Ni(II)-(E9A-HypB + SlyD), Zn(II)-E9A-HypB, and Zn(II)-SlyD achieved harmonic rejection by detuning one monochromator crystal to 50% peak intensity. Incident and transmitted x-rays were measured using a nitrogen-filled ionization chamber. Fluorescence was measured using a 30 element germanium detector.49 Samples were maintained at ~10 K using a liquid helium flow cryostat (Oxford Instruments). The energy for each scan was calibrated to reference nickel and zinc foils, assuming a lowest-energy inflection point of 8331.6 eV and 9665 eV respectively. A minimum of 8 scans for each sample were collected.
XAS data analysis and reduction were performed by using the EXAFSPAK suite of computer programs (George, G. N. (2001) EXAFSPAK: http://ssrl.slac.stanford.edu/exafspak.html). Data of repeated scans were averaged after sweeps and channels were reviewed for inconsistencies and calibrated using the lowest energy inflection point of the appropriate metal foil. A polynomial pre-edge function and a subsequent polynomial spline were used to extract the EXAFS oscillations χ(k). Erroneous data points that resulted from monochromator glitches were removed. Fourier transforms were phase corrected using theoretical phase functions from the largest EXAFS component. The Fourier transforms and EXAFS oscillations χ(k) were quantitatively analyzed using EXASPAK with ab initio theoretical phase and amplitude parameters from the program FEFF, version 8.25.50, 51 Scattering for second coordination sphere C and N atoms from imidazole rings were used but, for clarity, not included in fit tables. No smoothing, filtering, or similar operations were performed on the data.
Molecular Modeling
Density functional theory (DFT) calculations were performed using Gaussian 0952 on SHARCNET to examine HypB HAS nickel and zinc geometries. Calculations used the Becke three-parameter hybrid functional combined with Lee-Yang-Parr correlation functional (B3LYP)53, 54 and the Dunning correlation-consistent polarized double zeta basis set (cc-pVDZ).55 All complexes were assumed to have a singlet ground state. All computations were performed using the self-consistent reaction field solvation models (SCRF) to accommodate potential solvent effects. Up to 3 water molecules were included in our models to occupy any vacant coordination sites.
Nickel titration and competition assay
Nickel was titrated into apo-protein in the presence or absence of 1 mM EGTA and incubated overnight in an anaerobic glovebox at 4 °C in protein buffer to reach equilibrium. Ni(II)-HypB complex formation was monitored by an increase in absorbance at 320 nm. The apparent dissociation constant was calculated using a custom DynaFit45 script using a dissociation constant for Ni(II)-EGTA, adjusted for a pH of 7.6, of 5.21 ×10−11 M,56 and an ε320 for Ni(II)-HypB of 7255 M−1 cm−1.23
Hydrogenase assays
Anaerobic E. coli cultures were grown in supplemented TYEP media (10 g/L tryptone, 5 g/L yeast extract, 12 g/L K2HPO4, 3 g/L KH2PO4, 1 μM sodium molybdate, 1 μM sodium selenite, 30 mM sodium formate, 0.8% glycerol, and 100 μM arabinose), with 100 mg/L ampicillin where appropriate, at 37 °C for 18 hr after inoculation with 1% (v/v) of an overnight aerobic culture grown in LB media. The MC4100 strain was used as a wild-type control and DHBP cells (MC4100 ΔhypB) were used as a negative control. To test the impact of the E9A mutation, DHBP cells were transformed with either pBAD24-hypB or pBAD24-E9A-hypB plasmid. Following growth, cells were harvested by centrifugation, washed with cold 100 mM potassium phosphate, pH 7.6, and resuspended in cold 100 mM potassium phosphate, pH 7.6, supplemented with 200 μM PMSF and 1 mM DTT. The suspended cells were sonicated on ice, and the lysates were separated from the cellular debris by centrifugation for 20 minutes at 21000 g. The cell lysates were either immediately used or stored at −80 °C.
Total hydrogenase activity of crude cell lysates was calculated by measuring the hydrogen-dependent reduction of benzyl viologen.57 Reactions were prepared inside an anaerobic glovebox (95% N2 and 5% H2) and contained within a septum-sealed cuvette during the reaction. Activity was measured in units/mg of total protein, where one unit of activity corresponds to 1 μmol of benzyl viologen reduced/minute. The amount of reduced benzyl viologen was quantified by measuring the electronic absorption at 600 nm by using a UV/Visible spectrophotometer (Agilent 8453) and an extinction coefficient of 7400 M−1 cm−1. Total protein concentrations were determined by BCA protein assays (Pierce) with bovine serum albumin as a standard. The assay was completed in biological triplicates.
Expression levels of WT-HypB and E9A-HypB were confirmed by western blot. Proteins were resolved on 12.5% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). The blots were probed with an anti-HypB polyclonal rabbit antibody raised against purified HypB peptide (sequence LGEKHKVAVLSVTEGEDK, Cedarlane Labs, Burlington, Canada) at a 1:1000 dilution and then secondary goat anti-rabbit (Bio-Rad) antibodies conjugated to horseradish peroxidase at a dilution of 1:30000. Enhanced chemiluminescence (SuperSignal West Pico Chemiluminescence, Pierce) was used for detection.
Results & Discussion
The HAS of HypB binds zinc
The HypB HAS at the N-terminus of the protein is necessary for the maturation of [NiFe]-hydrogenase in E. coli and binds nickel in vitro,23, 25 however the selectivity of metal binding to this site is not known. A small peptide based on the N-terminal sequence of HypB was previously shown to bind zinc,32 but zinc binding to the HAS of the full-length protein had not been demonstrated. Purified HypB is loaded with nickel in the HAS, as previously demonstrated,23, 25 so nickel was first removed by incubation in an anaerobic glovebox (95% N2 and 5% H2) with excess EDTA as well as TCEP, to ensure all cysteines were reduced. Apo-HypB was desalted to remove the metal and EDTA, followed by the addition of ZnSO4 and incubation in an anaerobic glovebox at 4°C overnight. Unbound zinc was removed by gel filtration chromatography and zinc binding was monitored by ESI-MS. The reconstructed mass spectrum revealed a 1:1 protein:zinc complex of Zn(II)-HypB at 31,492 Da (Fig. S1). Quantitative zinc loading of the HypB HAS was not observed by ESI-MS, which could be due to incomplete loading or due to zinc dissociation during buffer exchange or during ionization, as observed during studies of the zinc-loaded peptide maquette.32 To determine if quantitative zinc binding was achieved in solution, the amount of zinc bound to HypB after desalting was measured with a colorimetric indicator PAR, which revealed that HypB bound 0.97 ± 0.07 equivalents of zinc. A competition experiment between PAR and apo-HypB for zinc was used to measure the affinity for zinc in the HAS (Fig. S2), revealing an apparent dissociation constant of (2.2 ± 0.6)×10−11 M, which indicated that zinc also binds tightly to the HAS of E. coli HypB. The conclusion that the zinc was bound to the HAS, rather than the G-domain, was made on the basis of the tighter apparent dissociation constant compared to that of zinc binding in the G-domain22 as well as XAS spectroscopy (vide infra).
SlyD blocks release of zinc from HAS of HypB
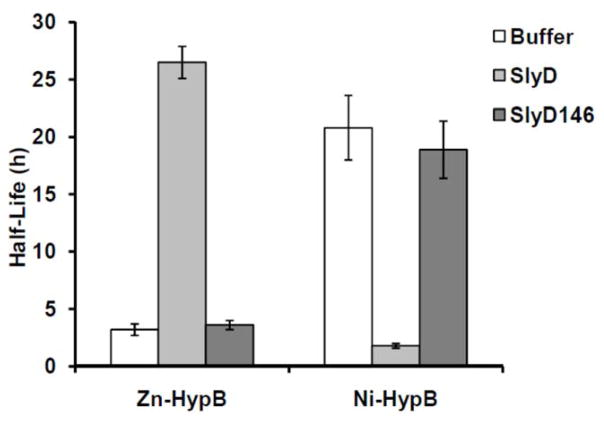
Given the strong nickel affinity of the HypB HAS, it is likely that there is some means to stimulate dissociation of the nickel ion so that it can be available for subsequent steps in the [NiFe]-hydrogenase biosynthetic pathway. Biochemical experiments revealed that SlyD can act as such a trigger, and experiments with a truncated form of SlyD suggested that acceleration of metal release from HypB is a key component of the role of SlyD during hydrogenase maturation.40, 43, 58 However, the selectivity of this process was not known. To examine the metal selectivity of the impact of SlyD on HypB, nickel and zinc release from HAS of HypB to PAR, which serves as a reporter for the amount of metal released from the protein,58 in the absence or presence of SlyD was monitored by electronic absorption spectroscopy. The data were fit to single-order exponential decay curves and the half-lives were extracted (Fig. 1, Fig. S3). SlyD was found to stimulate the release of nickel from the HAS of HypB, as previously reported,42 but SlyD inhibited the release of zinc. Chemical cross-linking experiments confirmed that Zn(II)-HypB interacts with SlyD to a similar extent as Ni(II)-HypB (Fig. S4). Since the activation of nickel release from HypB by SlyD is greater when HypB is loaded with GDP,40 zinc release experiments were performed in the presence of GDP, but the cofactor did not alter the rate of release of zinc (data not shown). Furthermore, the impact of a truncated SlyD lacking the C-terminal tail, SlyD146, was examined because this protein is unable to stimulate the release of nickel but still forms a complex with HypB, as shown by previous cross-linking experiments.42 Unlike the full-length protein, SlyD146 is unable to block zinc release from HypB (Fig. 1), indicating that the C-terminal MBD is necessary for the impact of SlyD on zinc-loaded HypB.
Figure 1.
Metal release from Zn(II)-HypB and Ni(II)-HypB. HypB (5 μM) loaded with zinc or nickel was incubated with 100 μM PAR in the absence or presence of full-length 50 μM SlyD or SlyD146. Metal release from HypB was monitored every 5 minutes by measuring the absorbance at 495 nm due to the metal-PAR2 complex. The half-lives were calculated by fitting the data as single-exponential decays. The data represent the averages from three independent experiments and the error bars indicate ± 1 standard deviation.
Metal release from HypB can also be followed by ESI-MS,40 and in these experiments the chelator EGTA was used as a metal sink. The data generated from ESI-MS experiments were complementary to those of the electronic absorption assay because free and bound HypB were directly observable. For these experiments, zinc release was monitored over 4 hr by comparing the ratio of the peak intensities of Zn(II)-HypB and apo-HypB. It is worth noting that Zn(II) bound to SlyD was not observed at any time point, indicating that zinc was not transferred from Zn(II)-HypB to SlyD (data not shown). In agreement with the results of the solution assays with PAR, the ESI-MS data revealed that SlyD blocks zinc release from HypB and that SlyD146 had no effect on Zn(II)-HypB (Fig. S5).
SlyD does not modify zinc or nickel coordination at the HAS of HypB
It is possible that SlyD stimulates the release of nickel and blocks the release of zinc from the HypB HAS by inducing changes in the coordination environments of the two metals. Therefore, X-ray absorption spectroscopy (XAS) was used to monitor the primary coordination spheres of nickel or zinc bound to the HypB HAS in the presence of SlyD. The XAS of just Ni(II)-HypB was consistent with previous analysis: the near-edge spectrum had a 1s-4pz pre-edge feature at 8336 eV that is typical of Ni(II) square-planar complexes (Fig. S6), and the EXAFS fit well to a S3/N coordination environment (Table S1, Fig. S6).24, 25 The XAS data of Ni(II)-(HypB + SlyD146) were very similar to those of HypB alone (Fig. S7), indicating that complex formation with SlyD146 does not alter the HAS of HypB, consistent with the results of the metal release assays.
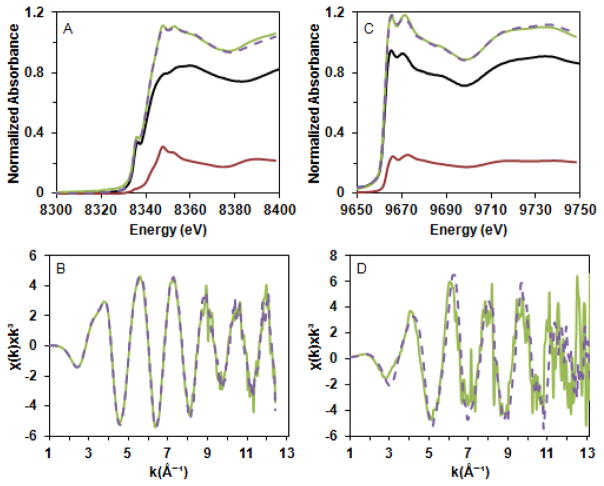
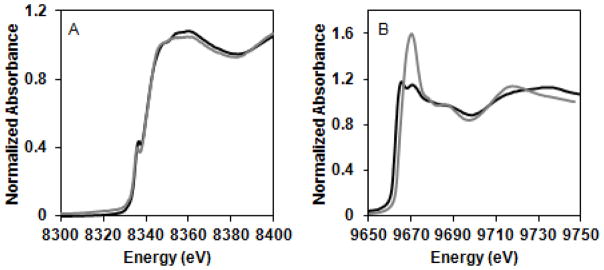
The near-edge spectrum of Ni(II)-(HypB + SlyD) exhibits distinct features compared to that of Ni(II)-HypB alone (Fig. 2). One possible explanation is that Ni(II)-(HypB + SlyD) is a mixture of Ni(II)-HypB and Ni(II)-SlyD rather than nickel in a modified site. Consistent with this hypothesis, the near-edge spectrum of Ni(II)-(HypB + SlyD) and the EXAFS region could be reconstructed from a combination of 79 % Ni(II)-HypB and 21% Ni(II)-SlyD by minimizing mean squared difference (Fig. 2). There are small differences between the EXAFS data and the reconstructed curve, so it is possible that the two proteins together have a subtle effect on the secondary coordination sphere of nickel; however, higher resolution data and multiple replicates would be needed to make this conclusion. It is also possible that cysteine side chains from SlyD replace those of HypB as nickel ligands in a ternary complex, resulting in very similar XAS data. This ligand exchange scenario is not likely given that SlyD lacking all cysteine residues is still capable of accelerating nickel release from HypB.40 Altogether, these data suggest that SlyD does not dramatically impact the local coordination environment of nickel ligated by the HypB HAS.
Figure 2.
Reconstruction of HypB + SlyD X-ray absorption spectra. The near-edge (A) and k3 weighted EXAFS (B) of Ni(II)-(HypB + SlyD) (green line) were reconstructed (dashed line) from those of the individual components using a composition of 79% HypB (black line) and 21% SlyD (red line). The near-edge (C) and k3 weighted EXAFS (D) of Zn(II)-(HypB + SlyD) (green line) were reconstructed (dashed line) from those of the individual components using a composition of 80% HypB (black line) and 20% SlyD (red line).
XAS was used to interrogate the coordination environments of Zn(II)-HypB and Zn(II)-SlyD, which have not yet been structurally characterized. The zinc K-edge spectrum of Zn(II)-HypB exhibits a white line intensity of 1.18 and two observable maxima, indicative of a four coordinate geometry composed of a mixture of sulfur and nitrogen or oxygen ligation (Fig. S8).59, 60 For consistency with previously published fits,24, 25 and between nickel and zinc fits, nitrogen was used since EXAFS fitting analysis cannot discriminate between nitrogen and oxygen. The EXAFS data fit well to both S3/N and S2/N2 coordination (Fig. S8, Table S1), but considering the proximity of the three cysteines at the metal-binding site and the sulfur Debye-Waller factors,61 a S3/N coordination is more likely.
SlyD can bind multiple zinc ions,39 but less than a single equivalent of zinc was added to the protein in an attempt to focus on the first event of zinc binding that is most applicable to this study. The Zn(II)-SlyD near-edge spectrum has a white line intensity of 1.23 and two maxima of similar intensity indicative of mixed sulfur and nitrogen and/or oxygen ligation in a 4-coordinate environment (Fig. S9). The EXAFS exhibits several distinctive oscillations, and the Fourier transform has intense features at R+Δ = 3–4, typical of histidine coordination (Fig. S9). The best fit to the EXAFS is a S2/His2 coordination (Fig. S9, Table S1). This single site model contrasts with the spectrum of Ni(II)-SlyD, which was best fit to a mixture of several sites.37 However, the possibility that zinc occupies more than one type of site with an average His2Cys2 coordination cannot be ruled out.
The XAS spectra of Zn(II)-HypB and Zn(II)-SlyD were used as a context for the analysis of the Zn(II)-(HypB+SlyD) mixture to probe for any SlyD induced changes in zinc coordination at the HypB HAS. It was possible to reconstruct the Zn(II)-(HypB+SlyD) spectrum using 80.5% Zn(II)-HypB and 19.5% Zn(II)-SlyD (Fig. 2), similar to the analysis of Ni(II)-(HypB + SlyD). Furthermore, analysis of Zn(II)-(HypB+SlyD146) revealed a similar spectrum to that of Zn(II)-HypB (Fig. S10). Collectively, the XAS of these mixtures indicate that SlyD does not induce significant changes in the metal coordination environment of the HypB HAS loaded with either nickel or zinc.
Molecular modeling of HypB HAS
The XAS analysis suggested that both nickel and zinc bound to the HypB HAS access similar types of ligands, but this experiment is limited because it cannot be used to identify the ligands or distinguish between nitrogen and oxygen coordination. To provide further insight into the zinc coordination at the HypB HAS, molecular modeling on nickel and zinc complexes with the B9 peptide was performed by using density functional theory (DFT). The B9 peptide (CTTCGCGEG) is comprised of the 9 N-terminal amino acids of the fully matured HypB with the N-terminal methionine removed (residues 2–10), and contains all the nickel ligands of the HAS of HypB.24, 32
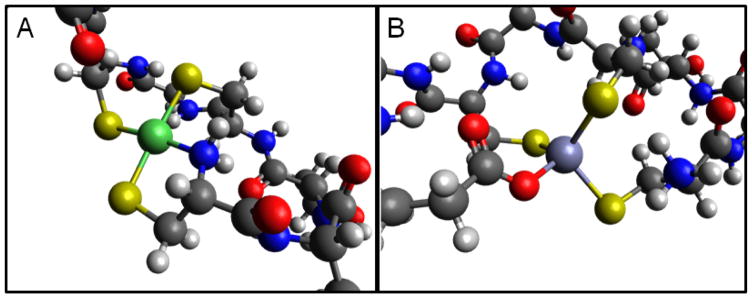
The optimized geometry of nickel-loaded B9 revealed a square planar coordination in a robust minimum (Fig. 3), starting from various geometries, consistent with the spectroscopic analysis of full-length protein. When the same ligands were used for zinc, either starting from a tetrahedral or square planar coordination environment, the geometry was optimized to a distorted tetrahedral environment. Inspection of this site revealed that this distortion was caused by the N-terminal cysteine residue that forms a 5-membered ring that contains the metal, cysteinyl sulfur and terminal amine, and imposes an N-metal-S bond angle of ~88°. While this structure would not exert significant strain in a square planar environment, which has an ideal bond angle of 90°, it does impact the tetrahedral coordination environment (108.5°) preferred by d10 Zn(II) when four ligands are available in biological systems.62 Therefore, we performed another set of optimizations in which the terminal amine was fully protonated to prevent ligation. In this case, Zn(II)-B9 adopts a tetrahedral four coordinate environment, recruiting the carboxylate of Glu9 as the fourth ligand (Fig. 3). Similarly, if the terminal amine is not available Ni(II)-B9 also recruits Glu9 and adopts a distorted square planar geometry. These results suggested that Glu9 could be important for metal binding.
Figure 3.
DFT optimized geometries of B9 zinc and nickel complexes (A) Nickel binds in a 4-coordinate square-planar geometry through 3 cysteinyl sulfurs and 1 N-terminal amine nitrogen. (B) Zinc binds in a 4-coordinate tetrahedral geometry through 3 cysteinyl sulfurs and 1glutamate oxygen. These putative optimized geometries were selected based on XAS spectroscopy results and mutagenesis experiments.
E9A-HypB nickel coordination is unchanged
To test whether Glu9 of HypB contributes to metal binding, as suggested by the results of the molecular modeling, an E9A-HypB mutant was created and the metal binding properties were examined. CD spectroscopy of E9A-HypB confirmed that the mutant and WT-HypB have similar secondary structures (data not shown).
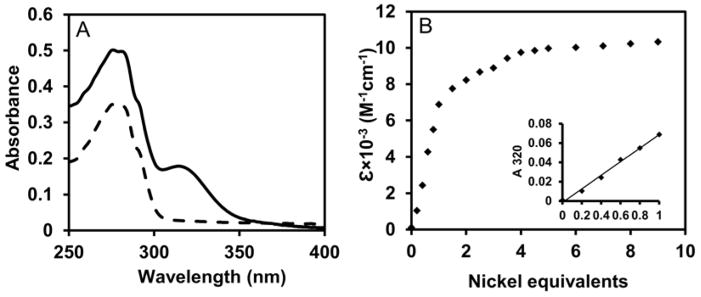
The stoichiometry of nickel binding was determined by titrating nickel into a solution of apo-E9A-HypB and monitoring the change in absorbance at 320 nm, characteristic of Cys→ Ni(II) LMCT.63 A linear increase in absorbance was observed upon addition of up to 1 equivalent of nickel, yielding an ε320 extinction coefficient of (6.9 ± 0.2)× 103 M−1 cm−1 (Fig 4B, inset), similar to WT-HypB. Further titration of E9A-HypB with Ni(II) resulted in a nonlinear increase in absorbance at 320 nm with a final extinction coefficient of approximately twice that of the first site (Fig. 4B), due to filling of the weaker G-domain site.23
Figure 4.
Electronic absorption spectroscopy of nickel binding to E9A-HypB. (A) The spectra of apo-E9A-HypB (dash line) and Ni(II)-E9A-HypB (solid line) with nickel loaded in the HAS. (B) Titration of apo-E9A-HypB to with nickel. (Inset) Linear titration curves with up to one equivalent of nickel, such as that shown, yielded an ε320 of (6.9 ± 0.2) × 103 M−1cm−1 (n=3).
The affinity of nickel binding to E9A-HypB was determined by titrating the protein with the metal ion in the presence of EGTA, and the fractional saturation of nickel in the HypB HAS was calculated by using the determined extinction coefficient. The apparent dissociation constants measured were (6.0± 0.6) × 10−14 M for WT-HypB and (9 ± 4) ×10−14 M for E9A-HypB (Fig. S11). Both values are similar to previously reported numbers for the wild-type protein,23 and they indicate that the high-affinity nickel site is intact in E9A-HypB. Furthermore, XAS analysis of Ni(II)-E9A-HypB revealed that the near-edge spectrum (Fig. 5) and EXAFS (Fig. S12) of Ni(II)-E9A-HypB are very similar to those of Ni(II)-loaded WT-HypB, providing additional evidence that there was little change in the nickel coordination site upon mutating Glu9.
Figure 5.
X-ray absorption spectroscopy of E9A-HypB. (A) The Ni(II) near-edge spectra of Ni(II)-HypB (black) and Ni(II)-E9A-HypB (grey) were obtained with protein incubated with 0.7 equivalents of nickel. Both spectra exhibit a 1s-4pz pre-edge feature at 8336 eV. (B) Zn(II) near-edge spectra of Zn(II)-HypB (black) and Zn(II)-E9A-HypB (grey) were obtained with protein incubated with 0.7 equivalents of zinc.
It is also possible that Glu9 could have a role in SlyD stimulated release of nickel. To test this model, nickel release from Ni(II)-E9A-HypB to PAR was monitored in the presence or absence of SlyD by using electronic absorption spectroscopy (Fig. S13). Ni(II)-E9A-HypB had t1/2 =16 ± 2 hr without SlyD and a t1/2 = 3 ± 2 hr with SlyD, similar to WT-HypB. Complex formation between Ni(II)-E9A-HypB and SlyD was confirmed with cross-linking experiments (Fig. S14). These results suggest that Glu9 is not required for recognition of Ni(II)-HypB by SlyD or coordination of nickel during the metal-release process.
E9A-HypB zinc coordination is weakened
In contrast to nickel, incubation of E9A-HypB with one equivalent of zinc followed by gel filtration chromatography and metal analysis using PAR revealed only 0.27 ± 0.12 equivalents of zinc, much less than the WT-HypB protein that bound 0.97 ±0.07 equivalents of zinc. Furthermore, zinc binding could not be detected by ESI-MS (data not shown), indicating that zinc binding to the HAS of E9A-HypB site was disrupted. E9A-HypB metal release experiments with SlyD did not reveal a stimulating or blocking effect (Fig. S13). However, it may not be possible to draw any conclusions from these data because of the poor zinc loading of E9A-HypB in these experiments. Preliminary competition experiments between E9A-HypB and PAR for zinc indicate that the zinc affinity is weakened by at least 10-fold (data not shown), giving an apparent dissociation constant consistent with a previously published zinc affinity for the B7-peptide of the HAS that lacks the Glu9 residue.32
XAS analysis of the zinc-loaded E9A-HypB complex was conducted, and the XAS of Zn(II)-E9A-HypB was decidedly different from that of the wild-type protein. This result confirms that, as suggested by the results of the computational and solution experiments, Glu9 is a zinc-binding residue. The near-edge spectrum of the mutant protein had a white line intensity of 1.59 (Fig. 5), which indicates an increase in coordination number, likely from 4 to 5,59, 60 and the edge energy (first maxima from first-derivative plot) shifted 1.1 eV from 9662.9 eV to 9664.0 eV. Photoreduction can cause a shift in edge energy, but no energy shift was observed over 8 successive sweeps (data not shown). Changes in edge energy could also be caused by changes in geometry, as found in isoelectronic Ga(III) small molecule complexes.64 This explanation is supported by the EXAFS spectrum of Zn(II)-E9A-HypB, which was fit best to a S3/N2 coordination (Table S1, Fig. S15). DFT calculations suggested that if the terminal amine was neutral, zinc adopted a 5-coordinate square pyramidal geometry with water in the axial position (supplemental information), a geometry consistent with the EXAFS fit. However, it is possible that in the context of the full-length protein Zn(II)-E9A-HypB recruits other residues that are not included in the peptide maquette. The near-edge spectrum of Zn(II)-E9A-HypB was also distinct from previously published spectrum of Zn(II) in the HypB G-domain site,25 indicating that the attenuated E9A-HypB HAS still bound zinc tighter than the G-domain. These results confirm that Glu9 is directly involved in zinc coordination at the E9A-HypB HAS.
E9A-HypB is active in E. coli hydrogenase biosynthesis
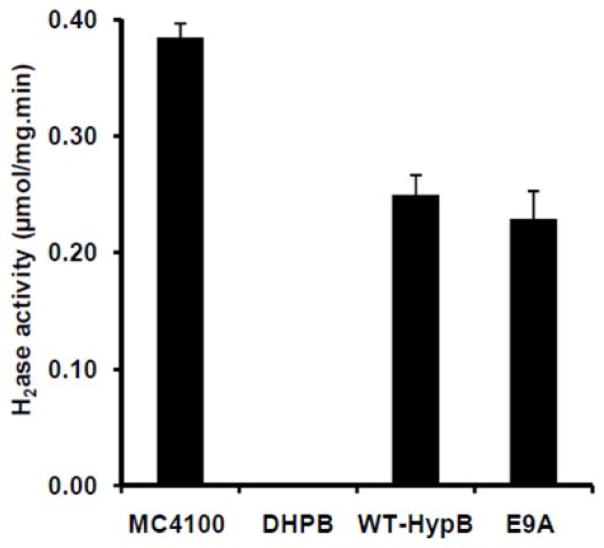
Given that Glu9 of HypB contributes to zinc binding but not nickel binding, the ability of E9A-HypB to function in E. coli hydrogenase production was examined to test if zinc coordination to Glu9 in the HypB HAS contributes to hydrogenase biosynthesis. Hydrogenase activity was measured in cell lysates from anaerobically grown DHBP (MC4100 ΔhypB) E. coli bearing the pBAD24-hypB or pBAD24-E9A-hypB plasmids. Western blot analysis of crude cell lysates revealed similar levels of expression of HypB and E9A-HypB (data not shown). MC4100 and DHBP strains were used as positive and negative controls, respectively. As expected, no significant hydrogenase activity was detected in DHPB cells (Fig. 6). Expression of either WT-HypB or E9A-HypB from the pBAD vector restored similar levels of activity, demonstrating that Glu9, and thus zinc coordination at the N-terminal metal-binding site, is not required for the hydrogenase biosynthesis activity of HypB.
Figure 6.
Hydrogenase activities. The E. coli strains MC4100, DHBP (MC4100 ΔhypB), DHPB transformed with pBAD24-hypB plasmid (WT-HypB), and DHPB transformed with pBAD24-E9A-hypB plasmid (E9A) were grown anaerobically. The hydrogenase activities of crude cell extracts were measured by monitoring the reduction of benzyl viologen in the presence of hydrogen gas. The results represent the average values from three biological replicates and the error bars indicate one standard deviation.
Conclusions
HypA, HypB, and SlyD facilitate nickel insertion into the [NiFe] hydrogenase precursor protein, and as a critical part of this process they must selectively deliver nickel while ensuring that other metals are not mistakenly incorporated into the enzyme active site. In this report, the metal selectivity of the N-terminal metal-binding site of HypB and the impact of the partner protein SlyD were investigated. Spectroscopic, computational, and in vivo experiments indicate that zinc binds tightly to HypB but through a coordination sphere that is distinct from nickel. Analysis of a mutant HypB with disrupted zinc binding supports the model that the HAS of HypB functions as a nickel site during hydrogenase biosynthesis. Furthermore, the fidelity of this function is likely modulated through the impact of SlyD, which selectively stimulates release of nickel and blocks release of zinc from HypB. These results add to the growing body of work that demonstrates that the metal selectivity of metallochaperone activity is not dictated by specific metal binding, but instead is achieved through allosteric coupling, often via partner protein complexes, as observed for example in several studies of the HypA-HypB complex.20–22
Previous analysis provided evidence that the HAS of HypB binds nickel with sub-picomolar affinity through a S3/N square planar site.23, 25 The work here confirmed this assignment, ruling out Glu9 as a ligand to nickel, and established that Glu9 contributes to the mid-picomolar binding affinity of zinc to HypB. The E9A mutation caused a decrease in the strength of zinc binding to the HAS of HypB (preliminary data suggests at least 10-fold weaker) and therefore widened the affinity gap between nickel and zinc to at least three orders of magnitude in favor of nickel, a surprisingly large difference considering the Irving-Williams series.18 One possible explanation for this gap in metal affinities of E9A-HypB, suggested by the results of the molecular modeling, stems from the terminal cysteine ligation that constrains the N-metal-S bond angle to ~88°, which would favour the square-planar geometry of four-coordinate Ni(II) complexes but induce strain into a tetrahedral site. This factor, along with the loss of a uniquely zinc ligand in Glu9, would significantly impact the zinc affinity relative to the strength of nickel binding at the N-terminal location of HypB.
The HAS of HypB is a putative nickel site because removal of the metal-binding residues at this site produces a hydrogenase-deficient phenotype in E. coli that was complemented by supplementation of the growth media with extra nickel.25 However, metal homeostasis networks are often connected, such that dysregulation of one metal network impacts other transition metals.65–67 Along these lines, it was possible that zinc binding to the HAS of HypB was a functional property; for example it could serve the nickel pathway by scrubbing the local environment of zinc and allow the highly conserved but weaker G-domain site to deliver nickel. However, even though the E9A-HypB mutant has weakened zinc affinity, the protein is fully competent during hydrogenase biosynthesis, suggesting that the HypB HAS does not function as a zinc site in vivo to a measurable extent with these growth conditions. This result raises the question of why E. coli HypB requires two nickel sites. The HypB G-domain transfers nickel to HypA, which interfaces directly with the hydrogenase precursor protein,21, 22, 31, 68 outlining a pathway for nickel delivery. So the role of another nickel site, especially one that has a much tighter affinity, is not evident. It is possible that the two sites are required under different growth conditions, such that the HAS serves as a nickel reserve that can be quickly mobilized, with the assistance of SlyD, during conditions that trigger rapid up regulation of hydrogenase production or nickel starvation. This hypothesis will be examined in future studies.
It has been demonstrated that zinc can inhibit hydrogenase maturation in vitro,69 but which step in the pathway was affected and whether this process is mitigated in vivo was not clear. A notable finding of this work is that SlyD not only enhances nickel release from HypB but blocks zinc release, indicating that complex formation between HypB and SlyD provides a step that checks the identity of the metal bound to the HAS. At equilibrium conditions, nickel would displace any zinc bound to the HypB HAS, on the basis of the relative apparent dissociation constants, however it is unlikely that these particular equilibria play a role on biologically relevant time scales given the tight affinities of both metals. Instead, SlyD serves to selectively mobilize kinetically trapped nickel bound to the HAS of HypB while ensuring that any zinc loaded erroneously would not be passed on to the subsequent steps in the pathway. In the solution experiments with the purified proteins, nickel released from HypB does not move to the MBD of SlyD but is captured by a small molecule chelator. In E. coli, it is possible that the nickel released from the HAS of HypB is used to fill the hydrogenase precursor protein, either directly or via other accessory proteins. This impact of SlyD provides an explanation for how the tight HAS of HypB could contribute to a physiological pathway. Spectroscopic analysis revealed that SlyD does not directly modify the primary coordination environment of HypB, so it is likely that SlyD activates metal release upon sensing which metal is loaded through the conformation resultant from the metal-specific coordination geometry. This mechanism, as well as other questions discussed, will be addressed in future work.
Supplementary Material
Significance to Metallomics Statement.
The biosynthesis of metalloenzymes often requires the activity of metallochaperones to selectively deliver the cognate metal(s). In this study, we examined two accessory proteins that deliver nickel to the bimetallic active site of [NiFe]-hydrogenase in E. coli, HypB and SlyD. The results demonstrate that metal selectivity is achieved through the cooperative activity of the two proteins together. This study provides a critical piece of information to the understanding of [NiFe]-hydrogenase maturation, suggesting a molecular mechanism for nickel selectivity, and contributes to a growing body of work illuminating the mechanisms by which metallochaperones accomplish a key step in metalloenzyme maturation.
Acknowledgments
We thank Michael D. Jones for help with XAS data collection, Kelly Summers for advice on XAS data analysis, and Karina A. Baksh for helpful suggestions. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET:www.sharcnet.ca) and Compute/Calcul Canada. This work was supported in part by funding from NSERC and CIHR.
Footnotes
Electronic supplementary information is available.
References
- 1.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 2.Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr Opin Chem Biol. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macomber L, Hausinger RP. Mechanisms of nickel toxicity in microorganisms. Metallomics. 2011;3:1153–1162. doi: 10.1039/c1mt00063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Zamble DB. Nickel Homeostasis and Nickel Regulation: An Overview. Chem Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Zhang F, Zhou P, Zhang C. Functional role of metalloproteins in genome stability. Front Biol. 2016;11:119–131. [Google Scholar]
- 6.Sydor AM, Zamble DB. Metallomics and the Cell. Springer; 2013. pp. 375–416. [DOI] [PubMed] [Google Scholar]
- 7.Bleackley MR, MacGillivray RTA. Transition metal homeostasis: from yeast to human disease. BioMetals. 2011;24:785–809. doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- 8.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeer-Wanklyn CJ, Zamble DB. Microbial nickel: cellular uptake and delivery to enzyme centers. Curr Opin Chem Biol. 2017;37:80–88. doi: 10.1016/j.cbpa.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Lubitz W, Ogata H, Ruüdiger O, Reijerse E. Hydrogenases. Chem Rev. 2014;114:4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 11.Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 12.Maier R. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem Soc Trans. 2005;33:83–85. doi: 10.1042/BST0330083. [DOI] [PubMed] [Google Scholar]
- 13.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HLT. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci USA. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benoit SL, Miller EF, Maier RJ. Helicobacter pylori stores nickel to aid its host colonization. Infect Immun. 2013;81:580–584. doi: 10.1128/IAI.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacasse MJ, Zamble DB. [NiFe]-hydrogenase maturation. Biochemistry. 2016;55:1689–1701. doi: 10.1021/acs.biochem.5b01328. [DOI] [PubMed] [Google Scholar]
- 16.Pinske C, Sawers R. Anaerobic formate and hydrogen metabolism. EcoSal Plus. 2016 doi: 10.1128/ecosalplus.esp-0011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forzi L, Sawers RG. Maturation of [NiFe]-hydrogenases in Escherichia coli. BioMetals. 2007;20:565–578. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- 18.Irving HMNH, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747. [Google Scholar]
- 19.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe S, Kawashima T, Nishitani Y, Kanai T, Wada T, Inaba K, Atomi H, Imanaka T, Miki K. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc Natl Acad Sci USA. 2015;112:7701–7706. doi: 10.1073/pnas.1503102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas CD, Ngu TT, Kaluarachchi H, Zamble DB. Metal transfer within the Escherichia coli HypB-HypA complex of hydrogenase accessory proteins. Biochemistry. 2013;52:6030–6039. doi: 10.1021/bi400812r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacasse MJ, Douglas CD, Zamble DB. The mechanism of selective nickel transfer From HypB to HypA, E. coli [NiFe]-hydrogenase accessory proteins. Biochemistry. 2016 doi: 10.1021/acs.biochem.6b00706. [DOI] [PubMed] [Google Scholar]
- 23.Leach MR, Sandal S, Sun H, Zamble DB. Metal binding activity of the Escherichia coli hydrogenase maturation factor HypB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- 24.Chung KC, Cao L, Dias AV, Pickering IJ, George GN, Zamble DB. A high-affinity metal-binding peptide from Escherichia coli HypB. J Am Chem Soc. 2008;130:14056–14057. doi: 10.1021/ja8055003. [DOI] [PubMed] [Google Scholar]
- 25.Dias AV, Mulvihill CM, Leach MR, Pickering IJ, George GN, Zamble DB. Structural and biological analysis of the metal sites of Escherichia coli hydrogenase accessory protein HypB. Biochemistry. 2008;47:11981–11991. doi: 10.1021/bi801337x. [DOI] [PubMed] [Google Scholar]
- 26.Gasper R, Scrima A, Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J Biol Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 27.Sydor AM, Lebrette H, Ariyakumaran R, Cavazza C, Zamble DB. Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB. J Biol Chem. 2014;289:3828–3841. doi: 10.1074/jbc.M113.502781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier T, Jacobi A, Sauter M, Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai F, Ngu TT, Kaluarachchi H, Zamble DB. Relationship between the GTPase, metal-binding, and dimerization activities of E. coli HypB. J Biol Inorg Chem. 2011;16:857–868. doi: 10.1007/s00775-011-0782-y. [DOI] [PubMed] [Google Scholar]
- 30.Blokesch M, Rohrmoser M, Rode S, Böck A. HybF, a zinc-containing protein involved in NiFe hydrogenase maturation. J Bacteriol. 2004;186:2603–2611. doi: 10.1128/JB.186.9.2603-2611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan Chung KC, Zamble DB. Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase. J Biol Chem. 2011;286:43081–43090. doi: 10.1074/jbc.M111.290726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas CD, Dias AV, Zamble DB. The metal selectivity of a short peptide maquette imitating the high-affinity metal-binding site of E. coli HypB. Dalton Trans. 2012;41:7876–7878. doi: 10.1039/c2dt30132f. [DOI] [PubMed] [Google Scholar]
- 33.Wülfing C, Lombardero J, Plückthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem. 1994;269:2895–2901. [PubMed] [Google Scholar]
- 34.Roof WD, Horne SM, Young KD, Young R. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 35.Kovermann M, Schmid FX, Balbach J. Molecular function of the prolyl cis/trans isomerase and metallochaperone SlyD. Biol Chem. 2013;394:965–975. doi: 10.1515/hsz-2013-0137. [DOI] [PubMed] [Google Scholar]
- 36.Hottenrott S, Schumann T, Plückthun A, Fischer G, Rahfeld JU. The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J Biol Chem. 1997;272:15697–15701. doi: 10.1074/jbc.272.25.15697. [DOI] [PubMed] [Google Scholar]
- 37.Kaluarachchi H, Sutherland DE, Young A, Pickering IJ, Stillman MJ, Zamble DB. The Ni(II)-binding properties of the metallochaperone SlyD. J Am Chem Soc. 2009;131:18489–18500. doi: 10.1021/ja9081765. [DOI] [PubMed] [Google Scholar]
- 38.Martino L, He Y, Hands-Taylor KLD, Valentine ER, Kelly G, Giancola C, Conte MR. The interaction of the Escherichia coli protein SlyD with nickel ions illuminates the mechanism of regulation of its peptidyl-prolyl isomerase activity. FEBS J. 2009;276:4529–4544. doi: 10.1111/j.1742-4658.2009.07159.x. [DOI] [PubMed] [Google Scholar]
- 39.Kaluarachchi H, Siebel JF, Kaluarachchi-Duffy S, Krecisz S, Sutherland DEK, Stillman MJ, Zamble DB. Metal selectivity of the Escherichia coli nickel metallochaperone, SlyD. Biochemistry. 2011;50:10666–10677. doi: 10.1021/bi2014882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaluarachchi H, Zhang JW, Zamble DB. Escherichia coli SlyD, more than a Ni(II) reservoir. Biochemistry. 2011;50:10761–10763. doi: 10.1021/bi201590d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JW, Butland G, Greenblatt JF, Emili A, Zamble DB. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J Biol Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- 42.Leach MR, Zhang JW, Zamble DB. The Role of Complex Formation between the Escherichia coli Hydrogenase Accessory Factors HypB and SlyD. J Biol Chem. 2007;282:16177–16186. doi: 10.1074/jbc.M610834200. [DOI] [PubMed] [Google Scholar]
- 43.Kaluarachchi H, Altenstein M, Sugumar SR, Balbach J, Zamble DB, Haupt C. Nickel binding and [NiFe]-hydrogenase maturation by the metallochaperone SlyD with a single metal-binding site in Escherichia coli. J Mol Biol. 2012;417:28–35. doi: 10.1016/j.jmb.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 44.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 45.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 46.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 47.Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 48.George MJ. XAS-Collect: a computer program for X-ray absorption spectroscopic data acquisition. J Synchrotron Radiat. 2000;7:283–286. doi: 10.1107/S090904950000683X. [DOI] [PubMed] [Google Scholar]
- 49.Cramer SP, Tench O, Yocum M, George GN. A 13-element Ge detector for fluorescence EXAFS. Nucl Instr Meth Phys Res A. 1988;266:586–591. [Google Scholar]
- 50.Mustre de Leon J, Rehr JJ, Zabinsky SI, Albers RC. Ab initio curved-wave x-ray-absorption fine structure. Phys Rev B. 1991;44:4146–4156. doi: 10.1103/physrevb.44.4146. [DOI] [PubMed] [Google Scholar]
- 51.Rehr JJ, Mustre de Leon J, Zabinsky SI, Albers RC. Theoretical x-ray absorption fine structure standards. J Am Chem Soc. 1991;113:5135–5140. [Google Scholar]
- 52.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark MJ, Heyd J, Brothers EN, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision D.01. Gaussian, Inc; Wallingford, CT, USA: 2009. [Google Scholar]
- 53.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 54.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 55.Dunning TH. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J Chem Phys. 1989;90:1007–1023. [Google Scholar]
- 56.Fahrni CJ, O’Halloran TV. Aqueous coordination chemistry of quinoline-based fluorescence probes for the biological chemistry of zinc. J Am Chem Soc. 1999;121:11448–11458. [Google Scholar]
- 57.Ballantine SP, Boxer DH. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol. 1985;163:454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leach MR, Zamble DB. Metallocenter assembly of the hydrogenase enzymes. Curr Opin Chem Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Giachini L, Veronesi G, Francia F, Venturoli G, Boscherini F. Synergic approach to XAFS analysis for the identification of most probable binding motifs for mononuclear zinc sites in metalloproteins. J Synchrotron Radiat. 2010;17:41–52. doi: 10.1107/S090904950904919X. [DOI] [PubMed] [Google Scholar]
- 60.Hennig C, Hallmeier KH, Zahn G, Tschwatschal F, Hennig H. Conformational Influence of Dithiocarbazinic Acid Bishydrazone Ligands on the Structure of Zinc(II) Complexes: A Comparative XANES Study. Inorg Chem. 1999;38:38–43. [Google Scholar]
- 61.Clark-Baldwin K, Tierney DL, Govindaswamy N, Gruff ES, Kim C, Berg J, Koch SA, Penner-Hahn JE. The limitations of X-ray absorption spectroscopy for determining the structure of zinc sites in proteins. When is a tetrathiolate not a tetrathiolate? J Am Chem Soc. 1998;120:8401–8409. [Google Scholar]
- 62.Rulíšek L, Vondrášek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 63.Jenkins RM, Singleton ML, Almaraz E, Reibenspies JH, Darensbourg MY. Imidazole-containing (N3S)-Ni(II) complexes relating to nickel containing biomolecules. Inorg Chem. 2009;48:7280–7293. doi: 10.1021/ic900778k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das U, Camacho-Bunquin J, Zhang G, Gallagher JR, Hu B, Cheah S, Schaidle JA, Ruddy DA, Hensley JE, Krause TR. Organometallic model complexes elucidate the active gallium species in alkane dehydrogenation catalysts based on ligand effects in Ga K-edge XANES. Catal Sci Tech. 2016;6:6339–6353. [Google Scholar]
- 65.Nies D, Grass G. Transition Metal Homeostasis. EcoSal Plus. 2009 doi: 10.1128/ecosalplus.5.4.4.3. [DOI] [PubMed] [Google Scholar]
- 66.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J Biol Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danielli A, Scarlato V. Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol Rev. 2010;34:738–752. doi: 10.1111/j.1574-6976.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe S, Sasaki D, Tominaga T, Miki K. Structural basis of [NiFe] hydrogenase maturation by Hyp proteins. Biol Chem. 2012;393:1089–1100. doi: 10.1515/hsz-2012-0197. [DOI] [PubMed] [Google Scholar]
- 69.Magalon A, Blokesch M, Zehelein E, Böck A. Fidelity of metal insertion into hydrogenases. FEBS Lett. 2001;499:73–76. doi: 10.1016/s0014-5793(01)02525-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.