Abstract
Protein arginine methylation regulates diverse functions of eukaryotic cells, including gene expression, the DNA damage response, and circadian rhythms. We showed that arginine residues within the third intracellular loop of the human D2 dopamine receptor, which are conserved in the DOP-3 receptor in the nematode Caenorhabditis elegans, were methylated by protein arginine methyl-transferase 5 (PRMT5). By mutating these arginine residues, we further showed that their methylation enhanced the D2 receptor–mediated inhibition of cyclic adenosine monophosphate (cAMP) signaling in cultured human embryonic kidney (HEK) 293T cells. Analysis of prmt-5–deficient worms indicated that methylation promoted the dopamine-mediated modulation of chemosensory and locomotory behaviors in C. elegans through the DOP-3 receptor. In addition to delineating a previously uncharacterized means of regulating GPCR (heterotrimeric guanine nucleotide–binding protein–coupled receptor) signaling, these findings may lead to the development of a new class of pharmacological therapies that modulate GPCR signaling by changing the methylation status of these key proteins.
INTRODUCTION
Dopamine plays an important role in neuronal and behavioral plasticity in diverse species (1–7). Signaling by this neuromodulator contributes to cognition, learning, motivation, reward, and motor control (8). Moreover, defects in dopamine signaling are linked to Parkinson’s disease, schizophrenia, and drug addiction (9–11). Understanding the mechanisms by which aberrant signaling contributes to the pathology of these diseases remains the focus of intense research, as does the search for approaches to modulate dopaminergic signaling to treat patients with these devastating conditions.
Mammalian dopamine receptors are GPCRs [heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptors], which are divided into two classes: the D1-like receptors (including D1 and D5) and the D2-like receptors (including D2, D3, and D4). The D1-like receptors generally signal through the G proteins Gαs or Gαolf to activate adenylyl cyclase and increase the intracellular abundance of the second messenger cyclic adenosine monophosphate (cAMP), whereas their D2-like counter-parts signal through Gαi and Gαo to inhibit adenylyl cyclase and decrease cAMP production (12). However, it is now appreciated that dopamine receptors also function in networks to control the activity of pathways not involving adenylyl cyclase (8, 12).
In the nematode Caenorhabditis elegans, as in vertebrates, dopamine activates GPCR signaling pathways (1, 2). C. elegans have one D1-like receptor (DOP-1), two D2-like receptors (DOP-2 and DOP-3), and one invertebrate-specific D1-like receptor (DOP-4) (1, 2). Dopamine also modulates a wide range of C. elegans behaviors. For example, animals lacking the D2-like receptor DOP-3 fail to reduce their rate of locomotion when they encounter a bacterial food source (13, 14); this “basal slowing” response is dependent upon endogenous dopamine signaling (15). In addition, dopamine modulates context-dependent behavioral responses of C. elegans to environmental stimuli (16–20).
One critical means of regulating signal transduction, including that by GPCRs, is through the posttranslational modification of proteins. Although phosphorylation remains the best-studied modification, arginine methylation is increasingly recognized as an important regulator of protein function. Protein arginine methylation is catalyzed by a family of enzymes termed protein arginine methyltransferases (PRMTs) (21). The human genome encodes nine such proteins (PRMT1 to PRMT9), as well as potentially two others (PRMT10 and PRMT11) (21). Type I PRMTs transfer either one or two methyl groups from S-adenosyl-L-methionine (SAM) to form mono-methyl arginine (MMA) or asymmetric dimethylarginine (ADMA), respectively. Type II PRMTs form MMA and symmetric dimethylarginine (SDMA). Type III PRMTs form only MMA. Each of the methylarginine modifications (MMA, ADMA, and SDMA) has distinct biological effects, and their dysregulation correlates with the etiology of chronic lung and kidney diseases, contributes to the toxicity of specific mutations that are linked to amyotrophic lateral sclerosis, and causes hypertension (22, 23). Among the type II PRMTs in eukaryotes, PRMT5 is the most conserved, and it methylates arginines in glycine- and arginine-rich motifs, in proline-, glycine-, and methionine-rich motifs, and in the absence of any recognizable motif (24, 25). PRMT5 influences gene expression, the DNA damage response, circadian rhythms, and germ cell development and pluripotency (23, 26).
Although a number of PRMT5 substrates have been identified (27–29), to our knowledge, this enzyme has never been found in association with a GPCR. However, in 2003, a large-scale proteomics analysis showed that a specific anti-SDMA antibody reacted with a number of GPCRs, suggesting a potential role for a type II PRMT in regulating this receptor class (30). Yet, direct evidence has remained elusive, and the functional importance of methylation in the regulation of GPCR signaling has not been demonstrated. Through a bioinformatics analysis, we identified 300 human GPCRs with a total of 583 predicted methylation motifs (RGG or RXR) within their intra-cellular domains. Almost 70% of these sites are conserved in the corresponding receptor sequences from other mammalian species, and seven of these sites (including that of the D2 receptor) are conserved from humans to C. elegans. Given the direct clinical relevance of dopamine and the broad evolutionary conservation of the predicted methylation motif in D2-like receptors, we focused on the role of arginine methylation in regulating signaling by these receptors. We showed that human PRMT5 methylated the third intracellular loop of the human D2 receptor in vitro and that mutating the conserved arginines within this region attenuated the D2-mediated inhibition of cAMP signaling in cultured human cells. Using C. elegans behavior as a readout for nervous system function, we found that PRMT-5 promoted dopaminergic signaling through the C. elegans D2-like receptor DOP-3 in vivo and that changing the predicted arginine methylation target sites in DOP-3 reduced its ability to regulate C. elegans behavior. Together, our data identify a previously uncharacterized mechanism by which PRMTs regulate signal transduction, and they reveal GPCRs that are functionally regulated by an arginine-methylating enzyme.
RESULTS
Bioinformatics analysis identifies predicted GPCR methylation motifs
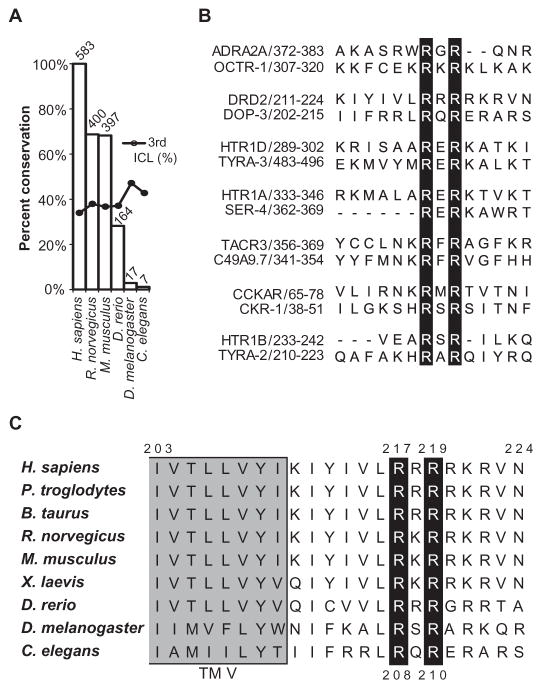
To identify the cohort of GPCRs that contain putative arginine methylation motifs, we analyzed the sequences of the intracellular domains of 737 annotated human GPCRs in the National Center for Biotechnology Information (NCBI) HomoloGene database. Because GPCRs are seven-pass transmembrane proteins, we reasoned that functional arginine methylation motifs could reside within any of the three intracellular loops or the C-terminal tail of the receptors. In total, we found 583 intracellular RGG or RXR motifs, and these were located in 300 of the human receptors within this data set (Fig. 1A and tables S1 and S2). Almost 70% of these motifs were conserved in the sequences of the homologous rat and mouse receptors (400 and 397 receptors, respectively), and ~1% (7) were conserved in the nematode C. elegans (Fig. 1, A and B, and tables S1 and S2). With regard to the localization of the predicted methylation motifs within the GPCRs, about 34% of those in the human GPCRs are within the third intracellular loop (Fig. 1A and tables S1 and S3).
Fig. 1. Predicated methylation motifs within the intracellular domains of GPCRs.
(A) Plot indicating the conservation of arginine methylation motifs across species. Among 737 annotated human GPCRs in the NCBI HomoloGene database, 300 contain a total of 583 predicted arginine methylation motifs (RGG or RXR). The height of each bar represents the percentage conservation of these sites between the human receptors and their homologs in the indicated species, and the number above each bar indicates the total number of predicted motifs conserved in each species. Dots indicate the percentage of motifs that are located within the third intracellular loop (ICL). (B) Alignments indicating conservation of the seven human and C. elegans GPCRs that contain conserved predicted arginine methylation motifs, as generated with the MUSCLE (multiple sequence comparison by log-expectation) algorithm (48). The human GPCR sequences are on top, and the corresponding C. elegans receptor sequences are aligned below. Amino acid positions are indicated after the receptor names. Black bars highlight the conserved arginine residues. (C) Alignment indicating conservation of residues among the D2 receptor homologs. The Arg217 and Arg219 residues of the human D2 receptor and their counterparts in other species lie within a highly conserved RXR motif, in which X can be any amino acid residue. These arginines are conserved from the C. elegans D2-like dopamine receptor DOP-3 to the human D2 receptor. The gray shading indicates the end of transmembrane domain five (TM V).
Given that invertebrate sequences may not be comprehensively annotated for homology to the human sequences and could therefore be missed in this analysis, we also generated a human–C. elegans homology data set by assembling the annotated human homologs of every C. elegans GPCR identified by Gene Ontology (GO). In analyzing these 565 human and 179 C. elegans GPCRs, we identified an additional 57 C. elegans receptors that contain conserved putative arginine methylation motifs (tables S1 and S4). Analysis of the more inclusive human–C. elegans homology data set indicated that 42% of the conserved sites within the C. elegans receptors are found within the third intracellular loop (Fig. 1A and tables S1 and S5).
PRMT5 methylates the human D2 receptor
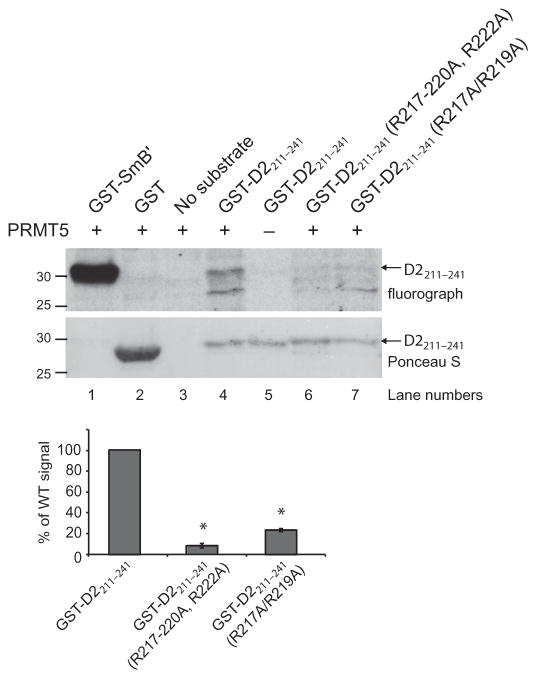
Given that the third intracellular loop of the D2 receptor contains a predicted PRMT5 methylation motif, which is conserved across diverse species (Fig. 1C), we tested the potential of human PRMT5 to methylate the human D2 receptor using an in vitro methylation assay. A recombinant fragment of the third intracellular loop of D2 [corresponding to amino acid residues 211 to 241 fused to glutathione S-transferase (GST)] was methylated in a PRMT5- dependent manner (Fig. 2, top). D2 receptor methylation was markedly decreased when all of the arginine residues within this region were replaced with alanines (R217–220A, R222A) and when only the two arginine residues (Arg217 and Arg219) that are conserved from human D2 to C. elegans DOP-3 were replaced with alanines (R217A/R219A) (Fig. 2, top). Quantification of the bands from three independent experiments revealed that less than 25% of the wild-type D2211–241 signal was present when either mutant was used as a substrate (Fig. 2, bottom). These data indicate that the third intracellular loop of the D2 receptor acts as a substrate for PRMT5 in vitro and suggest that Arg217 and Arg219 are key methylation sites within this region.
Fig. 2. PRMT5 methylates the human D2 receptor in vitro.
The indicated GST-tagged wild-type (WT) and mutant recombinant fragments of the third intracellular loop (amino acid residues 211 to 241) of the human D2 receptor were used in an in vitro methylation assay with active recombinant human PRMT5 as described in Materials and Methods. The PRMT5 substrate GST-SmB′ was used as a positive control, whereas GST alone served as a negative control. Top: Fluorograph shows that the WT GST-D2211–241 fragment was methylated in a PRMT5-dependent manner. The degrees of methylation of GST-D2211–241(R217–220A, R222A) and GST-D2211–241(R217A/R219A) were 10 and 24%, respectively, of that of the WT fragment. Ponceau S staining of the polyvinylidene difluoride (PVDF) membrane was performed to demonstrate equivalent loading of receptor fragments. Molecular mass markers (kD) are indicated on the left. Bottom: Quantification of the degree of methylation of the receptor fragments based on densitometric analysis of fluorographs. Data are means ± SEM of three independent experiments. *P = 0.000001.
Conserved arginines promote D2 receptor function in human cells
The extent to which Arg217 and Arg219 contribute to human D2 receptor function was tested by comparing signaling by the hemagglutinin (HA)–tagged wild-type D2 receptor (long isoform D2L) and the D2(R217A/R219A) mutant receptor stably expressed in human embryonic kidney (HEK) 293T cells (Fig. 3). Both flow cytometric and immunofluorescence microscopic analyses with an anti-HA antibody confirmed that mutating Arg217 and Arg219 did not alter the cell surface abundance of the receptor (Fig. 3A).
Fig. 3. Conserved arginines contribute to the function of D2 receptors in cultured human cells.
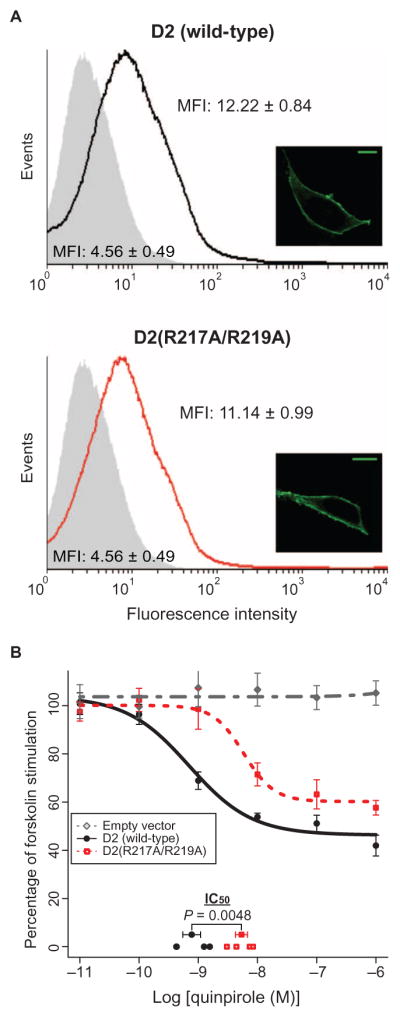
(A) HEK 293T cell lines stably expressing N-terminal HA-tagged WT D2 (top) or D2(R217A/R219A) mutant (bottom) receptors were analyzed by flow cytometry with an anti-HA antibody. Histograms show the analysis of 50,000 cells per sample and are representative of three independent experiments. Shaded gray histograms show background fluorescence of cells stably transfected with empty plasmid. Inset: Cell lines were analyzed by immunofluorescence microscopy with an anti-HA antibody. Images are representative of three experiments. Scale bars, 10 μm. (B) HEK 293T cells stably expressing empty plasmid (gray), N-terminal HA-tagged WT D2 receptors (black), or D2(R217A/R219A) mutant receptors (red) were treated for 30 min with the indicated concentrations of quinpirole in the presence of 20 μM forskolin. The amount of cAMP in each sample was measured using the HitHunter competitive immunoassay (DiscoveRx). Data are means ± SEM of four independent experiments, each performed in triplicate. Log IC50 (half maximal inhibitory concentration) values for each biological replicate are plotted on the x axis, together with the mean ± SEM of the log IC50 for each receptor variant. HA-D2(R217A/R219A) (log IC50 = −8.27 ± 0.10) shows a decrease in quinpirole-mediated inhibition of forskolin-stimulated cAMP accumulation when compared to HA-D2 (log IC50 = −9.11 ± 0.15) (P = 0.0048). MFI, mean fluorescence intensity.
The ability of the D2 receptor agonist quinpirole to dampen forskolin-stimulated cAMP generation in HEK 293T cells (31, 32) expressing either the HA-tagged human wild-type D2 receptor or D2(R217A/R219A) mutant receptor was assessed with the HitHunter cAMP competitive immunoassay (DiscoveRx). Whereas treatment of cells expressing the wild-type D2 receptor with quinpirole effectively blocked the forskolin-stimulated accumulation of cAMP (log IC50 = −9.11 ± 0.15), this ability was diminished in the D2(R217A/R219A)-expressing cells (log IC50 = −8.27 ± 0.10) (Fig. 3B). These data suggest that Arg217 and Arg219 promote efficient D2 receptor–mediated inhibition of cAMP production.
To further support a role for arginine methylation in the facilitation of D2 receptor–mediated signaling, we overexpressed PRMT5 in the HEK 293T cells expressing the HA-tagged wild-type human D2 receptor and again assessed the ability of quinpirole to dampen forskolin-stimulated cAMP accumulation. Cells overexpressing PRMT5 more effectively blocked cAMP accumulation in response to quinpirole (log IC50 = −9.53 ± 0.07) than did cells containing only endogenous PRMT5 (log IC50 = −9.20 ± 0.04) (fig. S1). This facilitation of D2 receptor–mediated signaling occurred despite a small, but statistically significant, decrease in the cell surface abundance of the D2 receptor in cells overexpressing PRMT5 (fig. S1).
PRMT5 methylates C. elegans DOP-3
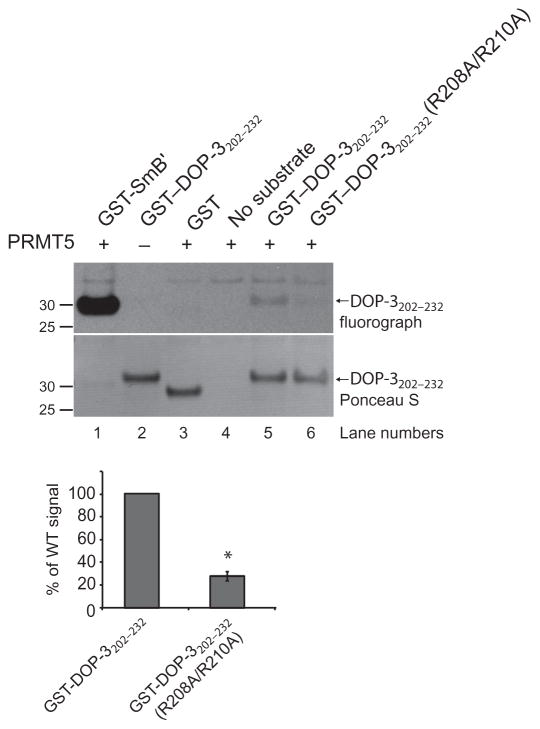
Before proceeding with animal studies, we confirmed that the C. elegans DOP-3 receptor was also methylated by PRMT5, similar to human D2. A recombinant fragment of the third intracellular loop of DOP-3 (amino acid residues 202 to 232 fused to GST) was methylated in a PRMT5-dependent manner (Fig. 4, top). The extent of methylation of the DOP-3 receptor was markedly decreased when the two conserved arginine residues were replaced with alanines (R208A/R210A) (Fig. 4, top). Quantification of the bands from three independent experiments revealed that 26% of that of wild-type DOP-3202–232 signal was present when this mutant was used as substrate (Fig. 4, bottom). These data establish the third intracellular loop of the DOP-3 receptor as a substrate for PRMT5 in vitro and suggest that Arg208 and Arg210 are key sites of methylation within this region.
Fig. 4. PRMT5 methylates C. elegans DOP-3 in vitro.
The indicated GST-tagged WT and mutant recombinant fragments of the third intracellular loop (amino acid residues 202 to 232) of the C. elegans DOP-3 receptor were used in an in vitro methylation assay with active, recombinant human PRMT5 as described in Materials and Methods. The PRMT5 substrate GST-SmB′ was used as a positive control, whereas GST alone served as a negative control. Top: Fluorograph shows that the WT GST–DOP-3202–232 fragment was methylated in a PRMT5-dependent manner. The degree of methylation of GST–DOP-3202–232(R208A/R210A) was 26% of that of the WT fragment. Ponceau S staining of the PVDF membrane was performed to demonstrate equivalent loading of receptor fragments. Molecular mass markers (kD) are indicated on the left. Bottom: Quantification of the degree of methylation of the receptor fragments based on densitometric analysis of fluorographs. Data are means ± SEM of three independent experiments. *P = 0.000055.
C. elegans lacking PRMT-5 are hypersensitive to dilute octanol
Mice lacking PRMT5 exhibit embryonic lethality, which has precluded whole-animal analysis of the physiological roles of this methyltransferase (33). In contrast, C. elegans prmt-5 mutants are viable (34), which enables in vivo studies that are not possible in mammals.
The sensitivity of C. elegans to environmental stimuli is dynamically regulated by their nutritional status, which influences signaling levels of biogenic amines, including dopamine (2, 35). The response of C. elegans to the aversive odorant octanol is scored as the amount of time it takes an animal to initiate backward locomotion when presented with a hair dipped in octanol (36, 37). When animals are assayed in the absence of food (a bacterial lawn), 100% octanol is detected by three pairs of sensory neurons in the head: the ASH, AWB, and ADL neurons (18, 37, 38). Among these, only the ASH neurons detect dilute (30%) octanol and, notably, they do so irrespective of the feeding status of the animal (18, 37, 38). Previous studies showed that animals lacking the tyrosine hydroxylase CAT-2, which is required for dopamine biosynthesis, are hypersensitive to dilute octanol; cat-2 mutant animals respond more quickly to the dilute stimulus than do wild-type animals (17, 20). In addition, animals specifically lacking dopamine signaling through the D2-like dopamine receptor DOP-3 are hypersensitive in their response to both 100% and dilute octanol (16).
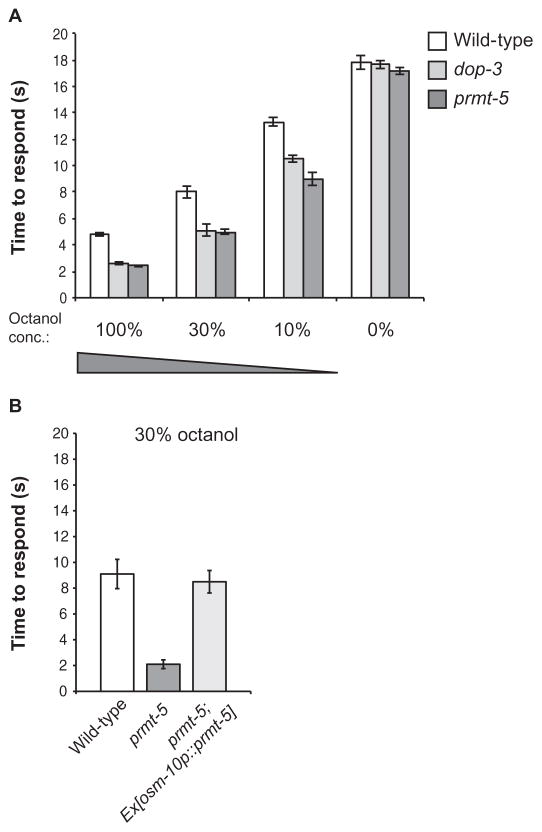
To determine whether protein arginine methylation contributed to the transduction or regulation of dopamine-modulated sensory signaling in vivo, we assessed the behavioral response of C. elegans lacking PRMT-5 function to the aversive odorant 1-octanol. These animals phenocopied the dop-3 mutant animals by responding more quickly than wild-type animals to all of the concentrations of octanol tested (Fig. 5A). Similar to the DOP-3 rescue experiments (16), restoring PRMT-5 in the ASH neurons [with the ASH cell–selective osm-10 promoter (36)] was sufficient to dampen the hypersensitivity to octanol. Although the osm-10 promoter also drives expression in the PHA and PHB tail neurons and weak expression in the ASI head neurons, these neurons are not involved in the detection of octanol (36). Indeed, prmt-5 mutant animals expressing the osm-10p::prmt-5 transgene responded similarly to wild-type animals when exposed to dilute (30%) octanol (Fig. 5B). These data suggest that PRMT-5 plays a role in promoting dopamine signaling in the ASH sensory neurons.
Fig. 5. Loss of C. elegans PRMT-5 function results in enhanced octanol avoidance.
(A) Effect of prmt-5 mutation on the timing of the octanol avoidance response. The times that it took WT, dop-3 mutant, and prmt-5 mutant C. elegans to respond to the presence of the indicated concentrations of octanol were determined as described in Materials and Methods. Animals were assayed 10 to 20 min after transfer to “off food” plates lacking OP50 Escherichia coli. Loss of prmt-5 function phenocopies loss of dop-3 at each concentration tested. P = 0.17 for 100% octanol, P = 0.84 for 30% octanol, and P = 0.13 for 10% octanol when comparing the responses of dop-3 and prmt-5 mutant animals. (B) Effect on octanol sensitivity of the expression of PRMT-5 in the ASH sensory neurons. WT and prmt-5 mutant animals were compared to prmt-5 mutant animals expressing prmt-5 driven by the osm promoter in ASH neurons in the time taken to respond (s) to the presence of 30% octanol. The combined data from three transgenic lines are shown. All data are means ± SEM. P = 0.7 when comparing WT animals to prmt-5 mutant animals expressing prmt-5 in the ASH neurons. Allele used: prmt-5(gk357). WT: the N2 WT strain. n ≥ 40 animals tested for each strain over at least three independent experiments.
C. elegans PRMT-5 contributes to the regulation of locomotion by both exogenous and endogenous dopamine
Dopamine modulates locomotor behavior in C. elegans by activating DOP-1 and DOP-3 (D1- and D2-like receptors, respectively) in the cholinergic motor neurons. Although the exposure of wild-type animals to exogenous dopamine causes paralysis, dop-3 mutant animals are resistant to this effect (13, 14). In addition, dop-3 mutant animals fail to reduce their rate of locomotion when they encounter a bacterial food source (13, 14); this basal slowing response is dependent upon signaling by endogenous dopamine (15). In both dopamine-induced paralysis and basal slowing, the function of DOP-3 is antagonized by DOP-1 (13, 14).
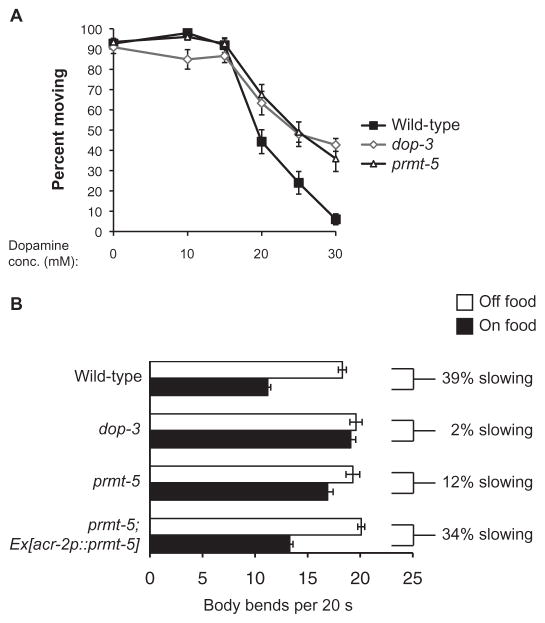
To establish whether loss of PRMT-5 function in C. elegans also phenocopied the loss of DOP-3 in locomotion, as it did for the aversive response to octanol, we first investigated whether prmt-5 mutant animals were resistant to dopamine-induced paralysis. Indeed, the locomotor behavior of prmt-5 and dop-3 mutant animals was indistinguishable across a broad range of dopamine concentrations (10 to 30 mM); both mutants exhibited resistance to paralysis induced by exogenously applied dopamine (Fig. 6A). Similarly, prmt-5 mutant animals displayed a diminished basal slowing response, which is mediated by endogenous dopamine (Fig. 6B). Whereas wild-type animals slowed their rate of locomotion by 39% on average when they encountered food, prmt-5 mutant animals slowed by only 12% (Fig. 6B). To determine whether, similar to DOP-3 (13, 14), PRMT-5 regulated dopamine-modulated locomotor behavior by acting in the cholinergic motor neurons, we used the acr-2 promoter (39) to restore PRMT-5 function in the prmt-5 mutant animals. Whereas prmt-5 mutant animals had a defect in basal slowing, prmt-5 animals expressing acr-2p::prmt-5 displayed a basal slowing response that was similar to that of wild-type animals (Fig. 6B). Together, these results suggest that, similar to the DOP-3 receptor, C. elegans PRMT-5 affects dopamine signaling in both chemosensation and locomotion.
Fig. 6. C. elegans PRMT-5 is required for efficacious dopamine-mediated paralysis and locomotor behavior.
(A) Effect of prmt-5 mutation on the dopamine-induced paralysis phenotype. The percentages of animals that were moving 20 min after transfer to plates containing the indicated concentrations of dopamine are shown for each genotype. Data are means ± SEM of ≥88 animals of each genotype. P > 0.1 when comparing prmt-5 and dop-3 mutant animals in terms of their responses to concentrations of dopamine from 15 to 30 mM. (B) Effect of prmt-5 mutation and its rescue in cholinergic motor neurons on the basal slowing response. Locomotion rates of the indicated C. elegans strains in the absence (white bars) and presence of HB101 E. coli (black bars) are given as the number of body bends per animal per 20-s time period. Animals lacking PRMT-5 function have a diminished basal slowing response. P = 0.00001 when comparing WT and prmt-5 mutant animals. P = 0.09 when comparing WT animals to prmt-5 mutant animals expressing prmt-5 in cholinergic motor neurons under the control of the acr-2 promoter. n ≥ 9 animals, 5 consecutive intervals each, for a total of ≥45 separate measurements. For the rescue experiment, the combined data of three independent transgenic lines are shown. n = 12 transgenic animals, 5 consecutive intervals each, for a total of 60 separate measurements under each condition. Alleles used: dop-3(vs106) and prmt-5(gk357). WT: the N2 WT strain.
Conserved arginines promote DOP-3 function in C. elegans
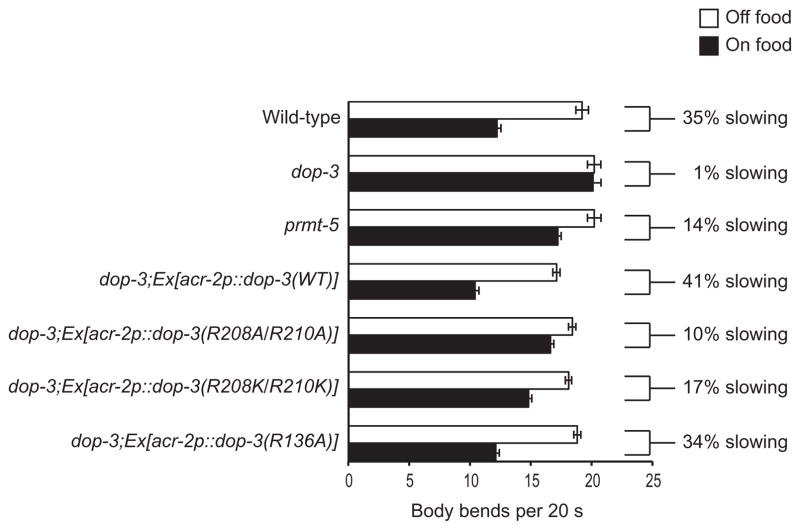
The residues Arg208 and Arg210 of the DOP-3 receptor in C. elegans correspond to the residues Arg217 and Arg219 of the human D2 receptor, respectively, in the putative RXR methylation motif within the third intracellular loop of the receptor (Fig. 1). To determine whether these conserved residues contributed to DOP-3 function in vivo, we generated the DOP-3(R208A/R210A) mutant receptor by site-directed mutagenesis. The expression of acr-2p::dop-3(R208A/R210A) failed to restore the basal slowing response of dop-3 mutant animals when a bacterial food source was encountered (Fig. 7). Similarly, when both arginines were mutated to lysines to preserve the positive charges at these positions, and when acr-2p::dop-3(R208K/R210K) was expressed in dop-3 mutant animals, these animals were likewise defective in their basal slowing response (Fig. 7). In both cases, the percentages by which the animals slowed (10 and 17%, respectively) were similar to that of animals lacking PRMT-5 (14%). Only one other arginine in the DOP-3 receptor is a predicted target for methylation (Arg136), and it lies within the second intracellular loop. However, this residue is not conserved in the human D2 receptor, and when the DOP-3(R136A) mutant receptor was expressed in dop-3 mutant animals, it rescued the basal slowing response similarly to the expression of wild-type DOP-3 in dop-3 mutants (Fig. 7). Together, these results suggest that evolutionarily conserved arginine residues within the predicted methylation motif of the third intracellular loop of D2-like dopamine receptors contribute to their signaling potential in vivo.
Fig. 7. DOP-3 residues Arg208 and Arg210 are required for a maximum basal slowing response.
Effect of mutating Arg208 and Arg210 to alanines or lysines on the basal slowing response. Locomotion rates of the indicated C. elegans strains in the absence (white bars) and presence of HB101 E. coli (black bars) are given as the number of body bends per animal per 20-s time period. Restoring WT DOP-3 fully rescued basal slowing. P = 0.4 when comparing WT animals to dop-3 mutant animals expressing dop-3 in cholinergic motor neurons under the control of the acr-2 promoter. dop-3 animals expressing DOP-3(R208A/R210A) or DOP-3(R208K/R210K) displayed a partially defective basal slowing response, similar to prmt-5 mutant animals. P = 0.3 and P = 0.2 when comparing prmt-5 mutant animals to dop-3 animals expressing DOP-3(R208A/R210A) or DOP-3(R208K/R210K), respectively. Alleles used: dop-3(vs106) and prmt-5(gk357). WT: the N2 WT strain. n ≥ 6 animals, 5 consecutive intervals each, for a total of ≥30 separate measurements. For the rescue experiments, the combined data for three or more independent transgenic lines are shown. n ≥ 12 transgenic animals, 5 consecutive intervals each, for a total of ≥60 separate measurements for each condition.
DISCUSSION
Understanding the mechanisms that regulate GPCR activity is crucial to manipulating their signaling for therapeutic benefit. One regulatory mechanism that is key to the functional diversity of many signaling proteins is posttranslational modification, such as arginine methylation by PRMTs. However, a major challenge in studying the physiological roles of PRMTs in vivo is that knockout mice have been generated for only seven PRMT family members, and in the case of three of these (including PRMT5), knockout results in embryonic lethality (21). Here, we took advantage of the fact that C. elegans prmt-5 loss-of-function mutants are viable (34), and used this system to identify adult animal phenotypes resulting from the loss of this PRMT.
Here, we provided evidence that arginine methylation of GPCRs contributes to their function. Our bioinformatics analysis identified putative arginine methylation motifs within the intracellular domains of human GPCRs and revealed that many of these are conserved across species (Fig. 1 and tables S1 to S5). Among the most conserved sites is a predicted RXR methylation motif in D2-like dopamine receptors. This motif is conserved from the C. elegans DOP-3 receptor to the human D2 receptor (Fig. 1C), a human D2 receptor fragment containing this motif was methylated by human PRMT5 in vitro (Fig. 2), and the conserved arginine residues within this motif (Arg217 and Arg219) enhanced D2 receptor function in cultured human cells (Fig. 3B).
We further showed a role for C. elegans PRMT-5 in D2-like (DOP-3) receptor–modulated behaviors in vivo. Similar to dop-3 mutant animals (13, 14, 16), animals lacking PRMT-5 displayed hypersensitivity to octanol (Fig. 5), an impaired basal slowing response (Fig. 6B), and resistance to paralysis by exogenously applied dopamine (Fig. 6A). Notably, conserved arginine residues that constitute the predicted methylation motif within the third intracellular loop of DOP-3 (Arg208 and Arg210) were required for its efficient methylation in vitro (Fig. 4) and also contributed to DOP-3 function in live animals (Fig. 7).
Together, these results led us to propose that arginine methylation promotes D2-like dopamine receptor signaling and that this mechanism of receptor regulation is conserved between nematodes and humans. Moreover, our finding that several hundred mammalian GPCRs contain predicted methylation sites within their cytoplasmic domains (Fig. 1A and tables S1 and S2) suggests that methylation may broadly regulate GPCR signaling in a previously unappreciated manner.
Our results also have implications for a possible mechanism by which arginine methylation could influence receptor signaling. The third intra-cellular loop of the D2 receptor is important for receptor function because it interacts with several signaling and regulatory proteins (40). For example, this region is the primary site of interaction with Gα proteins. In particular, the N-terminal section of the third intracellular loop of the human D2 receptor is sufficient to interact in a complex with Gαi1 and couples to Gαi/o proteins to inhibit adenylyl cyclase activity in cell membrane preparations (41–43); Arg217 and Arg219 are located within the Gα interaction domain. The addition of a methyl group to an arginine residue can remove a hydrogen bond donor and decrease the electrostatic surface potential at the residue, resulting in a change in size and hydrophobicity that can affect its interaction with binding partners (21). Therefore, methylation of the D2 receptor has the potential to regulate the interaction between the receptor and the Gα protein directly or the activation of the G protein by the receptor. Alternatively, it is possible that arginine methylation modulates interactions between the D2 receptor and accessory regulatory proteins that also bind to this region (40) or modulates agonist binding.
Targeting enzymes that catalyze the posttranslational modification of proteins has proven to be a useful therapeutic strategy in treating diseases, as exemplified by the many pharmacological agents that inhibit the activity of histone deacetylases. Similarly, GPCRs are the targets of almost 40% of all marketed pharmaceuticals (44). However, even with the large number of D2 receptor antagonists used to treat schizophrenia, they are not optimally effective in treating all of the symptoms associated with the disorder, and their application is often limited because of side effects (45). In addition, although dopamine receptor agonists are used in the treatment of Parkinson’s disease, treatment with L-DOPA remains the most effective course of action. However, its effectiveness diminishes over time, and its use in treatment is associated with the onset of severe motor complications (46). Thus, new approaches to treat diseases associated with dopaminergic dysfunction are critically needed. The findings described herein have the potential to facilitate the development of a new generation of treatments based on manipulating GPCR methylation status not only for D2 receptor–linked neuropsychiatric disorders but also for the treatment of diseases ranging from cancer to chronic heart failure that are also associated with aberrant GPCR signaling.
MATERIALS AND METHODS
Bioinformatics analysis
The amino acid sequences of 1655 human GPCRs were obtained from the GPCR database (www.gpcr.org/7tm/). This starting set of GPCRs was filtered to remove duplicates and was then used to query the NCBI RefSeq protein database, which resulted in a list of 963 nonredundant human GPCR accession numbers. These accession numbers were then used to locate NCBI HomoloGene database records, and GPCRs conserved in Mus musculus, Rattus norvegicus, Danio rerio, Drosophila melanogaster, and C. elegans were identified. TMHMM (transmembrane hidden Markov model) v2.0c (47) was used (with the default model parameters) to predict the locations of the transmembrane domains. All putative GPCRs that were not predicted to contain seven transmembrane domains were removed from the data set, resulting in a total of 737 human GPCRs in the final data set that was used for alignment and motif searching. For each GPCR, the human protein sequence was aligned to a homologous sequence with MUSCLE v3.8.31 (48), and within each alignment, the intracellular loops of each GPCR were searched for the putative methylation motifs RGG and RXR. The entire analysis workflow was automated with Perl (script available upon request). A more inclusive human–C. elegans homology data set was obtained with GPCR sequences from WormBase (www.wormbase.org). C. elegans GPCRs were identified with GO records, and the WormBase-annotated human homologs for each C. elegans GPCR were used to query the NCBI RefSeq database. This resulted in the identification of 565 nonredundant human GPCRs that were potential homologs of 179 C. elegans GPCRs. This data set was analyzed with TMHMM (47) to predict transmembrane domains, the sequences were aligned with MUSCLE (48), and the alignments were searched for RGG and RXR motifs as described earlier.
In vitro methylation assay
The GST fusion proteins used as substrates for methylation were expressed in the E. coli strain Rosetta DE3 (Novagen) and were purified with glutathione Sepharose beads (GE Healthcare) according to the manufacturer’s protocol. In vitro methylation assays were performed in a total volume of 20 μl with 5 μg of substrate, 210 ng of recombinant human PRMT5 complex (Active Motif), and 5.5 μCi of S-[methyl-3H]adenosyl-L-methionine (55 to 85 Ci/mmol; PerkinElmer) in 1× methylation buffer [150 mM NaCl, 50 mM tris-HCl (pH 8), 1 mM EDTA]. Reactions were incubated at 37°C for 8 hours, resolved by SDS–polyacrylamide gel electrophoresis, and then transferred to PVDF membranes. The membranes were sprayed with EN3HANCE (PerkinElmer) three times at 10-min intervals before being exposed to Kodak BioMax MS film with a BioMax TranScreen LE Intensifying Screen at −80°C for 2 weeks and subsequently were developed. Band intensities were quantified with ImageJ software (49) and were normalized according to gel loading.
Cell culture and viral transduction
HEK 293T cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies), 1 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified incubator with 5% atmospheric CO2. Vesicular stomatitis virus glycoprotein (VSV-G)–pseudotyped lentiviruses were produced in HEK 293T cells by the traditional calcium phosphate precipitation method using 6 μg of pMD2.G, 25 μg of psPAX2, and 30 μg of the appropriate lentiviral construct: pCS-MCS, pCS-MCS-HA-DRD2, or pCS-MCS-HA-DRD2(R217A/R219A). The viruses were then used to transduce HEK 293T cells, and stable cell lines were selected with puromycin (1 μg/ml; Sigma). All experiments were performed in cultured cells that were between passages 3 and 5 after retrieval from frozen stocks.
Flow cytometric analysis
HEK 293T cells stably expressing HA-tagged wild-type D2 receptor or the D2(R217A/R219A) mutant receptor were washed with ice-cold phosphate-buffered saline (PBS), harvested by trituration, and fixed with 1% para-formaldehyde (PFA; USB, Affymetrix) for 20 min at room temperature. The cells were then washed three times with PBS, 0.1% bovine serum albumin (BSA) and incubated with an anti-HA antibody (6E2, Cell Signaling) diluted 1:250 in PBS, 0.1% BSA for 30 min at room temperature. Samples were washed three times with PBS, 0.1% BSA, before incubation with Alexa Fluor 488–conjugated goat anti-mouse antibody (0.5 μg/ml; Life Technologies) for 60 min at room temperature. The cells were then washed three times with PBS, 0.1% BSA and passed through a nylon sieve. Surface labeling was measured with a FACSCalibur flow cytometer. Data analysis was performed with Cyflogic software (CyFlo Ltd.). The Student’s two-tailed t test was used for statistical analysis.
Immunofluorescence microscopy
HEK 293T cells stably expressing HA-tagged wild-type D2 or the D2(R217A/R219A) mutant receptor were plated at 50% confluency onto coverslips coated with poly-L-lysine in a six-well plate. Twenty-four hours later, the cells were washed with PBS and fixed with 4% PFA at room temperature for 15 min. Coverslips were then washed three times with PBS for 2 min each and incubated in blocking solution (goat serum diluted 1:125 in PBS) for 30 min without rocking. The blocking solution was then replaced with anti-HA antibody (6E2, Cell Signaling) diluted 1:125 in blocking solution. Coverslips were incubated at room temperature for 60 min followed by three washes with PBS and incubation with Alexa Fluor 488–conjugated goat anti-mouse antibody (Life Technologies) for 30 min. The coverslips were mounted onto slides with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories). Slides were dried overnight in the dark at 4°C. Imaging was performed with a Zeiss LSM 710 confocal microscope.
Whole-cell cAMP assay
HEK 293T cells stably expressing HA-tagged D2 or D2(R217A/R219A) were plated at a density of 10,000 cells per well onto a clear-bottomed, poly-L-lysine–coated 96-well plate (Corning). Forty-eight hours later, the cells were incubated with 20 μM forskolin (Sigma) together with increasing concentrations of quinpirole (Sigma) in serum-free medium for 30 min at 37°C. The abundance of cAMP in each sample was measured with the HitHunter competitive immunoassay (DiscoveRx) in accordance with the manufacturer’s protocol. Luminescence was measured with a FlexStation 3 Reader (Molecular Devices). The data were analyzed in R software (3.1.1) with the drc package (2.3–96). Dose-response curves were fitted to each individual biological replicate sample using a four-parameter logistic nonlinear regression model. The empty vector control curve was fitted with a linear regression model. The log10 of the IC50 values from each biological replicate were then compared by the Student’s two-tailed t test (n = 4 biological replicates).
C. elegans strains
C. elegans strains were maintained under standard conditions on NGM (nematode growth medium) agar plates seeded with OP50 E. coli bacteria. The strains used in this study include N2 Bristol wild-type, FG129 prmt-5(gk357), and LX703 dop-3(vs106).
C. elegans transgenic strains
For prmt-5 and dop-3 rescue experiments, pJM67 elt-2::gfp plasmid (50 ng/μl) (50) was used as the co-injection marker together with either the prmt-5 or dop-3 rescuing plasmids (50 ng/μl).
C. elegans behavioral assays
Well-fed young adult animals were used for analysis, and all behavioral assays were performed on at least two separate days, in parallel with controls. Behavioral assays were performed as previously described (13, 20, 36, 37). Dopamine (hydrochloride complex) was purchased from Sigma. All data are presented as means ± SEM. The Student’s two-tailed t test was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank J. Benovic and P. Sengupta for feedback on this manuscript and F. Bachand, D. Chase, and J. Côte for technical advice. We thank D. Chase, J. Côte, A. Fire, Y. Jin, Y. Kohara (National Institute of Genetics, Japan), M. Koelle, S. Suo, the Caenorhabditis Genetics Center, and S. Mitani and the Japanese National Bioresource Project for the Nematode for reagents. We are also grateful to F. Bachand, J. Benovic, J. Côte, R. Komuniecki, and P. Sengupta for helpful discussions.
Funding: This work was supported by the Ellison Medical Foundation (AG-NS-0380-07 to D.M.F.), the National Science Foundation (MCB-0917896 to D.M.F. and MCB-1051350 to M.C.Y.), and the NIH (R21MH101386-01A1 to D.M.F.). The Flow Cytometry Facility is funded by the NIH (National Cancer Institute core grant P30CA16056).
Footnotes
Author contributions: M.C.Y. and D.M.F. jointly conceived the study and oversaw experiments; N.L., C.A.J., M.C.K., M.C.Y., and D.M.F. designed and performed the experiments; C.A.J. and D.M.F. performed statistical analyses; C.A.J. performed the bioinformatics analysis for methylation motifs; M.-S.L., P.L., and S.T.A. generated lentivirus and the stable HEK 293T cell lines; J.F.W., B.B., K.L.M., and S.D.C. aided in the design and execution of experiments; N.L., M.C.Y., and D.M.F. wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
www.sciencesignaling.org/cgi/content/full/8/402/ra115/DC1
Materials and Methods
Fig. S1. Overexpression of PRMT5 promotes D2 receptor function in cultured human cells.
Table S1. Bioinformatics summary.
Table S2. Predicted arginine methylation motifs in the intracellular domains of human GPCRs and their conservation in GPCRs of other species, as identified by HomoloGene.
Table S3. Predicted arginine methylation motifs in the third intracellular domain of human GPCRs and their conservation in GPCRs of other species, as identified by HomoloGene.
Table S4. Predicted arginine methylation motifs in the intracellular domains of GPCRs in the human–C. elegans data set generated by GO analysis.
Table S5. Predicted arginine methylation motifs in the third intracellular domain of GPCRs in the human–C. elegans data set generated by GO analysis.
REFERENCES AND NOTES
- 1.McDonald PW, Jessen T, Field JR, Blakely RD. Dopamine signaling architecture in Caenorhabditis elegans. Cell Mol Neurobiol. 2006;26:593–618. doi: 10.1007/s10571-006-9003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chase DL, Koelle MR. The C. elegans Research Community, editor. WormBook. WormBook; Pasadena, CA: 2007. [Google Scholar]
- 3.Schafer WR. Addiction research in a simple animal model: The nematode Caenorhabditis elegans. Neuropharmacology. 2004;47:123–131. doi: 10.1016/j.neuropharm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Wolf FW, Heberlein U. Invertebrate models of drug abuse. J Neurobiol. 2003;54:161–178. doi: 10.1002/neu.10166. [DOI] [PubMed] [Google Scholar]
- 5.Waddell S. Dopamine reveals neural circuit mechanisms of fly memory. Trends Neurosci. 2010;33:457–464. doi: 10.1016/j.tins.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmiter RD. Dopamine signaling in the dorsal striatum is essential for motivated behaviors: Lessons from dopamine-deficient mice. Ann N Y Acad Sci. 2008;1129:35–46. doi: 10.1196/annals.1417.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovinger DM. Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology. 2010;58:951–961. doi: 10.1016/j.neuropharm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 9.Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 10.Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: Version III—The final common pathway. Schizophr Bull. 2009;35:549–562. doi: 10.1093/schbul/sbp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. doi: 10.1016/s0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 12.Neve KA, Seamans JK, Trantham-Davidson H. Dopamine receptor signaling. J Recept Signal Transduct Res. 2004;24:165–205. doi: 10.1081/rrs-200029981. [DOI] [PubMed] [Google Scholar]
- 13.Chase DL, Pepper JS, Koelle MR. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci. 2004;7:1096–1103. doi: 10.1038/nn1316. [DOI] [PubMed] [Google Scholar]
- 14.Allen AT, Maher KN, Wani KA, Betts KE, Chase DL. Coexpressed D1- and D2-like dopamine receptors antagonistically modulate acetylcholine release in Caenorhabditis elegans. Genetics. 2011;188:579–590. doi: 10.1534/genetics.111.128512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 16.Ezak MJ, Ferkey DM. The C. elegans D2-like dopamine receptor DOP-3 decreases behavioral sensitivity to the olfactory stimulus 1-octanol. PLOS One. 2010;5:e9487. doi: 10.1371/journal.pone.0009487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wragg RT, Hapiak V, Miller SB, Harris GP, Gray J, Komuniecki PR, Komuniecki RW. Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. J Neurosci. 2007;27:13402–13412. doi: 10.1523/JNEUROSCI.3495-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci USA. 2004;101:15512–15517. doi: 10.1073/pnas.0403369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezcurra M, Tanizawa Y, Swoboda P, Schafer WR. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J. 2011;30:1110–1122. doi: 10.1038/emboj.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferkey DM, Hyde R, Haspel G, Dionne HM, Hess HA, Suzuki H, Schafer WR, Koelle MR, Hart AC. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron. 2007;53:39–52. doi: 10.1016/j.neuron.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 23.Wei H, Mundade R, Lange KC, Lu T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle. 2014;13:32–41. doi: 10.4161/cc.27353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branscombe TL, Frankel A, Lee J-H, Cook JR, Yang Z-h, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric di-methylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, Brown JWS, Cerdán PD, Borevitz JO, Mas P, Ceriani MF, Kornblihtt AR, Yanovsky MJ. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 27.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 29.Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 30.Boisvert FM, Côté J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Filtz TM, Artymyshyn RP, Guan W, Molinoff PB. Paradoxical regulation of dopamine receptors in transfected 293 cells. Mol Pharmacol. 1993;44:371–379. [PubMed] [Google Scholar]
- 32.Masri B, Salahpour A, Didriksen M, Ghisi V, Beaulieu JM, Gainetdinov RR, Caron MG. Antagonism of dopamine D2 receptor/β-arrestin 2 interaction is a common property of clinically effective antipsychotics. Proc Natl Acad Sci USA. 2008;105:13656–13661. doi: 10.1073/pnas.0803522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLOS Genet. 2009;5:e1000514. doi: 10.1371/journal.pgen.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart AC, Kass J, Shapiro JE, Kaplan JM. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 1999;19:1952–1958. doi: 10.1523/JNEUROSCI.19-06-01952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 38.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 39.Hallam S, Singer E, Waring D, Jin Y. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development. 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- 40.Fukunaga K, Shioda N. Novel dopamine D2 receptor signaling through proteins interacting with the third cytoplasmic loop. Mol Neurobiol. 2012;45:144–152. doi: 10.1007/s12035-011-8227-8. [DOI] [PubMed] [Google Scholar]
- 41.Bofill-Cardona E, Kudlacek O, Yang Q, Ahorn H, Freissmuth M, Nanoff C. Binding of calmodulin to the D2-dopamine receptor reduces receptor signaling by arresting the G protein activation switch. J Biol Chem. 2000;275:32672–32680. doi: 10.1074/jbc.M002780200. [DOI] [PubMed] [Google Scholar]
- 42.Voss T, Wallner E, Czernilofsky AP, Freissmuth M. Amphipathic α-helical structure does not predict the ability of receptor-derived synthetic peptides to interact with guanine nucleotide-binding regulatory proteins. J Biol Chem. 1993;268:4637–4642. [PubMed] [Google Scholar]
- 43.Malek D, Münch G, Palm D. Two sites in the third inner loop of the dopamine D2 receptor are involved in functional G protein-mediated coupling to adenylate cyclase. FEBS Lett. 1993;325:215–219. doi: 10.1016/0014-5793(93)81076-c. [DOI] [PubMed] [Google Scholar]
- 44.White A. Market Wired. IBC Life Sciences; Westborough, MA: 2005. Biotechnology and pharmaceutical companies take aim at promising drug targets. [Google Scholar]
- 45.Ginovart N, Shitij K. In: The Dopamine Receptors. Neve KA, editor. chap. 16. Humana Press; New York: 2010. pp. 431–477. [Google Scholar]
- 46.Gurevich EV, Gurevich VV. In: The Dopamine Receptors. Neve KA, editor. chap. 18. Humana Press; New York: 2010. pp. 525–584. [Google Scholar]
- 47.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting trans-membrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- 51.Sugiura M, Fuke S, Suo S, Sasagawa N, Van Tol HHM, Ishiura S. Characterization of a novel D2-like dopamine receptor with a truncated splice variant and a D1-like dopamine receptor unique to invertebrates from Caenorhabditis elegans. J Neurochem. 2005;94:1146–1157. doi: 10.1111/j.1471-4159.2005.03268.x. [DOI] [PubMed] [Google Scholar]
- 52.Goulet I, Gauvin G, Boisvenue S, Côté J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and sub-cellular localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- 53.Miyoshi H, Blömer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.