Abstract
Skin sensitization is a major environmental and occupational health hazard. Although many chemicals have been evaluated in humans, there have been no efforts to model these data to date. We have compiled, curated, analyzed, and compared the available human and LLNA data. Using these data, we have developed reliable computational models and applied them for virtual screening of chemical libraries to identify putative skin sensitizers. The overall concordance between murine LLNA and human skin sensitization responses for a set of 135 unique chemicals was low (R = 28-43%), although several chemical classes had high concordance. We have succeeded to develop predictive QSAR models of all available human data with the external correct classification rate of 71%. A consensus model integrating concordant QSAR predictions and LLNA results afforded a higher CCR of 82% but at the expense of the reduced external dataset coverage (52%). We used the developed QSAR models for virtual screening of CosIng database and identified 1061 putative skin sensitizers; for seventeen of these compounds, we found published evidence of their skin sensitization effects. Models reported herein provide more accurate alternative to LLNA testing for human skin sensitization assessment across diverse chemical data. In addition, they can also be used to guide the structural optimization of toxic compounds to reduce their skin sensitization potential.
Keywords: Skin sensitization, QSAR modeling, human data, virtual screening
INTRODUCTION
Allergic contact dermatitis (ACD) is a prevalent occupational and environmental disease with high impact on individual working ability and quality of life.1 ACD is caused by topical exposure to chemical allergens2 and their abundance in commerce demands that hazardous chemicals must be identified and replaced by “greener”, i.e., safer, alternatives.3,4 The adverse outcome pathway (AOP) of ACD is characterized by two phases: induction and elicitation of the immune response.2,5 After a chemical gains access to the viable epidermis, the first phase is initiated by chemical binding to skin proteins/peptides to form an immunogenic complex. The second phase is an inflammatory process mediated by allergen-specific T cells.5,6
Skin sensitization is commonly evaluated in humans using the human repeated insult patch test and its variations.7–11 The human maximization test12,13 was designed to be a sensitive assay, where the “maximization step” produces slightly irritated skin. In most countries, these methods face both ethical issues and scientific validity and are only used as to confirm safe doses.14,15
In parallel, common animal tests for skin sensitization include the guinea pig maximization test16 and the murine local lymph node assay (LLNA);17 the latter is regarded as the preferred in vivo test for evaluating skin sensitization within REACH18 and by various regulatory agencies, such as UK Health and Safety Executive19 and US EPA20. In this assay, skin sensitization is evaluated by quantifying lymphocyte proliferation, which is correlated with the extent of sensitization after a repeated exposure to a sensitizing substance. A variation of this assay, named the reduced local lymph node assay (rLLNA), was later proposed and found to reduce the number of animals used for testing by almost 40%.21,22 The LLNA showed good overall correlation with human skin sensitization.23–27 However, some studies have shown that LLNA EC3 (the dose that produces the stimulation index of three, the threshold for a positive response) values failed in several cases to predict human skin sensitization potency.28,29
Animal tests have been forbidden for cosmetic ingredients in Europe since 2009, followed by the ban on the sale of cosmetics tested in animals after March 2013 anywhere in the world.30 Some reports have further questioned the use of animals in the evaluation of human safety with respect to both ethical concerns and scientific relevance.31,32 Although animal testing is still considered crucial to the evaluation of chemical safety33, toxicity testing in the 21st century is moving toward greater understanding of the disease pathways at multiple biological levels, so as to develop alternative methods.34–36 Several in vitro tests have been proposed, but a single test most likely will not be able to predict human skin sensitization.37 A recent analysis made by a group of experts revealed that in chemico and in vitro assays correctly identified most of the compounds requiring activation, despite some divergence between the assays.38 This reinforces the importance of applying integrated testing strategies (ITS) to address multiple key steps of the adverse outcome pathway.39,40
Quantitative Structure-Activity Relationship (QSAR) modeling is a major computational approach used in medicinal chemistry and toxicology to design novel bioactive compounds or evaluate chemical safety, respectively. This approach employs statistical or machine learning techniques to establish predictive correlations between intrinsic chemical properties (chemical descriptors) and measured bioactivity or toxicity and the resulting models are used to forecast the respective target properties of novel or untested componds.41,42 Several studies have generated computational models to predict skin sensitization based on LLNA.43–61 As we have alluded to in our recent papers,62,63 most published QSAR models are not compliant with the best practices on model development and validation,41,64 and thus their reliability for assessing chemically-induced skin sensitization is not assured.
In an attempt to address the skin sensitization AOP, a few groups have built local QSAR models using mechanistic information.59,60 The biological mechanism of skin sensitization is very well defined, and there are several substructures associated with this defined mechanism of action.38 However, these models were built using small datasets owing to the small size of publicly available data and the high number of compounds lacking protein binding alerts for skin sensitization. For instance, Nandy and Roy56 developed regression-based QSAR models for skin sensitization; however, these models were built using a small dataset of 51 compounds that resulted in the limited model applicability domain. Another recent study compared two structural alert-based systems to predict skin sensitization (QSAR Toolbox and Toxtree) with LLNA and human data.65 The authors found that structural alerts could predict human data better than LLNA, concluding that in silico models should be preferably developed using human data. While we agree with this overall conclusion, we have also shown recently66 that structural alerts do not serve as reliable predictors as they typically select toxic compounds accurately, but often identify very large numbers of false positives. Nevertheless, one might benefit from the use of structural alerts to build models and some interesting and innovative approaches have been proposed. A recent study58 reported hybrid models that used structural alerts along with chemical descriptors. The model accuracy was 93%, with sensitivity of 98% and specificity of 85% for 269 chemicals. Another study proposed a hierarchical model where skin permeability is evaluated using Monte Carlo simulations, chemical reactive centers are determined with expert rules, and protein reactivity is predicted by the means of quantum-mechanical modeling.61 The authors reported conspicuously impressive results for predicting skin sensitization of an external set mostly composed by LLNA, Buehler’s test, and GMPT data: sensitivity as high as 87%, specificity as high as 100%, and balanced accuracy of 93%, which exceeded the reported67 concordance of 89% between LLNA and GMPT.
The challenge of rationally designing environmental chemicals lacking skin sensitization effects resonates strongly with the subject of green chemistry68 that has been gaining broad attention as part of chemical toxicology.69–71 Computational methods represent an attractive approach for the design of safer chemicals.72–75 Besides high accuracy, it is also critical that computational models predict biological effects of chemicals that are most relevant to human health. To the best of our knowledge, there have been no reports on the computational modeling of human skin sensitization data, which has the potential to increase the accuracy of early chemical evaluation, especially when used in combination with other strategies.40,76 Considering the need to design safer compounds that would be less prone to induce skin sensitization, and the difficulty in obtaining human data, the aims of this study were to analyze the concordance between the results of human tests and LLNA and develop robust and predictive QSAR models for skin sensitization based on human data. We further endeavored to benchmark the performance of QSAR vs. LLNA in predicting human skin sensitization and analyze the performance of QSAR vs. LLNA on clusters of structurally similar compounds to identify which approach is more efficient for various chemotypes.
MATERIALS AND METHODS
Datasets
Human skin sensitization (Dataset A)
The dataset used in this study was retrieved from the Appendix C, Annex III-2 of ICCVAM Test Method Evaluation Report: The Usefulness And Limitations of the Murine Local Lymph Node Assay For Potency Categorization Of Chemicals Causing Allergic Contact Dermatitis In Humans.28 This dataset has been revised and expanded by scientists from the Integrated Laboratory Systems (http://www.ils-inc.com/). All the corrections are highlighted in the electronic supplementary information (ESI) (ESI-A-Data_predictions.xlsx). For instance, tests for (chloro)methylisothiazolinone were actually done for its mixture with methylisothiazolinone, i.e., for Kathon; therefore, these entries have been changed to Kathon. Additionally, some tests were previously combined (they used the same concentration, but different vehicles) and have now been separated. The dataset originally consisted of 302 chemical records (every record refers to a chemical compound but because of the presence of duplicates, several records could describe the same compound) and associated human data. Skin sensitization potential was based on human DSA05 data (dose per skin area that produces a positive response in 5% of the tested population), since it has been shown to correspond best with the LLNA EC3 value, compared against no observed effect level (NOEL) or lowest observed effect level (LOEL) values.28
After curation, 135 unique substances were kept for this study. This list contained 26 inorganics or mixtures that were used for comparison with the LLNA (see the Analysis of structural duplicates section) but were not included in the modeling set because chemical descriptors could not be computed for such substances. Given the complexity of skin sensitization and the lack of additional data, we decided to keep compounds with both positive and negative results as skin sensitizers due to the weight of evidence of the data, since most of the conflicts were dose-related. The modeling set contained the remaining 109 compounds (63 sensitizers and 46 non-sensitizers) with defined chemical structure.
Murine skin sensitization (Dataset B)
ICCVAM (2011) provided LLNA data for the 135 unique substances present in the human dataset that was described above (Dataset A). However, we obtained LLNA data from a larger database compiled by the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM).77 This source contained information for 1,060 LLNA tests. After curation, all the LLNA data for the 135 unique substances present in the human dataset (Dataset A) were compiled. Only 109 of them have defined chemical structures and thus were used for the comparison of QSAR and LLNA in predicting human data.
CosIng database
CosIng is the European Commission database that includes information on cosmetic substances and ingredients (http://ec.europa.eu/growth/tools-databases/cosing/). We retrieved 5166 records, and after curation, 3964 unique chemical compounds were used for virtual screening. The initial analysis of the CosIng database revealed 76 chemicals already present in our modeling set including 38 sensitizers and 38 non-sensitizers. Thus, we have applied our QSAR models to predict the human skin sensitization potential for the remaining 3850 compounds.
Data curation
Chemical structures were retrieved from either Chemicalize (http://www.chemicalize.org/), ChemSpider (http://www.chemspider.com/), or SciFinder (https://scifinder.cas.org) databases using the Chemical Abstracts Service (CAS) registry numbers and chemical names. Chemicals were not considered for modeling or virtual screening if their structures were not available, but the respective biological data were used to analyze the concordance between human and LLNA results. The datasets were thoroughly curated according to the workflows developed by our group78–80. Briefly, structural normalization of specific chemotypes, such as aromatic and nitro groups, was performed using ChemAxon Standardizer (v. 15.10.12.0, ChemAxon, Budapest, Hungary, http://www.chemaxon.com). Inorganic salts, organometallic compounds, and mixtures were also removed. After structural standardization, the duplicates were identified using ISIDA Duplicates81 and HiT QSAR82.
Concordance analysis of human vs. LLNA data
Concordance analysis of LLNA vs. human data was conducted to verify the relevance of the LLNA to human outcomes. Compounds that were tested in both assays were analyzed in three different ways: binary (sensitizer vs. non-sensitizer; 109 compounds); multiclass (strong/extreme sensitizer, weak/moderate sensitizer, and non-sensitizer; 109 compounds); and continuous scales (the concentration in mol/m2; 52 compounds). All compounds were divided into several classes according to their potency in different assays. When using LLNA EC3 data, three classes were defined: strong/extreme sensitizers had LLNA EC3 less or equal to 2%; weak/moderate sensitizers had LLNA EC3 between 2% and 100%; and compounds with no EC3 values were defined as non-sensitizers. For human data, strong/extreme sensitizers were defined as compounds with DSA05 less or equal to 500 mol/m2; weak/moderate sensitizers had DSA05 values above 500 mol/m2; and compounds with no DSA05 values were defined as non-sensitizers. We then compared quantitative potency data for human and LLNA assays to detect the level of concordance. For this analysis, eleven human sensitizers were excluded, either because it was not possible to calculate the molar concentration (mixtures), or a compound was a non-sensitizer in LLNA.
Cheminformatics approaches
Cluster analysis
Chemical clusters were generated by the Sequential Agglomerative Hierarchical Non-overlapping method implemented in the ISIDA/Cluster software.81 Briefly, the software generates a dendrogram of the parent-child relationships between clusters and a heat map of the proximity matrix colored according to the pairwise chemical similarity between compounds. This method was applied to check the structural diversity of compounds in the dataset and to identify possible structure-activity relationships rules and trends in success/failure of predicting human skin sensitization by QSAR models and LLNA.
Molecular descriptors
GUSAR uses a combination of three types of descriptors: whole-molecule descriptors, QNA (Quantitative Neighborhoods of Atoms) descriptors83 and “biological” descriptors84 which represent multiple bioactivity predictions by the PASS (Prediction of Activity Spectra of Substances) software.85
QNA descriptors are defined by two functions, P and Q. The values for P and Q for each atom i are calculated as:
| (1) |
| (2) |
where the k are all other atoms in the molecule and
| (3) |
Here IP is the ionization potential and EA is the electron affinity for each atom, and C is the connectivity matrix for the molecule. Two-dimensional Chebyshev polynomials are used for approximating the functions P and Q over all atoms of the molecule.86
The whole-molecule descriptors used in GUSAR are topological length, topological volume, lipophilicity, number of positive charges, number of negative charges, number of hydrogen bond acceptors, number of hydrogen bond donors, number of aromatic atoms, molecular weight, and number of halogen atoms.84,87
The PASS biological descriptors are calculated using the PASS algorithm84, which predicts a wide range of biological outcomes including transporter protein binding, gene expression activities, and various mechanisms of action, totaling ~ 6400 “biological activities” at a mean prediction accuracy threshold of at least 95%. The output from PASS is the probability for each predicted outcome that the compound will be active (Pa), and the probability that it will be inactive (Pi). The difference between these two values (Pa−Pi) for a randomly selected subset of the predicted activities constituted a molecular descriptor.
RBF-SCR algorithm
In the RBF-SCR algorithm, the descriptors are weighted during the calculation of the radial basis functions (RBF) by the coefficients obtained from self-consistent regression (SCR). These coefficients reflect the contribution of each particular descriptor (variable) to the final equation for the given activity. The higher the absolute value of the coefficient, the greater its contribution. Self-consistent regression is implemented as a regularized least-squares method that can be formulated as:
| (4) |
where a is the vector of regression coefficients, n is the number of objects, yi is the response value of the ith object, m is the number of independent variables, xik is the value of the kth independent variable of the ith object, ak is the kth value of the regression coefficients, and vk is the kth value of the regularization parameters.
Thus, RBF-SCR can be expressed as the equation:
| (5) |
where a is taken from equation 4.
The RBF-SCR algorithm uses linear radial basis functions because they allow for modeling of diverse training sets with a high level of dissimilarity between the set’s objects. Thus, the salient features of the RBF-SCR method are: (a) the weights for each descriptor vector used for the calculation of RBF are based on that descriptor’s importance for the given activity as determined by SCR, and (b) linear basis functions are used for better description of diverse data sets.88
QSAR modeling
Binary QSAR models were developed and rigorously validated according to the best practices of QSAR modeling.64 Some QSAR models were developed with RBF-SCR algorithm (see previous section) implemented in the GUSAR software.89 We have followed a 5-fold external cross-validation procedure for the estimation of model’s predictive power.41 Here the full set of compounds with known experimental activity was divided into five subsets of equal size (external folds) using modified Kennard-Stone algorithm.90 For each fold, we selected ten models with the highest Correct Classification Rate (CCR, computed as the average of sensitivity and specificity of the model). Then, the models that passed the Y-randomization test91 were applied to the external set compounds to predict their experimental properties. Applicability domain (AD) is an important characteristic of any QSAR model.92,93 Herein, AD was estimated using three different approaches: similarity, leverage, and accuracy assessment. Since in GUSAR several internal models are developed for each fold, a compound is considered to be within the AD if it is found within it for at least one model.88 Once the predictivity of the developed models was validated, they were applied to predict the skin sensitization potential of the ingredients used in cosmetic products, retrieved from the CosIng database on January 26, 2016 (http://ec.europa.eu/growth/tools-databases/cosing/). The developed QSAR models are available in the ESI (ESI-B-Human_Skin_Sens_QSAR).
ChemoText Analysis
An in-house tool called ChemoText94 was used to validate the skin sensitization potential of "hits" identified by QSAR modeling. ChemoText is a graph database that is used to extract Medical Subject Headings (MeSH terms) and PubMed article IDs from MEDLINE (2015 version; 25 million articles). ChemoText, implemented as a web server (http://chemotext.mml.unc.edu/), analyzes MEDLINE to extract MeSH terms that define diseases, proteins, and chemicals. ChemoText is used to find instances in published scientific abstracts indexed in MEDLINE/PubMed where certain MeSH terms co-occur, i.e., are mentioned together in the abstract. MeSH terms associated with skin sensitization, namely "Dermatitis, Occupational", "Dermatitis, Contact", "Dermatitis, Allergy", were queried in ChemoText for co-occurring chemical names. If a "hit" (chemical) and a skin sensitization MeSH term were found to co-occur in an abstract, then the PubMed article ID was obtained. The skin sensitization potential of the chemical "hit" was then manually verified by inspection of the corresponding full-text article.
RESULTS AND DISCUSSION
Analysis of structural duplicates
Human skin sensitization (Dataset A)
Almost 50% of compounds from Dataset A (62 out 135) were associated with multiple records (numbering from two to twelve). The number of records and the outcome of all assays for these 62 compounds is shown in ESI (Table S1, ESI-C-Clusters_and_Tables.docx). Only 28 substances out of 62 had concordant outcomes for all the records. We were over-cautious to avoid any toxicant being considered as non-toxic; thus, all the compounds with at least one record labeling the compound as a sensitizer were considered sensitizers, according to the weight of evidence approach adapted to risk assessment95 and lack of additional data in the literature. Thus, 50 of the 62 substances were considered to be sensitizers. Several compounds with more than five records (e.g., hydroxycitronellal, phenylacetaldehyde, and cinnamyl alcohol,) had large divergence between annotations, revealing the lack of concordance among the available data from the human tests. It is known that there can be large inter-individual differences in response to a chemical exposure. However, since expected no-effect sensitization levels (NESILs) are determined at a dose below the sensitization threshold, much of this variability is already taken into account 96. Despite this high variability, we modeled these data (see QSAR modeling section) and analyzed the predictions for these compounds when they were in the external folds. Four substances (ylang ylang, oakmoss, treemoss, and (chloro)methylisothiazolinone [Kathon]) out of 34 compounds with divergent annotations (and thus considered as sensitizers) were mixtures excluded from the analysis. Interestingly, 22 of 30 remaining sensitizer compounds were predicted as sensitizers.
Murine skin sensitization (Dataset B)
The murine skin sensitization dataset had 653 records for the 135 compounds from Dataset A. We found that only 19 of the 76 compounds with multiple records (from 2 to 44) had a discordance between annotations (see Table S2 in ESI). Even though some compounds had divergent annotations, they usually represented a small fraction of the total number of records. This is in agreement with earlier studies that LLNA has a low inter-laboratory variance.62,97,98 In general, variability was higher when different vehicles are used.99,100
Human vs. LLNA data analysis
There is a common understanding that LLNA results correlate with human skin sensitization potential and, therefore, LLNA has been regarded as a reliable method to predict whether a chemical is expected to be a sensitizer or not.28,101 We decided to repeat the analysis made by ICCVAM several years ago (2011)28 but considering compounds with defined chemical structure only (removing mixtures and inorganics) in order to analyze the structure-activity relationship trends. We conducted the analysis at three levels: binary (sensitizer vs. non-sensitizer); multi-class potency (strong/extreme sensitizer, weak/moderate sensitizer, and non-sensitizer); and continuous potency (using the concentration in mol/m2 for 52 sensitizers). Table 1 shows the confusion matrix reflecting the level of concordance between human and LLNA binary data. The accuracy of using LLNA results to predict human data is estimated to have the Correct Classification Rate (CCR) of 63%, sensitivity of 83%, positive predictive value (PPV) of 67%, specificity of 43%, and negative predictive value (NPV) of 65%. The Pearson’s correlation (R) between qualitative data for these two endpoints was 28% only.
Table 1.
Human vs. LLNA experimental outcomes for 109 compounds with defined chemical structure.
| HUMAN | ||||
|---|---|---|---|---|
|
|
||||
| Non-sensitizer | Sensitizer | Total | ||
|
|
||||
| LLNA | Non-sensitizer | 20 | 11 | 31 |
| Sensitizer | 26 | 52 | 78 | |
|
|
||||
| Total | 46 | 63 | 109 | |
All compounds were divided into several classes according to their potency as described above in the Methods section. As can be seen in Table 2, among 46 human non-sensitizers, 24 were identified as weak and two as strong sensitizers in LLNA. Ten human weak sensitizers were non-sensitizers in LLNA, while 29 compounds were weak and four compounds were strong sensitizers. One strong human sensitizer is non-sensitizer, while twelve are weak and seven are strong sensitizers in LLNA. The Pearson’s correlation (R) between the multiclass data was 43% only.
Table 2.
Human vs. LLNA multi-class annotation of experimental results for 109 compounds with defined chemical structure.
| HUMAN | |||||
|---|---|---|---|---|---|
|
|
|||||
| Non-sensitizer | Weak | Strong | Total | ||
|
|
|||||
| LLNA | Non-sensitizer | 20 | 10 | 1 | 31 |
| Weak | 24 | 29 | 12 | 65 | |
| Strong | 2 | 4 | 7 | 13 | |
|
|
|||||
| Total | 46 | 43 | 20 | 109 | |
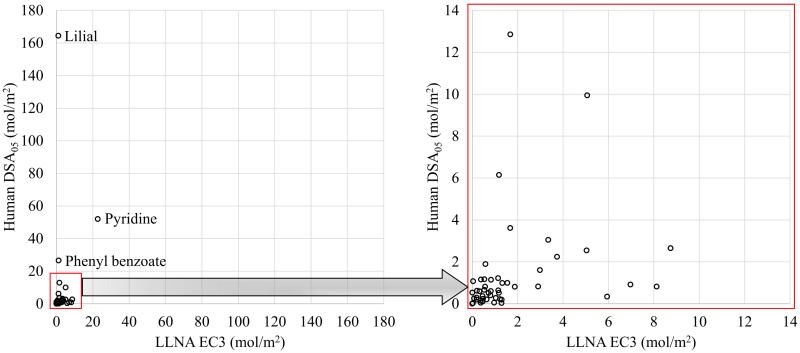
A relative potency was compared for all compounds that were sensitizers for both human and LLNA. List of figure captions
Figure 1 shows the distribution of sensitizers of human DSA05 vs. LLNA EC3, both expressed in (mol/m2). The coefficient of determination (R2) for the 52 sensitizers was very low (R2=0.05). Removing three outliers (lilial, pyridine, and phenyl benzoate), as shown in List of figure captions
Figure 1.
Human DSA05 vs. LLNA EC3 for all 53 sensitizers (y = 0.3111x + 0.8259; R² = 0.08) – Left Panel; and 50 sensitizers (y = 1.4383x + 2.961; R² = 0.05) remained after exclusion of three outliers (lilial, pyridine, and phenyl benzoate) – Right Panel.
Figure 1 B, provided no significant improvement (R2=0.08).
The high DSA05 values for lilial (164.48 mol/m2), pyridine (51.96 mol/m2), and phenyl benzoate (26.5 mol/m2) indicate that they are weak sensitizers.28 For lilial, the LOEL of 29,528 μg/cm2 (14.48 mol/m2) was also the NOEL in a separate test with fewer subjects. Thus, testing more subjects elicited an infrequent sensitization response for lilial. Two additional tests at lower concentrations, with NOEL of 3,750 μg/cm2 (1.84 mol/m2) and 4,125 μg/cm2 (2.02 mol/m2), yielded no sensitizing effects. ICCVAM (2011) reports one test each for pyridine and phenyl benzoate with LOELs of 34,483 μg/cm2 (43.65 mol/m2) and 9,448 μg/cm2 (4.77 mol/m2), respectively. The high DSA05 values for these substances reflect the fact that there are low sensitization rates in the tested populations at relatively high doses. This seems to be related to the high inter-individual variability of human tests.96
Cluster analysis
In this section, we present the detailed analysis of most interesting clusters of structurally similar chemicals. A complimentary analysis is available in the ESI (ESI-C-Clusters_and_Tables.docx). The dataset used in this study was small (109 chemicals) but structurally diverse. The main goal of the cluster analysis was to identify small groups of structurally similar compounds, assess whether all chemicals within each cluster have, as expected, similar skin sensitization effect, same mechanism of action, and compare the predictive performance of LLNA and QSAR models for these compounds. Compounds were clustered using ISIDA fragment descriptors and the hierarchical algorithm of ISIDA/Cluster as described in the Methods section. The resulting dendrogram and the associated distance matrix for Dataset A are shown in Figure 2. The dendrogram revealed 23 clusters containing 3-8 structurally similar compounds each. The information about all 23 clusters including chemical names, SMILES, associated QSAR predictions, and LLNA and human test outcomes is available in the ESI (ESI-D-Clusters.pdf). The summary of clusters showing the number of compounds correctly predicted by QSAR and LLNA with respect to the human data is shown in Table 4.
Figure 2.
Results of cluster analysis of 109 compounds with human skin sensitization data. (A) Heatmap and (B) dendrogram of the distance matrix, both colored according to structural similarity (blue/violet = similar; yellow/red = dissimilar).
Table 4.
Summary of cluster analysis showing the number of compounds correctly predicted by QSAR and LLNA when compared to the human data.
| Sensitizers | Non-sensitizers | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Cluster |
Human
data |
QSAR |
LLNA
data |
Human
data |
QSAR |
LLNA
data |
| 1 | 4 | 2 | 3 | 4 | 4 | 1 |
| 2 | 4 | 4 | 3 | 2 | 0 | 2 |
| 3 | 2 | 1 | 2 | 4 | 4 | 0 |
| 4 | 5 | 3 | 4 | 0 | - | - |
| 5 | 3 | 2 | 3 | 4 | 3 | 2 |
| 6 | 4 | 1 | 4 | 2 | 2 | 1 |
| 7 | 4 | 2 | 3 | 2 | 2 | 2 |
| 8 | 1 | 0 | 1 | 3 | 3 | 0 |
| 9 | 3 | 1 | 2 | 1 | 0 | 0 |
| 10 | 3 | 2 | 3 | 1 | 1 | 1 |
| 11 | 2 | 2 | 2 | 2 | 0 | 1 |
| 12 | 3 | 1 | 3 | 1 | 1 | 1 |
| 13 | 3 | 2 | 3 | 1 | 1 | 1 |
| 14 | 2 | 0 | 1 | 5 | 5 | 0 |
| 15 | 1 | 0 | 1 | 2 | 1 | 1 |
| 16 | 0 | - | - | 3 | 1 | 2 |
| 17 | 3 | 3 | 0 | 0 | - | - |
| 18 | 3 | 3 | 3 | 0 | - | - |
| 19 | 5 | 5 | 5 | 1 | 0 | 1 |
| 20 | 3 | 3 | 3 | 2 | 1 | 1 |
| 21 | 1 | 0 | 1 | 4 | 4 | 4 |
| 22 | 3 | 3 | 3 | 0 | - | - |
| 23 | 1 | 1 | 0 | 2 | 2 | 0 |
|
| ||||||
| Total | 63 | 41 | 53 | 46 | 35 | 21 |
Three clusters (Clusters 17, 14, and 9, see Table 5) were of the particular interest because all compounds in these clusters belonged to the same chemical class. The three aminoglycosides (neomycin, streptomycin, and kanamycin) present in Cluster 17 were human sensitizers accurately predicted by QSAR but they produced a false negative response in LLNA. Cluster 14 contained four damascone derivatives. All compounds in this cluster were non-sensitizers in humans except for δ-damascone, which was the only compound mispredicted by both our models and LLNA. All of its highly similar analogs (damascone, trans-α-damascone, and trans-β-damascone) were non-sensitizers, which explains why the QSAR model produced a false negative response for this compound. It should be noted that this chemical is described as “may cause sensitization by skin contact” (http://www.thegoodscentscompany.com/data/rw1006961.html) so its classification as a sensitizer may be inconclusive. Cluster 9 is formed by terpenes. It has been shown that terpenes are not allergenic themselves, but they oxidize when in contact with air to produce allergenic compounds.102 Thus, the potency of skin sensitization depends on how much oxidation impurities terpenes contain. This fact explains why QSAR models have confusing predictions for this chemical class.
Table 5.
Human data vs. LLNA results vs. external QSAR predictions within selected chemical clusters
LLNA also has a high disagreement with human data for non-sensitizers present in Cluster 1. All the non-sensitizers were correctly predicted by QSAR, although one of the non-sensitizers (resorcinol) has been recently labeled as a sensitizer. This compound is used at high levels in hair dyes and skin preparations, but is not considered dangerous, since it has low frequency of human sensitization. Benzyl alcohol (sensitizer) also had wrong concordance with LLNA and it was mispredicted by QSAR. Similar to outliers in previous sections, benzyl alcohol as a high DSA05 of 48.67 μg/cm2 (45.06 mol/m2) and a NOEL of 5,906 μg/cm2 (5.47 mol/m2). The very low skin sensitization potency may be the reason for wrong prediction by our QSAR model. LLNA also showed a poor concordance for non-sensitizers in Clusters 3 and 8 (mispredicted all four and three compounds, respectively). LLNA failed to predict all of the cinammyl derivatives in Cluster 8; α-amylcinnamyl alcohol was the only human sensitizer and the only compound mispredicted by QSAR.
LLNA has shown full concordant response with human data in six out of 23 clusters (Clusters 10, 12, 13, 18, 19, and 21). Out of these six clusters, only Cluster 10 (terpenoids) and Cluster 21 (short-chain alcohols), had more than three compounds belonging to the same chemical class. LLNA showed higher predictivity of human data then QSAR for all of these clusters except 18. LLNA also predicted all the three coumarins in Cluster 6 correctly, while QSAR failed to predict 3,4-dihydrocoumarin (human sensitizer). The non-sensitizer 6-methylcoumarin was correctly predicted by both QSAR and LLNA. In our previous study,62 the initial data source21 labeled coumarin as non-sensitizer. In a newer report used in the present work,28 a positive LLNA response was reported for coumarin, although the original reference103 showed that pure coumarin does not induce skin sensitization in mice and it is also well tolerated in humans. In our previous analysis,62 3,4-dihydrocoumarin was the only LLNA sensitizer within the cluster of coumarins. Further investigation suggested that 3,4-dihydrocoumarin is a prohapten, i.e., it requires a biotransformation to cause skin sensitization.104
QSAR and LLNA in the prediction of human skin sensitization
The statistical characteristics of QSAR models are summarized in Table 3. QSAR models were built using QNA, biological, and whole-molecule descriptors combined with radial basis function interpolation and self-consistent regression.88 We selected ten best models for each fold; these included only QNA and whole-molecule descriptors such as logP, topological length of the molecule, etc. No biological descriptors were selected. Consensus QSAR models presented higher predictivity of human data than LLNA. Although LLNA had both higher sensitivity (83% vs. 65%) and NPV (69% vs. 61%), the developed QSAR models outperformed LLNA in terms of overall accuracy, i.e., CCR (71% vs. 63%), PPV (79% vs. 67%), and specificity (76% vs. 43%).
Table 3.
Statistical characteristics of LLNA results vs. external QSAR predictions (5-fold external cross-validation) for predicting human skin sensitization.
| Model | CCR | Sensitivity | PPV | Specificity | NPV | Coverage |
|---|---|---|---|---|---|---|
| QSAR model | 0.71 | 0.65 | 0.79 | 0.76 | 0.61 | 1.00 |
| LLNA result | 0.63 | 0.83 | 0.67 | 0.43 | 0.65 | 1.00 |
|
| ||||||
| Combined (QSAR + LLNA) | 0.82 | 0.88 | 0.90 | 0.76 | 0.72 | 0.52 |
Despite many efforts in the last decade toward the development of alternative methods for evaluating skin sensitization potential of chemicals,62,105–110 LLNA is still considered essential for the evaluation of skin sensitization potency in compounds that lack human data.101 Although the results obtained both in a recent study29 and in this work have shown that in certain cases LLNA does not correlate well with human potency, it certainly contributes valuable information towards skin sensitization categorization.28 On the other hand, due to the prohibition of animal tests for cosmetics research in Europe, there is a strong need to develop alternative test methods.
Overall, the results presented in Table 3 show that our models could predict human skin sensitization with higher overall accuracy than LLNA. Notably, very high sensitivity but very low specificity implies that LLNA merely classifies the majority of molecules as sensitizers. Although the high sensitivity of this test is important from a point of view of regulatory precaution, it may ultimately lead to the withdrawal of many potentially useful and harmless compounds from the development because of false sensitization alerts. At the same time, one can see that developed QSAR models have high CCR to predict skin sensitization of new compounds lacking LLNA data or containing the chemotypes for which LLNA fails (see Cluster analysis section).
The ultimate goal of any method for evaluating skin sensitization is to provide an accurate assessment of the potential risk of a chemical with respect to human safety.111 Due to the high inter-individual variability of human tests, multiple sources of exposure must be evaluated for quantitative risk assessment of skin sensitization.96 It has been shown that consensus prediction typically affords models of higher accuracy.91,112,113 Thus, we decided to combine QSAR predictions and LLNA assessment to predict human skin sensitization. Under this scenario, only cases when QSAR and LLNA agreed with each other were considered; cases when QSAR and LLNA had contradictory outcomes were treated as inconclusive and were discarded, leading to a coverage of 52% (see Table 3). Although the combined model could make predictions for only about half of all compounds, the prediction accuracy was much higher, i.e., CCR increased from 71 to 82%.
These results reveal that concordant predictions by LLNA and QSAR of the human skin sensitization are of the highest confidence, but at the expense of the reduced coverage, i.e., the models could not provide estimates for all compounds of interest. Nevertheless, our studies suggest that for the most reliable but conservative assessment, the consensus approach should be used preferably. However, when the entire chemical dataset of interest needs to be assessed for potential human skin sensitization effects, the QSAR models built with the currently available human data should be chosen over LLNA for the higher accuracy. Our models could be used in combination with other strategies, as proposed earlier.40,76 Considering the small size of the data, it was not feasible to build local models based on the mechanism of action. Nevertheless, it has been shown that, usually, there are no improvements in model accuracy when using local over global QSAR models.114,115 Thus, the QSAR model presented here affords a powerful alternative to current animal and in vitro approaches for assessing skin sensitization effect of chemicals in human.
Structural optimization of chemicals
Cheminformatics analyses can be used to determine structural features associated with toxicity in order to design new compounds with improved properties, e.g., less toxic.66 In the spirit of molecular design oriented by green chemistry, we illustrate herein an example of structural optimization for reduced toxicity, using experimental data and predictions derived from the developed models (Figure 3). Starting from phenyl benzoate, a known human skin sensitizer, several structurally similar compounds, most likely sharing the same properties of interest, could be revealed by clustering, and then evaluated for skin sensitization potential using QSAR. As can be seen in Figure 3, all chosen structurally similar chemicals were predicted to be non-sensitizers and thus could serve as a potential alternative to phenyl benzoate. In this example, three compounds similar to phenyl benzoate have human and LLNA data (benzyl salicylate, benzyl cinnamate, and benzyl benzoate). All three compounds are non-sensitizers in humans and yet they show false positive results in LLNA. Despite lacking both human and LLNA data, the remaining compounds shown in Figure 3 were predicted as non-sensitizers based on structurally similarity and QSAR modeling results. Next, through structural interpretation of the QSAR model and clustering, one can propose that the toxic potential of phenyl benzoate could be reduced through an increase of the linker length to the benzene rings and/or an addition of a hydroxyl group in the ortho-position on the benzene ring. Phenyl benzoate acts as an acyl transfer agent, reacting with the sulfur of the cysteine residues. The proposed modifications may result in less reactive compounds because they lead to a reduced partial charge on the carbon atom of the carboxylate group, which hinders the expulsion of the oxygen from the tetrahedral intermediate.116 This hypothesis is supported by the analysis of the atomic contributions derived by the interpretation of the developed models (Figure 4). These contributions are illustrated by different colors: red – increasing the sensitization potential; blue – decreasing the sensitization potential; and green – no significant contribution. Green arrows represent higher confidence of the prediction. This higher confidence is caused by the absence of fragments that increase the sensitization potential. Other proposed compounds contain such fragments; however, their collective contribution is overpowered by the negative contributions of other fragments resulting in the absence of the sensitization potential for respective molecules. As seen from Figure 4, the carbon of the carboxylate is slightly darker for benzyl salicylate and benzyl benzoate, and much greener for benzyl cinnamate, indicating that this atom was predicted to have lower influence on the skin sensitization potential than the same carbon of phenyl benzoate. In other compounds, although the carbon of carboxylate was predicted to positively contribute to the activity, the contributions of the structural neighborhood into the sensitization potential is much lower than in phenyl benzoate. This analysis provides another evidence that structural alerts alone are not sufficient to flag a compound as toxic.66 This information could be used for designing new compounds preserving the specific desired properties of phenyl benzoate but removing its skin sensitization action.
Figure 3.
Example of structural transformation of human skin sensitizer phenyl benzoate into various non-sensitizers using developed models.
Figure 4.
Example of interpretation of QSAR models visualized as color-coding according to atom contributions in change of sensitization potential: red – sensitization increase; blue – sensitization decrease; green – no significant contribution. Green arrows represent higher confidence of the prediction. This information helps to guide structural transformation of human skin sensitizer phenyl benzoate into various non-sensitizers (see Figure 3).
Virtual screening of ingredients found in cosmetic products
We applied the developed QSAR models to the European Commission database for information on cosmetic substances and ingredients (CosIng) in order to identify possible skin sensitizers. Using the QSAR model developed with GUSAR, 1061 compounds were predicted as sensitizers. Since CAS numbers of chemicals in the CosIng database may correspond to mixtures, we regenerated all possible names and CAS numbers for the curated database. Using ChemoText,94 we queried three MeSH terms associated with skin sensitization (see Methods) for instances when CosIng "hits" predicted by QSAR models were mentioned in the literature. We confirmed 17 chemicals as skin sensitizers (see Table S3 in ESI). It should be noted that not all chemicals/chemical names are indexed in MEDLINE. Therefore, many possible "hits" could not be verified using ChemoText. The list containing all the predicted sensitizers is available in Supplemental Materials.
CONCLUSIONS
We have collected and curated the largest publicly available dataset of 135 substances (109 defined chemical structures) tested in both LLNA and human assays. We have conducted a variability and concordance analysis between human and LLNA data using substances with multiple test results. This analysis showed that human data have higher variability than the LLNA data and that the concordance between human and LLNA was relatively low (CCR = 28-43%; see Table 1 and Table 2).
We have developed validated and externally predictive QSAR models for skin sensitization using the available human data and benchmarked their performance in comparison with LLNA results. Our skin sensitization models, compared to LLNA, showed lower sensitivity (65% vs. 83%), but higher CCR (71% vs. 63%) and much higher specificity (76% vs. 43%). The analysis of positive prediction values showed that our models had a higher probability (79 % vs. 67%) to predict new sensitizers correctly. More importantly, the conservative combination of QSAR and LLNA (i.e., only for compounds with concordant predictions by both methods) outperformed each of the contributing approaches by up to 11% as evaluated by CCR or PPV but at the expense of significantly reduced (by nearly 50%) chemical coverage. Thus, our results confirm the importance of combining experimental and computational methods for most accurate conservative assessment of chemical safety for humans but also suggest that QSAR models built with human data afford significantly higher accuracy of predicting human effects than murine LLNA.
We have employed hierarchical clustering to examine if similar chemicals also have similar human sensitization data as generally expected. Indeed, for some clusters, the expected trend was observed, but there were also inconsistencies, which could be explained by inter-individual differences in the human data. For three largest clusters of similar compounds (aminoglycosides, damascones, and terpenes), LLNA showed poor concordance with human data, whereas QSAR predictions were highly accurate (Table 4). Both LLNA and QSAR showed almost random prediction accuracy for terpenes, which could be explained by the known facts that the skin effects are caused by metabolites of these compounds rather than by compounds themselves.
In summary, the analysis conducted in this study was important to benchmark advantages and drawbacks of both LLNA and QSAR models to predict the human skin sensitization effects of chemicals with the available data. Our models could be employed for identifying putative sensitizers as the first step of a multi-tiered testing strategy. For instance, virtual screening of the CosIng database including 3,964 chemicals with the QSAR models built in this study identified 1061 potential skin sensitizers that may be candidates for targeted testing. The ChemoText search of additional sensitization data in the scientific literature identified 17 new sensitizers that were absent in our dataset. All were predicted as sensitizers by our QSAR model serving as additional proof of model accuracy. Note that LLNA data for the same compounds were unavailable, which is another illustration of the advantages of QSAR models, that require the knowledge of chemical structure only, over LLNA. Further efforts are needed to identify additional high-quality human data by both more pervasive literature and digital sources search of substances with known effect on human or by additional experimental testing. Rebuilding our models with additional data will increase their predictive power and reliability. Summarizing, our findings provide a strong argument in favor of using in silico models as a strong alternative to both animal and human testing for skin sensitization. All curated datasets, predictions, chemical clusters, and the models developed in this study have been made publicly available in the ESI. GUSAR is a proprietary software; thus, one should purchase it in order to develop or execute our model. However, we will make and share predictions free of charge for any compound of interest upon request.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by NIH (GM096967), FAPEG (grant 201310267001095), and CNPq (grant 400760/2014-2). J.S. is supported the NIEHS, NIH under Contract No. HHSN273201500010C to ILS in support of NICEATM. ILS staff provide technical support for NICEATM, but do not represent NIEHS, NTP, or the official positions of any federal agency. This article may be the work product of an employee or group of employees of the NIEHS, NIH, or other organizations; however, the statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NIEHS, NIH, the United States government, or other organizations. The use of commercial product names is for comparative purposes only and does not constitute endorsement by any of the authors, organizations, or agencies. The authors express sincere gratitude to Drs. Vladimir Poroikov, Dmitri Filimonov, and Alexey Zakharov for providing the GUSAR Software.
Footnotes
CONFLICT OF INTERESTS
The authors declare no actual or potential conflict of interests.
REFERENCES
- 1.Macan J, Rimac D, Kežić S, Varnai VM. Dermatology. 2013;227:321–329. doi: 10.1159/000354763. [DOI] [PubMed] [Google Scholar]
- 2.Hennino A, Vocanson M, Chavagnac C, Saint-Mezard P, Dubois B, Kaiserlian D, Nicolas J. An. Bras. Dermatol. 2005;80:335–347. doi: 10.1586/1744666X.1.1.75. [DOI] [PubMed] [Google Scholar]
- 3.Schulte PA, McKernan LT, Heidel DS, Okun AH, Dotson GS, Lentz TJ, Geraci CL, Heckel PE, Branche CM. Environ. Heal. 2013;12:31. doi: 10.1186/1476-069X-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins T. Green Chem. 2003;5:G51–G52. [Google Scholar]
- 5.OECD The adverse outcome pathway for skin sensitisation initiated by covalent binding to proteins part 1: scientific evidence, OECD Enviroment, Health and Safaty Publications. 2012 http://search.oecd.org/officialdocuments/displaydocumentpdf/?cote=env/jm/mono(2012)10/part1&doclanguage=en, (accessed 8 February 2016)
- 6.Fyhrquist N, Lehto E, Lauerma A. Curr. Opin. Allergy Clin. Immunol. 2014;14:430–435. doi: 10.1097/ACI.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 7.Marzulli FN, Maibach HI. Contact Dermatitis. 1976;2:1–17. doi: 10.1111/j.1600-0536.1976.tb02972.x. [DOI] [PubMed] [Google Scholar]
- 8.Kligman AM. J. Invest. Dermatol. 1966;47:369–374. doi: 10.1038/jid.1966.158. [DOI] [PubMed] [Google Scholar]
- 9.Griffith JF. Toxicol. Appl. Pharmacol. 1969;14:90–102. doi: 10.1016/0041-008x(69)90101-x. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz L, Peck SM. J. Natl. Assoc. Chirop. 1946;36:7–16. [PubMed] [Google Scholar]
- 11.Politano VT, Api AM. Regul. Toxicol. Pharmacol. 2008;52:35–38. doi: 10.1016/j.yrtph.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Kligman AM. J. Invest. Dermatol. 1966;47:393–409. doi: 10.1038/jid.1966.160. [DOI] [PubMed] [Google Scholar]
- 13.Kligman AM, Epstein W. Contact Dermatitis. 1975;1:231–239. doi: 10.1111/j.1600-0536.1975.tb05389.x. [DOI] [PubMed] [Google Scholar]
- 14.Basketter DA. Cutan. Ocul. Toxicol. 2009;28:49–53. doi: 10.1080/15569520902938032. [DOI] [PubMed] [Google Scholar]
- 15.Kimber I, Basketter DA, Berthold K, Butler M, Garrigue JL, Lea L, Newsome C, Roggeband R, Steiling W, Stropp G, Waterman S, Wiemann C. Toxicol. Sci. 2001;59:198–208. doi: 10.1093/toxsci/59.2.198. [DOI] [PubMed] [Google Scholar]
- 16.Magnusson B, Kligman AM. J. Invest. Dermatol. 1969;52:268–276. doi: 10.1038/jid.1969.42. [DOI] [PubMed] [Google Scholar]
- 17.Basketter DA, Evans P, Fielder RJ, Gerberick GF, Dearman RJ, Kimber I. Food Chem. Toxicol. 2002;40:593–598. doi: 10.1016/s0278-6915(01)00130-2. [DOI] [PubMed] [Google Scholar]
- 18.European Union Off. J. Eur. Union. 2007:3–280. [Google Scholar]
- 19.Cockshott A, Evans P, Ryan CA, Gerberick GF, Betts CJ, Dearman RJ, Kimber I, Basketter DA. Hum. Exp. Toxicol. 2006;25:387–394. doi: 10.1191/0960327106ht640oa. [DOI] [PubMed] [Google Scholar]
- 20.EPA Health Effects Test Guidelines: OPPTS 870.2600 Skin Sensitization. 2003 https://www.regulations.gov/#!documentDetail;D=EPA-HQ-OPPT-2009-0156-0008, (accessed 9 June 2016)
- 21.ICCVAM The reduced murine local lymph node assay: an alternative test method using fewer animals to assess the allergic contact dermatitis potential of chemicals and products. 2009 http://ntp.niehs.nih.gov/iccvam/docs/immunotox_docs/LLNA-LD/TMER.pdf, (accessed 15 July 2015)
- 22.Roberts DW. Regul. Toxicol. Pharmacol. 2015;71:437–443. doi: 10.1016/j.yrtph.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Basketter DA, Balikie L, Dearman RJ, Kimber I, Ryan CA, Gerberick GF, Harvey P, Evans P, White IR, Rycroft RJ. Contact Dermatitis. 2000;42:344–348. doi: 10.1034/j.1600-0536.2000.042006344.x. [DOI] [PubMed] [Google Scholar]
- 24.Ryan CA, Gerberick GF, Cruse LW, Basketter DA, Lea L, Blaikie L, Dearman RJ, Warbrick EV, Kimber I. Contact Dermatitis. 2000;43:95–102. doi: 10.1034/j.1600-0536.2000.043002095.x. [DOI] [PubMed] [Google Scholar]
- 25.Gerberick GF, Robinson MK, Ryan C. a., Dearman RJ, Kimber I, Basketter D. a., Wright Z, Marks JG. Am. J. contact Dermat. 2001;12:156–161. doi: 10.1053/ajcd.2001.23926. [DOI] [PubMed] [Google Scholar]
- 26.Griem P, Goebel C, Scheffler H. Regul. Toxicol. Pharmacol. 2003;38:269–290. doi: 10.1016/j.yrtph.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Schneider K, Akkan Z. Regul. Toxicol. Pharmacol. 2004;39:245–255. doi: 10.1016/j.yrtph.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.ICCVAM Usefulness and limitations of the murine local lymph node assay for potency categorization of chemicals causing allergic contact dermatitis in humans. 2011 http://ntp.niehs.nih.gov/pubhealth/evalatm/test-method-evaluations/immunotoxicity/llna-potency/tmer/index.html, (accessed 9 February 2015)
- 29.Api AM, Basketter D, Lalko J. Cutan. Ocul. Toxicol. 2014;34:1–5. doi: 10.3109/15569527.2014.979425. [DOI] [PubMed] [Google Scholar]
- 30.European Commission On the animal testing and marketing ban and on the state of play in relation to alternative methods in the field of cosmetics, Communication from the commision to the european parliament and the council. 2013 http://ec.europa.eu/consumers/sectors/cosmetics/files/pdf/animal_testing/com_at_2013_en.pdf, (accessed 9 February 2016)
- 31.Hartung T. ALTEX. 2013;30:275–291. doi: 10.14573/altex.2013.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey J, Thew M, Balls M. Altern. Lab. Anim. 2014;42:181–199. doi: 10.1177/026119291404200306. [DOI] [PubMed] [Google Scholar]
- 33.Mangipudy R, Burkhardt J, Kadambi VJ. Regul. Toxicol. Pharmacol. 2014;70:439–441. doi: 10.1016/j.yrtph.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Adler S, Basketter D, Creton S, Pelkonen O, van Benthem J, Zuang V, Andersen KE, Angers-Loustau A, Aptula A, Bal-Price A, Benfenati E, Bernauer U, Bessems J, Bois FY, Boobis A, Brandon E, Bremer S, Broschard T, Casati S, Coecke S, Corvi R, Cronin M, Daston G, Dekant W, Felter S, Grignard E, Gundert-Remy U, Heinonen T, Kimber I, Kleinjans J, Komulainen H, Kreiling R, Kreysa J, Leite SB, Loizou G, Maxwell G, Mazzatorta P, Munn S, Pfuhler S, Phrakonkham P, Piersma A, Poth A, Prieto P, Repetto G, Rogiers V, Schoeters G, Schwarz M, Serafimova R, Tähti H, Testai E, van Delft J, van Loveren H, Vinken M, Worth A, Zaldivar J-M. Arch. Toxicol. 2011;85:367–485. doi: 10.1007/s00204-011-0693-2. [DOI] [PubMed] [Google Scholar]
- 35.Langley G, Austin CP, Balapure AK, Birnbaum LS, Bucher JR, Fentem J, Fitzpatrick SC, Fowle JR, Kavlock RJ, Kitano H, Lidbury BA, Muotri AR, Peng S-Q, Sakharov D, Seidle T, Trez T, Tonevitsky A, van de Stolpe A, Whelan M, Willett C. Environ. Health Perspect. 2015;123:268–272. doi: 10.1289/ehp.1510345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop PL, Manuppello JR, Willett CE, Sandler JT. Environ. Health Perspect. 2012:1631–1639. doi: 10.1289/ehp.1104666. C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reisinger K, Hoffmann S, Alépée N, Ashikaga T, Barroso J, Elcombe C, Gellatly N, Galbiati V, Gibbs S, Groux H, Hibatallah J, Keller D, Kern P, Klaric M, Kolle S, Kuehnl J, Lambrechts N, Lindstedt M, Millet M, Martinozzi-Teissier S, Natsch A, Petersohn D, Pike I, Sakaguchi H, Schepky A, Tailhardat M, Templier M, van Vliet E, Maxwell G. Toxicol. Vitr. 2015;29:259–270. doi: 10.1016/j.tiv.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Casati S, Aschberger K, Asturiol D, Basketter D, Dimitrov S, Dumont C, Karlberg A-T, Lepoittevin J-P, Patlewicz G, Roberts DW, Worth A. Ability of non-animal methods for skin sensitisation to detect pre- and pro-haptens: Report and recommendations of an EURL ECVAM expert meeting, EUR 27752 EN. 2016 [Google Scholar]
- 39.MacKay C, Davies M, Summerfield V, Maxwell G. ALTEX. 2013;30:473–486. doi: 10.14573/altex.2013.4.473. [DOI] [PubMed] [Google Scholar]
- 40.Strickland J, Zang Q, Kleinstreuer N, Paris M, Lehmann DM, Choksi N, Matheson J, Jacobs A, Lowit A, Allen D, Casey W. J. Appl. Toxicol. 2016 doi: 10.1002/jat.3281. Epub ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz’min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Gasteiger J, Richard A, Tropsha A. J. Med. Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dearden JC. Int. J. Quant. Struct. Relationships. 2016;1:1–44. [Google Scholar]
- 43.Estrada E, Patlewicz G, Chamberlain M, Basketter D, Larbey S. Chem. Res. Toxicol. 2003;16:1226–1235. doi: 10.1021/tx034093k. [DOI] [PubMed] [Google Scholar]
- 44.Fedorowicz A, Zheng L, Singh H, Demchuk E. Int. J. Mol. Sci. 2004;5:56–66. [Google Scholar]
- 45.Fedorowicz A, Singh H, Soderholm S, Demchuk E. Chem. Res. Toxicol. 2005;18:954–969. doi: 10.1021/tx0497806. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Fedorowicz A, Singh H, Soderholm SC. J. Chem. Inf. Model. 2005;45:952–964. doi: 10.1021/ci050049u. [DOI] [PubMed] [Google Scholar]
- 47.Miller MD, Yourtee DM, Glaros AG, Chappelow CC, Eick JD, Holder AJ. J. Chem. Inf. Model. 2005;45:924–929. doi: 10.1021/ci050018z. [DOI] [PubMed] [Google Scholar]
- 48.Roberts DW, Patlewicz G, Dimitrov SD, Low LK, Aptula AO, Kern PS, Dimitrova GD, Comber MIH, Phillips RD, Niemelä J, Madsen C, Wedebye EB, Bailey PT, Mekenyan OG. Chem. Res. Toxicol. 2007;20:1321–1330. doi: 10.1021/tx700169w. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Tseng YJ, Pan D, Liu J, Kern PS, Gerberick GF, Hopfinger AJ. Chem. Res. Toxicol. 2007;20:114–128. doi: 10.1021/tx6002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan H, Huang J, Cao C. Int. J. Mol. Sci. 2009;10:3237–3254. doi: 10.3390/ijms10073237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golla S, Madihally S, Robinson RL, Gasem KAM. Toxicol. In Vitro. 2009;23:454–465. doi: 10.1016/j.tiv.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 52.Chaudhry Q, Piclin N, Cotterill J, Pintore M, Price NR, Chrétien JR, Roncaglioni A. Chem. Cent. J. 2010;4(Suppl 1):S5. doi: 10.1186/1752-153X-4-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gunturi SB, Theerthala SS, Patel NK, Bahl J, Narayanan R. SAR QSAR Environ. Res. 2010;21:305–335. doi: 10.1080/10629361003773955. [DOI] [PubMed] [Google Scholar]
- 54.Lu J, Zheng M, Wang Y, Shen Q, Luo X, Jiang H, Chen K. J. Comput. Aided. Mol. Des. 2011;25:885–893. doi: 10.1007/s10822-011-9472-7. [DOI] [PubMed] [Google Scholar]
- 55.Nandy A, Kar S, Roy K. Mol. Simul. 2013;39:432–441. [Google Scholar]
- 56.Nandy A, Kar S, Roy K. SAR QSAR Environ. Res. 2013;24:1009–1023. doi: 10.1080/1062936X.2013.821422. [DOI] [PubMed] [Google Scholar]
- 57.Nandy A, Kar S, Roy K. Mol. Simul. 2014;40:261–274. [Google Scholar]
- 58.Asturiol D, Casati S, Worth A. Toxicol. In Vitro. 2016;36:197–209. doi: 10.1016/j.tiv.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Roberts DW, Aptula AO, Patlewicz G. Chem. Res. Toxicol. 2006;19:1228–1233. doi: 10.1021/tx060102o. [DOI] [PubMed] [Google Scholar]
- 60.Dearden JC, Hewitt M, Roberts DW, Enoch SJ, Rowe PH, Przybylak KR, Vaughan-Williams GD, Smith ML, Pillai GG, Katritzky AR. Chem. Res. Toxicol. 2015;28:1975–1986. doi: 10.1021/acs.chemrestox.5b00197. [DOI] [PubMed] [Google Scholar]
- 61.Kostal J, Voutchkova-Kostal A. Chem. Res. Toxicol. 2016;29:58–64. doi: 10.1021/acs.chemrestox.5b00392. [DOI] [PubMed] [Google Scholar]
- 62.Alves VM, Muratov EN, Fourches D, Strickland J, Kleinstreuer N, Andrade CH, Tropsha A. Toxicol. Appl. Pharmacol. 2015;284:262–272. doi: 10.1016/j.taap.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alves VM, Muratov EN, Fourches D, Strickland J, Kleinstreuer N, Andrade CH, Tropsha A. Toxicol. Appl. Pharmacol. 2015;284:273–280. doi: 10.1016/j.taap.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tropsha A. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 65.Urbisch D, Honarvar N, Kolle SN, Mehling A, Ramirez T, Teubner W, Landsiedel R. Toxicol. Vitr. 2016;34:194–203. doi: 10.1016/j.tiv.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 66.Alves VM, Muratov EN, Capuzzi SJ, Politi R, Low Y, Braga RC, Zakharov AV, Sedykh A, Mokshyna E, Farag S, Andrade CH, Kuz’min VE, Fourches D, Tropsha A. Green Chem. 2016;18:4348–4360. doi: 10.1039/C6GC01492E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson SE, Siegel PD, Meade BJ. J. Allergy. 2011;2011:424203. doi: 10.1155/2011/424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anastas P, Eghbali N. Chem. Soc. Rev. 2010;39:301–312. doi: 10.1039/b918763b. [DOI] [PubMed] [Google Scholar]
- 69.Zimmerman JB, Anastas PT, Miller GW. Toxicol. Sci. 2014;141:4–5. doi: 10.1093/toxsci/kfu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunn PJ. Chem. Soc. Rev. 2012;41:1452–1461. doi: 10.1039/c1cs15041c. [DOI] [PubMed] [Google Scholar]
- 71.DeVito SC. Green Chem. 2016;18:4332–4347. [Google Scholar]
- 72.Maertens A, Anastas N, Spencer PJ, Stephens M, Goldberg A, Hartung T. ALTEX. 2014;31:243–249. doi: 10.14573/altex.1406181. [DOI] [PubMed] [Google Scholar]
- 73.Naven R, Louise-May S. Hum. Exp. Toxicol. 2015;34:1304–1309. doi: 10.1177/0960327115605440. [DOI] [PubMed] [Google Scholar]
- 74.Gramatica P, Cassani S, Sangion A. Green Chem. 2016 [Google Scholar]
- 75.Melnikov F, Kostal J, Voutchkova-Kostal A, Zimmerman JB, Anastas PT. Green Chem. 2016 [Google Scholar]
- 76.Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts DW, Mekenyan O. Regul. Toxicol. Pharmacol. 2014;69:529–545. doi: 10.1016/j.yrtph.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 77.ICCVAM Evaluations of non-animal skin sensitization test methods and testing strategies. 2013 http://ntp.niehs.nih.gov/go/40500, (accessed 15 July 2015)
- 78.Fourches D, Muratov E, Tropsha A. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fourches D, Muratov E, Tropsha A. Nat. Chem. Biol. 2015;11:535–535. doi: 10.1038/nchembio.1881. [DOI] [PubMed] [Google Scholar]
- 80.Fourches D, Muratov E, Tropsha A. J. Chem. Inf. Model. 2016;56:1243–1252. doi: 10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov’ev V, Hoonakker F, Tetko I, Marcou G. Curr. Comput. Aided-Drug Des. 2008;4:191–198. [Google Scholar]
- 82.Kuz’min VE, Artemenko AG, Muratov EN. J. Comput. Aided. Mol. Des. 2008;22:403–421. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 83.Filimonov DA, Zakharov AV, Lagunin AA, Poroikov VV. SAR QSAR Environ. Res. 2009;20:679–709. doi: 10.1080/10629360903438370. [DOI] [PubMed] [Google Scholar]
- 84.Lagunin A, Zakharov A, Filimonov D, Poroikov V. Mol. Inform. 2011;30:241–250. doi: 10.1002/minf.201000151. [DOI] [PubMed] [Google Scholar]
- 85.Lagunin A, Filimonov D, Zakharov A, Xie W, Huang Y, Zhu F, Shen T, Yao J, Poroikov V. QSAR Comb. Sci. 2009;28:806–810. [Google Scholar]
- 86.Zakharov AV, Varlamova EV, Lagunin AA, Dmitriev AV, Muratov EN, Fourches D, Kuz’min VE, Poroikov VV, Tropsha A, Nicklaus MC. Mol. Pharm. 2016;13:545–556. doi: 10.1021/acs.molpharmaceut.5b00762. [DOI] [PubMed] [Google Scholar]
- 87.Zakharov AV, Lagunin AA, Filimonov DA, Poroikov VV. Chem. Res. Toxicol. 2012;25:2378–2385. doi: 10.1021/tx300247r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zakharov A, Peach ML, Sitzmann M, Nicklaus MC. J. Chem. Inf. Model. 2014 doi: 10.1021/ci400737s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Filimonov DAA, Zakharov AVV, Lagunin AAA, Poroikov VVV. SAR QSAR Environ. Res. 2009;20:679–709. doi: 10.1080/10629360903438370. [DOI] [PubMed] [Google Scholar]
- 90.Braga RC, Alves VM, Silva MFB, Muratov E, Fourches D, Tropsha A, Andrade CH. Curr. Top. Med. Chem. 2014;14:1399–1415. doi: 10.2174/1568026614666140506124442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuz’min VE, Muratov EN, Artemenko AG, Varlamova EV, Gorb L, Wang J, Leszczynski J. QSAR Comb. Sci. 2009;28:664–677. [Google Scholar]
- 92.Gadaleta D, Mangiatordi GF, Catto M, Carotti A, Nicolotti O. Int. J. Quant. Struct. Relationships. 2016;1:45–63. [Google Scholar]
- 93.Mathea M, Klingspohn W, Baumann K. Mol. Inform. 2016;35:160–180. doi: 10.1002/minf.201501019. [DOI] [PubMed] [Google Scholar]
- 94.Baker NC, Hemminger BM. J. Biomed. Inform. 2010;43:510–519. doi: 10.1016/j.jbi.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burton GA, Chapman PM, Smith EP. Hum. Ecol. Risk Assess. An Int. J. 2002;8:1657–1673. [Google Scholar]
- 96.Basketter D, Safford B. Regul. Toxicol. Pharmacol. 2016;74:105–116. doi: 10.1016/j.yrtph.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 97.Scholes EW, Basketter DA, Sarll AE, Kimber I, Evans CD, Miller K, Robbins MC, Harrison PT, Waite SJ. J. Appl. Toxicol. 1992;12:217–222. doi: 10.1002/jat.2550120312. [DOI] [PubMed] [Google Scholar]
- 98.ICCVAM and NICEATM The murine local lymph node assay: a test method for assessing the allergic contact dermatitis potential of chemicals/compounds. 1999 http://ntp.niehs.nih.gov/iccvam/docs/immunotox_docs/llna/llnarep.pdf, (accessed 15 July 2015)
- 99.Hoffmann S. ALTEX. 2015;32:379–383. doi: 10.14573/altex.1505051. [DOI] [PubMed] [Google Scholar]
- 100.Dumont C, Barroso J, Matys I, Worth A, Casati S. Toxicol. Vitr. 2016;34:220–228. doi: 10.1016/j.tiv.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 101.Basketter DA, Alépée N, Ashikaga T, Barroso J, Gilmour N, Goebel C, Hibatallah J, Hoffmann S, Kern P, Martinozzi-Teissier S, Maxwell G, Reisinger K, Sakaguchi H, Schepky A, Tailhardat M, Templier M. Dermat. contact, atopic, Occup. drug. 2014;25:11–21. doi: 10.1097/DER.0000000000000003. [DOI] [PubMed] [Google Scholar]
- 102.Matura M, Sköld M, Börje A, Andersen KE, Bruze M, Frosch P, Goossens A, Johansen JD, Svedman C, White IR, Karlberg A-T. Contact Dermatitis. 2005;52:320–328. doi: 10.1111/j.0105-1873.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 103.Vocanson M, Goujon C, Chabeau G, Castelain M, Valeyrie M, Floc’h F, Maliverney C, Gard A, Nicolas JF. Int. Arch. Allergy Immunol. 2006;140:231–238. doi: 10.1159/000093248. [DOI] [PubMed] [Google Scholar]
- 104.Gerberick GF, Vassallo JD, Bailey RE, Chaney JG, Morrall SW, Lepoittevin J-P. Toxicol. Sci. 2004;81:332–343. doi: 10.1093/toxsci/kfh213. [DOI] [PubMed] [Google Scholar]
- 105.Ramirez T, Mehling A, Kolle SN, Wruck CJ, Teubner W, Eltze T, Aumann A, Urbisch D, Van Ravenzwaay B, Landsiedel R. Toxicol. Vitr. 2014;28:1482–1497. doi: 10.1016/j.tiv.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 106.van der Veen JW, Rorije E, Emter R, Natsch A, van Loveren H, Ezendam J. Regul. Toxicol. Pharmacol. 2014;69:371–379. doi: 10.1016/j.yrtph.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 107.Urbisch D, Mehling A, Guth K, Ramirez T, Honarvar N, Kolle S, Landsiedel R, Jaworska J, Kern PS, Gerberick F, Natsch A, Emter R, Ashikaga T, Miyazawa M, Sakaguchi H. Regul. Toxicol. Pharmacol. 2014;71:337–351. doi: 10.1016/j.yrtph.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 108.EURL ECVAM Recommendation on the Direct Peptide Reactivity Assay (DPRA) 2013 https://eurl-ecvam.jrc.ec.europa.eu/eurl-ecvam-recommendations/eurl-ecvam-recommendation-on-the-direct-peptide-reactivity-assay-dpra, (accessed 29 January 2015)
- 109.Ashikaga T, Yoshida Y, Hirota M, Yoneyama K, Itagaki H, Sakaguchi H, Miyazawa M, Ito Y, Suzuki H, Toyoda H. Toxicol. In Vitro. 2006;20:767–773. doi: 10.1016/j.tiv.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 110.Ade N, Martinozzi-Teissier S, Pallardy M, Rousset F. J. Immunotoxicol. 2006;3:189–197. doi: 10.1080/15476910600978038. [DOI] [PubMed] [Google Scholar]
- 111.Basketter D, White I, McFadden J, Kimber I. Hum. Exp. Toxicol. 2015;34:1222–1230. doi: 10.1177/0960327115601760. [DOI] [PubMed] [Google Scholar]
- 112.Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko IV. J. Chem. Inf. Model. 2008;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- 113.Wang XS, Tang H, Golbraikh A, Tropsha A. J. Chem. Inf. Model. 2008;48:997–1013. doi: 10.1021/ci700404c. [DOI] [PubMed] [Google Scholar]
- 114.Sheridan RP. J. Chem. Inf. Model. 2014;54:1083–1092. doi: 10.1021/ci500084w. [DOI] [PubMed] [Google Scholar]
- 115.Helgee EA, Carlsson L, Boyer S, Norinder U. J. Chem. Inf. Model. 2010;50:677–689. doi: 10.1021/ci900471e. [DOI] [PubMed] [Google Scholar]
- 116.Roberts DW, Patlewicz G, Kern PS, Gerberick F, Kimber I, Dearman RJ, Ryan C. a., Basketter D. a., Aptula AO. Chem. Res. Toxicol. 2007;20:1019–1030. doi: 10.1021/tx700024w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.