Abstract
Neuroblastoma (NB) is one of the most frequently occurring cancerous tumors in children. The current grading evaluations for patients with this disease require pathologists to identify certain morphological characteristics with microscopic examinations of tumor tissues. Thanks to the advent of modern digital scanners, it is now feasible to scan cross-section tissue specimens and acquire whole-slide digital images. As a result, computerized analysis of these images can generate key quantifiable parameters and assist pathologists with grading evaluations. In this study, image analysis techniques are applied to histological images of haematoxylin and eosin (H&E) stained slides for identifying image regions associated with different pathological components. Texture features derived from segmented components of tissues are extracted and processed by an automated classifier group trained with sample images with different grades of neuroblastic differentiation in a multi-resolution framework. The trained classification system is tested on 33 whole-slide tumor images. The resulting whole-slide classification accuracy produced by the computerized system is 87.88%. Therefore, the developed system is a promising tool to facilitate grading whole-slide images of NB biopsies with high throughput.
Keywords: Quantitative image analysis, Microscopy images, Neuroblastoma prognosis, Grade of differentiation, Multi-resolution pathological image analysis, Machine learning
1. Introduction
Peripheral neuroblastic tumors (pNTs) are a group of embryonal tumors of the sympathetic nervous system, and include neuroblastoma (NB), ganglioneuroblastoma, and ganglioneuroma category [1]. Each year, more than 600 children and adolescents are diagnosed with pNTs in the United States, and it comprises about 8–10% of all childhood cancers [1,2]. Of all the cancer categories in pNTs, NB, com-posed of neoplastic neuroblasts in various maturation grades with no or limited Schwannian stromal development, is the most common tumor that affects children ranging from newly born infants to teenagers.
According to the International Neuroblastoma Pathology Classification System (the Shimada system), NB can be further classified into three categories, namely undifferentiated (UD), poorly differentiated (PD), and differentiating (D) subtype, based on the grade of differentiation [3]. A simplified classification tree diagram of this recommended classification system is shown in Fig. 1. NB, with different grades, usually has unique pathological characteristics and micro-texture features [4]. Representative tumors of the three differentiation grades are shown in Fig. 2. Typical features of these subtypes can be briefly summarized as follows:
Tumors in the UD subtype often present such features as small to medium-sized NB cells, thin cytoplasm, none-to-few neurites, round to elongated nuclei, and the salt and pepper appearance of chromatin with or without prominent nucleoli.
As for PD cases, the typical rosette formations and/or clearly recognizable neurites are observed in tumor tissues.
Tumors in the D subtype contain > 5% of D neuroblasts characterized by nuclear and cytoplasmic enlargement; an eccentrically located nucleus containing a single prominent nucleolus in most cases; and the increased ratio of the diameter of the cell to that of the nucleus (typically > 2).
Fig. 1.
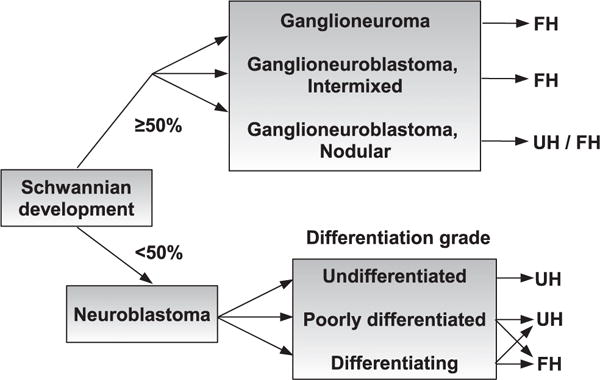
A simplified tree diagram of the International Neuroblastoma Pathology Classification (the Shimada system), where UH represents “unfavorable histology” and FH stands for “favorable histology”.
Fig. 2.
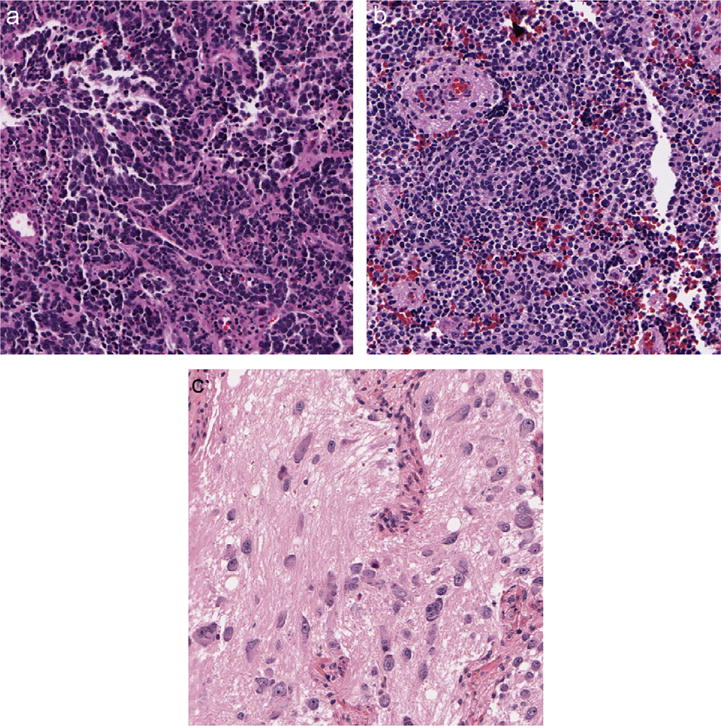
Typical tissue images associated with the three differentiation grades: (a) undifferentiated grade, (b) poorly-differentiated grade, and (c) differentiating grade.
It is usually the case that the more differentiated tumors are, the less aggressively they behave. As a result, patients with tumors of higher grades of differentiation may have better chances to survive. In clinical practice, treatments for cases with different neuroblastic grades are quite different. For this reason, an accurate grading of a NB sample is crucial to make an appropriate choice of treatment plans.
In current clinical practice, differentiation grading is made with visual examinations of tumors by pathologists under the microscope. There are several weaknesses associated with visual evaluations. First of all, it is often time-consuming and cumbersome for pathologists to review a large number of slides in practice. Secondly, visual evaluations can be subject to unacceptable inter- and even intra-reviewer variations. A recent study reports that there is a 20% discrepancy between central and institutional reviewers [5]. Thirdly, for practical reasons, pathologists often sample slide regions to be examined, making the whole process subject to sampling bias. However, this may lead to erroneous results for tumors exhibiting strong heterogeneity.
To overcome these weaknesses rooted in the visual evaluation process, several computerized methods that automate the image analysis procedures are being developed with promising initial results [6–8]. However, to the best of our knowledge, no research work, so far, has been devoted to developing a computer-aided classification methodology that automates the process of classifying NB whole-slide images in accordance with the grade of differentiation. In this study, we propose an image analysis framework that integrates intensive computer vision and machine learning techniques for the purpose of grading NB images. Within this system, an image hierarchy consisting of multiple image resolution levels is established for each given tumor image. Furthermore, the system dynamically changes the image resolution level at which it proceeds with sequential image analysis steps. At each image resolution level, every image is first segmented into four cytological components using an automated image segmentation method. Discriminating features extracted from segmented image regions are then used to classify each image into one of the three grading classes by a family of classifiers. The resulting decisions are next combined using a two-step classifier combining mechanism. Each classification decision is first evaluated with a confidence measure that indicates the degree of agreements across different classifiers. Based on the evaluation results, the proposed system either stops its analysis process or continues with further investigations by including more image details.
2. Methods
2.1. Image acquisition
In this study, all NB tumor slides are collected from Nationwide Children’s Hospital in accordance with an Institutional Review Board (IRB) protocol. According to the protocols commonly used in the Children’s Oncology Group, these tissue slides are cut at a thickness of 5 μm and soaked in paraffin at the preparation stage. Each NB slide in the dataset is prepared using a dual staining procedure in which haematoxylin and eosin (H&E) are used to increase the visual contrasts among different cytological components. After being stained with H&E, each thin tissue slide is then fixed on a scanning bed and digitized using ScanScope T2 digitizer (Aperio, San Diego, CA) at 40× magnification, allowing for clear visualization of tumor architectures. The resulting whole-slide images are quite large with their sizes up to 40 GB. Due to the limited hardware storage capability, the resulting digital images are compressed following the JPEG compression standards at approximately a 1:40 compression ratio. After the compression, the typical image sizes can vary from 1 to 4 GB. To make the image analysis more tractable, we partition each histology slide image into multiple non-overlapping image tiles of the size 512 × 512 in pixels, rather than requiring our classification system to work on the whole-slide image. Another benefit of breaking down whole-slide images into tiles is that we can make full use of the distributed computational infrastructure. The parallel implementation details will be discussed in Section 2.3.
2.2. Image dataset
The image dataset used in this study consists of 36 NB cases, covering all three subtypes of neuroblastic grading. All tumor slides are selected in such a way that they are good representatives of different grade subtypes and contain a sufficiently large number of cytological components of interest in the tissue regions. In our study, the training dataset consists of 389 image tiles of the size 512×512 in pixels, equally selected at random from three representative cases (one from each subtype). The remaining 33 case images from the dataset are used for the testing purpose. The images in our database are evaluated by an experienced pathologist who visually categorized them into three distinctive differentiation grades. The average storage size of testing slides is about 20 GB before compression, which approximately corresponds to 27,400 image tiles of 512 × 512 pixels in size.
2.3. Software and hardware
The developed classification algorithm and the graphical user interface are designed using MATLAB (The MathWorks, Inc., Natick, MA). All experimental evaluations of our work are carried out on a 64-node cluster with Linux OS owned by the Department of Biomedical Informatics at The Ohio State University. Each node of the cluster is equipped with dual 2.4 GHz Opteron 250 processors, 8 GB of DDR400 RAM with 1 GB dimms and a 250 GB SATA hard disk. The computation infrastructure is designed with a master–client architecture in which a master application and multiple client applications work in a collaborative pattern [9]. For each computation task, one master node is responsible for partitioning the tumor slide images into image tiles with a fixed size and distributing data to clients for further processing in a round-robin fashion. Each client keeps local copies of the assigned image tiles and initiates a local MATLAB application to analyze the cached image tiles with the developed classification algorithm. Once the automated image analysis process ends, the master node is, again, in charge of collecting classification results from client nodes and re-assembles them in order before it produces the grading classification results over the whole-slide images.
2.4. Multi-resolution paradigm
Multi-resolution analysis has shown its power in many computer-aided diagnosis (CAD) systems, as CAD systems usually involve processing a large volume of medical image data with prohibitive computational costs. In the work presented by Liang and Page [10], they addressed the problem of demanding computations by adjusting the weights in the neural network with a multi-resolution strategy. Yu et al. took the similar idea and used a hierarchical clustering method to obtain the coarse-to-fine classifiers from a clustering tree [11]. Furthermore, a multi-resolution classification model was proposed by He et al. who classified data using the support vector machines (SVM) in a multi-resolution classification model [12]. In another histopathological image analysis application, Doyle et al. performed pixel-wise Bayesian binary classification at each image resolution level to produce the likelihood scenes from selected regions of interest [8].
Our CAD system for discriminating the grade of neuroblastic differentiation makes full use of the multi-resolution principle in that the system emulates the way pathologists examine histology slides with different magnifications. In accordance with the coarse-to-fine strategy, the developed classification system begins analyzing images at the lowest image resolution level. Processing at higher resolution levels is only invoked when the classification performance associated with the lower resolution level is not sufficient.
At each image resolution level, a complete and automated analysis pipeline is followed. As shown in Fig. 3, the sequence of processes include image–tile generation, establishment of the multi-resolution hierarchy, color conversion, clustering-based segmentation, feature construction, feature selection, dimensionality reduction, multi-classification, classifier combination, and performance evaluation [13].
Fig. 3.
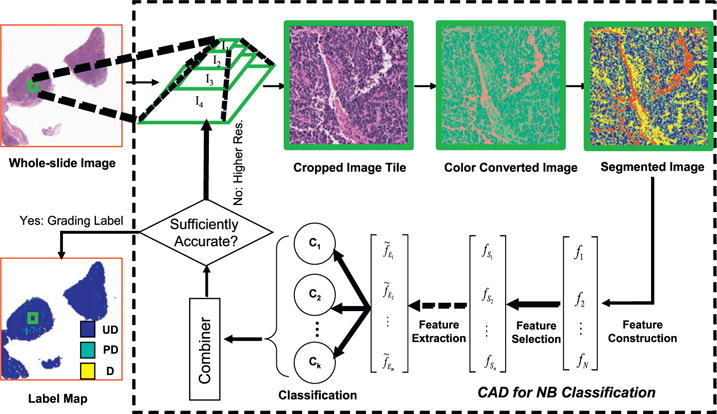
Flowchart of the developed image processing system. A whole-slide image of a neuroblastoma tumor with its size of 59, 412×64, 990 in pixels (13, 932 μm×15, 183μm) is processed with steps involving color conversion, image segmentation, feature construction–selection–extraction, classification, and classifier aggregation, where the scalar N, n, and m satisfy N ⩾ n ⩾ m. Additionally, k indicates the number of independent classifiers used in the system.
As the initial step of the whole process, each image tile analyzed by the computerized system is decomposed into a set of image representations in such a way that the lower resolution image contains most of the relevant image details from higher resolution levels.
Let us denote IL[n, m] as the input image tile at the full resolution, where L is the number of resolution hierarchies. The image versions with a sequence of resolutions can be represented by
| (1) |
where Il[n, m] is the image that is down-sampled for L−l times from the full resolution copy IL[n, m]; and are the integer fields associated with row and column directions at the revolution level l; Nl and Ml, respectively, designate the row number and the column number of Il[n, m].
For each down-sampling process, we follow such a method that the output image is a non-aliasing version in the spatial frequency domain of the next higher resolution tile. This process can be mathematically expressed as
| (2) |
where and . In Eq. (2), Fl[k, s] is the two-dimensional discrete Fourier transform of Il[n, m]. Although there is no particular reason to claim that Il[n, m] has a limited bandwidth, we can in general run an ideal window filter and truncate its bandwidth in the spatial frequency domain:
| (3) |
The ideal low pass filter Hl[k, s] is defined as
| (4) |
where α(≤ 1) is the size reduction factor. In our application, α = 0.5. Therefore, the resulting down-sampled image can be obtained by taking the inverse Fourier transform of the non-zero response area in Hl[k, s].
The designed algorithm initially examines the image tiles at the lowest resolution that corresponds to the lowest optical magnification under the microscope. The lower-resolution image is scaled by a factor of two on each image dimension, thus a quarter as large as the image tile of the next higher resolution level. In our tests, a four layered multi-resolution hierarchy is built up, with {(512×512), (256×256), (128 × 128), (64 × 64) as the set of tile sizes from the highest to the lowest resolution, respectively. An example of image resolution hierarchy with its resolution decreased from the bottom to the top level is shown in Fig. 4, where L = 4.
Fig. 4.
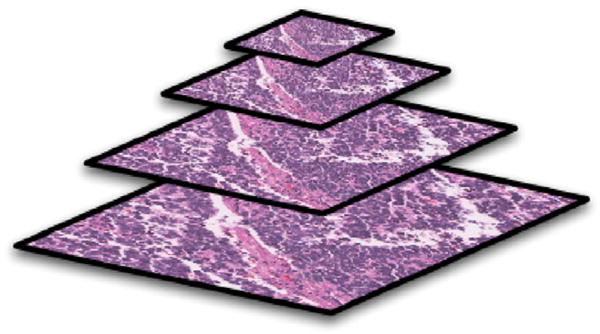
A typical example of the multi-resolution representation hierarchy with image sizes scaled by half at each dimension from bottom to top resolution level.
Since the lowest resolution images are the smallest ones in size within the image representation hierarchy, it requires the least amount of time to process these images. However, if the images at lower resolution levels do not contain sufficient image details, the computerized system automatically switches to work on images at the next higher resolution. As a result, the dynamic change across images of different resolution levels is an analogy to the way a pathologist adjusts the microscope magnification based on the amount of details needed to analyze a particular portion of a tumor slide.
2.5. Image segmentation
At each resolution level, each image is segmented into multiple cytological components necessary for further analysis. Although there are large variations of tissue architectures in images of NB samples having different differentiation grades, five salient components (nuclei, cytoplasm, neuropil, red blood cells (RBCs), and background) can usually be discerned. Additionally, relatively discriminating color contrasts enhanced by the H&E staining process provide us many useful image clues. For example, nuclei and cytoplasm regions are stained with blue-purple in color while regions with pink and red hues suggest neuropils. As a result, it is promising to develop a clustering-based segmentation analysis that identifies different cytological components in a well-formulated feature space. Guided by these ideas, we develop a novel segmentation method, namely EMLDA [14], that works in a feature space constructed with combined color and entropy information extracted from the RGB and the La*b* image channels.
The La*b* color space is developed by the Commission Internationale d’Eclairage (CIE) [15]. When it is compared to other color spaces such as HSI, YIQ, and YUV, it presents the desired color perceptual uniformity that allows the use of Euclidean distance metric rational. The La*b* color space is also a good choice in terms of its ability to represent luminance and chrominance information separately. By its definition, channel L carries the information for the light intensity while color information is contained in a* and b* components. As a supplement, three entropy statistics computed with a 9 × 9 window shifting across the R, G, and B image components are used to enrich the feature vector.
Our novel segmentation approach integrates the Fisher–Rao criterion into the generic expectation–maximization algorithm and iteratively partitions data in the resulting feature space in such a way that the data associated with different classes can be separated as much as possible. This process is repeated until the Fisher–Rao criterion, the ratio of the sum of squared between-class distances to the sum of squared within-class distances, converges to its maxima.
Suppose C components are to be segmented from a given image. Let us denote as the dataset in a p-dimensional feature space. The expectation and maximization step of the EMLDA method can then be summarized as follows.
-
E step (expectation):
Find the optimal projection matrix:
where J(V|θ) is the Fisher–Rao criterion to be maximized. V* is a p×s matrix consisting of s discriminant vectors as its columns, where s ≤C−1. In addition, θ, is the labelling configuration determined from the previous step, while SB and SW are the between- and within-class scatter matrices that are symmetric and positive-definite [16].(5) -
M step (maximization):
The matrix V* maximizing the Fisher–Rao criterion J(V|θ) is composed of s column-wise discriminant feature vectors onto which the set of data X are projected. By , the projected data are mapped to a lower dimensional space where the data associated with the C classes can be best discriminated.
Next, find:
| (6) |
Where Θ = {1, 2,…, C} is the label set; mi and are the means of class i in the original and reduced dimensional feature space that are related by . After finding the labels for all data points, we substitute θ with and repeat steps (1) and (2) again, until J(V*|θ) converges to a local maximum.
One beauty associated with this method is that one can simply skip the feature normalization step due to the following theorem.
Theorem 1
Feature normalization by linear scaling to unit range does not change the end classification result with EMLDA.
Proof of Theorem 1
Suppose the mapping functions are linear scaling transformations that map each feature x to a specific unit range, i.e., , where M = diag(m11,…,mpp) and D = (d1⋯dp)T, then the between-class scatter matrix in the transformed data space is
| (7) |
where m is the overall mean and ni is the number of samples of class i.
Similarly, we have the within-class scatter matrix in the transformed data space as Furthermore, we have
| (8) |
where .
Eq. (8) reveals the fact that J and can be simultaneously maximized when . The projected and the mean in the lower dimension space spanned by the discriminant vectors for X = {x|x ε Rp} are xTV*and , while those associated with are
| (9) |
i.e., the projected data derived from X and are related by a simple translation. Therefore, the end classification results in X and feature space are conserved to be the same. ■
In our study, we define four classes: nuclei, cytoplasm, neuropils, and background (i.e., C = 4). Given the fact that RBCs may occupy a large area within image tiles, a simple yet efficient threshold-based method is used to identify the RBC regions before the iterations are initiated. As a result, the number of defined classes is kept as low as possible, which contributes to the reduction of time costs. A set of segmentation results on a typical image tile associated with the UD grade are shown in Fig. 5, which verifies the effectiveness of this segmentation approach.
Fig. 5.
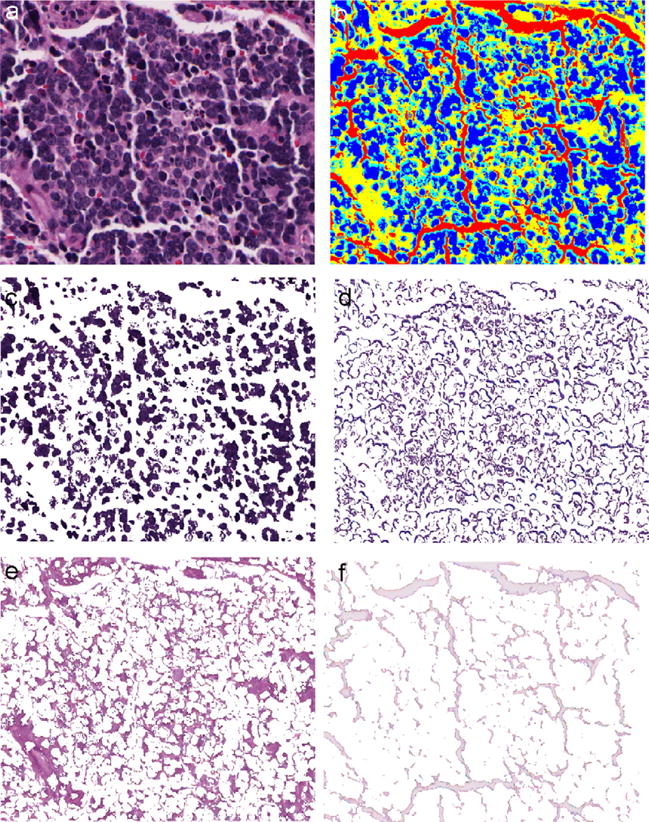
The segmented components in a typical image from undifferentiated subtype are shown. (a) Original image. (b) Partitioned image shown in colors with nuclei in blue, cytoplasm in cyan, neuropil in yellow, red blood cells in gray, and background in red. (c) Nuclei component. (d) Cytoplasm component. (e) Neuropil component. (f) Background component.
2.6. Feature construction
Although pathologists tend to rely heavily on morphologic features such as nuclear size and cellularity, a large amount of information derived from textures of different histological structures is integrated into their decision procedures in an implicit way [17]. By creating the feature vector, it is our intention to input into the computerized system information that helps discriminate different pathological components effectively. Therefore, in our implementation, all features are derived from color information and textural patterns as they depend less on the segmentation accuracy.
In our study, local statistical measures are computed only from the segmented regions associated with cytoplasm and neuropils identified by the proposed segmentation algorithm in that they usually bear texture patterns considerably distinct across tumor tissues of different grades. Associated with cytoplasm and neuropil regions, four textural Haralick features [18] are computed: the entropy, mean, and variance of the range of values within a local neighborhood, and the homogeneity degree of the co-occurrence matrix in L, a* and b* channels. As a result, a feature vector composed of 24 elements is constructed as a symbolic representation of each image tile. For comparison, summaries of the set of features used by pathologists and those employed in the computerized system are reported in Tables 1 and 2, where two apparent distinctions can be observed.
Table 1.
Features used by pathologists in the visual grading process
| Features | Description | Category |
|---|---|---|
| Neuropil | The degree of presence of neuropil | No/minimal/sparse/moderate/prominent |
| Cell cellularity | Number of cells per HPF (high power field) | Low/intermixed/high/intermediate |
| Nuclear size | Variation of nuclear size | Variable/uniform/pleomorphic |
| Nuclear shape | Shape regularity | Round-to-oval/pleomorphic |
| Mitotic karyorrhectic index | Number of tumor cells in mitosis and karyorrhexis | Low/intermediate/high |
| Mitotic rate | Number of mitoses in 10 contiguous HPFs at 400× magnification | Low/high |
| Calcification | Presence of dense basophilic clumps or amorphous granular material | Yes/no |
Table 2.
Statistical features derived from L, A*, and B* image channels used by the developed computerized system in the automated classification process
| Features | Description |
|---|---|
| Entropy | A measure of the uncertainty of the local pattern from the information theory |
| Mean of the range of values | The mean value of the differences between the maximum and minimum values within neighborhoods across the image |
| Variance of the range of values | The variance of the differences between the maximum and minimum values within neighborhoods across the image |
| Homogeneity | A statistical value that measures the uniformity of a neighborhood |
Pathologists tend to use morphological and pathological characteristics in their prognosis, while the computer system prefers using statistical features.
The characteristics used by pathologists are qualitative, as opposed to the quantitative features utilized by the computer system.
2.7. Feature selection and classification
Although we use a vector of features to characterize the texture patterns of cytological structures, it is not the case, in general, that each extracted feature component contributes to the characterization of the texture patterns. More importantly, they may not increase the classification accuracy in equal proportions. Some features may contain far more discriminating information than others. By contrast, some other features may contribute less in improving the classification accuracy.
The choice of the most discriminating subset of features is not only conducive to substantial reduction in computational complexity, but it also leads to improved classification accuracies. In fact, if more than the necessary number of features are used, the classification performance can deteriorate due to the “peaking phenomenon” [16]. As a result, we are interested in seeking out the best subset of features that yield the best classification accuracy and have the least number of members. In our application, the subset of features sufficient for classifying well-organized data are determined by a popular feature selection technique, namely the sequential floating forward selection (SFFS) procedure [19]. The SFFS procedure is an upgraded version of the sequential selection procedures with back-tracking mechanisms, such as plus l − take away r algorithm. In addition to these methods, the number of forward and backtracking steps (i.e., l and r) in SFFS are dynamically adjusted rather than fixed values set in advance. In other words, SFFS allows a dynamic number of features that have been selected to be removed in a dynamic number of posterior steps. As a result, SFFS is very efficient and effective even on problems of high dimensionality with non-monotonic feature selection criterion functions.
Since our proposed multi-resolution hierarchy has four resolution levels, the optimal subset of features needs to be determined with training image tiles from each resolution layer. This is due to the fact that neither histopathological nor statistical characteristics of image tiles across different resolution levels are necessarily the same. In practice, they could be considerably distinct from each other. As a result, the ensuing subset of selected features may not be the same when computed at different resolutions. After the optimal subsets of features extracted from the training data associated with different resolutions are obtained in the training phase, we can establish the new feature vectors with only those feature components selected in the training process and classify the feature data in a lower dimensional feature space in the testing stage.
Furthermore, the feature selection process is carried out in combination with different classifiers over all the training data in an offline pattern. To achieve a good classification performance, multiple classifiers were employed: K-nearest neighbor (KNN), linear discriminant analysis (LDA) & KNN, LDA & nearest mean (NM), correlation LDA (CORRLDA) [20] & KNN, CORRLDA & NM, LDA & Bayesian and SVM [21] with a linear kernel.
KNN is a very intuitive classifier that assumes observations associated with the same class label are close to each other measured by some metric in a feature space. Assuming {x1, x2,…, xk} is a set consisting of the nearest K observations, under the distance}metric d(.,.), to the given data x whose class label is to be determined, the class·label of x is the majority vote of the nearest K neighbors. Likewise, NM classifier assigns data to the class associated with the closest class mean. Unlike the non-parametric classifiers, Bayesian classifier can reach the optimal recognition result given that all underlying true class-conditional probability density functions are known. Another useful classifier widely used n machine learning process is known as SVM that can deal with non-linearly separable cases by mapping data from a lower dimensional space to a higher dimensional one where data becomes linearly separable.
In addition to the four classifiers, two feature extraction methods, i.e., LDA and CORRLDA, are used in combination with the classifiers in reducing the dimensionality of the feature space. LDA aims at finding the best subspace where the between-class variance is maximized while the within-class variance is minimized. However, it does not necessarily guarantee the minimum Bayes error of the given data distributions when the eigenvectors associated with the largest eigenvalues of the between-class scatter matrix have a high correlation with the most principal eigenvectors of the within-class scatter matrix. By contrast, CORRLDA is a method to select the most discriminative eigenvectors of the within-class scatter matrix that is often singular. Rather than picking up the most principle eigenvectors, it keeps the eigenvectors that are most correlated to the bases spanning the range space of the between-class scatter matrix. For more detailed discussions on these methods, readers are referred to [16,20,21].
As different feature extraction approaches and classifiers are optimal under different assumptions, no single combination of one specific dimensionality reduction technique and one fixed classifier can guarantee the best feature representation and the resulting minimum Bayes error in the absence of assumptions on feature distributions. As a result, no general algorithm exists that can always achieve superior recognition rates to those of others under all possible circumstances. However, one feasible way to boost the system classification performance is to use a collection of classifiers, rather than a single one. It has been shown that the overall error rate decreases monotonically as more classifiers are used in a system, as long as each individual classifier has an error rate less than random guessing [22]. As each classifier has its own feature regions where it yields the best performance, a combination of classifiers can achieve higher classification accuracies in theory. As a result, the ensemble classification system comprising seven classifiers is developed for the decision-making component. Since the seven classifiers included in this classification group present different characteristics and various classification mechanisms, aggregation of these classifiers can help improve the end recognition performances.
2.8. Classifier combiner
To make full use of the group of classifiers, a combination strategy is proposed to combine those decisions made by the collection of classifiers in a parallel pattern. Although a number of architectures for classifier combination have been proposed, such as the dynamic classifier selection (DCS), classifier hierarchical concatenation (CHC), and serial combination (SC), we choose to aggregate the outputs of the multiple classifiers with a straightforward two-step classifier combining mechanism that consists of a voting and weighting procedure. This is due to the fact that a sum rule-based combination paradigm outperforms the others in general [23]. The combining mechanism can be described in the following two steps.
Step 1: The combiner evaluates the outputs of all K classifiers (K = 7 in this work) and produces a final decision θ* that refers to the decisions supported by the majority of the K classifiers:
| (10) |
where Ψ(i) is the number of votes for the ith class collected from the K classifiers and C is the number of classes (C = 3 in this work).
Step 2: We next evaluate the confidence degree of the voted classification result at the current resolution level. Since the combination scheme of the sum rule is usually superior to the others, the confidence degree in this study is defined as the sum of weights assigned to classifiers that concur with the combiner. The corresponding weight assigned to each classifier, in each resolution level, is computed by normalizing the priori classification accuracies of all the classifiers over the training data with the leave-one-out validation process. The higher the priori classification accuracy one classifier has, the more biased it is weighted. If the sum of weights of the classifiers whose decisions concur with each other is greater than a given threshold, the ensemble decision supported by the majority of classifiers is accepted as an end result. Otherwise, the system switches to work on the image version at the next higher resolution level where the same sequence of image processing steps are followed again.
In summary, the hypothesis test and the resulting decision rule can be written as
H0: classification result is good enough; quit the process;
- H1: go to the next higher resolution level for classification;
where(11)
In Eq. (11), wl(i) is the priori recognition rate of the classifier i at resolution level l; δi,j is the Kronecker delta function; θ(i) is the class label decided by the classifier i; γl,l+1 is the threshold of the confidence measure with which the system decides whether or not it needs to proceed with further analysis from image resolution l to l + 1.
3. Results
Before the developed system can achieve reasonably good grading accuracy, it needs to be well trained. A brief outline of the training–testing process is shown in Fig. 6, where solid and dashed arrows indicate the steps executed online and offline, respectively. In the offline training stage, multiple statistical learning models are used to enrich the system knowledge base with the ground-truth given by an experienced pathologist. As the computer system is trained with more and more typical patterns, it starts producing outputs similar to those made by the pathologist. After the training process is finished, testing data can then be passed into the “educated” system for evaluating its performance.
Fig. 6.
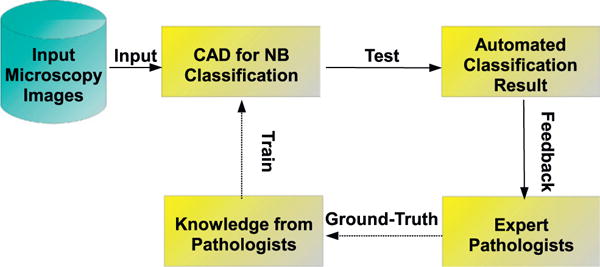
Flowchart of the developed classification system. The solid arrows indicate online steps and the dashed arrows indicate offline steps.
3.1. System training
For training purpose, a priori knowledge on the architectural patterns of NB with different neuroblastic differentiations is derived from a training dataset. It consists of 387 image tiles extracted from three representative whole-slide images. Achieving a good separation of the features from the training tiles of different grades, the training set is considered a good representation of the underlying discriminating information. In order to visualize how well the training data can be separated in an appropriate feature space, we show in Fig. 7 the scatter plots of training features associated with the four resolution levels in a two-dimensional feature space obtained with a combined use of the dimension reduction technique of LDA and the classifier of KNN (K = 5). The resulting plots illustrated in Fig. 7 show that the data cloud of each class is relatively compact and well apart from the others. This considerably contributes to a good grading accuracy in the following classification process.
Fig. 7.
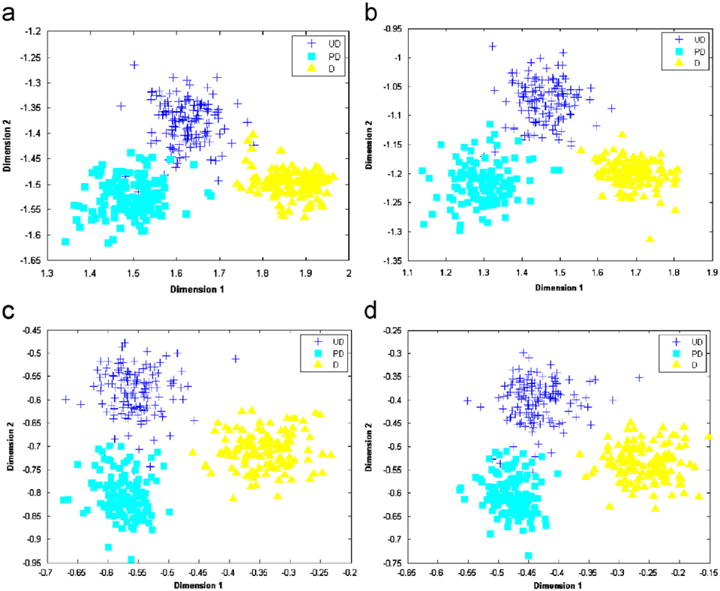
Scatter plots of the training features in a two-dimensional feature space associated with four resolution levels (from the lowest to highest) are shown in (a)–(d). These features are selected and extracted when LDA and the classifier of KNN (K = 5) are used.
In Table 3, we report the numbers of selected features associated with different choices of classifiers on each image resolution level over the training data using the leave-one-out cross-validation method. In this cross-validation approach, all but one sample of training data is used for training purpose. The sample left out is then used for testing with the trained classifiers. This is repeated for all possible ways of leaving one training sample out (i.e., leaving out one sample selected in a cyclical pattern).
Table 3.
When our system is trained with the training data using the leave-one-out cross-validation method, the numbers of features selected using the SFFS procedure for different classifiers are presented
| Feature extraction and classification | Resolution level
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| KNN | 9 | 7 | 7 | 6 |
| LDA&KNN | 16 | 13 | 10 | 11 |
| LDA&NM | 10 | 16 | 11 | 9 |
| CORRLDA&KNN | 8 | 8 | 6 | 6 |
| CORRLDA&NM | 4 | 9 | 2 | 6 |
| LDA&BAYESIAN | 17 | 16 | 11 | 11 |
| SVM | 3 | 6 | 10 | 5 |
It can be noticed from Table 3 that the resulting numbers of selected features associated with different classifiers vary considerably. For example, numbers of selected features with LDA&BAYESIAN are much larger than those associated with CORRLDA&NM. Although there are no direct clues that can help explain the differences, some educated explanations can be given by a careful look at the working criteria different feature extraction approaches take and natures various classifiers exhibit. Again, the example of the comparison be-tween LDA&BAYESIAN and CORRLDA&NM is used for illustration. It is well known that LDA tries to maximize the ratio of the between-class to the within-class distance. Therefore, a larger set of features allow LDA a larger degree of freedom in finding the best projection vectors. CORRLDA, in contrast, computes eigenvectors of the within-class scatter matrix and only cares about those most correlated to the bases that span the range space of the between-class scatter matrix. This results in its preference over smaller sets of features, as the selection of a small number of features helps increase the degree of correlation. Furthermore, Bayesian classifier is a parametric classifier and thereby usually requires a relatively larger set of features for a better estimation of its parameters. Unlike Bayes, NM is a non-parametric classification machine that demands a higher ratio of the number of samples to the dimensionality of the feature space. As a result, LDA&BAYESIAN selects a larger sets of features than CORRLDA&NM across all resolution levels.
Table 4 presents the corresponding classification accuracies using the leave-one-out cross-validation method. It can be concluded with Table 4 that, in general, higher resolution levels yield better classification accuracies, although they require longer processing time.
Table 4.
When our system is trained with the training data using the leave-one-out cross-validation method, the resulting tile-level classification accuracies with the use of the SFFS procedure for different classifiers are presented
| Feature extraction and classification | Resolution level
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| KNN | 94.32% | 94.57% | 95.35% | 97.16% |
| LDA&KNN | 98.97% | 98.19% | 98.45% | 99.48% |
| LDA&NM | 98.19% | 98.45% | 98.19% | 98.97% |
| CORRLDA&KNN | 96.12% | 95.35% | 97.67% | 98.19% |
| CORRLDA&NM | 94.32% | 95.35% | 95.61% | 97.16% |
| LDA&BAYESIAN | 98.71% | 98.19% | 98.71% | 99.22% |
| SVM | 98.72% | 97.44% | 98.72% | 97.44% |
3.2. System testing
To evaluate the generalization ability of the developed system to new clinical data, we carry out experiments on an independent testing dataset comprising 33 whole-slide images: 10 from UD, 10 from PD, and 13 from D subtypes.
In our experiments, we process images with the most critical confidence threshold setting:
where Ω5 is the confidence configuration label;γl,l+1 is the confidence threshold with which the system decides whether or not it is necessary to move from resolution level l to l + 1 (see Eq. (11)).
The grading accuracies on the whole-slide level can be summarized in Table 5 where the accuracies of identifying UD, D, and PD cases are presented, in addition to the overall grading accuracy of 87.88%.
Table 5.
Classification accuracies of the whole-slide experiments are shown
| Confidence configuration | Slide-level classification accuracy
|
|||
|---|---|---|---|---|
| Undifferentiated | Differentiating | Poorly differentiated | Overall | |
| Ω5 | 90.00% | 84.62% | 90.00% | 87.88% |
In addition to the ensemble results reported in Table 5, results of three representative slides (one from each grading class) are also presented to illustrate the experimental results in more details. The description of the characteristics of the three typical slides is reported in Table 6.
Table 6.
Neuroblastic differentiation grades of typical three testing slides diagnosed by pathologists and their whole-slide image sizes are presented
| Slide no. | Differentiation grade | Image size (uncompressed) (GB) |
|---|---|---|
| 1 | Undifferentiated | 10.8 |
| 2 | Poorly differentiated | 15.5 |
| 3 | Differentiating | 28.5 |
Besides the most critical confidence threshold setting Ω5 used for slide-based evaluations, four other configurations for the classifier combiner are also created for testing these three slides. Each one represents a different strategy of manipulating the dynamic resolution changes in our multi-resolution grading system. These five configurations, each consisting of three threshold values, are summarized in Table 7.
Table 7.
Thresholds γl,l+1∀l =1, 2, 3 with which we conduct the hypothesis tests for evaluating the confidence degrees of the voted classification results at resolution level l are presented for the five different confidence configurations (i.e., Ω1–Ω5)
| Confident threshold | Different configuration settings
|
||||
|---|---|---|---|---|---|
| Ω1 | Ω2 | Ω3 | Ω4 | Ω5 | |
| γ1,2 | 0.7 | 0.8 | 0.9 | 0.9 | 1.0 |
| γ2,3 | 0.8 | 0.8 | 0.8 | 0.9 | 1.0 |
| γ3,4 | 0.9 | 0.8 | 0.7 | 0.9 | 1.0 |
In Fig. 8, the image tile-level classification accuracies associated with the confidence configurations Ω1–Ω5 are shown for the slides from Nos. 1 to 3, as presented in Table 6. For better visual effects, it is worth mentioning that the vertical scale of Fig. 8(a) begins from 92%. Generally speaking, the larger the threshold (i.e., a value between 0 and 1), the more difficult it becomes to satisfy the system at each resolution level. As a result, a better grading accuracy can be expected, but at the cost of longer execution time.
Fig. 8.
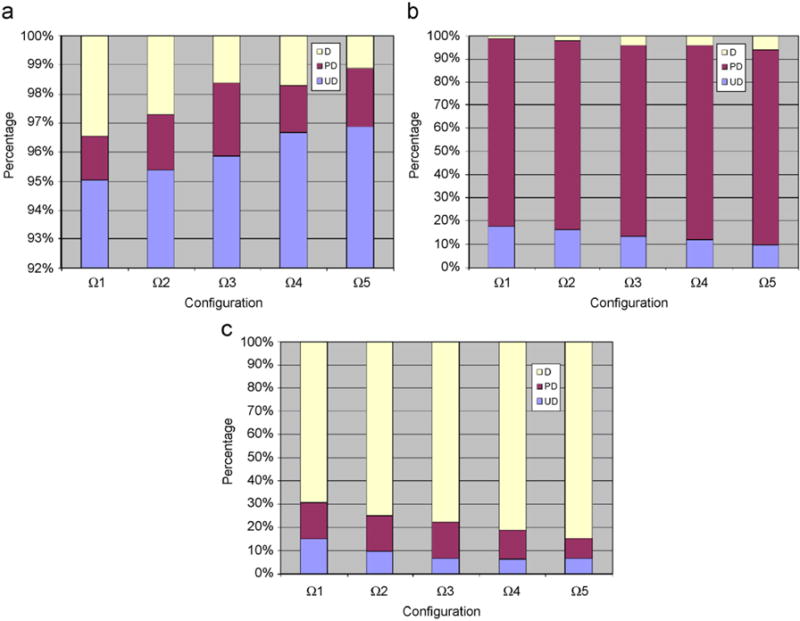
Percentages of image tiles graded as undifferentiated, poorly differentiated, and differentiating for the three testing slides (i.e., Nos. 1–3 in Table 6) are shown in (a)–(c), respectively. For each slide, classification results associated with confidence configurations Ω1–Ω5 in Table 7 are presented, respectively. For better visual effects, the vertical axis of (a) starts with 92%.
To illustrate classification results of the three typical tumor slides, we assign to each image tile (512×512 in size) a color that represents its grade of neuroblastic differentiation identified by our system. We name the resulting image representations as classification maps, as shown in Fig. 9(b), (e), and (h). With these classification maps, we are able to recognize quickly how tumors of different grades are spatially distributed. In our application, the colors blue, cyan, and yellow are used for image tiles classified as UD, PD, and D, respectively. Likewise, a gray level is assigned to each image tile as an indicator of the resolution level at which the accuracy of the grading evaluation process convinces the system. The resulting images, consisting of resolution levels at which final grading decisions are made, are named decision level maps, as shown in Fig. 9(c), (f), and (i). The gray level of each pixel indicates the decision level of the corresponding image tile.
Fig. 9.
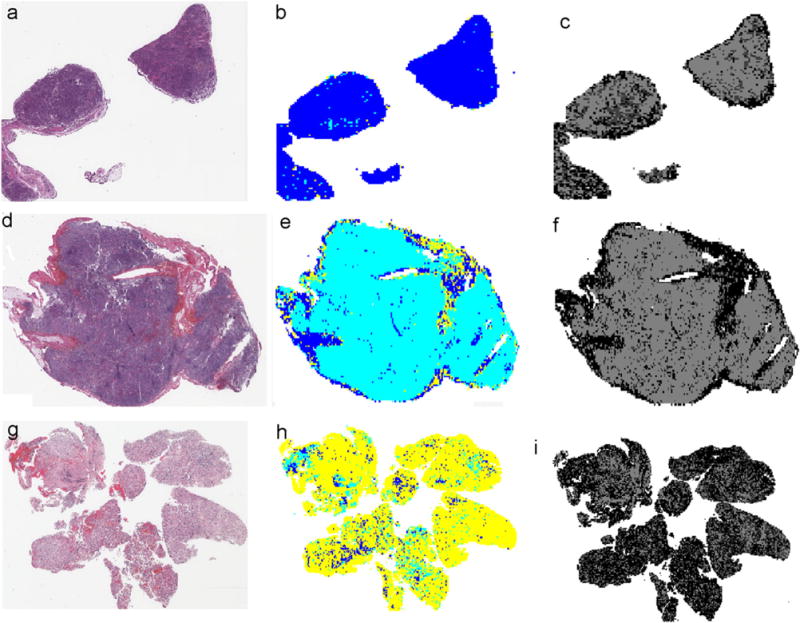
Image tile-level classification results of three typical tumor slides using configuration Ω5 are shown. (a), (d), and (g) are the H&E stained whole-slide images classified as undifferentiated, poorly differentiated, and differentiating class by an experienced pathologist. (b), (e), and (h) are the corresponding classification maps where the colors blue, cyan, and yellow represent undifferentiated, poorly differentiated, and differentiating class identified by the developed computer system. (c), (f), and (i) are the associated decision level maps where the intensity of each pixel represents the resolution level at which each 512 × 512 image tile is eventually classified. Darker intensities indicate higher resolution levels.
4. Discussions
The developed system is a useful tool that pathologists and clinicians can use in diagnosing the grade of NB. Although decision accuracy of the computerized system is promising in our tests, it should be emphasized that the role of the computerized system should always be limited to that of a second reader or pre-screener who can make consistent decisions on NB classification in parallel with expert pathologists. In other words, the computerized methodology is not intended to replace the pathologist, but to support the conventional prognostic procedure. Coincidentally, one similar paradigm of having a computerized system as a second reader for diagnosis in the field of radiology has been shown to be safe and effective [24].
Additionally, the computerized system is consistent and does not suffer from certain human-reader limitations even experienced pathologists may encounter. One other benefit of this system is that it provides a relatively generic methodology that can be modified for a vast number of practical applications involving pathological image analysis components. Furthermore, given the fact that the selected features exhibit the helpful information for class discrimination, this proposed generic multi-resolution paradigm can be also tailored to other medical image analysis systems with minor efforts.
Although a large number of CAD and prognosis systems have been proposed in the literature [7,25], our system has multiple advantages over these systems:
The whole-slide image dataset is analyzed. The storage size of a typical H&E stained image before being compressed is around 25 GBs, which gives rise to the following two challenges that we have to overcome: the demanding computational complexity and the large-scale data storage. These two difficulties in our application are solved with a generalizable computational infrastructure for grid-based biomedical image processing applications developed within the Department of Biomedical Informatics at The Ohio State University [9]. Within this service-based infrastructure, multiple developers can have access to a common image data and code repository where a pool of different resources can be shared with the public. This is specifically designed for very large-scale image processing projects that require pipelined processing capabilities. In this system, the images are automatically de-clustered by a central coordinator. Then, the remaining slave nodes follow the processing sequence and analyze the assigned image blocks cropped by the central coordinator. Finally, the image processing results, either saved in texts or as an image, are collected and assembled in compliance with the de-cluster order at the central coordinator.
Color images are analyzed. Instead of analyzing gray level images, discriminating information derived from three color channels were jointly used in our application. In addition to combining the color with the entropy information in constructing features, we apply feature selection and extraction techniques to the established feature pool, resulting in the most discriminating features that lead to promising grading accuracies.
A multi-resolution schema is used. An automated dynamic change in the image resolution is devised in the developed system to eliminate from the analysis as many superfluous computations as possible. By emulating the way pathologists examine tumor slides, the computerized system is designed to work in an efficient way that produces much savings of computational resources.
Since neuroblastic differentiation is the key for the correct protocol assignment, we will further improve the developed system in our future work. Since the higher-order decision information may contribute to either a more efficient or accurate global labelling configuration, it is desirable to develop a labelling scheme that takes into account decision information from adjacent image tiles. Expansion into an even larger group of features is another part of work we will explore in future study, as only color and texture information is used in our current implementation. We will also investigate the way to group the best training dataset, since data generalization is a critical problem that may impact overall performance significantly. With all these aspects improved, it is promising to expect even better classification accuracy sufficient for the clinical requirements in practice.
5. Conclusions
This article presents an automated grading system for the quantitative analysis of the histological images of the H&E stained NB cross-sections. To emulate the way pathologists assess resected specimens, the complete image analysis pipeline is designed within a multi-resolution paradigm. When tested on 33 whole-slide images, the overall classification accuracy of the system is 87.88%. The developed algorithm chooses to work on images of the lowest resolution level where sufficient image details are retained for the grading analysis. On each resolution of the whole image hierarchy, crucial statistical features needed for accurate evaluations are constructed from the segmented image regions where the most discriminating information resides. Systematic selection of the best subset of features not only improves the system efficiency but also the ensuing grading accuracy. Moreover, a collection of classifiers is used for exploring different areas of the feature space, which mimics the scenario when multiple pathologists are available within a prognosis panel. The resulting competitive classification accuracy and high throughput performance suggests that the developed system is promising for the grading assessments of NB.
Biographies
JUN KONG received the B.S. in Information and Control and M.S. in Electrical Engineering from Shanghai Jiao Tong University, Shanghai, China, in 2001 and 2004. He is currently a Ph.D. student in the Department of Electrical and Computer Engineering at Ohio State University, Columbus, OH. His research interests include computer vision, machine learning, and pathological image analysis.
OLCAY SERTEL received his B.S. from Yildiz Technical University, Istanbul, Turkey, and his M.Sc. degree from Yeditepe University, Istanbul, Turkey, in 2004 and 2006, both in Computer Engineering. He is currently a Ph.D. student at the Department of Electrical and Computer Engineering and working as a Graduate Research Associate at the Department of Biomedical Informatics at The Ohio State University, Columbus, OH. His research interests include computer vision, image processing and pattern recognition with applications in medicine.
HIROYUKI SHIMADA is currently an Associate Professor of Clinical in Children’s Hospital Los Angeles and Department of Pathology and Laboratory Medicine at The University of Southern California. He is the Pathologist-of-Record for the Children’s Cancer Group (CCG) Neuroblastoma Studies. He is also a core member of International Neuroblastoma Pathology Committee and chairing workshops and other activities. In 1999 this Committee established the International Neuroblastoma Pathology Classification (the Shimada system) by adopting my original classification published in 1984. His research interests include investigating morphological characteristics of pediatric tumors.
KIM L. BOYER received the BSEE (with distinction), MSEE, and Ph.D. degrees, all in Electrical Engineering, from Purdue University in 1976, 1977, and 1986, respectively. Since 1986, he has been with the Department of Electrical and Computer Engineering, The Ohio State University, where he holds the rank of a Professor. In 1986, he founded the Signal Analysis and Machine Perception Laboratory at Ohio State. He is a fellow of the IEEE and a fellow of IAPR. He is a member of the Governing Board for the International Association for Pattern Recognition and Chair of the IEEE Computer Society Technical Committee on Pattern Analysis and Machine Intelligence. Dr. Boyer’s research interests include all aspects of computer vision and medical image analysis, including perceptual organization, structural analysis, graph theoretical methods, stereopsis in weakly constrained environments, optimal feature extraction, large model bases, and robust methods.
METIN N. GURCAN received his M.Sc. degree in Digital Systems Engineering from UMIST, Manchester, England and his Ph.D. degree in Electrical and Electronics Engineering from Bilkent University, Ankara, Turkey. He is working as an Assistant Professor at the Department of Biomedical Informatics at The Ohio State University since 2006. Dr. Gurcan’s research interests include image analysis and understanding, computer vision with applications to medicine.
JOEL H. SALTZ is a Professor and Chair of the Department of Biomedical Informatics, Professor in the Department of Computer Science and Engineering at The Ohio State University (OSU), Davis Endowed Chair of Cancer at OSU, and a Senior Fellow of the Ohio Supercomputer Center. He received his M.D. and Ph.D. degrees in Computer Science at Duke University. Dr. Joel Saltz has developed a rich set of middleware optimization and runtime compilation methods that target irregular, adaptive, and multi-resolution applications. Dr. Saltz is also heavily involved in the development of ambitious biomedical applications for high-end computers, very large-scale storage systems, and grid environments. He has played a pioneering role in the development of pathology virtual slide technology and has made major contributions to informatics applications that support point-of-care testing.
References
- 1.Goodman MJ, Gurney JG, Smith MA, Olshan AE. Sympathetic Nervous System Tumors. National Cancer Institute; 1999. Cancer incidence and survival among children and adolescents: United states surveillance, epidemiology, and end results (seer) program 1975–1995. http://seer.cancer.gov/ (Chapter IV) [Google Scholar]
- 2.Castleberry EP. Neuroblastoma. European Journal of Cancer. 1997;33:1430–1438. doi: 10.1016/s0959-8049(97)00308-0. [DOI] [PubMed] [Google Scholar]
- 3.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, Gastlebery RP. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86(2):364–372. [PubMed] [Google Scholar]
- 4.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B. Terminology and morphologic criteria of neuroblastic tumors: recommendation by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–363. [PubMed] [Google Scholar]
- 5.Teot LA, Khayat RSA, Qualman S, Reaman G, Parham D. The problems and promise of central pathology review: development of a standardized procedure for the children’s oncology group. Pediatric and Developmental Pathology. 2007;10:199–207. doi: 10.2350/06-06-0121.1. [DOI] [PubMed] [Google Scholar]
- 6.Ayres FJ, Zuffo MK, Rangayyan RM, Boag GS, Filho VO, Valente M. Estimation of the tissue composition of the tumor mass in neuroblastoma using segmented CT images. Medical and Biological Engineering and Computing. 2004;42:366–377. doi: 10.1007/BF02344713. [DOI] [PubMed] [Google Scholar]
- 7.Madabhushi A, Feldman M, Metaxas D, Tomasezweski J, Chute D. Automated detection of prostatic adenocarcinoma from high resolution ex vivo MRI. IEEE Transactions on Medical Imaging. 2005;24(12):1611–1625. doi: 10.1109/TMI.2005.859208. [DOI] [PubMed] [Google Scholar]
- 8.Doyle S, Madabhushi A, Feldman MD, Tomaszeweski JE. A boosting cascade for automated detection of prostate cancer. The 9th International Conference on MICCAI. 2006:504–511. doi: 10.1007/11866763_62. [DOI] [PubMed] [Google Scholar]
- 9.Cambazoglu BB, Sertel O, Kong J, Saltz J, Gurcan MN, Catalyurek UV. Efficient processing of pathological images using the grid: computer-aided prognosis of neuroblastoma. Proceedings of the IEEE International Conference on Challenges of Large Applications in Distributed Environments. 2007:35–41. [Google Scholar]
- 10.Liang Y, Page EW. Multiresolution learning paradigm and signal prediction. IEEE Transactions on Signal Processing. 1997;45(11):2858–2864. [Google Scholar]
- 11.Yu H, Yang J, Han J. Classifying large data sets using SVMs with hierarchical clusters. SIGKDD. 2003:306–315. [Google Scholar]
- 12.He Y, Zhang B, Li J. A new multiresolution classification model based on partitioning of feature space. IEEE International Conference on Granular Computing. 2005;2:462–467. [Google Scholar]
- 13.Kong J, Sertel O, Shimada H, Boyer K, Saltz J, Gurcan M. Computer-aided grading of neuroblastic differentiation: multi-resolution and multi-classifier approach. Proceedings of the IEEE International Conference on Image Processing. 2007:525–528. [Google Scholar]
- 14.Kong J, Shimada H, Boyer K, Saltz J, Gurcan M. Image analysis for automated assessment of grade of neuroblastic differentiation, in: Proceedings of the IEEE International Symposium on Biomedical Imaging: Macro to Nano. 2007:61–64. [Google Scholar]
- 15.Paschos G. Perceptually uniform color spaces for color texture analysis: an empirical evaluation. IEEE Transactions on Image Processing. 2001;10(6):932–937. [Google Scholar]
- 16.Fukunaga K. Introduction to Statistical Pattern Recognition. Academic Press; New York: 1990. [Google Scholar]
- 17.Ambros IM, Hata J, Joshi VV, Roald B, Dehner LP, Tüchler H, Pötschger U, Shimada H. Morphologic features of neuroblastoma (Schwannian stroma-poor tumors) in clinically favorable and unfavorable groups. Cancer. 2002;94(5):1574–1583. doi: 10.1002/cncr.10359. [DOI] [PubMed] [Google Scholar]
- 18.Haralick R, Shanmugam K, Dinstein I. Texture features for image classification, IEEE Transactions on Systems. Man and Cybernetics. 1996;3:610–621. [Google Scholar]
- 19.Pudil P, Ferri F, Novovicova J, Kittler J. Floating search methods for feature selection with nonmonotonic criterion functions. Proceedings of Computer Vision and Image Processing. 1994;2:279–283. [Google Scholar]
- 20.Zhu M, Martinez AM. Selecting principal components in a two-stage LDA algorithm. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition. 2006:132–137. [Google Scholar]
- 21.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20(3):273–297. [Google Scholar]
- 22.Hanse LK, Salamon P. Neural network ensembles. IEEE Transactions of Pattern Analysis and Machine Intelligence. 1990;12(10):993–1001. [Google Scholar]
- 23.Kittler J, Hatef M, Duin RPW, Matas J. On combining classifiers. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1998;20(3):226–239. [Google Scholar]
- 24.Burhenne LJW, Wood SA, D’Orsi CJ, Feig SA, Kopans DB, O’Shaughnessy KF, Sickles EA, Tabar L, Vyborny CJ, Castellino RA. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology. 2000;215:554–562. doi: 10.1148/radiology.215.2.r00ma15554. [DOI] [PubMed] [Google Scholar]
- 25.Esgiar AN, Naguib RNG, Sharif BS, Bennett MK, Murray A. Microscopic image analysis for quantitative measurement and feature identification of normal of cancerous colonic mucosa. IEEE Transactions on Information Technology in Biomedicine. 1998;2(3):197–203. doi: 10.1109/4233.735785. [DOI] [PubMed] [Google Scholar]


