The d3-GHR genetic variant polymorphism modulates GH responsiveness throughout life span and positively affects male longevity.
Keywords: growth hormone receptor, IGF-I, positive pleiotropy, longevity, Centenarians, d3-GHR
Abstract
Although both growth hormone (GH) and insulin-like growth factor 1 (IGF-1) signaling were shown to regulate life span in lower organisms, the role of GH signaling in human longevity remains unclear. Because a GH receptor exon 3 deletion (d3-GHR) appears to modulate GH sensitivity in humans, we hypothesized that this polymorphism could play a role in human longevity. We report a linear increased prevalence of d3-GHR homozygosity with age in four independent cohorts of long-lived individuals: 841 participants [567 of the Longevity Genes Project (LGP) (8% increase; P = 0.01), 152 of the Old Order Amish (16% increase; P = 0.02), 61 of the Cardiovascular Health Study (14.2% increase; P = 0.14), and 61 of the French Long-Lived Study (23.5% increase; P = 0.02)]. In addition, mega analysis of males in all cohorts resulted in a significant positive trend with age (26% increase; P = 0.007), suggesting sexual dimorphism for GH action in longevity. Further, on average, LGP d3/d3 homozygotes were 1 inch taller than the wild-type (WT) allele carriers (P = 0.05) and also showed lower serum IGF-1 levels (P = 0.003). Multivariate regression analysis indicated that the presence of d3/d3 genotype adds approximately 10 years to life span. The LGP d3/d3-GHR transformed lymphocytes exhibited superior growth and extracellular signal–regulated kinase activation, to GH treatment relative to WT GHR lymphocytes (P < 0.01), indicating a GH dose response. The d3-GHR variant is a common genetic polymorphism that modulates GH responsiveness throughout the life span and positively affects male longevity.
INTRODUCTION
Growth hormone (GH) and insulin-like growth factor (IGF) play a central role in development, differentiation, growth, and metabolism among divergent taxa (1). For instance, the absence of the IGF-1 gene is associated with poor prenatal growth both in rodents and in humans (2). The activities of IGF-1 are regulated by six IGF binding proteins (IGFBP1 to IGFBP6), which act as carrier and modulatory proteins for IGF-1 and are produced in a diverse array of tissues via complex regulatory processes. Among these six proteins, IGFBP3 is complexed to more than 90% of the circulating IGF-1 (3).
Dwarf individuals appear to live longer among many species (4, 5), suggesting a role for the GH/IGF-1 axis in modulating aging and life span. A considerable body of in vitro experimental evidence also suggests an important role for the IGF axis in human longevity and aging-related processes in a tissue-specific manner (3). Furthermore, several studies in selected human populations lend support on the role of this axis in health and life span. For instance, we have previously identified a cluster of functional mutations in the IGF-1 receptor in centenarians (6). We showed that Laron dwarfs, who are naturally short, have decreased prevalence of diabetes, cancer, and stroke, suggesting increased health span although life span in this small sample size cannot be determined accurately (7). Also, we previously established that centenarians with lower levels of IGF-1 had significantly longer survival (8). Clearly, individuals with severe GH deficiency have reduced life expectancy (9), suggesting that some GH is necessary for survival. On the other hand, interventional GH therapy in humans is commonly used to reverse age-related morbidities; hence, the kind of deficiency that will be most beneficial for health span and longevity needs to be further established.
GH is produced by the anterior pituitary gland and circulates in a pulsatile manner, with peaks during sleep. The production of GH is mediated by ghrelin and GH-releasing hormone and inhibited by somatostatin and IGF-1 feedback inhibition. The peaks of circulating GH are modulated by several factors, including age, health conditions, gender, and nutrition. GH production is decreased with age; however, it is never completely diminished (10). That said, there is accumulating evidence that GH may play a crucial role in modulating aging. Surprisingly, GH deficiency or diminished secretion has been linked to longevity phenotypes both in mice models and in humans with familial longevity (11, 12).
The GH receptor (GHR) gene is located on the short arm of chromosome 5 (p12-p13.1) and has nine coding exons (13). It consists of two common isoforms: (i) full-length GHR—flGHR (NM_000163) and (ii) a shorter form with a deletion of exon 3 in which exon 2 is spliced in frame to exon 4, resulting in the deletion of 22 amino acids within the N-terminal domain of the receptor—d3-GHR (14, 15). The allele frequencies of these isoforms among human populations range from 68 to 90% and 10 to 32% for flGHR and d3-GHR (15–17), respectively.
The effects of GHR isoforms on human health have provided mixed results. Likewise, results on the efficacy of human GH (hGH) therapy to d3-GHR human subjects are variable. Following the initial observation of a positive association between growth response and hGH therapy among d3-GHR carriers (16), both positive (16, 17) and no relationship (18, 19) have been reported, resulting in two meta-analyses showing modest positive dominant effects of the d3-GHR genotype for the response to GH in various etiologies of short stature (20, 21). In two Genome Wide Association Studies (GWAS) based on single-nucleotide polymorphisms (SNPs) (22, 23), the GHR locus showed association with final height. However, to our knowledge, the association of d3-GHR with final height has not been examined possibly because individuals with d3-GHR are expected to maintain normal GH action despite lower GH production, as hypothesized by Bougnères (24) and Pantel et al. (25). Hence, it is reasonable to hypothesize that increased GH sensitivity can also alter IGF-1 secretion (26, 27) and therefore regulate longevity (28, 29).
Given the potential role of the GH/IGF axis in longevity, we hypothesize that low IGF-1 levels will assure longevity of the d3-GHR carriers. To address this hypothesis, we genotyped d3-GHR locus in four human cohorts with long-lived participants, and we tested its association with longevity-related phenotypes and stature with a relatively common GHR variation.
RESULTS
Relationship between d3-GHR and phenotypic traits
d3-GHR and longevity
In Ashkenazi males, but not in females, a marked difference in allele frequency for the exon 3 deletion polymorphism (d3-GHR) was found between centenarian and control, as well as offspring and control groups. Whereas the male control group carried only 4% homozygote deletions, male offspring of centenarians and male centenarians carried 11 and 12%, respectively. Although these differences did not reach statistically significant levels among the offspring (P = 0.07), they crossed the significance threshold in centenarians (P < 0.05) versus control (Fig. 1). Note that the mean age difference between the centenarians and their offspring is about 30 years. We applied age trend analysis to test the relationship between the d3-GHR polymorphism in pooled control and centenarian’s data. The results showed a significant positive trend (P = 0.017), implying an enrichment of this allele with advancing age (Fig. 2A).
Fig. 1. Percentage of d3-GHR homozygosity among female (black) (F) and male (gray) (M) Ashkenazi centenarians (95F and 102M), offspring (113F and 110M), and controls (53F and 94M).
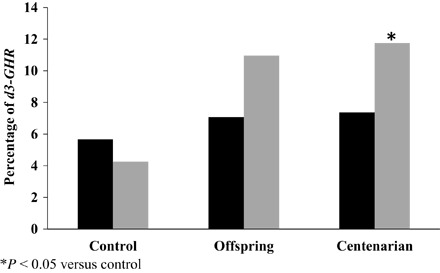
Fig. 2. d3-GHR age prevalence within and between cohorts.

Prevalence of d3-GHR homozygotes in relation to age groups in: (A) Ashkenazi Jew (AJ) (female and male of control and centenarian, n = 344) combined and (A1) split by gender (196 males and 148 females), (B) OOA 152 males, (C) 61 white males of the CHS, and (D) 61 French white males and (E) mega analysis of the four cohorts (470 males). In each of the cohorts, there is increased prevalence of the homozygote d3-GHR (*P < 0.05 for the trend).
We further validated these results in three independent cohorts—the Old Order Amish (OOA), Cardiovascular Health Study (CHS), and the French Long-Lived Study (FLLS). In the OOA, homozygosity for the d3-GHR increased with age among males, from 5% at 45 years of age to 21% at 75 years of age (P = 0.02) (Fig. 2B). In CHS, these differences showed similar trends; the homozygotes increased from 8.3% at 75 to 85 years of age to 26.5% at 86 to 95 years of age (P = 0.14; Fig. 2C). In the French cohort, the frequency of d3-GHR homozygotes increased from 6.45% at <100 years of age to 30% at >100 years of age (P = 0.02) (Fig. 2D). Finally, we conducted a mega analysis with males of the four cohorts in which d3-GHR homozygotes increased from 6.45% at <100 years of age to 30% at >100 years of age (P = 0.02) (Fig. 2E). These results demonstrate a consistent relationship between homozygosity for the d3-GHR deletion allele and longevity among the cohorts studied. However, this observation was limited only to males; the frequency of d3-GHR deletion homozygosity among females did not differ with age in any of the cohorts studied.
d3-GHR and height
Because d3-GHR has been shown to be associated with shorter stature (30), we investigated height among homozygotes for this deletion using a recessive model adjusted for gender, age, and group in our Ashkenazi Jew study cohorts (AJs). In the combined data set of AJ subjects, d3-GHR homozygotes across ages were taller compared to wild type (WT) and the heterozygotes by 1 inch {(67.3 inches versus 66.3 inches, P = 0.05; table S1), this phenomenon might be driven by the effect observed among centenarians [66.8 inches versus 64.7 inches, P = 0.05; table S1 (other validation cohorts did not have this phenotype)]}.
Relationships among d3-GHR, IGF-1, and IGFBPs
We further tested whether the d3-GHR polymorphism was related to the levels of IGF-1 and its IGFBPs. Previously, Suh et al. (6) demonstrated high IGF-1 levels and short stature among female offspring of centenarians and suggested a mechanism by which IGF-1 insensitivity could occur in females. However, this finding was not replicated among males in the offspring of centenarians. In contrast, the frequency of the homozygote GHR deletion polymorphism increased in males in all cohorts, suggesting gender specificity. Also, this increment was associated with low IGF-1 (tables S1 and S2 under dominant model). d3-GHR homozygote deletion carriers or those with heterozygote under dominant model showed significant low levels of IGFBP1 compared with WT, in offspring (table S3) and in allelic or dominant model among all groups (table S1), and control and offspring combined (table S4). Other IGFBPs did not reveal this relationship.
d3-GHR and GHBP
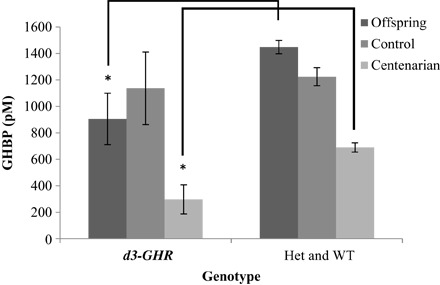
We further tested for association of the d3-GHR deletion with the levels of GHBP. All association tests revealed a significant inverse relationship between the carriers of the deletion and Growth Hormone Binding Protein (GHBP) levels (Fig. 3 and tables S1 to S4).
Fig. 3. Distribution of GHBP (in pM) among the centenarians (95F and 102M), their offspring (113F and 110M), and controls (53F and 94M), all of whom are of Ashkenazi (AJ) descent, in recessive model [homozygous GHR deletion of exon 3 (d3-GHR) versus heterozygote (Het) and WT combined], adjusted for gender and age (*P < 0.05).
d3-GHR and survival
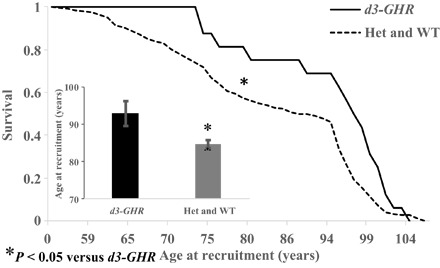
Of the 196 genotyped male subjects in the longevity cohort, 16 were homozygous for the deletion (>8% of total). The relationship between the d3-GHR deletion genotypes and mortality assessed through Kaplan-Meier survival function suggests that the probability of survival was significantly greater among the deletion carriers than the noncarriers (P = 0.01; Fig. 4). The survivors were 10 years older, on average, than the WT carriers, suggesting plausible beneficial effects of this polymorphism on age and longevity.
Fig. 4. Kaplan-Maier survival curves of 196 male AJ centenarians (n = 102) and control (n = 94) of d3-GHR in recessive model [homozygous GHR deletion of exon 3 (d3-GHR) versus heterozygote (Het) and WT combined].
Functional studies of d3-GHR polymorphism
To assess the functional impact of the deletion, we determined the in vitro effects of GH stimulation (100 ng/ml) on the rate of proliferation of transformed lymphocytes from male centenarians. In the basal state, lymphocytes from d3-GHR homozygous subjects had lower proliferation rates compared to WT homozygotes. However, in the presence of GH, we observed higher proliferation rates in d3 homozygotes (Fig. 5A). In addition, we assessed the phosphorylation of extracellular signal–regulated kinase (ERK) in transformed lymphocytes from male centenarians. Lymphocytes from d3-GHR homozygotes displayed lower basal activation of ERK compared to WT homozygotes. Conversely, higher activation of ERK was found in response to GH treatment (100 ng/ml), compared to WT homozygotes (P < 0.01) (Fig. 5B). No effect on the phosphorylation of signal transducers and activators of transcription 5 (STAT5) and AKT was evident (fig. S1), suggesting a unique, pathway-specific effect of the d3-GHR polymorphism on GH signaling with decreased constitutive activation yet enhanced GH-driven signaling. These results are compatible with reports on the growth of d3-GHR children showing an increased responsiveness to GH only in studies that use high doses of GH.
Fig. 5. Functional effect of d3-GHR carriers.
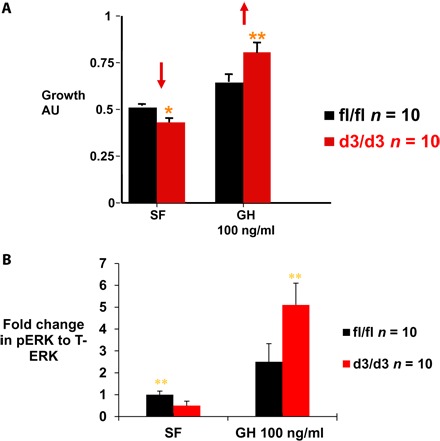
(A) In vitro effect of GH stimulation (100 ng/ml) on proliferation of transformed lymphocytes from male AJ centenarians. In the basal state [serum-free (SF)], lymphocytes from d3-GHR homozygous subjects have reduced proliferation rates; however, in the presence of GH, there is enhanced responsiveness and overall higher growth rates. This is compatible with reports in humans showing an increased responsiveness to GH only in studies that use high doses of GH. AU, arbitrary units. (B) Phosphorylation of ERK in homozygous WT and d3-GHR transformed lymphocytes from male AJ centenarians. Lymphocytes from d3-GHR homozygotes displayed significantly lower pERK levels under serum-free conditions but higher activation of ERK in response to GH treatment (100 ng/ml), compared to WT (fl/fl) carriers (**P < 0.01).
DISCUSSION
Although numerous genes have been shown to influence longevity (31), certain genes, including Igf1, appear to affect life span across diverse organisms, ranging from worms to humans (32). In humans, IGF-1 is associated with key biochemical pathways that regulate energy metabolism, immunity, inflammation, and imprinting and hence influence growth, development, and senescence from fetus to old age (33). Hence, it is reasonable to suggest that the IGFs exert pleiotropic effects on a wide range of cellular, physiological, and morphological variables throughout the life span of individuals. For instance, IGF-1 has been shown to prevent age-related myopathy through attenuation of myocyte death, repair muscle damage through regeneration of skeletal muscles in old animals, and up-regulate telomerase activity (34). Furthermore, as presented elsewhere, mutations in Igf1 have been shown to bring about a series of correlated responses: decreased IGF-1 levels, lower incidence of heart failure and mortality, and increased longevity in mice (35). Our results corroborate similar studies conducted on other cohorts. For instance, an inverse relationship between reduced IGF-1 levels and longevity has been reported on an Italian centenarian cohort (36) and in the Leiden 85-plus Study (37).
Similarly, GHR deficiency is associated with an unchanged life span but with a reduced incidence of cancer and diabetes (7). The phenotypic effects of GHR could be attributed to the direct activation of tyrosine kinase or indirect induction of IGF-1 (38), whose excess can be detrimental.
In centenarians, most IGF-1 regulation seems to respond to caloric and protein nutritional signals, not from GH. IGF-1 is not lower in carriers of d3-GHR during childhood, adolescence, and adulthood despite several reports showing that the GHR genotype may influence circulating IGF-1 under basal conditions. People with d3-GHR or fl-GHR alleles produce comparable amounts of circulating IGF-1 (16, 39–41). That said, we suspect people with d3-GHR alleles to have a decreased GH secretion. GH secretion by pituitary somatotropic cells is normally regulated by hypothalamic neuroendocrine mediators that are, at least partially, sensitive to feedback effects of IGF-1 or metabolic substrates. A higher level of GH transduction signaling through the d3-GHR would result in the following conditions: a greater GH impact on liver and the growth plate; and among other targets, increased IGF-1 generation, less feedback exerted on pituitary GH production, lower circulating GH levels, less binding to the d3-GHR–equipped cells, and reestablishment of a desirable level of GH action on relevant targets.
It appears that deletion of the GHR gene exon 3 might have originated from complex genomic events taking place after the emergence of Old World monkeys, followed by homologous recombination between two retro-elements in Homo sapiens. Thereafter, it spread throughout the human clades to be present now in approximately 25% of Caucasian chromosomes.
Strawbridge et al. (26) suggested that d3-GHR may offer some degree of protection against type 2 diabetes mellitus (T2DM) because the percent of individuals homozygous for the d3-GHR allele was only 7% in T2DM versus 27% in Normal Glucose Tolerance (NGT). The diminution of T2DM prevalence may increase longevity. This protective effect of the d3-GHR genotype is independent of IGF-1, because no significant differences of circulating IGF-1 were observed across the three d3-GHR genotypic groups. This is consistent with previous GWAS and studies that have not found that the GHR genotype contributes to the individual variation of circulating IGF-1 (42).
The mode of GH secretion has not yet been studied in centenarians. It is possible that people exposed lifelong to lower GH secretion live longer, because the activity of GH is involved with many physiological systems associated with longevity. In addition, we and others have reported an association between low IGF-1 and longevity (8, 37, 43). Similarly, several others (16, 39–41) have shown numerically higher IGF-1 levels in d3-GHR carriers, which did not reach significant levels because of absence of recessive model analysis or small sample size. Further, people with d3-GHR may have increased in number because of these advantages, including low GH secretion for a given IGF-1 level: If nutrition increases IGF-1 in these people, then they will keep their GH relatively low through a more efficient feedback. We should remember that GH–IGF-1 relationships are regulated to favor growth in young ages but fit other purposes in older ages, such as body composition, insulin secretion, and energy flux. Finally, GHRKO mice demonstrated longer life expectancy and lower IGF-1 levels compared with WT (44). Following this line of thought, we suggest that people with d3-GHR should maintain normal GH action despite less GH being produced. The inverse relationship between low IGF-1 and increased d3-GHR prevalence (as indicated in this paper) supports the notion that d3-GHR is favorable to longevity.
The positive association between d3-GHR homozygote frequencies and longevity in the three distinct populations with different demographic histories found in our study agrees with the earlier reports that reduced GH/IGF signaling may be involved in modulating human longevity and could signify a broader phenomenon. The positive relationship between the d3-GHR homozygote frequency and longevity, as well as height, suggests that decreased levels of GHR expression in homozygotes may have favorable effects on longevity. Further, whereas in most reports d3-GHR homozygote carriers are associated with short or no difference in stature, Audi et al. (19) demonstrated a tendency of the d3-GHR homozygote carriers to be taller, and Strawbridge et al. (26) reported significant 2+-inches-taller normal glucose-tolerant d3/d3 subjects in an additive model compared with the other genotypes. This general trend was further confirmed through cell culture studies, in which cells carrying GHR homozygote exon 3 deletions displayed a significantly slower rate of growth and lower activation of ERK at baseline. However, with GH treatment, d3-GHR homozygous lymphocytes showed superior growth and ERK activation, relative to homozygous WT lymphocytes. This phenomenon could be explained by the positive relationship of GHBP with longevity among AJs, further suggesting the plausibility of a consistent relationship between d3-GHR variation and GH secretion from cellular to organism levels over the life course of individuals. These observations suggest that IGF-1 may exert positive pleiotropy (45) on many traits associated with healthy longevity. Our observation that IGFBP1 is higher in the d3-GHR subjects suggests that they are more insulin-sensitive (46).
Our study provides the first consistent evidence linking the GHR to human longevity. Although our study consists of four cohorts with a total of 841 subjects, the sample size ranged from 60 to 600; hence, it is generally low relative to the sample sizes used in many association studies. Mutations in GH-related genes may exert pleiotropic (45) effects throughout the life span of individuals, which appears to be a universal feature of most if not all major longevity assurance genes. Because IGFs are found across a wide range of organisms and perform similar functions among these, as suggested by Waddington (47), they could be viewed as “canalized” genes playing a vital role in the survival of organisms across taxa. Although the GHR deletion appears to show age-, cohort-, and gender-specific effects, we hypothesize that a combination of empirical and systems analyses would provide answers toward understanding the contextual effects of GH on longevity in relation to age, stage, and gender.
In summary, our findings suggest that the d3-GHR, as indicated by the distribution of homozygotes among the four populations examined, may be involved in modulating human longevity. These results may have implications in devising precision medicine strategies, such as GH-related interventional therapies in the elderly.
MATERIALS AND METHODS
Subjects and phenotyping
The Ashkenazi centenarian cohort (Longevity Genes Project)
The contemporary AJ population worldwide is descended from a founder population (estimated to be several thousand) originating in the 15th century. To a large extent, this population exhibits both cultural and genetic homogeneity. For these reasons, the AJ population has been successfully used in the discovery of many disease-associated genes (48). The AJ cohort in the present study, which consists of three groups: centenarians, their offspring, and controls (see below), was recruited as previously described (49–52). Most of the participants in this cohort were born in or moved to the United States before World War II. The ages of 197 male AJ centenarians with exceptional longevity were 95 or above, as verified from their passports. Offspring of the long-lived centenarians consisted of 223 subjects (49.3% male and 50.7% female) with a mean age of 68.4 years (range, 49 to 88 years). The control group, used for the purpose of comparison with the offspring group, consisted of 147 individuals (64% male and 36% female) recruited from two different sources: (i) spouses of the offspring (n = 65) and (ii) a group of independently recruited AJ from the Einstein Aging Study (n = 82) (53). In the control group, we excluded individuals if the parents lived to the age of >85 years. The mean age of the control group was 71 years (range, 43 to 92 years). Because there were no differences between any measured traits or GHR genotype frequencies between the two subgroups, they were combined and were treated as one control group. A detailed medical history questionnaire was administered, and a physical examination on the AJ was performed as previously described (49–51). Informed written consent was obtained from the participants in accordance with the policy of the Committee on Clinical Investigations of the Albert Einstein College of Medicine.
Independent (validation) cohorts
In addition to the 567 subjects from the AJ cohort, DNAs from the OOA, the CHS, and the FLLS were genotyped. Additional details of these cohorts are provided below.
Old Order Amish. The Amish Family Calcification Study (AFCS) was initiated in 2001 at the University of Maryland to identify the joint determinants of Bone Mineral Density (BMD) and vascular calcification specific to the Amish community of Lancaster County, PA. The Amish immigrated to the United States from Western Europe in the late 1700s and represent a genetically homogeneous founder population. The AFCS cohort consisted of 428 males and 627 females with an average age of 55 years (range, 25 to 98 years). Of these, 152 white males with a mean age of 56.4 years (range, 21 to 85 years) and d3-GHR genotypes were reported in this study (table S5A). The AFCS was approved by the University of Maryland Institutional Review Board (IRB), and written informed consent was obtained from all research participants.
Cardiovascular Health Study. This is a prospective population-based cohort study consisting of 5888 adult men and women who are 65 years and older. They were recruited from four field centers: Forsyth County, NC; Sacramento County, CA; Washington County, MD; and Pittsburgh, PA (54). Baseline examination for the original cohort, of whom 4925 or 95% self-identified their ethnicity as white, was performed over 1 year, beginning in May 1989. Of these, 61 white males with a mean age of 77 years (range, 65 to 94 years) and d3-GHR genotypes were reported in this study (table S5A). All clinical examinations/procedures were conducted under institutionally approved protocols for use of human subjects.
French Long-Lived Study. The FLLS included 325 French long-lived Caucasian male participants from 87 to 110 years old and 381 adult controls from 34 to 85 years old. Of these, 61 white males with a mean age of 94 years (range, 70 to 109 years) and d3-GHR genotypes were reported in this study (table S5A). Written informed consent was obtained from the study participants.
Phenotyping
Wide ranging phenotypic data encompassing physiological (lipids: cholesterol, triglyceride, and high-density lipoprotein and low-density lipoprotein cholesterol) (51), and IGF-1–related biomarkers (IGF-2, IGFBPs, insulin, and GHBP) (55), disease prevalence (myocardial infarction, stroke, diabetes, or cancer), and anthropometric (height and weight) (51) traits have been collected from the Ashkenazi centenarians, their offspring, and age-matched (to the offspring) controls (table S5B).
Genotyping
Genotyping of the d3-GHR polymorphism was performed using two consecutive polymerase chain reactions (PCRs) using the same genomic DNA sample, one with the G1-G2 set of primers followed by another with the G1-G3 set of primers (table S6). We used 50 ng of genomic DNA in a 50-μl PCR reaction mix consisting of 25-μl Amplitaq Gold PCR Master Mix (Applied Biosystems), 0.1 pM of each primer, and double-distilled water. The PCR was performed in a Bio-Rad instrument with the following conditions: 94°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 90 s, and a final extension at 72°C for 7 min. We then analyzed the PCR-amplified products by electrophoresis on a 1% agarose gel stained with ethidium bromide. The lengths of the PCR products for the G1-G2 and G1-G3 were 532 and 935 base pairs (bp), respectively. These bands were classified as follows: 935 bp–935 bp = homozygote for WT, 935 bp–532 bp = heterozygote, and 532 bp–532 bp = homozygote for the deletion.
Analysis of transformed lymphocytes
Blood samples obtained from subjects were rapidly processed at the General Clinical Research Center at the Albert Einstein College of Medicine for generating Epstein-Barr virus–transformed lymphocytes using the established methods. Informed written consent was obtained from all the participants, in accordance with the policy of the Committee on Clinical Investigations of the Albert Einstein College of Medicine. To assess the ability of GH to activate signal transduction via the GHR, we incubated lymphocytes harboring mutation and those without the mutation, as described (6). Growth assays on these cells were recorded, followed by total cell protein extraction from cell lysates. These were further subjected to SDS–polyacrylamide gel electrophoresis; immunoblotting with total- and phospho-AKT, STAT5 and ERK using antibodies from Cell Signaling; and densitometry. Results are means ± SD. Statistical significance was determined using unpaired t tests.
Statistical analysis
Data on serum triglycerides were ln-transformed for analysis and were back-transformed for presentation. Four comparisons of phenotypic data were performed on the AJ cohort: (i) within each group (that is, centenarian, offspring, and control—all of whom are of AJ descent); (ii) between centenarians, offspring, and controls; (iii) between offspring and controls; and (iv) between centenarians and controls. All analyses were adjusted for age and sex. We applied the most frequently used models in association studies: allelic (carriers of the deletion versus none), dominant (WT versus heterozygote and deletion homozygote), additive (WT versus heterozygote versus deletion homozygote), and recessive (deletion homozygote versus WT and heterozygote) in each comparison (tables S1 to S4).
Analysis of genotype data
Genotypes were checked for Mendelian consistency using the PedCheck software (56) before analysis. Mendelian errors were resolved or removed before analysis. Allele frequencies were calculated by gene counting, and all the genotypes conformed to Hardy-Weinberg expectations. We evaluated the association between SNP genotype and phenotype (for example, IGF levels or height) under the additive or dominant models using a variance component approach. We modeled the probability that the subject was a case or control as a function of the individual’s age, sex, and genotype, conditional on the correlations in phenotype among relative pairs. Statistical analyses were performed using JMP 12.
Supplementary Material
Acknowledgments
We thank all the participants from the three cohorts used in this study. Funding: This study was funded in part by grants from the NIH [(P01AG021654 to N.B.), (1R01AG042188 to G.A.), and (1R01AG 034430 and 1P01AG034906 to P.C.)], the Nathan Shock Center of Excellence for the Biology of Aging (P30AG038072 to N.B.), the Glenn Center for the Biology of Human Aging (Paul Glenn Foundation Grant to N.B. and P.C.), and Diabetes Center (DK-20541 to N.B.). D.F. and P.B. were supported by INSERM with additional contribution from Association de Recherche sur le Diabète and an institutional grant from Novo Nordisk France. CHS was supported by contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-55222, N01 HC-15103, N01-HC-75150, and N01-HC-45133; grant number U01 HL080295 from the National Heart, Lung, and Blood Institute; U19 AG023122 from the National Institute on Aging Longevity Consortium; and P30 AG021334 from the National Institute on Aging, Claude D. Pepper Older Americans Independence Centers; and grant R01 HL-071862 (to A.R.), with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of participating CHS investigators and institutions can be found at http://chs-nhlbi.org. Genotyping services for CHS were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the NIH to the Johns Hopkins University (contract number N01-HG-65403). Author contributions: D.B.-A. and D.R.G. performed data analysis and participated in drafting the manuscript. T.B., D.F., B.L., S.O., and L.S. were responsible for laboratory preparation and data acquisition. D.G. and A.R. performed data acquisition and helped with drafting the manuscript. BPS initial recruitment and data acquisition. P.D. and R.K. helped with drafting the manuscript. P.B., A.R.S., P.C., and N.B. coordinated and designed the studies and helped with drafting the manuscript. G.A. coordinated and designed the study, performed data analysis, and participated in drafting the manuscript. All authors read and approved the final manuscript. Competing interests: A.R.S. is an employee of Regeneron Pharmaceuticals. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The data and materials in this work can only be used in accordance with the filed University of Maryland IRB.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1602025/DC1
fig. S1. Phosphorylation of STAT5 and AKT in homozygous WT and d3-GHR transformed lymphocytes from male AJ centenarians.
table S1. Analysis (allelic, dominant, additive, and recessive models) of various variables (means ± SE), includes all AJs groups (n = 567) and adjusted for gender, age, and group.
table S2. Analysis (allelic, dominant, additive, and recessive) of various variables (means ± SE), includes AJ centenarian (C) and control (C) groups (n = 344) and adjusted for gender, age, and group.
table S3. Groups analysis of various variables (means ± SE) adjusted for gender and age within AJs.
table S4. Analysis (allelic, dominant, additive, and recessive models) of various variables (means ± SE), includes AJ offspring (O) and control (C) (n = 370) and adjusted for gender, age, and group.
table S5A. Genotyping efforts among the four cohorts (n = 841): AJ (n = 567, 56% female), OOA 152 males, CHS 61 males, and FLLS 61 males.
table S5B. Crude measurements of various variables (means ± SE) in the three AJs (n = 567) study groups.
table S6. PCR procedure using primers.
REFERENCES AND NOTES
- 1.Barbieri M., Bonafè M., Franceschi C., Paolisso G., Insulin/IGF-I-signaling pathway: An evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. 285, E1064–E1071 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Fu Q., Yu X., Callaway C. W., Lane R. H., McKnight R. A., Epigenetics: Intrauterine growth retardation (IUGR) modifies the histone code along the rat hepatic IGF-1 gene. FASEB J. 23, 2438–2449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberbauer A. M., The regulation of IGF-1 gene transcription and splicing during development and aging. Front. Endocrinol. 4, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J., Anzo M., Cohen P., Control of aging and longevity by IGF-I signaling. Exp. Gerontol. 40, 867–872 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Bartke A., Healthspan and longevity can be extended by suppression of growth hormone signaling. Mamm. Genome 27, 289–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh Y., Atzmon G., Cho M.-O., Hwang D., Liu B., Leahy D. J., Barzilai N., Cohen P., Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U.S.A. 105, 3438–3442 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guevara-Aguirre J., Balasubramanian P., Guevara-Aguirre M., Wei M., Madia F., Cheng C.-W., Hwang D., Martin-Montalvo A., Saavedra J., Ingles S., de Cabo R., Cohen P., Longo V. D., Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 3, 70ra13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milman S., Atzmon G., Huffman D. M., Wan J., Crandall J. P., Cohen P., Barzilai N., Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 13, 769–771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besson A., Salemi S., Gallati S., Jenal A., Horn R., Mullis P. S., Mullis P. E., Reduced longevity in untreated patients with isolated growth hormone deficiency. J. Clin. Endocrinol. Metab. 88, 3664–3667 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Höybye C., Christiansen J. S., Growth hormone replacement in adults—Current standards and new perspectives. Best Pract. Res. Clin. Endocrinol. Metab. 29, 115–123 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Bartke A., Pleiotropic effects of growth hormone signaling in aging. Trends Endocrinol. Metab. 22, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Spoel E., Jansen S. W., Akintola A. A., Ballieux B. E., Cobbaert C. M., Slagboom P. E., Blauw G. J., Westendorp R. G. J., Pijl H., Roelfsema F., van Heemst D., Growth hormone secretion is diminished and tightly controlled in humans enriched for familial longevity. Aging Cell 15, 1126–1131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godowski P. J., Leung D. W., Meacham L. R., Galgani J. P., Hellmiss R., Keret R., Rotwein P. S., Parks J. S., Laron Z., Wood W. I., Characterization of the human growth hormone receptor gene and demonstration of a partial gene deletion in two patients with Laron-type dwarfism. Proc. Natl. Acad. Sci. U.S.A. 86, 8083–8087 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palizban A. A., Radmansorry M., Bozorgzad M., Exon 3-deleted and full-length growth hormone receptor polymorphism frequencies in an Iranian population. Res. Pharm. Sci. 9, 489–494 (2014). [PMC free article] [PubMed] [Google Scholar]
- 15.Pantel J., Machinis K., Sobrier M.-L., Duquesnoy P., Goossens M., Amselem S., Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J. Biol. Chem. 275, 18664–18669 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos C., Essioux L., Teinturier C., Tauber M., Goffin V., Bougnères P., A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat. Genet. 36, 720–724 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Jorge A. A. L., Marchisotti F. G., Montenegro L. R., Carvalho L. R., Mendonca B. B., Arnhold I. J. P., Growth hormone (GH) pharmacogenetics: Influence of GH receptor exon 3 retention or deletion on first-year growth response and final height in patients with severe GH deficiency. J. Clin. Endocrinol. Metab. 91, 1076–1080 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Binder G., Baur F., Schweizer R., Ranke M. B., The d3-growth hormone (GH) receptor polymorphism is associated with increased responsiveness to GH in Turner syndrome and short small-for-gestational-age children. J. Clin. Endocrinol. Metab. 91, 659–664 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Audi L., Esteban C., Carrascosa A., Espadero R., Pérez-Arroyo A., Arjona R., Clemente M., Wollmann H., Fryklund L., Parodi L. A. Spanish SGA Study Group , Exon 3-deleted/full-length growth hormone receptor polymorphism genotype frequencies in Spanish short small-for-gestational-age (SGA) children and adolescents (n = 247) and in an adult control population (n = 289) show increased fl/fl in short SGA. J. Clin. Endocrinol. Metab. 91, 5038–5043 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Renehan A. G., Solomon M., Zwahlen M., Morjaria R., Whatmore A., Audi L., Binder G., Blum W., Bougnères P., Dos Santos C., Carrascosa A., Hokken-Koelega A., Jorge A., Mullis P. E., Tauber M., Patel L., Clayton P. E., Growth hormone receptor polymorphism and growth hormone therapy response in children: A Bayesian meta-analysis. Am. J. Epidemiol. 175, 867–877 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Wassenaar M. J. E., Dekkers O. M., Pereira A. M., Wit J. M., Smit J. W., Biermasz N. R., Romijn J. A., Impact of the exon 3-deleted growth hormone (GH) receptor polymorphism on baseline height and the growth response to recombinant human GH therapy in GH-deficient (GHD) and non-GHD children with short stature: A systematic review and meta-analysis. J. Clin. Endocrinol. Metab. 94, 3721–3730 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Zoledziewska M., Sidore C., Chiang C. W. K., Sanna S., Mulas A., Steri M., Busonero F., Marcus J. H., Marongiu M., Maschio A., Del Vecchyo D. O., Floris M., Meloni A., Delitala A., Concas M. P., Murgia F., Biino G., Vaccargiu S., Nagaraja R., Lohmueller K. E. UK10K Consortium, Timpson N. J., Soranzo N., Tachmazidou I., Dedoussis G., Zeggini E. Understanding Society Scientific Group, Uzzau S., Jones C., Lyons R., Angius A., Abecasis G. R., Novembre J., Schlessinger D., Cucca F., Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 47, 1352–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanktree M. B., Guo Y., Murtaza M., Glessner J. T., Bailey S. D., Onland-Moret N. C., Lettre G., Ongen H., Rajagopalan R., Johnson T., Shen H., Nelson C. P., Klopp N., Baumert J., Padmanabhan S., Pankratz N., Pankow J. S., Shah S., Taylor K., Barnard J., Peters B. J., Maloney C. M., Lobmeyer M. T., Stanton A., Zafarmand M. H., Romaine S. P. R., Mehta A., van Iperen E. P. A., Gong Y., Price T. S., Smith E. N., Kim C. E., Li Y. R., Asselbergs F. W., Atwood L. D., Bailey K. M., Bhatt D., Bauer F., Behr E. R., Bhangale T., Boer J. M. A., Boehm B. O., Bradfield J. P., Brown M., Braund P. S., Burton P. R., Carty C., Chandrupatla H. R., Chen W., Connell J., Dalgeorgou C., de Boer A., Drenos F., Elbers C. C., Fang J. C., Fox C. S., Frackelton E. C., Fuchs B., Furlong C. E., Gibson Q., Gieger C., Goel A., Grobbee D. E., Hastie C., Howard P. J., Huang G.-H., Johnson W. C., Li Q., Kleber M. E., Klein B. E. K., Klein R., Kooperberg C., Ky B., Lacroix A., Lanken P., Lathrop M., Li M., Marshall V., Melander O., Mentch F. D., Meyer N. J., Monda K. L., Montpetit A., Murugesan G., Nakayama K., Nondahl D., Onipinla A., Rafelt S., Newhouse S. J., Otieno F. G., Patel S. R., Putt M. E., Rodriguez S., Safa R. N., Sawyer D. B., Schreiner P. J., Simpson C., Sivapalaratnam S., Srinivasan S. R., Suver C., Swergold G., Sweitzer N. K., Thomas K. A., Thorand B., Timpson N. J., Tischfield S., Tobin M., Tomaszewski M., Verschuren W. M. M., Wallace C., Winkelmann B., Zhang H., Zheng D., Zhang L., Zmuda J. M., Clarke R., Balmforth A. J., Danesh J., Day I. N., Schork N. J., de Bakker P. I. W., Delles C., Duggan D., Hingorani A. D., Hirschhorn J. N., Hofker M. H., Humphries S. E., Kivimaki M., Lawlor D. A., Kottke-Marchant K., Mega J. L., Mitchell B. D., Morrow D. A., Palmen J., Redline S., Shields D. C., Shuldiner A. R., Sleiman P. M., Smith G. D., Farrall M., Jamshidi Y., Christiani D. C., Casas J. P., Hall A. S., Doevendans P. A., Christie J. D., Berenson G. S., Murray S. S., Illig T., Dorn G. W. II, Cappola T. P., Boerwinkle E., Sever P., Rader D. J., Reilly M. P., Caulfield M., Talmud P. J., Topol E., Engert J. C., Wang K., Dominiczak A., Hamsten A., Curtis S. P., Silverstein R. L., Lange L. A., Sabatine M. S., Trip M., Saleheen D., Peden J. F., Cruickshanks K. J., März W., O’Connell J. R., Klungel O. H., Wijmenga C., Maitland-van der Zee A. H., Schadt E. E., Johnson J. A., Jarvik G. P., Papanicolaou G. J. Hugh Watkins on behalf of PROCARDIS, Grant S. F. A., Munroe P. B., North K. E., Samani N. J., Koenig W., Gaunt T. R., Anand S. S., van der Schouw Y. T. Meena Kumari on behalf of the Whitehall II Study and the WHII 50K Group, Soranzo N., Fitzgerald G. A., Reiner A., Hegele R. A., Hakonarson H., Keating B. J., Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 88, 6–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bougnères P., The exon-3 deletion of the growth hormone receptor (GHR) gene still has a limited impact in clinical endocrinology. J. Clin. Endocrinol. Metab. 95, 56–59 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Pantel J., Grulich-Henn J., Bettendorf M., Strasburger C. J., Heinrich U., Amselem S., Heterozygous nonsense mutation in exon 3 of the growth hormone receptor (GHR) in severe GH insensitivity (Laron syndrome) and the issue of the origin and function of the GHRd3 isoform. J. Clin. Endocrinol. Metab. 88, 1705–1710 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Strawbridge R. J., Kärvestedt L., Li C., Efendic S., Östenson C. G., Gu H. F., Brismar K., GHR exon 3 polymorphism: Association with type 2 diabetes mellitus and metabolic disorder. Growth Horm. IGF Res. 17, 392–398 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Ouni M., Castell A.-L., Linglart A., Bougnères P., Genetic and epigenetic modulation of growth hormone sensitivity studied with the IGF-1 generation test. J. Clin. Endocrinol. Metab. 100, E919–E925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tapia P. C., RhoA, Rho kinase, JAK2, and STAT3 may be the intracellular determinants of longevity implicated in the progeric influence of obesity: Insulin, IGF-1, and leptin may all conspire to promote stem cell exhaustion. Med. Hypotheses 66, 570–576 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Rincon M., Muzumdar R., Atzmon G., Barzilai N., The paradox of the insulin/IGF-1 signaling pathway in longevity. Mech. Ageing Dev. 125, 397–403 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Park S. W., Lee S.-T., Sohn Y. B., Kim S. H., Cho S.-Y., Ko A.-r., Ji S.-T., Kwon J.-Y., Yeau S., Paik K.-H., Kim J.-W., Jin D.-K., A polymorphism in the growth hormone receptor is associated with height in children with Prader-Willi syndrome. Am. J. Med. Genet. A 155A, 2970–2973 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Tacutu R., Craig T., Budovsky A., Wuttke D., Lehmann G., Taranukha D., Costa J., Fraifeld V. E., de Magalhães J. P., Human Ageing Genomic Resources: Integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 41, D1027–D1033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longo V. D., Finch C. E., Evolutionary medicine: From dwarf model systems to healthy centenarians? Science 299, 1342–1346 (2003). [DOI] [PubMed] [Google Scholar]
- 33.C. Finch, The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Lifespans (Academic Press, 2007). [Google Scholar]
- 34.Anversa P., Aging and longevity: The IGF-1 enigma. Circ. Res. 97, 411–414 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Bartke A., Can growth hormone (GH) accelerate aging? Evidence from GH-transgenic mice. Neuroendocrinology 78, 210–216 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Bonafè M., Barbieri M., Marchegiani F., Olivieri F., Ragno E., Giampieri C., Mugianesi E., Centurelli M., Franceschi C., Paolisso G., Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: Cues for an evolutionarily conserved mechanism of life span control. J. Clin. Endocrinol. Metab. 88, 3299–3304 (2003). [DOI] [PubMed] [Google Scholar]
- 37.van der Spoel E., Rozing M. P., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J. M., Westendorp R. G. J., van Heemst D., Association analysis of insulin-like growth factor-1 axis parameters with survival and functional status in nonagenarians of the Leiden Longevity Study. Aging 7, 956–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks A. J., Waters M. J., The growth hormone receptor: Mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 6, 515–525 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Giavoli C., Ferrante E., Profka E., Olgiati L., Bergamaschi S., Ronchi C. L., Verrua E., Filopanti M., Passeri E., Montefusco L., Lania A. G., Corbetta S., Arosio M., Ambrosi B., Spada A., Beck-Peccoz P., Influence of the d3GH receptor polymorphism on the metabolic and biochemical phenotype of GH-deficient adults at baseline and during short- and long-term recombinant human GH replacement therapy. Eur. J. Endocrinol. 163, 361–368 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Jallad R. S., Trarbach E. B., Duarte F. H., Jorge A. A. L, Bronstein M. D., Influence of growth hormone receptor (GHR) exon 3 and -202A/C IGFBP-3 genetic polymorphisms on clinical and biochemical features and therapeutic outcome of patients with acromegaly. Pituitary 18, 666–673 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Glad C. A. M., Carlsson L. M. S., Melander O., Almgren P., Sjöström L., Nilsson S., Larsson I., Svensson P.-A., Johannsson G., The GH receptor exon 3 deleted/full-length polymorphism is associated with central adiposity in the general population. Eur. J. Endocrinol. 172, 123–128 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Alfred T., Ben-Shlomo Y., Cooper R., Hardy R., Cooper C., Deary I. J., Gaunt T. R., Gunnell D., Harris S. E., Kumari M., Martin R. M., Sayer A. A., Starr J. M., Kuh D., Day I. N. M. HALCyon Study Team , A multi-cohort study of polymorphisms in the GH/IGF axis and physical capability: The HALCyon programme. PLOS ONE 7, e29883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perice L., Barzilai N., Verghese J., Weiss E. F., Holtzer R., Cohen P., Milman S., Lower circulating insulin-like growth factor-I is associated with better cognition in females with exceptional longevity without compromise to muscle mass and function. Aging 8, 2414–2424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Junnila R. K., Duran-Ortiz S., Suer O., Sustarsic E. G., Berryman D. E., List E. O., Kopchick J. J., Disruption of the GH receptor gene in adult mice increases maximal lifespan in females. Endocrinology 157, 4502–4513 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Parsons P. A., Antagonistic pleiotropy and the stress theory of aging. Biogerontology 8, 613–617 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Rajwani A., Ezzat V., Smith J., Yuldasheva N. Y., Duncan E. R., Gage M., Cubbon R. M., Kahn M. B., Imrie H., Abbas A., Viswambharan H., Aziz A., Sukumar P., Vidal-Puig A., Sethi J. K., Xuan S., Shah A. M., Grant P. J., Porter K. E., Kearney M. T., Wheatcroft S. B., Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes 61, 915–924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. H. Waddington, The Strategy of Genes (George Allen & Unwin, 1957). [Google Scholar]
- 48.Lancaster J. M., Carney M. E., Futreal P. A., BRCA 1 and 2–A genetic link to familial breast and ovarian cancer. Medscape Womens Health 2, 7 (1997). [PubMed] [Google Scholar]
- 49.Atzmon G., Gabriely I., Greiner W., Davidson D., Schechter C., Barzilai N., Plasma HDL levels highly correlate with cognitive function in exceptional longevity. J. Gerontol., Ser. A 57, M712–M715 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Atzmon G., Schechter C., Greiner W., Davidson D., Rennert G., Barzilai N., Clinical phenotype of families with longevity. J. Am. Geriatr. Soc. 52, 274–277 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Barzilai N., Atzmon G., Schechter C., Schaefer E. J., Cupples A. L., Lipton R., Cheng S., Shuldiner A. R., Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA 290, 2030–2040 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Atzmon G., Rincon M., Schechter C. B., Shuldiner A. R., Lipton R. B., Bergman A., Barzilai N., Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLOS Biol. 4, e113 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verghese J., Lipton R. B., Hall C. B., Kuslansky G., Katz M. J., Buschke H., Abnormality of gait as a predictor of non-Alzheimer’s dementia. N. Engl. J. Med. 347, 1761–1768 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Fried L. P., Borhani N. O., Enright P., Furberg C. D., Gardin J. M., Kronmal R. A., Kuller L. H., Manolio T. A., Mittelmark M. B., Newman A., O’Leary D. H., Psaty B., Rautaharju P., Tracy R. P., Weiler P. G., The Cardiovascular Health Study: Design and rationale. Ann. Epidemiol. 1, 263–276 (1991). [DOI] [PubMed] [Google Scholar]
- 55.Wan J., Atzmon G., Hwang D., Barzlai N., Kratzsch J., Cohen P., Growth hormone receptor (GHR) exon 3 polymorphism status detection by dual-enzyme-linked immunosorbent assay (ELISA). J. Clin. Endocrinol. Metab. 98, E77–E81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connell J. R., Weeks D. E., PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1602025/DC1
fig. S1. Phosphorylation of STAT5 and AKT in homozygous WT and d3-GHR transformed lymphocytes from male AJ centenarians.
table S1. Analysis (allelic, dominant, additive, and recessive models) of various variables (means ± SE), includes all AJs groups (n = 567) and adjusted for gender, age, and group.
table S2. Analysis (allelic, dominant, additive, and recessive) of various variables (means ± SE), includes AJ centenarian (C) and control (C) groups (n = 344) and adjusted for gender, age, and group.
table S3. Groups analysis of various variables (means ± SE) adjusted for gender and age within AJs.
table S4. Analysis (allelic, dominant, additive, and recessive models) of various variables (means ± SE), includes AJ offspring (O) and control (C) (n = 370) and adjusted for gender, age, and group.
table S5A. Genotyping efforts among the four cohorts (n = 841): AJ (n = 567, 56% female), OOA 152 males, CHS 61 males, and FLLS 61 males.
table S5B. Crude measurements of various variables (means ± SE) in the three AJs (n = 567) study groups.
table S6. PCR procedure using primers.


