Hyperpolarized micro-NMR nondestructively analyzes metabolic activities in scant cells.
Keywords: Hyperpolarization, Nuclear magnetic resonance, Cancer metabolism, metabolic flux, Leukemia stem cells, microfluidics
Abstract
Metabolic reprogramming is widely considered a hallmark of cancer, and understanding metabolic dynamics described by the conversion rates or “fluxes” of metabolites can shed light onto biological processes of tumorigenesis and response to therapy. For real-time analysis of metabolic flux in intact cells or organisms, magnetic resonance (MR) spectroscopy and imaging methods have been developed in conjunction with hyperpolarization of nuclear spins. These approaches enable noninvasive monitoring of tumor progression and treatment efficacy and are being tested in multiple clinical trials. However, because of their limited sensitivity, these methods require a larger number of cells, on the order of 107, which is impractical for analyzing scant target cells or mass-limited samples. We present a new technology platform, a hyperpolarized micromagnetic resonance spectrometer (HMRS), that achieves real-time, 103-fold more sensitive metabolic analysis on live cells. This platform enables quantification of the metabolic flux in a wide range of cell types, including leukemia stem cells, without significant changes in viability, which allows downstream molecular analyses in tandem. It also enables rapid assessment of metabolic changes by a given drug, which may direct therapeutic choices in patients. We further advanced this platform for high-throughput analysis of hyperpolarized molecules by integrating a three-dimensionally printed microfluidic system. The HMRS platform holds promise as a sensitive method for studying metabolic dynamics in mass-limited samples, including primary cancer cells, providing novel therapeutic targets and an enhanced understanding of cellular metabolism.
INTRODUCTION
Altered metabolism in cancer, first discovered as a mitochondrial defect in the 1920s (1), is now recognized as one of the hallmarks of cancer; cancer cells can reprogram their metabolism to promote cellular growth and proliferation (2–4), adapt nutrient- or oxygen-depleted environments (5–8), and even escape immune surveillance (9–11). A common phenotype is an increased glucose uptake, which can be metabolized through glycolysis, regardless of oxygen availability. Known as aerobic glycolysis or the “Warburg effect,” this pathway provides metabolic intermediates critical for numerous biosynthetic processes, conferring proliferative advantages (4), and acidifies the tumor microenvironment, promoting metastasis to other organs (12, 13). Aerobic glycolysis is thus closely correlated with poor prognosis (14–16). Considering that the glycolytic rate, or “flux,” represents the metabolic activity of energetic reactions at any given moment derived from glucose, its analysis may describe how cancer cells respond to environmental changes, such as nutrient availability or drug treatment, in real time (9, 17).
Nuclear magnetic resonance (NMR) spectroscopy and imaging techniques have been developed in conjunction with hyperpolarization of nuclear spins for real-time analysis of metabolism in intact cells or organs. Dynamic nuclear polarization (DNP), in particular, is an emerging technique that achieves hyperpolarization by transferring polarization from unpaired electrons in a stable organic free radical to neighboring nuclei, demonstrating more than a 10,000-fold enhancement of NMR sensitivity (18). Using 13C-enriched metabolites (for example, [1-13C]pyruvate or [U-13C]glucose, which are NMR-active nuclei) as hyperpolarized molecules, the DNP-NMR technique can measure multiple metabolic pathways with minimal toxicity (17, 19, 20), and its application to cancer diagnosis is undergoing multiple clinical trials. However, this method requires numerous cells of interest (order of 107), because of its limited sensitivity, and analytical application of DNP-NMR has been challenging with mass-limited samples, including primary cancer cells, stem cells, or organoids.
Here, we report a rapid and sensitive NMR platform designed for metabolic flux analysis on a small number of cells. Termed a hyperpolarized micromagnetic resonance spectrometer (HMRS), the system is (i) equipped with a dual-tuned microcoil circuit for sensitive detection of 13C molecules, (ii) optimized to process a small mass of biological sample, and (iii) integrated with a microfluidic system for high-throughput analysis. The HMRS system can quantify glycolytic flux with a 1000-fold higher sensitivity than conventional DNP-NMR approaches. We applied this system to quantify metabolic flux in leukemia stem cells (LSCs) in vitro and to assess drug treatment response before any changes in cell viability or proliferation have occurred. Furthermore, we extended this platform to the analysis of more than 10 samples of hyperpolarized molecules in one experiment, demonstrating its potential as a powerful system to study metabolic flux.
RESULTS
Hyperpolarized micromagnetic resonance spectrometer
Figure 1A illustrates the fundamental difference in glycolytic metabolism between cancer and normal cells. Pyruvate, a three-carbon intermediate product of glycolysis, is predominantly converted to lactate by lactate dehydrogenase (LDH; EC 1.1.2.4) in the cytoplasm or to acetyl–coenzyme A by pyruvate dehydrogenase (EC 1.2.4.1) in the mitochondria. LDH is typically overexpressed in cancer cells (4) and drives the conversion from pyruvate to lactate. In normal cells, pyruvate is oxidized to a greater degree and used for adenosine triphosphate generation in the mitochondria. When [1-13C]pyruvate enters cells directly through the monocarboxylate transporters, it enters the intracellular pool of pyruvate and is metabolized, producing higher [1-13C]lactate in cancer cells as compared to normal cells (fig. S1) (21, 22). Hyperpolarization of [1-13C]pyruvate then provides a means of probing this last step of glycolysis noninvasively with a marked enhancement of MR signal.
Fig. 1. HMRS platform.
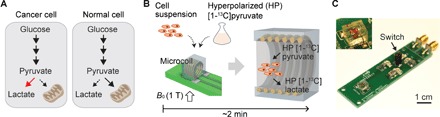
(A) Schematic of glycolytic metabolism in cancer and normal cells. (B) Schematic of HMRS assay. 13C-enriched pyruvate is hyperpolarized and mixed with cell suspension. Once it is loaded into the microcoil, 13C NMR acquisition starts in a 1.05-T magnetic field. (C) Optical picture of the microcoil probe in the HMRS. The mechanical switch was used for changing the resonance mode of the microcoil. Inset shows the microcoil embedded in a polydimethylsiloxane (PDMS) block. Net volume inside the coil is 2 μl.
Figure 1B shows the HMRS assay scheme. Samples (cell suspension) are mixed with hyperpolarized [1-13C]pyruvate and then loaded into the microcoil (diameter, 1.4 mm; volume, 2 μl ) in the HMRS system (Fig. 1C). Once the microcoil probe is moved into the center of the magnetic field (B0, 1.05 T), the NMR signal acquisition is started. Because pyruvate and lactate have relatively long spin-lattice relaxation times, T1, at 1.05 T (71 and 43 s, respectively) (23), the hyperpolarization states of these molecules do not decay significantly during the metabolic reactions and signal acquisition (24). It is important to note that the whole process of analyzing metabolic flux is completed within 2 min (Fig. 1B). The microcoil probe circuit was implemented to resonate at the Larmor frequencies of 1H and 13C in our permanent magnet (44.69 and 11.24 MHz, respectively). In general, a permanent magnet provides a cost-effective solution for MR research, but it suffers from temporal and spatial inhomogeneity of a magnetic field. We addressed these problems with the 1H resonance mode in the microcoil; the drift and inhomogeneity were diagnosed and calibrated with first-order shimming in the 1H resonance mode. The resonance mode was then switched from 1H mode to 13C mode immediately before 13C NMR experiments (Fig. 1C and fig. S2).
The HMRS system acquired NMR spectra every 4 s with a 30° radio frequency (RF) pulse and quantified them on the basis of the peak area. Utilizing 50,000 UOK262 cells (a renal cell carcinoma) (25), we observed that the lactate peak was detected from the first NMR spectrum (Fig. 2, A and B). The positioning of the micro-NMR probe into the center of the magnetic field (B0) required a delay of 30 s before signal acquisition, because of the mechanical characteristics of the magnet system (nanoScan, Mediso). Therefore, the lactate peak in the first spectrum indicates that the cancer cells in the microcoil had already begun converting hyperpolarized pyruvate and generating lactate within the delay. Also, the slower decay of the lactate signal compared to the pyruvate demonstrated that the pyruvate-to-lactate conversion continued during the measurements (Fig. 2, B and C); hyperpolarized signals decay exponentially with a decay constant of 1/T1, and the fact that the lactate (T1 of 43 s) decayed slower than the pyruvate (T1 of 71 s) indicated that the former was generated and the latter was consumed (fig. S3).
Fig. 2. Analysis of metabolic flux using HMRS.
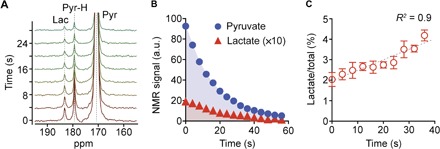
(A) NMR spectra of hyperpolarized metabolites from 50,000 UOK262 cells. Each spectrum was acquired every 4 s with a 30° RF pulse. Lac, lactate; Pyr-H, pyruvate hydrate; Pyr, pyruvate. (B) Plot of the NMR signals of pyruvate and lactate peaks shown in (A). Each signal was quantified from the integral of each area. a.u., arbitrary units. (C) Plot of the ratio of lactate signal to total signal (sum of pyruvate, pyruvate hydrate, and lactate signal) with time. All measurements were performed in duplicate. Error bars show the SD.
Because the 13C NMR signal of pyruvate is significantly larger than that of lactate and pyruvate hydrate, the increase in 13C lactate signal relative to 13C total signal (pyruvate + pyruvate hydrate + lactate) can represent the conversion rate constant of pyruvate to lactate, kPL (fig. S4). The overall conversion rate from pyruvate to lactate in the HMRS assay, therefore, can be assumed to be the product of kPL and the initial amount of pyruvate, [Pyr]t=0, loaded into the system (28 × 10−9 mol in 2 μl). We defined the overall conversion rate (kPL × [Pyr]t=0) as a metabolic flux metric ξ.
Analysis of metabolic flux in live cells with a 1000-fold higher sensitivity
To assess the sensitivity of our approach, we performed titration experiments with UOK262 cells, demonstrating detection sensitivity down to 104 cancer cells with a linear response to cell numbers (R2 > 99%; Fig. 3A). Conventional hyperpolarized NMR techniques typically require much larger number of cells (>107) for metabolic flux analysis, whereas the HMRS platform required on the order of 104 cells of interest. This 1000-fold higher sensitivity is the key advantage of the HMRS system and was achieved with the miniaturization of the coil design. The signal-to-noise ratio (SNR) of NMR signals depends on the magnetic field induced by unit current flowing through the coil (26, 27); thus, a smaller coil generates a higher SNR than a larger one with the same amount of target molecules (fig. S5). Another contributing factor for a higher SNR is the filling factor of the target molecule. Our microcoil, embedded into a polymer block, allowed the sample to fill the entire coil, maximizing the filling factor (≈1) for NMR detection (28).
Fig. 3. Sensitive and nondestructive analysis of glycolytic flux with HMRS.
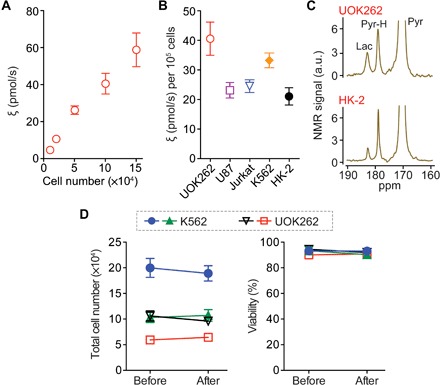
(A) Titration data of the flux metric ξ of UOK262 cells. (B) Profiling of the flux metric in five different cell lines: UOK262 (kidney cancer), U87 (glioblastoma), Jurkat (acute T cell leukemia), K562 (CML), and HK-2 (kidney). (C) NMR spectra of hyperpolarized metabolites acquired from malignant cancer cells (UOK262) and nonmalignant ones (HK-2). For each HMRS assay, 105 cells were used. (D) Comparison of viability before and after HMRS assays. Two cancer cell lines (K562 and UOK262) were tested. The “Before” measurement was conducted 15 min before the HMRS assay, and the “After” measurement was conducted 15 min after the HMRS assay. All measurements were performed in duplicate. Error bars show the SD.
We profiled five different cell lines, UOK262, U87, Jurkat, K562, and HK-2 (Fig. 3B). As expected, UOK262 showed approximately twofold higher flux when compared to benign renal tubular cells, HK-2 (40.6 and 21.0 pmol/s, respectively), which is well matched to previous studies (Fig. 3C) (22, 29). We further assessed the cell viability before and after the HMRS assay with two cancer cell lines, UOK262, as a representative of solid cancer, and K562, a chronic myeloid leukemia (CML) cell line, as a representative of liquid cancer. The cell loss was negligible during the experiments, and their viability was not significantly different (P value = not significant; Fig. 3D and fig. S6), demonstrating another advantage of the HMRS platform: nondestructive analysis of metabolic flux.
Quantification of metabolic flux in LSCs
LSCs, defined by their ability to initiate and re-establish malignancy upon transplantation, are more resistant to conventional therapeutic regimens as compared to bulk leukemia populations (30–32). Considering that distinct metabolic features confer the survival benefits to cancer cells, understanding reliance on specific metabolic pathways has the potential to illuminate more effective therapeutic strategies to target cancer stem cells. With this in mind, we took advantage of the increased sensitivity of the HMRS platform to quantify metabolic flux in primary LSCs, previously not possible using conventional approaches.
LSCs in acute myeloid leukemia (AML) driven by the MLL-AF9 oncogene are of particular interest because MLL-AF9 is related to deregulated expression of Myc (33, 34), which potentially mediates metabolic reprogramming in cancer (35). LSCs, collected from the bone marrow in MLL-AF9 AML mice, were sorted on the basis of the surface protein c-Kit (CD117) and assayed rapidly within 24 hours noninvasively (Fig. 4, A and B) (36). Intriguingly, LSCs (c-KitHi) demonstrated a nearly twofold higher flux than leukemia non–stem cells (c-KitLo) (Fig. 4C). Given that our approach is nondestructive, we were able to further assay the status of c-Myc in these primary cells by immunoblot and confirmed that c-KitHi cells also exhibited high expression levels of Myc (fig. S7) (33, 34). Although further experiments are necessary to characterize the control of metabolism by c-Myc in primary c-KitHi LSCs, this work using hyperpolarized substrates provides quantitative evidence of differential metabolism in the LSC compartment.
Fig. 4. Quantification of metabolic flux in LSCs.
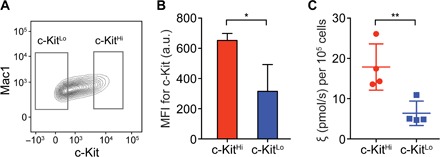
(A) Representative plot of flow cytometry of leukemia cells from a mouse with MLL-AF9 AML. The leukemia cells, collected from a mouse bone marrow, were sorted using the gates indicated in the plot. (B) Median fluorescence intensity of c-Kit in the LSCs (c-KitHi) and leukemia non–stem cells (c-KitLo) after 20 hours in media. MFI, mean fluorescence intensity. *P = 0.0281. (C) Profiling of the flux metric ξ in the leukemia cells. **P = 0.0045.
Rapid quantitative assessment of drug treatment response
Because metabolic changes can be induced by anticancer drug treatments before major clinicopathological changes occur (37), any metabolic changes related to the pathway can be an early indicator of the treatment efficacy. With this premise in mind, we applied the HMRS platform for rapid assessment of drug-treatment efficacy, based on the advantages of high sensitivity and nondestructiveness for metabolic flux analysis.
Imatinib (Gleevec), a breakpoint cluster region–Abelson murine leukemia viral oncogene homolog (BCR-ABL) tyrosine kinase inhibitor developed for CML treatment, has been shown to affect glycolytic activity in cancer cells (38, 39). We reasoned that glycolytic flux analysis on K562 cells with imatinib treatment would be an ideal model to assess time-dependent changes in drug action. After 24 hours of drug treatment, the rate of cell proliferation was slightly decreased, and the cell viability was not significantly changed at concentrations as high as 1 μM imatinib (Fig. 5, A and B, and fig. S8). However, using the HMRS, we found that the lactate peak decreased by approximately 75% after 24 hours of 1 μM imatinib treatment (Fig. 5, C and D). By assessing different time points, we found that the drug effect could be detected as early as 3 hours after treatment (Fig. 5, E and F). This reduction in the glycolytic flux with drug treatment was attributed to multiple factors, and we analyzed the protein levels in the signaling pathways, targeted by imatinib treatment, and the concentration of endogenous cofactors NADH [reduced form of NAD+ (nicotinamide adenine dinucleotide)] and NAD+. The phosphorylation level of the target protein, BCR-ABL, decreased even at 0.25 μM imatinib treatment, which led to down-regulation of the PI3K/AKT/mTOR (phosphoinositide 3-kinase/AKT/mammalian target of rapamycin) pathway (40, 41) and the key metabolic enzyme LDH-A (fig. S9). Intriguingly, the concentration of NADH, the cofactor for the pyruvate-to-lactate conversion, decreased significantly with the treatment (fig. S10). The mechanism by which BCR-ABL inhibition affects the NADH level has not been elucidated comprehensively, and this requires further study. Contrary to the flux analysis, the analysis of metabolite concentrations (pool sizes) using a conventional high-field NMR spectrometer showed much slower response to treatment (Fig. 5G). This significant difference between the analyses of flux and pool sizes can be explained as follows: The flux indicates the metabolic activity at a given moment, but the pool size indicates the integration of the metabolic activity over a certain period. Imatinib starts to affect metabolic activities in K562 cells as early as 3 hours after treatment, and the decreased glycolytic activity begins to change the lactate pool size, which is large (on the order of millimolar). In our assay, it required about 12 hours to make a significant change in the pool size. These results demonstrate another advantage of the HMRS flux analysis: rapid assessment of treatment efficacy.
Fig. 5. Rapid assessment of drug treatment response with HMRS.
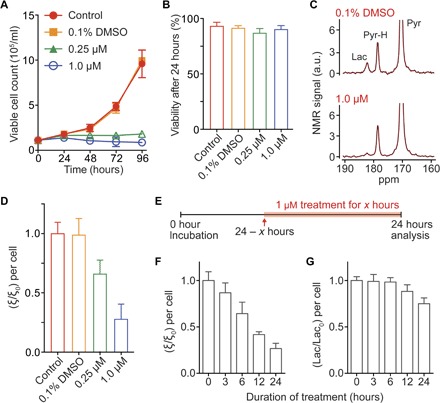
(A) Measurement of K562 cell counts after imatinib (Gleevec) treatment. (B) Viability of K562 cells after 24 hours of treatment. (C) NMR spectra of hyperpolarized metabolites acquired by the HMRS after 24 hours of treatment. (D) Plot of normalized metric ξ/ξ0 after treatment. The metric ξ0 was calculated from nontreated K562 cells (control). (E) Schematic of metabolic analysis with different treatment durations. The dose of imatinib was set to 1 μM. (F) Plot of normalized metric ξ/ξ0 with different durations. The metric ξ0 was calculated from nontreated K562 cells (duration was 0 hour). (G) Plot of normalized lactate concentrations from cell culture media with different durations. Each lactate signal was acquired by a conventional high-field NMR spectrometer. All measurements were performed in triplicate. Error bars show the SD.
Multiple analyses in a single dissolution
Although current approaches provide a means of rapidly measuring metabolic fluxes, there is a great need for developing high-throughput approaches. To address this need, we further advanced the HMRS platform to enable high-throughput analysis of hyperpolarized molecules. The DNP method provides greater than 20% polarization for 13C nuclei, but it has some limitations in throughput: (i) It takes more than an hour for hyperpolarization with relatively expensive chemical compounds, and (ii) the hyperpolarized state decays to thermal equilibrium rapidly with RF excitation pulses, which allows for only one NMR experiment. There is a great need for approaches that enable a high-throughput analysis with the DNP-NMR method. We hypothesized that multiple NMR analyses could be performed if intact hyperpolarized molecules could be supplied constantly into the microcoil in a time frame proportional to the T1 relaxation time of the hyperpolarized molecule.
We designed a three-dimensionally (3D) printed microreservoir to hold samples for multiple experiments (up to 100 μl) and connected it to the inlet of the microcoil channel (Fig. 6A). The channel outlet was connected to a syringe pump (PH2000, Harvard Apparatus) with a pulling rate of 5 μl/s. Figure 6B describes the assay scheme of high-throughput analysis from a single dissolution; (i) the syringe pump pulled the sample (Z1) to the microcoil region, followed by 13C NMR signal acquisition; (ii) after the acquisition, the syringe pump pulled the intact sample (Z2) into the microcoil region; and (iii) another 13C NMR signal acquisition started. Integrated with the microreservoir, the HMRS platform successfully conducted 12 analyses of the hyperpolarized spin states from a single dissolution (Fig. 6, C and D, and fig. S11). All 12 analyses were fit to a single decay model with a T1 of 67 s and an RF pulse of 60° flip angle, which yielded an R2 of 99.5 ± 0.4% and a root mean square error (RMSE) of 3.07 ± 1.05% between analyses (Fig. 6D). Moreover, we extended this approach to the multiple analyses of glycolytic flux in cancer cells from a single dissolution (Fig. 6E). Our integrated HMRS platform, separating the sample reservoir from the microcoil where RF pulses were applied, achieved multiple analyses from a single dissolution of hyperpolarized molecules. This analytical capability shows the potential of the HMRS for high-throughput flux analysis in cells.
Fig. 6. HMRS integrated with a microfluidic system for multiple analyses.
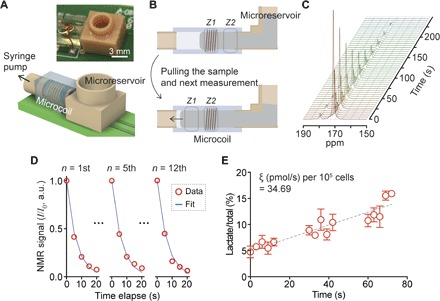
(A) Schematic of the integrated HMRS platform. (B) Assay schematic. (C) Sequential spectra of hyperpolarized pyruvate collected from multiple analyses. (D) Quantification and comparison of NMR signals from 12 analyses. NMR signal in each analysis (I) was normalized by the first signal of the same analysis (I0). All data sets were fitted to a single model of decay of hyperpolarized spin states. The 12 analyses yielded an R2 of 99.5 ± 0.4% and an RMSE of 3.07 ± 1.05%. (E) Multiple analyses of glycolytic flux from a single dissolution of hyperpolarized pyruvate. Each analysis consists of five NMR acquisitions (15 s in total), followed by a 15-s interval to stabilize the sample after movement. The overall glycolytic flux metric ξ was 34.69 (pmol/s) per 105 cells, which was well matched to the ξ of K562 (33.24 ± 2.51) presented in Fig. 3B. The measurements were performed in triplicate. Error bars show the SD.
DISCUSSION
Aerobic glycolysis or Warburg effect is a unique metabolic feature of cancer cells, closely related to multiple oncogenic signaling pathways. Because the glycolytic flux represents the activity of the major metabolic pathway at any given moment, it has emerged as a potent biomarker for cancer diagnosis and treatment response monitoring. Conventional techniques, such as optical microscopy or liquid chromatography–mass spectrometry (LC-MS), have been developed for metabolism research, but they are impractical for analyzing metabolic fluxes in mass-limited samples. Optical microscopy, which can provide sensitivity at single-cell resolution (42, 43), is limited in detectable metabolic pathways, and fluorescent labels can distort metabolic reactions unexpectedly. LC-MS, the most widely used method (44, 45), requires extended periods of incubation time with labeled molecules, and a large number of target cells are needed for flux analyses, because it is a destructive approach (fig. S12). There has been a great need for methods that could perform rapid flux analyses in mass-limited samples, including cancer stem cells or primary cells, and the HMRS system presented here could be a promising solution for that.
The developed HMRS system has several advantages over conventional techniques for metabolic flux analysis. First, its detection modality is based on MR, which makes a flux analysis completely nondestructive, allowing repeated measurements as well as downstream molecular analyses using other techniques. Second, its miniaturized detection coil increases the filling factor of the target molecules and enhances the sensitivity. Third, by exploiting the DNP hyperpolarization technique, it monitors metabolic reactions in real time without any signal averaging. Last, it can perform multiple analyses with an integrated microfluidic system, enabling the high-throughput system for analyzing hyperpolarized molecules as well as the potential to integrate this platform into other high-throughput approaches.
On the basis of its technical advancement, the HMRS platform quantitatively profiled metabolic fluxes in LSCs and assessed therapeutic responses in cancer cells much earlier than any changes in viability, hardly achievable using conventional methods. This work opens new opportunities for stem cell research where rapid and sensitive analytical capabilities are critical; this platform can be used to investigate how stem cells metabolically respond to external stimuli, such as drug treatment. Further studies with drugs targeting metabolic differences in LSCs and hematopoietic stem cells will lead to more effective therapeutic strategies to cure cancer. The HMRS platform can also be applicable for rapid metabolic flux analysis in clinically relevant samples, such as biopsy specimens or patient-derived organoids, in a continuous manner to assess treatment response.
Technical optimizations could further advance the HMRS system. For the analysis of multiple metabolic pathways (for example, pyruvate-to-lactate, pyruvate-to-bicarbonate, or pyruvate-to-alanine) in a smaller number of cells, there are multiple strategies: (i) A smaller coil for NMR signal acquisition can be implemented. A coil with a diameter of <1.4 mm could provide a higher sensitivity, but precise loading of the sample into a submicroliter space within a limited time frame is not trivial. One strategy to overcome this challenge is to integrate a micropneumatic actuator into the HMRS, which could move liquid sample rapidly and precisely into a microcoil. (ii) High-order shimming coils can be designed to make a magnetic field more homogeneous. The current system was equipped with first-order shimming coils, which provided a limited homogeneity of magnetic field (0.14 parts per million full width at half maximum) in the microcoil circuit region. Because the NMR signal is proportional to the number of target nuclei experiencing the same magnetic field, a more homogeneous magnetic field leads to narrower NMR peaks and a higher SNR. Adding second- and third-order shimming coils into the permanent magnet will improve the homogeneity of the magnetic field (46), thereby leading to the enhanced sensitivity of the HMRS. (iii) A selective RF pulse can be applied for signal acquisition. The current system acquired the NMR spectra using a single RF pulse, which excited all the metabolic substrates and products at the same time. A selective pulse designed to excite the products (for example, lactate) only can leave the hyperpolarized spin states of substrates (for example, pyruvate) relatively intact (47–49). The benefit of slowly decaying hyperpolarized substrates will allow flux analysis for extended periods of time, providing an opportunity to monitor multiple metabolic pathways.
The recent advancement of hyperpolarized NMR technology has shed light on how to study metabolism in live cells or organs. The HMRS platform can expand the applications of the hyperpolarized NMR technology further, which will provide opportunities for rapid and sensitive exploration of metabolic dynamics in biologically relevant systems.
MATERIALS AND METHODS
Microcoil fabrication
Thin magnet wire [Belden 8042, 32 AWG (American wire gauge)] was wound five times around a metal rod (diameter, 1.4 mm), and it was embedded into PDMS (Dow Corning). After overnight PDMS curing at 80°C, the wire was pulled out, which formed a miniaturized solenoid coil (microcoil) with a microfluidic channel.
3D-printed microreservoir
We used a computer-aided design software (3ds MAX, Autodesk) and a 3D printer (Micro Plus Hi-Res, EnvisionTEC) to design and print a microreservoir. Its outlet (diameter, 1.4 mm) was designed to fit into the microfluidic channel, and its inlet (diameter, 10 mm) was designed for easy loading of the sample. Its maximum capacity was 100 μl.
Drug (imatinib) treatment on K562 cells
K562 cells were plated with a concentration of 100,000 cells/ml in 10 ml of complete RPMI medium. Complete RPMI medium (10 μl), dimethyl sulfoxide (DMSO), 0.25 mM imatinib in DMSO, and 1 mM imatinib in DMSO were loaded for the control sample, the 0.1% DMSO-treated sample, and the 0.25 and 1.0 μM imatinib-treated samples, respectively. Imatinib was purchased from Cayman Chemical. Cell number and viability were checked after 24 hours, and the cells were washed with complete RPMI medium once and prepared for hyperpolarization experiments.
Hyperpolarization using DNP
We used a SPINlab polarizer (GE) to polarize [1-13C]pyruvate. The preparation steps for polarization are as follows: (i) The 13C-enriched pyruvate sample was prepared with a stable organic radical: 15 mM AH-111501 (GE) was mixed in [1-13C]pyruvic acid (Sigma-Aldrich, 677175) thoroughly. (ii) Buffer solution for dissolution was prepared: 0.4 mM EDTA was added in 40 mM Trizma hydrochloride solution (Sigma-Aldrich, T2663). (iii) The pyruvate sample (100 μl) from step (i) and 20 ml of the buffer solution from step (ii) were loaded into the SPINlab polarizer (3.35 T, 0.98 K). After 90 min of polarization, the pyruvate sample was quickly dissolved into an ice-cold flask with 120 μl of 10 N sodium hydroxide solution (Fisher Scientific, SS255) to make the dissolved sample neutral (pH ~ 7.4) and reach 37°C faster. The dissolved pyruvate sample was added into the cell suspension with a ratio of 1:10, which made the concentration of pyruvate 14 mM.
Cell culture
K562 and Jurkat cells were grown in Gibco RPMI 1640 medium (Thermo Fisher Scientific, 11875-093). UOK262 and HK-2 cells were grown in Advanced Dulbecco’s modified Eagle’s medium/F-12 (Thermo Fisher Scientific, 12634-010). U87 cells were grown in Eagle’s minimum essential medium (American Type Culture Collection, 30-2003). All media were supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin.
Cell number and viability
Cell number and viability were measured using a Cellometer Mini Cell Counter (Nexcelom Bioscience); the cell suspension was mixed at a 1:1 ratio with the trypan blue solution before the measurement. For each measurement, the cell suspension was diluted so that its concentration would be within the 0.8 × 106 to 2 × 106/ml range.
Preparation of primary leukemia cells
Bones collected from leukemic mice, transplanted with MLL-AF9 transformed leukemic cells, were crushed in a sterile mortar in the addition of serum-free RPMI 1640 medium. The bone marrow leukemic cells were strained (70 μM Nylon strainer, Falcon), resuspended in red blood cell lysis buffer (Qiagen) to remove red blood cells, and washed with serum-free RPMI 1640 media. After centrifugation (15,000 rpm, 5 min), the cell pellet was resuspended in 2% FBS/RPMI medium and stained with Mac1-PacBlue and c-Kit–PeCy7 (34). The top 15% and lowest 20% of c-Kit cells were sorted as c-KitHi and c-KitLo cells, respectively, using a BD FACSAria III instrument. The c-KitHi and c-KitLo cells were cultured separately in 10% FBS RPMI medium with the addition of stem cell factor (10 ng/ml), interleukin-3 (IL-3) (10 ng/ml), and IL-6 (10 ng/ml). All cytokines were purchased from PeproTech. After a 20-hour incubation, hyperpolarization experiments were conducted with approximately 0.3 million cells loaded into the microcoil. All animal procedures were approved by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center (MSKCC).
Western blot analysis
K562 cells, washed with ice-cold phosphate-buffered saline (PBS) without Ca + Mg and concentrated by centrifugation (1400 rpm, 5 min), were lysed in radioimmunoprecipitation assay buffer (Thermo Scientific, 89901) containing protease and phosphatase inhibitors (Thermo Scientific, 78480 and 78420). The concentration of protein lysates was quantified using bicinchoninic acid assay. Protein lysates were resolved using NuPAGE precast gels and transferred to Novex polyvinylidene difluoride membrane (Thermo Fisher Scientific, LC2002), as described in the protocol. Target proteins in the membrane were immunoblotted with the following primary antibodies: BCR-ABL (Cell Signaling, 3902), phospho–BCR-ABL (Cell Signaling, 3901), lactate dehydrogenase A (LDHA) (Cell Signaling, 2012), phospho-LDHA (Cell Signaling, 8176), and β-actin (Cell Signaling, 8457). The horseradish peroxidase (HRP)–conjugated secondary antibodies (Santa Cruz Biotechnology, sc-2004) were then incubated with the membrane. With the chemiluminescent (enhanced chemiluminescence) HRP substrate (Thermo Fisher Scientific, 34080), protein bands were detected on x-ray film (Thermo Fisher Scientific, 34090).
Quantification of NADH/NAD+ from cell extracts
K562 cells, washed with ice-cold PBS without Ca + Mg and concentrated by centrifugation (1400 rpm, 5 min), were resuspended in methanol solution (80% in water) and kept at −80°C. After 24 hours, the supernatant of the mixture was collected, centrifuged (4000 rpm, 30 min), and dried using a solvent evaporator (EZ-2 Elite Genevac, SP Scientific). The dried material was resuspended in D2O with the internal standard solution and transferred to a high-field NMR tube. After an hour-long 1H NMR acquisition (Ultrashield Plus 14.1 T, Bruker), the spectrum was analyzed using Chenomx NMR Suite.
Supplementary Material
Acknowledgments
We thank V. Di Gialleonardo and L. Salamanca-Cardona for helping with the Western blot analysis and providing insightful comments on experimental details. Funding: This work was supported in part by U.S. NIH grants R01HL113156 (H.L.), R21CA205322 (H.L.), R00EB014328 (K.R.K.), and R21CA212958-01 (K.R.K.); Cancer Center Support Grant P30CA008748 (K.R.K.); and Department of Defense Ovarian Cancer Research Program Award W81XWH-14-1-0279 (H.L.) as well as the Center for Molecular Imaging and Nanotechnology at MSKCC. Author contributions: S.J., R.E., S.M.P., S.S.T., R.W., M.G.K., H.L., and K.R.K. designed the study and experiments; S.J., R.E., S.M.P., S.S.T., and J.A. performed experiments; S.J., R.E., J.A., S.M.P., M.G.K., H.L., and K.R.K. analyzed the data; and S.J., R.E., S.M.P., R.W., H.L., and K.R.K. wrote the manuscript. Competing interests: K.R.K. and S.J. are inventors on a pending patent application related to this work filed by MSKCC (PCT/US17/21355, filed 8 March 2017). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1700341/DC1
fig. S1. Schematic of [1-13C]pyruvate metabolism in cells.
fig. S2. Tuning/matching characteristic of the microcoil in the HMRS platform.
fig. S3. Metabolic flux measurement using the HMRS.
fig. S4. Mathematical analysis of the increased rate of lactate relative to total carbon (lactate + pyruvate + pyruvate hydrate) with time.
fig. S5. Comparison of NMR signals from three different coils.
fig. S6. Comparison of viability before and after HMRS assays.
fig. S7. Immunoblot analysis for c-Myc in LSCs (c-KitHi) and leukemia non–stem cells (c-KitLo).
fig. S8. Viability after imatinib treatment on K562 cells.
fig. S9. Changes in protein expression and phosphorylation levels in K562 cells after 24-hour imatinib treatment.
fig. S10. Changes in the NADH concentration after the imatinib treatment.
fig. S11. High-throughput analysis of hyperpolarized pyruvate.
fig. S12. Comparison of the HMRS technique with conventional approaches to study metabolism.
REFERENCES AND NOTES
- 1.Warburg O., Wind F., Negelein E., The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Commisso C., Davidson S. M., Soydaner-Azeloglu R. G., Parker S. J., Kamphorst J. J., Hackett S., Grabocka E., Nofal M., Drebin J. A., Thompson C. B., Rabinowitz J. D., Metallo C. M., Vander Heiden M. G., Bar-Sagi D., Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamphorst J. J., Nofal M., Commisso C., Hackett S. R., Lu W., Grabocka E., Vander Heiden M. G., Miller G., Drebin J. A., Bar-Sagi D., Thompson C. B., Rabinowitz J. D., Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller K. E., Tan I. S., Lee Y.-S., SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science 338, 1069–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J.-w., Tchernyshyov I., Semenza G. L., Dang C. V., HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Chang C.-H., Qiu J., O’Sullivan D., Buck M. D., Noguchi T., Curtis J. D., Chen Q., Gindin M., Gubin M. M., van der Windt G. J. W., Tonc E., Schreiber R. D., Pearce E. J., Pearce E. L., Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer K., Hoffmann P., Voelkl S., Meidenbauer N., Ammer J., Edinger M., Gottfried E., Schwarz S., Rothe G., Hoves S., Renner K., Timischl B., Mackensen A., Kunz-Schughart L., Andreesen R., Krause S. W., Kreutz M., Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood 109, 3812–3819 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Gottfried E., Kunz-Schughart L. A., Ebner S., Mueller-Klieser W., Hoves S., Andreesen R., Mackensen A., Kreutz M., Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107, 2013–2021 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Walenta S., Wetterling M., Lehrke M., Schwickert G., Sundfør K., Rofstad E. K., Mueller-Klieser W., High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 60, 916–921 (2000). [PubMed] [Google Scholar]
- 13.Rofstad E. K., Mathiesen B., Kindem K., Galappathi K., Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 66, 6699–6707 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Dimopoulos M. A., Barlogie B., Smith T. L., Alexanian R., High serum lactate dehydrogenase level as a marker for drug resistance and short survival in multiple myeloma. Ann. Intern. Med. 115, 931–935 (1991). [DOI] [PubMed] [Google Scholar]
- 15.Koukourakis M. I., Giatromanolaki A., Sivridis E., Bougioukas G., Didilis V., Gatter K. C., Harris A. L.; Tumour and Angiogenesis Research Group, Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 89, 877–885 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek G., Tse Y. F., Hu Z., Cox D., Buboltz N., McCue P., Yeo C. J., White M. A., DeBerardinis R. J., Knudsen E. S., Witkiewicz A. K., MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 9, 2233–2249 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues T. B., Serrao E. M., Kennedy B. W. C., Hu D.-E., Kettunen M. I., Brindle K. M., Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat. Med. 20, 93–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardenkjær-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thaning M., Golman K., Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day S. E., Kettunen M. I., Gallagher F. A., Hu D.-E., Lerche M., Wolber J., Golman K., Ardenkjaer-Larsen J. H., Brindle K. M., Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 13, 1382–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Keshari K. R., Sriram R., Van Criekinge M., Wilson D. M., Wang Z. J., Vigneron D. B., Peehl D. M., Kurhanewicz J., Metabolic reprogramming and validation of hyperpolarized 13C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate 73, 1171–1181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu S., Balakrishnan A., Bok R. A., Anderton B., Larson P. E. Z., Nelson S. J., Kurhanewicz J., Vigneron D. B., Goga A., 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 14, 131–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshari K. R., Sriram R., Koelsch B. L., Van Criekinge M., Wilson D. M., Kurhanewicz J., Wang Z. J., Hyperpolarized 13C-pyruvate magnetic resonance reveals rapid lactate export in metastatic renal cell carcinomas. Cancer Res. 73, 529–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tee S. S., DiGialleonardo V., Eskandari R., Jeong S., Granlund K. L., Miloushev V., Poot A. J., Truong S., Alvarez J. A., Aldeborgh H. N., Keshari K. R., Sampling hyperpolarized molecules utilizing a 1 Tesla permanent magnetic field. Sci. Rep. 6, 32846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keshari K. R., Wilson D. M., Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization. Chem. Soc. Rev. 43, 1627–1659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong W.-H., Sourbier C., Kovtunovych G., Jeong S. Y., Vira M., Ghosh M., Romero V. V., Sougrat R., Vaulont S., Viollet B., Kim Y.-S., Lee S., Trepel J., Srinivasan R., Bratslavsky G., Yang Y., Linehan W. M., Rouault T. A., The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 20, 315–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb A., Increasing the sensitivity of magnetic resonance spectroscopy and imaging. Anal. Chem. 84, 9–16 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Zalesskiy S. S., Danieli E., Blümich B., Ananikov V. P., Miniaturization of NMR systems: Desktop spectrometers, microcoil spectroscopy, and “NMR on a Chip” for chemistry, biochemistry, and industry. Chem. Rev. 114, 5641–5694 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Lee H., Yoon T.-J., Figueiredo J.-L., Swirski F. K., Weissleder R., Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc. Natl. Acad. Sci. U.S.A. 106, 12459–12464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sriram R., Van Criekinge M., Hansen A., Wang Z. J., Vigneron D. B., Wilson D. M., Keshari K. R., Kurhanewicz J., Real-time measurement of hyperpolarized lactate production and efflux as a biomarker of tumor aggressiveness in an MR compatible 3D cell culture bioreactor. NMR Biomed. 28, 1141–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M. A., Dick J. E., A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Huntly B. J. P., Gilliland D. G., Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat. Rev. Cancer 5, 311–321 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Kreso A., Dick J. E., Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Somervaille T. C. P., Matheny C. J., Spencer G. J., Iwasaki M., Rinn J. L., Witten D. M., Chang H. Y., Shurtleff S. A., Downing J. R., Cleary M. L., Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 4, 129–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S.-M., Gönen M., Vu L., Minuesa G., Tivnan P., Barlowe T. S., Taggart J., Lu Y., Deering R. P., Hacohen N., Figueroa M. E., Paietta E., Fernandez H. F., Tallman M. S., Melnick A., Levine R., Leslie C., Lengner C. J., Kharas M. G., Musashi2 sustains the mixed-lineage leukemia–driven stem cell regulatory program. J. Clin. Invest. 125, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., Dang C. V., MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krivtsov A. V., Twomey D., Feng Z., Stubbs M. C., Wang Y., Faber J., Levine J. E., Wang J., Hahn W. C., Gary Gilliland D., Golub T. R., Armstrong S. A., Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Stroobants S., Goeminne J., Seegers M., Dimitrijevic S., Dupont P., Nuyts J., Martens M., van den Borne B., Cole P., Sciot R., Dumez H., Silberman S., Mortelmans L., van Oosterom A., 18FDG-positron emission tomography for the early prediction of response in advanced soft tissue sarcoma treated with imatinib mesylate (Glivec®). Eur. J. Cancer 39, 2012–2020 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Barnes K., McIntosh E., Whetton A. D., Daley G. Q., Bentley J., Baldwin S. A., Chronic myeloid leukaemia: An investigation into the role of Bcr-Abl-induced abnormalities in glucose transport regulation. Oncogene 24, 3257–3267 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Klawitter J., Anderson N., Klawitter J., Christians U., Leibfritz D., Eckhardt S. G., Serkova N. J., Time-dependent effects of imatinib in human leukaemia cells: A kinetic NMR-profiling study. Br. J. Cancer 100, 923–931 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kharas M. G., Janes M. R., Scarfone V. M., Lilly M. B., Knight Z. A., Shokat K. M., Fruman D. A., Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J. Clin. Invest. 118, 3038–3050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorski T., Kanakaraj P., Nieborowska-Skorska M., Ratajczak M. Z., Wen S. C., Zon G., Gewirtz A. M., Perussia B., Calabretta B., Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 86, 726–736 (1995). [PubMed] [Google Scholar]
- 42.Walsh A. J., Cook R. S., Sanders M. E., Aurisicchio L., Ciliberto G., Arteaga C. L., Skala M. C., Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 74, 5184–5194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szulczewski J. M., Inman D. R., Entenberg D., Ponik S. M., Aguirre-Ghiso J., Castracane J., Condeelis J., Eliceiri K. W., Keely P. J., In vivo visualization of stromal macrophages via label-free FLIM-based metabolite imaging. Sci. Rep. 6, 25086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., Rabinowitz J. D., Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayers J. R., Torrence M. E., Danai L. V., Papagiannakopoulos T., Davidson S. M., Bauer M. R., Lau A. N., Ji B. W., Dixit P. D., Hosios A. M., Muir A., Chin C. R., Freinkman E., Jacks T., Wolpin B. M., Vitkup D., Vander Heiden M. G., Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson W. A., Electrical current shims for correcting magnetic fields. Rev. Sci. Instrum. 32, 241–250 (1961). [Google Scholar]
- 47.Larson P. E. Z., Kerr A. B., Chen A. P., Lustig M. S., Zierhut M. L., Hu S., Cunningham C. H., Pauly J. M., Kurhanewicz J., Vigneron D. B., Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J. Magn. Reson. 194, 121–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C., Harrison C., Jin E. S., Chuang D. T., Sherry A. D., Malloy C. R., Merritt M. E., DeBerardinis R. J., Simultaneous steady-state and dynamic13C NMR can differentiate alternative routes of pyruvate metabolism in living cancer cells. J. Biol. Chem. 289, 6212–6224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen A. P., Cunningham C. H., Single voxel localization for dynamic hyperpolarized 13C MR spectroscopy. J. Magn. Reson. 258, 81–85 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/6/e1700341/DC1
fig. S1. Schematic of [1-13C]pyruvate metabolism in cells.
fig. S2. Tuning/matching characteristic of the microcoil in the HMRS platform.
fig. S3. Metabolic flux measurement using the HMRS.
fig. S4. Mathematical analysis of the increased rate of lactate relative to total carbon (lactate + pyruvate + pyruvate hydrate) with time.
fig. S5. Comparison of NMR signals from three different coils.
fig. S6. Comparison of viability before and after HMRS assays.
fig. S7. Immunoblot analysis for c-Myc in LSCs (c-KitHi) and leukemia non–stem cells (c-KitLo).
fig. S8. Viability after imatinib treatment on K562 cells.
fig. S9. Changes in protein expression and phosphorylation levels in K562 cells after 24-hour imatinib treatment.
fig. S10. Changes in the NADH concentration after the imatinib treatment.
fig. S11. High-throughput analysis of hyperpolarized pyruvate.
fig. S12. Comparison of the HMRS technique with conventional approaches to study metabolism.


