Abstract
The discovery of new genetic determinants of inherited skin disorders has been instrumental to the understanding of epidermal function, differentiation, and renewal. Here, we show that mutations in KDSR (3-ketodihydrosphingosine reductase), encoding an enzyme in the ceramide synthesis pathway, lead to a previously undescribed recessive Mendelian disorder in the progressive symmetric erythrokeratoderma spectrum. This disorder is characterized by severe lesions of thick scaly skin on the face and genitals and thickened, red, and scaly skin on the hands and feet. Although exome sequencing revealed several of the KDSR mutations, we employed genome sequencing to discover a pathogenic 346 kb inversion in multiple probands, and cDNA sequencing and a splicing assay established that two mutations, including a recurrent silent third base change, cause exon skipping. Immunohistochemistry and yeast complementation studies demonstrated that the mutations cause defects in KDSR function. Systemic isotretinoin therapy has achieved nearly complete resolution in the two probands in whom it has been applied, consistent with the effects of retinoic acid on alternative pathways for ceramide generation.
Keywords: KDSR, skin, erythrokeratoderma, ichthyosis, genome sequencing, inversion, splicing, TSC10, isotretinoin, ceramide
Main Text
Mendelian disorders of cornification (MEDOCs) are severe skin disorders that feature localized or generalized scaling and redness and can be associated with significant morbidity and mortality. Genetic investigation has revealed MEDOC-causing mutations in over 50 genes; despite diverse function, they share a common pathobiologic feature of increased transepidermal water loss. Included among MEDOCs are progressive symmetric erythrokeratoderma (PSEK [MIM: 133200], which shows symmetrically distributed hyperkeratotic and erythematous plaques on the face, buttocks, and groin and palmoplantar keratoderma in about half of affected individuals) and erythrokeratoderma variabilis (which exhibits similar features in conjunction with transient patches of erythema). Both disorders display substantial phenotypic heterogeneity, and cutaneous involvement can vary markedly from localized to generalized disease, even within kindreds. Previously described Mendelian forms of erythrokeratoderma are usually dominant and often result from heterozygosity for a de novo mutation. These include erythrokeratoderma variabilis (MIM: 133200; caused by heterozygosity for a mutation in GJB3 [MIM: 603324], GJB4 [MIM: 605425], or GJA1 [MIM: 121014], which encode connexins 31, 30.3, and 43, respectively1, 2, 3), Vohwinkel syndrome (MIM: 604117; caused by heterozygosity for a mutation in LOR [MIM: 152445], encoding loricrin4), and a syndrome of erythrokeratoderma, ichthyosis, and cardiomyopathy (EKC syndrome [MIM: 615821], caused by heterozygosity for a missense mutation in a single domain of DSP [MIM: 125647], encoding desmoplakin5).
We recruited a large cohort of 750 kindreds with disorders of cornification, including subjects with ichthyosis and erythrokeratoderma phenotypes. The Yale Human Investigation Committee approved the study in accordance with institutional and national ethical standards, and subjects provided verbal and written informed consent. We screened probands for 48 genes in which mutations cause MEDOCs via targeted amplification and high-throughput sequencing. Subjects without pathogenic mutations identified in this gene panel make up a subset that facilitates the discovery of new genetic causes of MEDOCs, typically via exome sequencing.3, 5
In the course of exome sequencing of the MEDOC discovery cohort, two probands described by their referring physicians as having an unusual keratinization or PSEK-like disorder (and unaffected parents) were each found to be compound heterozygous for a deleterious mutation and a silent mutation in the same gene, KDSR (MIM: 136440; GenBank: NM_002035.2 and NP_002026.1; Tables S1 and S2 and Figures S1A and S1B). Subject 429 is heterozygous for the previously unreported mutation c.164_166delAAG (p.Gln55_Gly56delinsArg), and subject 101 is heterozygous for the previously unreported splice-site mutation c.256−2A>C. Both subjects are heterozygous for the silent mutation c.879G>A (p.Gln293Gln) (minor allele frequency 0.00003).
The silent mutation shared by both probands is at the last base of exon 9. Because guanine is highly conserved at this position,6 we hypothesized that this mutation might alter splicing. We used skin tissue from subject 429 to examine KDSR transcripts, which revealed an in-frame deletion of exon 9, leading to p.Gln260_Gln293del (Figures 1A and S1C). Using a splicing assay, we also found that c.256−2A>C results in an in-frame deletion of exon 4, leading to p.Val86_Gln107del (Figures 1B and S1D).
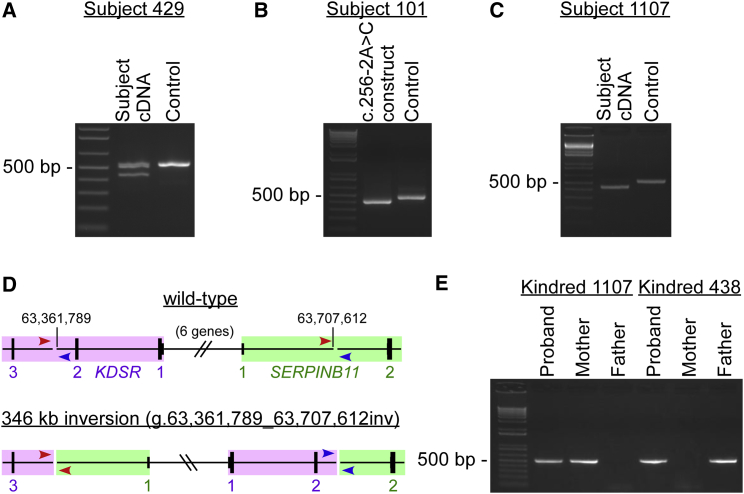
Figure 1.
KDSR Mutations in PSEK Subjects Include Single-Nucleotide Changes That Affect Splicing and a 346 kb Inversion That Abolishes Expression
(A) Reverse transcription and amplification of a portion of KDSR RNA spanning exons 6–10 from a wild-type control sample produced a band of the expected size (543 bp, right lane), whereas RNA from subject 429, a subject heterozygous for a substitution at the last base of exon 9 (c.879G>A), also produced a smaller band (441 bp, left lane) in which exon 9 had been skipped (r.778_879del [p.Gln260_Gln293del]). Corresponding Sanger sequences are shown in Figure S1C.
(B) Reverse transcription and amplification of a portion of KDSR RNA generated by a wild-type KDSR construct expressed in HEK cells produced a single product including exons 3–5 (450 bp, middle lane), whereas RNA from a KDSR construct with the exon 4 splice acceptor mutation found in subject 101 (c.256−2A>C) produced a smaller band (384 bp, left lane) in which exon 4 had been skipped (r.256_321del [p.Val86_Gln107del]). Corresponding Sanger sequences are shown in Figure S1D.
(C) When the same experiment shown in (A) was performed with RNA from subject 1107, who, like subject 429, is heterozygous for the KDSR c.879G>A mutation (causing skipping of exon 9), all amplification products lacked exon 9, demonstrating that his other KDSR allele harbors a mutation that either also affects exon 9 splicing or ablates expression of KDSR entirely.
(D) Genome sequencing of DNA from subject 1107 revealed a 346 kb inversion on chromosome 18 (g.63,361,789_63,707,612inv), which flips the genomic sequence between intron 2 of KDSR and intron 1 of SERPINB11, as shown. KDSR is in purple, and SERPINB11 is in green; there are six additional genes between these (not shown). Amplification of genomic DNA across the boundaries of the inversion with the red or blue primer set (arrows) should produce a product only when the inversion is present.
(E) Amplification of genomic DNA with primers shown in blue in (D) produced the expected PCR product (533 bp) with DNA from subject 1107 and his mother and with DNA from subject 438 and her father, confirming that both probands are compound heterozygous for this inversion and another deleterious KDSR mutation (one inherited from each parent). The same result was obtained with the primers shown in red in (D) (665 bp, not shown). Corresponding Sanger sequences for both primer sets are shown in Figures S1E and S1F.
Ongoing exome sequencing of subjects new to our MEDOC cohort subsequently revealed two probands, each born to unaffected parents, with single inherited KDSR mutations (Tables S1 and S2). Subject 1107 is heterozygous for the mutation shown to result in p.Gln260_Gln293del, and subject 438 is heterozygous for the previously unreported mutation c.557A>T (p.Tyr186Phe). Although exome sequencing did not show any other KDSR mutations in these two probands, examination of aligned reads with the Integrative Genomics Viewer (IGV)7 revealed that an intronic SNP (c.108+166C>T, rs62098681, minor allele frequency 0.01) was most likely present in both subjects, despite low exome coverage. In both kindreds, this SNP was inherited from the parent in whom exome sequencing did not reveal a damaging KDSR mutation (Tables S2 and S3). Because the minor allele frequency is too high to be plausibly pathogenic for this disorder but also too infrequent for this to be a coincidental observation, we hypothesized that the SNP was a marker for another KDSR mutation that is shared by both subjects but is not observable by exome sequencing.
To investigate the possibility of an intronic mutation affecting KDSR, we obtained a skin biopsy from subject 1107 to examine KDSR transcripts. Although subject 1107 is heterozygous for the mutation that causes exon 9 skipping, exon 9 was omitted in all KDSR transcripts detectable in this experiment (Figure 1C). This result indicated that the other allele harbored either an intronic mutation also affecting splicing of exon 9 or a mutation that abolishes production of KDSR transcripts altogether. Sequencing of the entire intronic sequence flanking exon 9 revealed no rare or novel mutations, effectively ruling out the former possibility.
Subsequent genome sequencing of subject 1107 revealed a novel 346 kb inversion on chromosome 18 (g.63,361,789_63,707,612inv), which flips the genomic sequence flanked by intron 2 of KDSR (coded on the (−) strand) and intron 1 of SERPINB11 (a gene distal to KDSR and coded on the (+) strand) (Figures 1D and S2). This inversion replaces KDSR’s upstream promoter and 5,485 bp of gene sequence, including the 5′ UTR, start codon, and first two exons, with unrelated sequence. PCR with primers at the flanking boundaries confirmed the presence of this inversion in both subjects 1107 and 438. Similar amplification of parental DNAs affirmed that in both kindreds this previously unreported mutation is on the same allele as SNP rs62098681 (as predicted) and consequently that both subjects 1107 and 438 are compound heterozygous for deleterious KDSR mutations (Figures 1E, S1E, and S1F). The inversion abolishes expression of KDSR (Figures 1C and 1D). Although fusion transcripts of KDSR and SERPINB11 are theoretically possible, the consistent phenotype of subjects with and without the inversion strongly suggests that such transcripts do not significantly contribute to the phenotype and that disease pathobiology results from KDSR loss of function.
All four of the probands compound heterozygous for deleterious coding mutations in KDSR (Table 1) were found to exhibit a similar and previously undescribed skin phenotype (Figure 2). All presented either at birth or in the perinatal period with thickened red skin with vernix (subjects 429 and 101), thickened skin in the diaper area (subject 1107), or tight red skin with deep fissures and collodion membrane (subject 438). Erythema faded in infancy, and by 4 months all four probands developed well-demarcated, thickened, scaly skin lesions on the cheeks and periocular areas and erythema and thickening of the palms and soles. Most experienced thickened scaly plaques on the genitals, and subject 438 also had well-demarcated scaly plaques on the torso, legs, and arms. All reported exacerbation with cold weather. Subject 438’s sister, who died of pneumonia at 9 days of age, showed similar skin findings, consistent with inheritance of a recessive Mendelian disorder.
Table 1.
Compound Heterozygosity for Damaging KDSR Mutations in PSEK Subjects
| DNA Change | Exon(s) | Protein Change | Method(s) | Subject 429 | Subject 101 | Subject 1107 | Subject 438 |
|---|---|---|---|---|---|---|---|
| g.63,361,789_63,707,612inv | 1, 2 | expression lost | genome sequencing, cDNA analysis | – | – | maternal | paternal |
| c.164_166delAAG | 2 | p.Gln55_Gly56delinsArg | exome sequencing | paternal | – | – | – |
| c.256−2A>C | 4 | p.Val86_Gln107del | exome sequencing, splicing assay | – | maternal | – | – |
| c.557A>T | 6 | p.Tyr186Phe | exome sequencing | – | – | – | maternal |
| c.879G>A | 9 | p.Gln293Gln, p.Gln260_Gln293del | exome sequencing, cDNA analysis | maternal | paternal | paternal | – |
Shown for each mutation are the DNA change, affected exon(s), effect on the protein, method(s) of discovery, and inheritance in subjects (on the maternal or paternal allele). The genomic position is from the UCSC Genome Browser build hg38. Accession numbers for cDNA, protein, and exon positions are GenBank: NM_002035.2, NP_002026.1, and NG_028249.1, respectively.
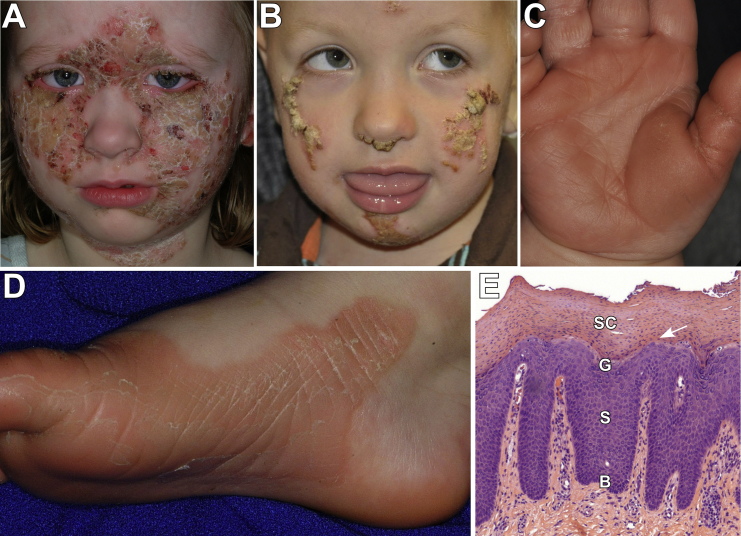
Figure 2.
Clinical and Histologic PSEK Features Due to KDSR Mutations
The face, palms, soles, and genitals are the most severely affected areas in all subjects.
(A) Image of the face of subject 101 shows well-demarcated pink-red plaques, with overlying thick yellow-white scale, which are prominent on the cheeks, central forehead, and neck. There are focal, denuded, red areas that showed no evidence of bacterial superinfection but could be the result of over-grooming.
(B) Subject 1107 has similar focal plaques with thick scale on the nose, cheeks, and chin.
(C and D) Images of the palm of subject 1107 (C) and sole of subject 101 (D) show erythematous palmoplantar hyperkeratosis with peeling scale.
(E) Histology of affected skin from the buttock of subject 1107 is remarkable for epidermal thickening (acanthosis), sparse to absent keratohyalin granules in the granular layer, and retention of nuclei in the stratum corneum (arrow). Epidermal layers are labeled as follows: B, basal layer; S, stratum spinosum; G, granular layer; SC, stratum corneum.
KDSR encodes 3-ketodihydrosphingosine reductase (KDSR), an enzyme in the ceramide synthesis pathway (Figure 3A). After palmitate and serine are condensed by serine palmitoyl transferase (SPT), KDSR reduces 3-ketodihydrosphingosine (KDS) to dihydrosphingosine (DHS), and either DHS or its hydroxylated product PHS is acylated by ceramide synthases (CERS1–CERS6) for the generation of ceramides.8, 9 Ceramides are central to cutaneous barrier function and are secreted by keratinocytes with cholesterol and free fatty acids to form ordered lipid lamellae as cells transition from the stratum granulosum to the stratum corneum.10 In addition to playing a role in barrier function, ceramides regulate cutaneous proliferation and differentiation.11 Recessive mutations in CERS3 (MIM: 615276) cause ichthyosis characterized by encasement in a collodion membrane at birth, which sheds in infancy to reveal whole-body erythroderma and fine or coarse scaling.12, 13 CERS3 mutations have been found to alter the ceramide content of epidermis and to lead to defects in cutaneous differentiation.13
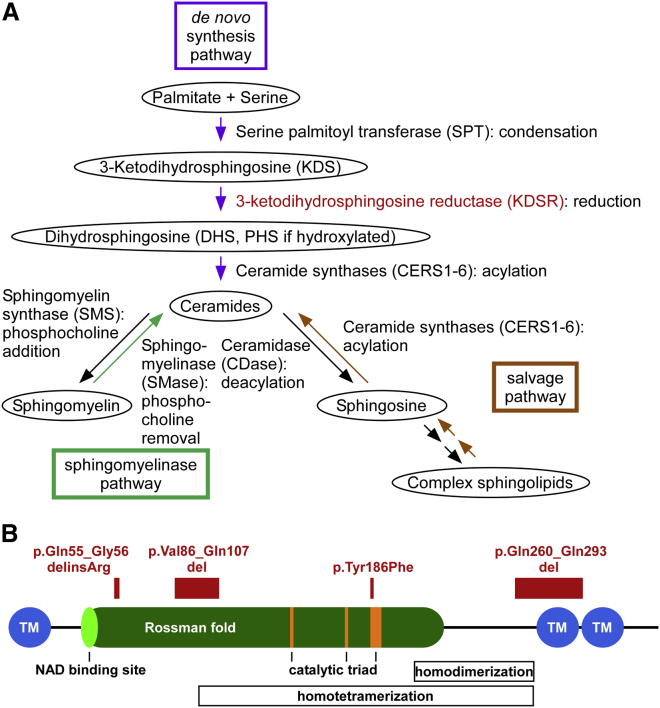
Figure 3.
KDSR Encodes 3-Ketodihydrosphingosine Reductase, an Enzyme in the Ceramide Synthesis Pathway
(A) The three major metabolic pathways capable of producing ceramides (the de novo, sphingomyelinase, and salvage pathways) are shown in purple, aqua, and brown, respectively. KDSR (in red) is critical to the creation of ceramides via de novo synthesis but is not intrinsic to the other two pathways.
(B) The domain structure of KDSR and the locations of mutations in PSEK subjects are shown. The TyrXXXLys, Asn, and Ser residues that form the canonical catalytic triad are in orange, the putative ThrGlyXXXGlyxGly NAD binding site and Rossman fold are in light and dark green, respectively, and the putative transmembrane domains are in blue. Putative homodimerization and homotetramerization domains are indicated with boxes. Residues that are substituted or deleted as a result of mutations in PSEK subjects are in red.
The domain structure of KDSR is homologous to that of other reductase enzymes (Figure 3B). The canonical TyrXXXLys reductase active site in KDSR is at amino acids 186–190. Conserved Asn and Ser residues at amino acids 145 and 173, respectively, form the canonical catalytic triad within a larger hydrophilic domain that extends from residues 22 to 270. A structurally conserved NAD-binding Rossman fold (an alpha-beta fold with a central beta sheet) extends from the putative NAD binding site ThrGlyXXXGlyXGly at amino acids 38–45 to residue 222. There are predicted homodimer and homotetramer interfaces at amino acids 208–268 and 98–268, respectively.14 Immunofluorescence studies have shown that KDSR localizes to the endoplasmic reticulum (ER), and proteinase K digestion has demonstrated that the large hydrophilic domain faces the cytosol.14, 15 There are putative transmembrane domains at residues 1–21, 271–291, and 294–314.14
The KDSR variants p.Val86_Gln107del and p.Gln260_Gln293del most likely have profound effects on protein structure. Both affect residues within the hydrophilic enzymatic domain. The former occurs within the NAD-binding domain and the homotetramer interface, and the latter is within both the homodimer and homotetramer interfaces and would omit one of the putative transmembrane domains, thereby affecting membrane topology at the C-terminal end of the protein. The p.Gln55_Gly56delinsArg variant alters highly conserved residues within the hydrophilic enzymatic domain. The p.Tyr186Phe variant affects the active-site tyrosine, which is completely conserved in both orthologs and paralogs and is the most conserved residue in reductase proteins (Figures 3B and S3). SIFT16 and PolyPhen17 predict that p.Tyr186Phe is highly damaging with scores of 0 and 1, respectively.
To investigate the consequence of KDSR mutations in the skin, we immunostained affected tissue from subject 1107 with antibodies to KDSR and to keratin 14 (KRT14, a marker of basal keratinocytes), keratin 10 (KRT10, a marker of suprabasal keratinocyte differentiation), and filaggrin (FLG, a marker of terminal differentiation in the granular layer of the epidermis). KDSR immunostaining intensity and localization were similar between normal tissue and affected tissue from this subject (Figures 4A and 4D). Although programs of epidermal differentiation can be grossly disrupted in some disorders of keratinization, including expansion of basal cell markers and loss of differentiation markers,18, 19 affected tissue from subject 1107 showed basal distribution of KRT14 and suprabasal distribution of KRT10, as was found in normal tissue (Figures 4B, 4C, 4F, and 4G). Notably, despite histologic absence of a granular layer (Figure 2E), affected tissue from subject 1107 showed expansion of filaggrin immunostaining (Figures 4D and 4H). Together, these results suggest a defect in keratinocyte terminal differentiation. Similar findings have been seen in tissue from affected individuals with widespread recessive congenital ichthyosis due to CERS3 mutations, which also affect ceramide synthesis.12, 13
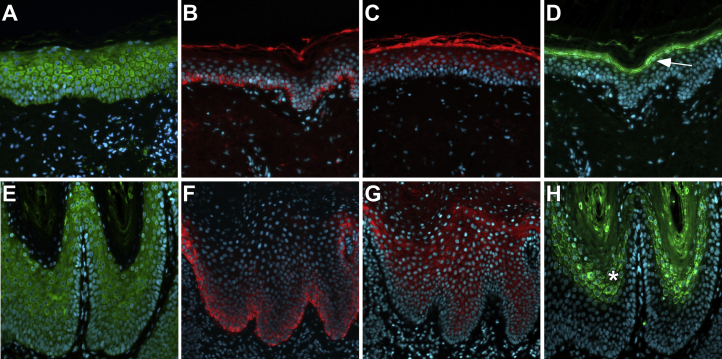
Figure 4.
Filaggrin Distribution Is Expanded in Affected PSEK Tissue
Tissue from a normal donor and affected tissue from subject 1107 was immunostained with primary antibodies to KDSR, KRT14, KRT10, and FLG (Santa Cruz Biotechnology sc-366781, sc-53253, sc-53252, and sc-25897, respectively) and corresponding Cy2 (green) or Cy3 (red) secondary antibodies. DAPI was used as a nuclear counterstain (blue).
(A and E) Positive immunostaining for KDSR (green) was observed in all layers of the epidermis in normal (A) and affected (E) tissue.
(B and F) Positive immunostaining for KRT14 (red) was limited to the basal layer of the epidermis in normal (B) and affected (F) tissue.
(C and G) Positive immunostaining for KRT10 (red), a marker of epidermal differentiation, was limited to the suprabasal epidermis in normal (C) and affected (G) tissue. Normal tissue showed typical autofluorescence of the stratum corneum.
(D and H) Positive immunostaining for FLG (green), a marker of the granular layer of the epidermis, was tightly restricted in normal tissue (D, arrow) but expanded within affected tissue (H, asterisk).
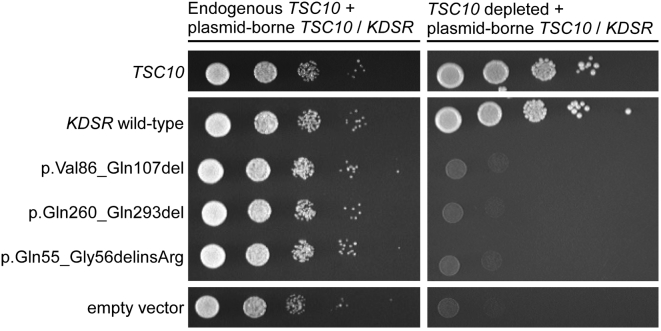
It has been previously shown that expression of wild-type human KDSR rescues the growth defect in yeast null for the orthologous TSC10, which otherwise cannot grow without supplementation with KDSR product DHS or PHS.14 To demonstrate the effect of the KDSR mutations observed in our subjects on the function of KDSR, we expressed both wild-type and mutant forms of human KDSR, including all of the mutant forms found in our initial two subjects, in yeast with TSC10 under a galactose-inducible and glucose-repressible conditional promoter. As expected, wild-type KDSR complemented the depletion of endogenous Tsc10, but the mutant forms of KDSR failed to complement, demonstrating their detrimental effect on KDSR function (Figures 5 and S4). We did not test the two other mutations identified. The inversion abolishes expression of KDSR, and the active-site Tyr186 has been characterized in other studies. This site is crucial to reductase function in both KDSR and paralogous members of the large reductase family of enzymes. Tyr-to-Phe substitutions at this site in several other reductases abolish enzymatic activity in vitro.23, 24, 25 Additionally, a temperature-sensitive p.Tyr186Phe KDSR variant expressed in Tsc10-null yeast failed to complement at 37°C.15
Figure 5.
Mutant KDSR Does Not Complement Depletion of Endogenous Yeast TSC10
A yeast strain with galactose-inducible and glucose-repressible expression of the KDSR ortholog TSC10 was created,20, 21, 22 and depletion of endogenous TSC10 was verified (Figure S4). Ten-fold serial dilutions of yeast expressing plasmid-borne mutant forms of KDSR in the presence or absence of endogenous yeast TSC10 showed severe growth defects in the absence of endogenous TSC10, whereas both yeast TSC10 and wild-type KDSR fully complemented the loss of endogenous yeast TSC10. The empty vector served as a negative control.
In complex organisms, three major pathways are capable of producing ceramides (Figure 3A). The de novo pathway generates ceramides “from scratch” via smaller building-block molecules, whereas the other pathways release ceramides via breakdown of larger molecules, either sphingomyelin from cell membranes (sphingomyelinase pathway) or sphingolipids that are converted to sphingosine and then acylated to form ceramides (salvage pathway).8, 9, 26 KDSR is critical to the creation of ceramides via de novo synthesis but is not intrinsic to the other two pathways.
Two of the probands (subjects 429 and 438) have been treated with a systemic retinoic acid derivative (isotretinoin), which remarkably resulted in nearly complete resolution of their scale and erythema (Figure S5). A third subject (1107) has been advised to start systemic isotretinoin therapy as a result of this work. Interestingly, retinoic acid has been shown to increase sphingosine acylation and to upregulate sphingomyelinase.27, 28, 29 These effects serve to stimulate the ceramide salvage pathway and sphingomyelinase pathway, which produce ceramides independent of KDSR (Figure 3A). This suggests that retinoic acid therapy is effective, at least in part, by compensating for a genetic defect in the ceramide de novo synthesis pathway via pharmacologic induction of alternative pathways for ceramide generation.
The identification of four unrelated probands compound heterozygous for previously unreported or extremely rare damaging mutations in KDSR, all of whom exhibit a unique skin disorder with well-demarcated symmetric scaling, erythema, and mild palmoplantar keratoderma, provides definitive evidence that recessive mutations in KDSR cause progressive symmetric erythrokeratoderma. This discovery underscores the central importance of ceramides in epithelial differentiation. The finding that systemic retinoids lead to nearly complete resolution of the skin findings in two subjects, most likely in part through activation of the salvage pathway for ceramide synthesis, provides an important therapeutic opportunity. Finally, RNA analysis and genome sequencing were essential to the detection of pathogenic mutations. Three of the four subjects are heterozygous for a silent third base change, for which cDNA sequencing was required to establish its damaging effect on the transcript, and two of the four are heterozygous for an inversion undetectable by exome sequencing. Consequently, at least one of the mutations in each of the four subjects would have been undetected by standard methods of exome sequencing and filtering. This work highlights the importance of comprehensive genetic evaluation in the study of severe, rare disorders such as erythrokeratoderma.
Acknowledgments
We thank the study subjects, their families, and the health-care professionals whose participation made this work possible. We also thank Erin Loring, Carol Nelson-Williams, Irina Tikhonova, Christopher Castaldi, Kaya Bilguvar, James Knight, and Bill Rizzo for technical assistance. This work was supported in part by a Clinician Scientist Development Award from the Doris Duke Charitable Foundation to K.A.C., the Foundation for Ichthyosis and Related Skin Types, and the National Institutes of Health (R01 AR068392 to K.A.C., R01 GM115710 to S.J. Baserga, and the Yale Center for Mendelian Genomics U54 HG006504).
Published: June 1, 2017
Footnotes
Supplemental Data include five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.003.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Database of Genomic Variants, http://dgv.tcag.ca/dgv/app/home
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
ExonPrimer, https://ihg.helmholtz-muenchen.de/ihg/ExonPrimer.html
Genome Analysis Toolkit (GATK), https://software.broadinstitute.org/gatk/
Integrative Genomics Viewer (IGV), http://software.broadinstitute.org/software/igv/
OMIM, https://www.omim.org/
SNPmasker, http://bioinfo.ebc.ee/snpmasker/
UCSC Genome Browser, https://genome.ucsc.edu/index.html
Variant Effect Predictor, http://useast.ensembl.org/info/docs/tools/vep/index.html
Supplemental Data
References
- 1.Richard G., Smith L.E., Bailey R.A., Itin P., Hohl D., Epstein E.H., Jr., DiGiovanna J.J., Compton J.G., Bale S.J. Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis. Nat. Genet. 1998;20:366–369. doi: 10.1038/3840. [DOI] [PubMed] [Google Scholar]
- 2.Macari F., Landau M., Cousin P., Mevorah B., Brenner S., Panizzon R., Schorderet D.F., Hohl D., Huber M. Mutation in the gene for connexin 30.3 in a family with erythrokeratodermia variabilis. Am. J. Hum. Genet. 2000;67:1296–1301. doi: 10.1016/s0002-9297(07)62957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyden L.M., Craiglow B.G., Zhou J., Hu R., Loring E.C., Morel K.D., Lauren C.T., Lifton R.P., Bilguvar K., Paller A.S., Choate K.A., Yale Center for Mendelian Genomics Dominant De Novo Mutations in GJA1 Cause Erythrokeratodermia Variabilis et Progressiva, without Features of Oculodentodigital Dysplasia. J. Invest. Dermatol. 2015;135:1540–1547. doi: 10.1038/jid.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maestrini E., Monaco A.P., McGrath J.A., Ishida-Yamamoto A., Camisa C., Hovnanian A., Weeks D.E., Lathrop M., Uitto J., Christiano A.M. A molecular defect in loricrin, the major component of the cornified cell envelope, underlies Vohwinkel’s syndrome. Nat. Genet. 1996;13:70–77. doi: 10.1038/ng0596-70. [DOI] [PubMed] [Google Scholar]
- 5.Boyden L.M., Kam C.Y., Hernández-Martín A., Zhou J., Craiglow B.G., Sidbury R., Mathes E.F., Maguiness S.M., Crumrine D.A., Williams M.L. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum. Mol. Genet. 2016;25:348–357. doi: 10.1093/hmg/ddv481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 7.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannun Y.A., Obeid L.M. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 9.Kihara A. Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides. Prog. Lipid Res. 2016;63:50–69. doi: 10.1016/j.plipres.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Borodzicz S., Rudnicka L., Mirowska-Guzel D., Cudnoch-Jedrzejewska A. The role of epidermal sphingolipids in dermatologic diseases. Lipids Health Dis. 2016;15:13. doi: 10.1186/s12944-016-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida Y. Ceramide signaling in mammalian epidermis. Biochim. Biophys. Acta. 2014;1841:453–462. doi: 10.1016/j.bbalip.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckl K.M., Tidhar R., Thiele H., Oji V., Hausser I., Brodesser S., Preil M.L., Onal-Akan A., Stock F., Müller D. Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J. Invest. Dermatol. 2013;133:2202–2211. doi: 10.1038/jid.2013.153. [DOI] [PubMed] [Google Scholar]
- 13.Radner F.P., Marrakchi S., Kirchmeier P., Kim G.J., Ribierre F., Kamoun B., Abid L., Leipoldt M., Turki H., Schempp W. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 2013;9:e1003536. doi: 10.1371/journal.pgen.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kihara A., Igarashi Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 2004;279:49243–49250. doi: 10.1074/jbc.M405915200. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S.D., Gable K., Han G., Borovitskaya A., Selby L., Dunn T.M., Harmon J.M. Tsc10p and FVT1: topologically distinct short-chain reductases required for long-chain base synthesis in yeast and mammals. J. Lipid Res. 2009;50:1630–1640. doi: 10.1194/jlr.M800580-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAleer M.A., Pohler E., Smith F.J., Wilson N.J., Cole C., MacGowan S., Koetsier J.L., Godsel L.M., Harmon R.M., Gruber R. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J. Allergy Clin. Immunol. 2015;136:1268–1276. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirza H., Kumar A., Craiglow B.G., Zhou J., Saraceni C., Torbeck R., Ragsdale B., Rehder P., Ranki A., Choate K.A. Mutations Affecting Keratin 10 Surface-Exposed Residues Highlight the Structural Basis of Phenotypic Variation in Epidermolytic Ichthyosis. J. Invest. Dermatol. 2015;135:3041–3050. doi: 10.1038/jid.2015.284. [DOI] [PubMed] [Google Scholar]
- 20.Mumberg D., Müller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Albalat R., González-Duarte, Atrian S. Protein engineering of Drosophila alcohol dehydrogenase. The hydroxyl group of Tyr152 is involved in the active site of the enzyme. FEBS Lett. 1992;308:235–239. doi: 10.1016/0014-5793(92)81282-q. [DOI] [PubMed] [Google Scholar]
- 24.Obeid J., White P.C. Tyr-179 and Lys-183 are essential for enzymatic activity of 11 beta-hydroxysteroid dehydrogenase. Biochem. Biophys. Res. Commun. 1992;188:222–227. doi: 10.1016/0006-291x(92)92373-6. [DOI] [PubMed] [Google Scholar]
- 25.Ensor C.M., Tai H.H. Bacterial expression and site-directed mutagenesis of two critical residues (tyrosine-151 and lysine-155) of human placental NAD(+)-dependent 15-hydroxyprostaglandin dehydrogenase. Biochim. Biophys. Acta. 1994;1208:151–156. doi: 10.1016/0167-4838(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 26.Kitatani K., Idkowiak-Baldys J., Hannun Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal. 2008;20:1010–1018. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalén A., Borchardt R.A., Bell R.M. Elevated ceramide levels in GH4C1 cells treated with retinoic acid. Biochim. Biophys. Acta. 1992;1125:90–96. doi: 10.1016/0005-2760(92)90160-w. [DOI] [PubMed] [Google Scholar]
- 28.Riboni L., Prinetti A., Bassi R., Caminiti A., Tettamanti G. A mediator role of ceramide in the regulation of neuroblastoma Neuro2a cell differentiation. J. Biol. Chem. 1995;270:26868–26875. doi: 10.1074/jbc.270.45.26868. [DOI] [PubMed] [Google Scholar]
- 29.Kraveka J.M., Li L., Bielawski J., Obeid L.M., Ogretmen B. Involvement of endogenous ceramide in the inhibition of telomerase activity and induction of morphologic differentiation in response to all-trans-retinoic acid in human neuroblastoma cells. Arch. Biochem. Biophys. 2003;419:110–119. doi: 10.1016/j.abb.2003.08.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.