Abstract
Sperm motility is vital to human reproduction. Malformations of sperm flagella can cause male infertility. Men with multiple morphological abnormalities of the flagella (MMAF) have abnormal spermatozoa with absent, short, coiled, bent, and/or irregular-caliber flagella, which impair sperm motility. The known human MMAF-associated genes, such as DNAH1, only account for fewer than 45% of affected individuals. Pathogenic mechanisms in the genetically unexplained MMAF remain to be elucidated. Here, we conducted genetic analyses by using whole-exome sequencing and genome-wide comparative genomic hybridization microarrays in a multi-center cohort of 30 Han Chinese men affected by MMAF. Among them, 12 subjects could not be genetically explained by any known MMAF-associated genes. Intriguingly, we identified compound-heterozygous mutations in CFAP43 in three subjects and a homozygous frameshift mutation in CFAP44 in one subject. All of these recessive mutations were parentally inherited from heterozygous carriers but were absent in 984 individuals from three Han Chinese control populations. CFAP43 and CFAP44, encoding two cilia- and flagella-associated proteins (CFAPs), are specifically or preferentially expressed in the testis. Using CRISPR/Cas9 technology, we generated two knockout models each deficient in mouse ortholog Cfap43 or Cfap44. Notably, both Cfap43- and Cfap44-deficient male mice presented with MMAF phenotypes, whereas the corresponding female mice were fertile. Our experimental observations on human subjects and animal models strongly suggest that biallelic mutations in either CFAP43 or CFAP44 can cause sperm flagellar abnormalities and impair sperm motility. Further investigations on other CFAP-encoding genes in more genetically unexplained MMAF-affected individuals could uncover novel mechanisms underlying sperm flagellar formation.
Keywords: CFAP43, CFAP44, CFAP65, cilia, CGH, flagella, male infertility, motility, sequencing, sperm
Introduction
Male infertility is one of the major issues of human health.1 Abnormalities in sperm flagellar morphology can impair sperm motility and function.2 Individuals with multiple morphological abnormalities of the flagella (MMAF) mostly present with a combination of sperm flagellar malformations, which include absent, short, coiled, bent, and/or irregular-caliber flagella and can be assessed by light microscopy.2, 3, 4 Furthermore, unassembled sperm fibrous sheaths and a lack of central microtubules and/or dynein arms have been observed by transmission electron microscopy.3, 4 Men with MMAF have been previously reported as having dysplasia of fibrous sheath (DFS) and short tails.5, 6 The term MMAF was proposed in 2014 to provide a clear definition of the phenotypes in the affected individuals.3 The incidence of MMAF or DFS has not been reported yet but was previously estimated to be higher than that of primary ciliary dyskinesia (PCD [MIM: 244400]; 1 per 10,000–20,000 births).6, 7, 8 As a result of severe flagellar abnormalities, MMAF impairs sperm motility and even leads to total immotility. No success of spontaneous pregnancy or conventional in vitro fertilization has been previously reported for MMAF, but individuals with MMAF have had a good prognosis after intracytoplasmic sperm injection.9
Previous studies on genetic mutants in model organisms have revealed genetic contributions to flagellar motility defects.2, 10, 11, 12, 13, 14 So far, mutations in only three genes, AKAP4 (MIM: 300185), CCDC39 (MIM: 613798), and DNAH1 (MIM: 603332), have been formally identified to cause MMAF in humans.2, 3, 15, 16 Among them, biallelic DNAH1 mutations in MMAF have been recurrently identified across studies and account for 28%–44% of MMAF cases.3, 17, 18 However, the genetic causes or molecular mechanisms in the other half of MMAF cases are still unclear, indicating the genetic heterogeneity of MMAF.
In this study, we recruited 30 Han Chinese men with MMAF from multiple centers in China. We conducted whole-exome sequencing (WES) to identify point mutations and indels potentially responsible for human MMAF. Because some copy-number variations (CNVs, including deletions and duplications) cannot be readily detected by WES,19, 20 we also employed high-density oligonucleotide-based comparative genomic hybridization (CGH) microarrays for genome-wide CNV analysis in the MMAF cases that cannot be resolved by point and indel mutations alone. Remarkably, biallelic mutations in CFAP43 (also known as WDR96) and CFAP44 (WDR52) were identified in 4 of 12 (33%) genetically unexplained MMAF-affected individuals in this study. CFAP43 and CFAP44, encoding cilia- and flagella-associated proteins (CFAPs), are specifically or preferentially expressed in the testis. We also generated two knockout models for mouse orthologs Cfap43 and Cfap44. Both mouse models mimicked the human MMAF phenotypes. Notably, we observed a homozygous nonsense mutation in another CFAP-encoding gene, CFAP65 (CCDC108 [MIM: 614270]), in one subject from a consanguineous family. All together, our findings strongly suggest that both CFAP43 and CFAP44 are associated with MMAF and that their biallelic mutations can impair sperm motility and cause male infertility.
Material and Methods
Study Participants
30 Han Chinese men with MMAF were enrolled at multiple centers in China, including Suzhou Hospital (affiliated with Nanjing Medical University), Yantai Yuhuangding Hospital (affiliated with Qingdao University), the First Affiliated Hospital of Nanjing Medical University, Shanghai General Hospital, and Jinghua Hospital of Shenyang Eastern Medical Group. Parental consanguinity was self-reported by three individuals (P001, P014, and P019). Parental DNA samples were available for 9 of 30 MMAF-affected subjects. The parenthood of these nine trios was confirmed by the EX20 kit (AGCU ScienTech Incorporation). This work was approved by the institutional review boards of the School of Life Sciences at Fudan University, Suzhou Hospital, and the other participating institutions. Informed consent was obtained from each subject.
Semen Analysis and Sperm Morphological Study
Semen samples of human subjects were collected by masturbation after 2–7 days of sexual abstinence and examined after liquefaction for 30 min at 37°C. Semen volume and sperm concentration and motility were evaluated according to the World Health Organization (WHO) guidelines. The semen analysis was replicated at least twice. Semen samples of the mouse models were collected from the epididymis, diluted in 1 mL sperm rinse (10101, Vitrolife), and examined after incubation for 30 min at 37°C. Semen was analyzed with a computer-assisted analysis system (Cyto-S, VideoTesT). At least four 8-week-old male C57BL/6 mice were studied for each of three groups: wild-type, Cfap43-deficient, and Cfap44-deficient mice.
Sperm morphology was analyzed on slides after staining by a modified Papanicolaou method. At least 200 spermatozoa were examined. The percentages of morphologically normal and abnormal spermatozoa were evaluated according to the WHO guidelines. The morphological abnormalities of sperm flagella were classified into five categories: (1) absent, (2) short, (3) coiled flagella, (4) angulation, and (5) irregular caliber.3 Irregular caliber was defined as big meshes occupying the space of the tail structures. One spermatozoon was classified in only one morphological category according to its major flagellar abnormality.4
WES and Data Processing
WES was performed on all 30 subjects with MMAF. Genomic DNAs were extracted from peripheral-blood samples of the subjects and their available parents with the DNeasy Blood and Tissue Kit (QIAGEN). The Agilent SureSelectXT Human All Exon Kit was employed to enrich the human exome. Next-generation sequencing was conducted with the Illumina HiSeq X-TEN platform at CloudHealth Genomics.
Reads were aligned to the human genome reference assembly (UCSC Genome Browser hg19) with the Burrows-Wheeler Aligner.21 We employed Picard software to remove PCR duplicates and evaluate the quality of variants by attaining effective reads, effective base, average coverage depth, and 90×–120× coverage ratio. Single-nucleotide variants (SNVs) and indels were called and analyzed by the Genome Analysis Toolkit.22 The SNVs with read depths less than 4× were filtered out. Then we used ANNOVAR for functional annotation with OMIM, Gene Ontology, KEGG Pathway, SIFT, PolyPhen-2, MutationTaster, and the Exome Aggregation Consortium (ExAC) Browser.23, 24, 25, 26, 27, 28, 29 The candidate pathogenic mutations and their parental origins were verified by Sanger sequencing.
Array-Based CGH
For the MMAF-affected subjects who could not be explained by indel or point mutations alone, we conducted CNV analysis by using Agilent high-density oligonucleotide-based CGH microarrays. Some experimental details of the array-based CGH (aCGH) assay have been previously described.30, 31 The test DNA of each subject and the human male reference DNA (Promega) were fragmented by both AluI and RsaI digestion. DNA was labeled with the Agilent SureTag DNA Labeling Kit. Different fluorescence dyes were used for DNA labeling of each test DNA (Cy5-dUTP) and the reference DNA (Cy3-dUTP). Each labeled test DNA was hybridized together with the labeled reference DNA onto Agilent 1× 1M human CGH microarrays. DNA processing, microarray handling, and data processing were conducted according to the Agilent oligonucleotide CGH protocol (version 6.0). The genomic CNVs were analyzed with Agilent Genomic Workbench software. For CNV verification, long-range PCR was conducted to amplify CNV breakpoints with TaKaRa LA Taq polymerase, and the product was analyzed by Sanger sequencing.30
Generation of Gene-Deficient Models in Mouse Orthologs Cfap43 and Cfap44
The frameshift mutations were generated in mouse Cfap43 and Cfap44 with CRISPR/Cas9 technology.32, 33, 34 The guide RNAs were designed against exon 22 of Cfap43 and exon 15 of Cfap44 in accordance with the positions of pathogenic mutations in individuals P003 and P002, respectively. The Cas9 and single-guide RNA (sgRNA) pX458 plasmid was obtained from Addgene (plasmid 48138). The sgRNAs were synthesized, annealed, and ligated to the pX458 plasmid, which was digested with BbsI. The pX458 plasmid harboring corresponding sgRNA was transfected into embryonic stem cells with Lipofectamine 3000 Reagent according to the manufacturer’s instructions. 24 hr after transfection, the cells expressing EGFP were separated with flow cytometry and plated on dishes. Single colonies were picked and expanded for genotyping. C57BL/6 female mice were superovulated and mated with wild-type C57BL/6 male mice. Blastocysts were collected and injected with embryonic stem cells carrying genetic modifications. After a short in vitro culture, the injected blastocysts were transferred into pseudopregnant female mice. The frameshift mutations in Cfap43 and Cfap44 were identified in founder mice and their offspring by PCR and Sanger sequencing. This study was carried out in accordance with the recommendation of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Quantitative RT-PCR
Total RNAs were extracted from mouse testis with the Rneasy MiNi Kit (QIAGEN) and treated with 5 U RNase-free DNAase I (TaKaRa) at 25°C for 10 min. Approximately 0.3 μg total RNAs were converted into cDNAs with SuperScript III reverse transcriptase (Invitrogen) and oligo (dT) primers (TaKaRa). The cDNAs were individually diluted 10-fold to be used as templates for the subsequent real-time fluorescence quantitative PCR with AceQ qPCR SYBR Green Master Mix (Vazyme). Mouse Gapdh was used as an internal control. Cfap43 and Cfap44 mRNA expression was quantified according to the 2−ΔΔCt method.
Histological Analysis of Mouse Tissues
Fresh mouse testicular and epididymal tissues were fixed in Bouin’s solution and 4% paraformaldehyde, respectively, for over 24 hr, embedded in paraffin, and sectioned at 4 μm intervals for the generation of tissue slides. After deparaffinization, slides were stained with hematoxylin and eosin via standard methods for visualization of structures.
Electron Microscopy Evaluation
For scanning electron microscopy (SEM) assay, the sperm specimens were immersed in 2.5% glutaraldehyde for 4 hr at 4°C, rinsed in 0.1 mol/L phosphate buffer for 30 min, and post-fixed in 1% osmic acid for 1 hr at 4°C. After being rinsed thoroughly in the same buffer for 30 min, the specimens were progressively dehydrated with an ethanol and isoamyl acetate gradient and dried by a CO2 critical-point dryer (Eiko HCP-2, Hitachi). Afterward, the specimens were mounted on aluminum stubs, sputter-coated by an ionic sprayer meter (Eiko E-1020, Hitachi), and analyzed by SEM (Stereoscan 260) under an accelerating voltage of 20 kV.
For transmission electron microscopy (TEM), the sperm cells were fixed routinely. After being embedded in Epon 812, ultrathin sections were stained with uranyl acetate and lead citrate and observed and photographed by TEM (TECNAI-10, Philips) with an accelerating voltage of 80 kV. For quantification of axonemal anomalies by TEM, at least 50 flagella with cross sections and several longitudinal sections were counted.
Mouse testicular tissues were fixed, embedded, and stained in the same manner as the sperm cells and were observed by TEM (HT 7700, Hitachi) with an accelerating voltage of 120 kV. Given that spermatogenesis was impaired in both Cfap43- and Cfap44-deficient male mice, quantitative assessment of sperm flagella was not available.
Results
Biallelic Mutations in CFAP43 and CFAP44, Encoding CFAPs
The workflow of our MMAF genetic analysis using WES and aCGH assays is shown in Figure S1. Because men with MMAF are infertile, the pathogenic variants responsible for MMAF cannot be frequent in human populations. Therefore, we excluded the genetic variants with allele frequencies ≥ 0.01 according to the ExAC Browser and 1000 Genomes Project.29, 35 Nonsense, frameshift, and essential splice-site variants were preferred. In addition, the missense variants predicted to be potentially deleterious simultaneously by SIFT, PoyPhen-2, and MutationTaster were also kept for further evaluation.26, 27, 28 Because an autosomal-recessive inheritance has been assumed for MMAF,2, 4 the genes with two alleles with loss-of-function mutations or potentially deleterious missense mutations in any MMAF-affected subject were ascertained. 18 of 30 (60%) MMAF-affected subjects fit the autosomal-recessive inheritance of the known human MMAF-associated genes, including DNAH1 (17 subjects) and CCDC39 (one subject) (Table S1).
In the remaining subjects with MMAF, we identified biallelic mutations in CFAP43, CFAP44, or CFAP65 in five subjects (Tables 1 and S2). Remarkably, these individuals accounted for 42% (5/12) of those with disease unexplained by previously known genes associated with human MMAF. All three genes encode CFAPs and are specifically or preferentially expressed in the testis according to data from ENCODE, FANTOM, and GTEx.36, 37, 38
Table 1.
Biallelic CFAP43 and CFAP44 Mutations Identified in the Subjects with MMAF
|
Subject |
|||||||
|---|---|---|---|---|---|---|---|
| P003 | P028 | P029 | P002 | ||||
| Gene | CFAP43 | CFAP43 | CFAP43 | CFAP44 | |||
| cDNA mutationa | c.2802T>A | c.4132C>T | c.253C>T | c.3945_4431del | c.386C>A | c.2802T>A | c.2005_2006delAT (homozygous) |
| Protein alteration | p.Cys934∗ | p.Arg1378∗ | p.Arg85Trp | p.Ile1316Leufs∗10 | p.Ser129Tyr | p.Cys934∗ | p.Met669Valfs∗13 |
| Mutation type | nonsense | nonsense | missense | exonic deletion, frameshift | missense | nonsense | frameshift |
| Allele frequency in ExAC Browser | 0 | 0 | 3.3 × 10−5 | 0 | 0 | 0 | 8.3 × 10−6 |
| Conservationb | |||||||
| Phastcons | 0.579 | 0.998 | 0.996 | NA | 1 | 0.579 | 0.644 |
| Phylop | 0.017 | 0.465 | 4.963 | NA | 5.249 | 0.017 | 3.461 |
| Functional Prediction | |||||||
| SIFT | NA | NA | damaging | NA | damaging | NA | NA |
| PolyPhen-2 | NA | NA | probably damaging | NA | probably damaging | NA | NA |
| MutationTaster | NA | NA | disease causing | NA | disease causing | NA | NA |
Abbreviation: NA, not available.
The accession numbers for CFAP43 and CFAP44 are GenBank: NM_025145.6 and NM_018338.3, respectively.
The Phastcons value is close to 1 when a nucleotide is conserved, and the predicted conserved sites are assigned positive scores by Phylop.
Five CFAP43 mutations were identified in three MMAF-affected subjects (P003, P028, and P029) (Figure 1 and Table 1). In subject P003 (II-1 in family 003 in Figure 1A), both CFAP43 mutations (GenBank: NM_025145.6) are nonsense: c.2802T>A (p.Cys934∗) and c.4132C>T (p.Arg1378∗). Notably, the c.2802T>A was recurrent in unrelated subject P029 (II-1 in family 029 in Figure 1A), in whom we also identified the second allele with the novel missense CFAP43 mutation c.386C>A (p.Ser129Tyr). This mutation was located at a conserved position of CFAP43 and was predicted to be potentially deleterious by all three bioinformatic tools: SIFT, PolyPhen-2, and MutationTaster (Figure 1 and Table 1).26, 27, 28 Another rare and potentially deleterious missense mutation in CFAP43, c.253C>T (p.Arg85Trp), was identified in subject P028 (II-1 in family 028 in Figure 1A). In addition to this point mutation revealed by WES, we also identified a 3.3-kb heterozygous deletion within CFAP43 in P028 by using aCGH and long-range PCR assays (Figures 1A and S2). This deletion eliminated exons 32–34 and caused a frameshift in CFAP43 (Figure S2 and Table 1).
Figure 1.
Biallelic Mutations in CFAP43 and CFAP44 Identified in Subjects with MMAF
(A) Biallelic CFAP43 mutations (M1–M5) were identified in three subjects with MMAF (P003, P028, and P029). M1 was recurrent in unrelated individuals P003 and P029. The parental origins of all CFAP43 mutations are shown.
(B) The positions of four point mutations in CFAP43 (M1–M3 and M5) are shown. The affected amino acid residues are conserved across species.
(C) The homozygous frameshift mutation in CFAP44 (M6) was identified in one subject with MMAF (P002). Both of his parents carried the heterozygous mutation.
All CFAP43 and CFAP44 mutations were verified by Sanger sequencing. Red arrows indicate the positions of point mutations, and red rectangles indicate the shifted sequences after deletion start points. Abbreviation: WT, wide-type.
The homozygous frameshift mutation c.2005_2006delAT (p.Met669Valfs∗13) in CFAP44 (GenBank: NM_018338.3) was identified in subject P002 (II-1 in Figure 1C) and was predicted to cause premature translational termination of CFAP44 (Table 1). All of the aforementioned biallelic mutations in CFAP43 and CFAP44 were confirmed to be inherited from the parents carrying heterozygous recessive mutations (Figure 1).
In affected individual P019 (II-1 in Figure S3A) from a consanguineous family, we identified the homozygous nonsense mutation c.5341G>T (p.Glu1781∗) in CFAP65 (GenBank: NM_194302.3). Furthermore, CNV analysis using a high-density CGH microarray did not identify any obvious deletion in CFAP65 (Figure S3C), suggesting that nonsense mutation c.5341G>T is preferentially a homozygous mutation rather than hemizygous. However, the parental DNA samples of P019 were not available for analysis of mutation origins. Therefore, this CFAP65 mutation was not included in our subsequent functional studies for MMAF.
To accurately assess the allele frequency of these candidate pathogenic mutations in CFAP43, CFAP44, and CFAP65, we investigated three control populations of Han Chinese ancestry: (1) 201 fertile Han Chinese men as a healthy control group, (2) 575 Han Chinese individuals (335 males and 240 females) as the first population control group, and (3) 208 Han Chinese individuals from the 1000 Genomes Project (phase 3) as the second population control group. Notably, none of the candidate pathogenic variants in CFAP43, CFAP44, and CFAP65 were identified in any of the three control populations (totaling 984 individuals with 1,968 alleles). Our observations suggest that the identified CFAP43, CFAP44, and CFAP65 mutations in our MMAF-affected subjects are rare (allele frequency < 0.001 in the Han Chinese).
We also studied the publically available genetic data of more than 100,000 individuals from the ExAC Browser and its newly released Genome Aggregation Database (gnomAD). No homozygous loss-of-function (nonsense, frameshift, or essential splice-site) mutations in CFAP43, CFAP44, or CFAP65 (canonical transcripts were used) were identified in gnomAD (Table S3). These observations suggest that the homozygous loss-of-function mutations in these genes are pathogenic.
MMAF Phenotypes in Subjects with Biallelic Mutations in CFAP43 or CFAP44
The detailed MMAF phenotypes in three subjects with CFAP43 mutations (P003, P028, and P029) and one with a CFAP44 homozygous mutation (P002) were ascertained through semen analysis and light and electron microscopy. All four subjects were shown to have severe to complete asthenospermia according to semen characteristics. No spermatozoa with progressive motility could be observed in any of the subjects (Table 2).
Table 2.
Semen Characteristics and Sperm Morphology in the Subjects Carrying Biallelic Mutations in CFAP43 and CFAP44
|
Subject |
||||
|---|---|---|---|---|
| P003 | P028 | P029 | P002 | |
| Gene | CFAP43 | CFAP43 | CFAP43 | CFAP44 |
| Semen Parameters | ||||
| Semen volume (mL) | 2.2–3.8 | 1.5–2.5 | 2.5–4.0 | 2.4–3.8 |
| Sperm concentration (106/mL) | 16.1–39.4 | 5.0–10.0 | 12.2–18.9 | 5.6–12.5 |
| Motility (%) | 0 | 2 | 1 | 0 |
| Progressive motility (%) | 0 | 0 | 0 | 0 |
| Sperm Morphology | ||||
| Normal flagella (%) | 20.5 | 3.0 | 7.5 | 0.5 |
| Absent flagella (%) | 8.0 | 11.0 | 18.5 | 42.5 |
| Short flagella (%) | 52.0 | 36.5 | 31.5 | 40.5 |
| Coiled flagella (%) | 9.5 | 39.0 | 25.0 | 5.0 |
| Angulation (%) | 8.0 | 3.5 | 8.5 | 4.0 |
| Irregular caliber (%) | 2.0 | 7.0 | 9.0 | 7.5 |
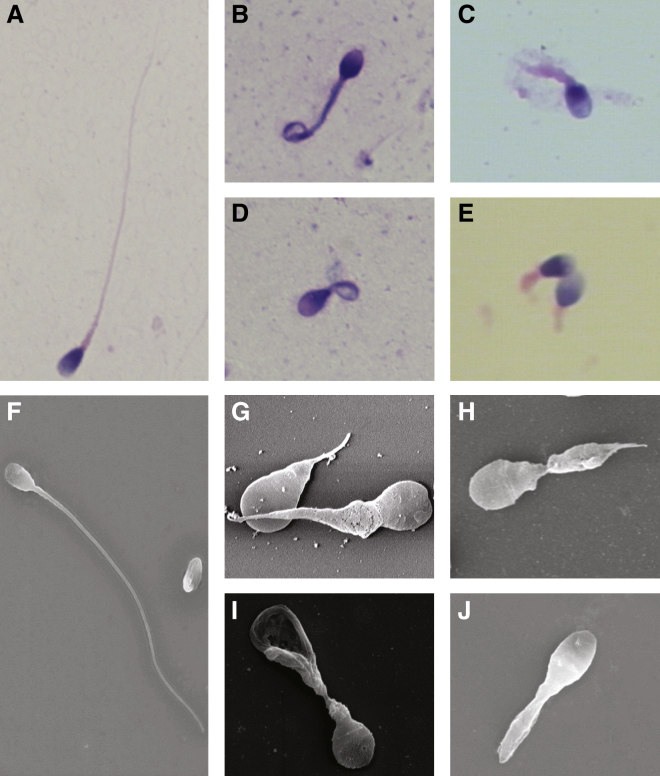
The morphological abnormalities of sperm flagella were first assessed by light microscopy, which identified severely distorted sperm flagella, including absent, short, coiled, bent, and irregular-caliber flagella (Figures 2B–2E). The affected spermatozoa with abnormal flagella accounted for 79.5%–99.5% of the spermatozoa analyzed in these individuals carrying biallelic mutations in CFAP43 or CFAP44 (Table 2), and short, coiled, and absent flagella were the most frequently observed. SEM was also used for studying sperm morphology, the results of which were consistent with those of light microscopy (Figures 2G–2J).
Figure 2.
Sperm Morphology in the Subjects Carrying Biallelic Mutations in CFAP43 or CFAP44
(A) Light microscopy shows a spermatozoon with normal morphology from a healthy man.
(B–D) Most spermatozoa of the subjects with biallelic CFAP43 mutations (B, P003; C, P028; D, P029) presented with morphological abnormalities of the sperm flagella, such as short and coiled flagella, flagella of irregular caliber, and other malformations diagnosed as MMAF.
(E) In individual P002, who carries homozygous frameshift mutations in CFAP44, most spermatozoa had short or absent flagella.
(F) SEM shows a spermatozoon with a normal flagellum.
(G–I) SEM shows that most spermatozoa of the subjects with biallelic CFAP43 mutations (G, P003; H, P028; I, P029) presented with absent, short, or coiled flagella and other MMAF phenotypes.
(J) The spermatozoa in CFAP44-deficient subject P002 were abnormal. One of the major abnormalities was short flagella. The results of SEM were consistent with those of light microscopy.
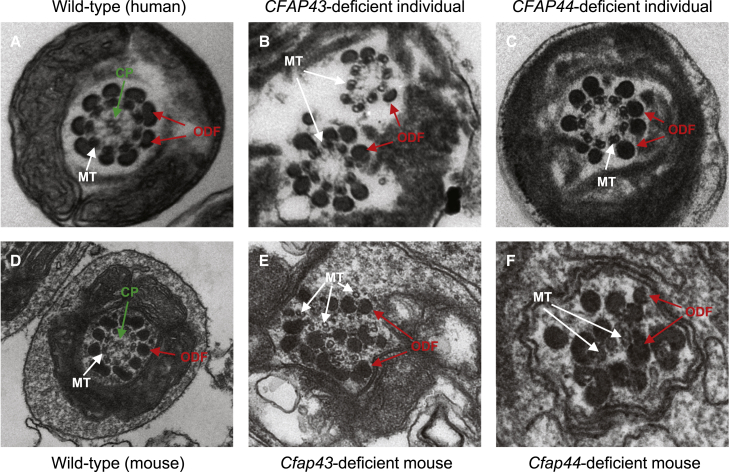
TEM observations on the above specimen further revealed that the spermatozoa from these four subjects presented with disorganization in fibrous sheaths and other axonemal and periaxonemal structures (Figures 3B and 3C). Cross sections showed hypertrophy and hyperplasia of fibrous sheaths. The central microtubules were absent (9+0) in most flagella, and various cytoskeletal components were distorted as well.
Figure 3.
Sperm Ultrastructures in MMAF-Affected Individuals with Mutations in CFAP43 and CFAP44
(A) The ultrastructure of a cross section in a normal spermatozoon from a healthy man. The typical “9+2” microtubule structure (nine peripheral microtubule doublets paired with nine outer dense fibers and the central pair of microtubules) is shown.
(B and C) In CFAP43-deficient (B) and CFAP44-deficient (C) subjects, most spermatozoa lacked the central pair of microtubules and had hyperplasia of fibrous sheaths.4 (B) Two axonemes in one section were probably due to a folded flagellum. The flagellum showed a “9+0” arrangement of microtubules and lacked the central pair of microtubules. (C) The cross section shows a thickened fibrous sheath and an absent central pair of microtubules. The outer dense fibers and peripheral microtubule are regularly arranged.
(D) TEM shows the typical “9+2” microtubule structure in a normal testicular spermatozoon from a wild-type male mouse.
(E and F) TEM analysis in Cfap43-deficient (E) and Cfap44-deficient (F) male mice shows a totally disorganized axoneme in a testicular spermatozoon. The central pair of microtubules are not apparent. The outer dense fibers and peripheral microtubules are misarranged.
Abbreviations: CP, central pair of microtubules (green arrows); MT, peripheral microtubule doublet (white arrows); and ODF, outer dense fiber (red arrows).
Cfap43- and Cfap44-Deficient Mouse Models Show MMAF Phenotypes
Frameshift mutations in mouse orthologs Cfap43 and Cfap44 were induced by CRISPR/Cas9 technology. For Cfap43, two frameshift mutations were generated: a 2-bp deletion and a 27-bp deletion plus a 10-bp insertion at the deletion interval (Figure S4). For Cfap44, a frameshift mutation was generated via the deletion of a 14-bp coding sequence (Figure S5). The levels of Cfap43 or Cfap44 transcripts in the testis were investigated by quantitative PCR assays. The Cfap43 mRNA level in Cfap43−/− mice was reduced to approximately 12% of that in wild-type mice, whereas the Cfap44 mRNA level in Cfap44−/− mice was reduced to approximately 16% of that in wild-type mice (Figure S6). This suggests nonsense-mediated mRNA decay triggered by premature translational termination. No commercial antibody was available for western blot analysis.
To investigate the fertility of Cfap43- and Cfap44-deficient mice, we bred both male and female knockout mice with wild-type mice. We found that both Cfap43- and Cfap44-deficient male mice were infertile, whereas female mice with corresponding mutants could generate offspring. No significant difference was observed in body and testis weights between wild-type and knockout male mice (Table 3). Semen characteristics of Cfap43- and Cfap44-deficient male mice were examined, and severe defects in sperm motility were observed in both mouse models (Table 3). No normal sperm flagella were observed in these knockout male mice. The primary morphological abnormalities in spermatozoa shown by light microscopy were short, coiled, and absent flagella (Figure 4 and Table 3). The flagellar ultrastructures of testicular sperm shown by TEM indicated a lack of the central pair of microtubules, which is consistent with the observations in human MMAF-affected subjects carrying biallelic mutations in CFAP43 or CFAP44. Furthermore, the nine peripheral microtubule doublets and outer dense fibers of the spermatozoa from knockout mice were scattered disorderly (Figures 3E and 3F).
Table 3.
Characteristics and Sperm Morphology in the Cfap43- and Cfap44-Deficient Male Mice
| Wild-Type Male Micea | Cfap43-Deficient Male Micea | Cfap44-Deficient Male Micea | |
|---|---|---|---|
| Body weight (g) | 24.2 (22.4–25.9) | 22.1 (20.5–24.5) | 24.7 (21.3–27.0) |
| Testis Weight | |||
| Left (g) | 0.093 (0.079–0.108) | 0.084 (0.078–0.094) | 0.091 (0.077–0.102) |
| Right (g) | 0.095 (0.074–0.117) | 0.089 (0.082–0.101) | 0.091 (0.072–0.108) |
| Semen Parameters | |||
| Sperm count (106)b | 6.8 (6.7–7.0) | 0.2 (0.1–0.4) | 0.4 (0.2–0.7) |
| Motility (%) | 76 (72–79) | 0 | 1 (0–2) |
| Progressive motility (%) | 65 (70–71) | 0 | 1 (0–2) |
| Sperm Morphology | |||
| Normal flagella (%) | 90.8 (89.0–94.0) | 0 | 0 |
| Absent flagella (%) | 1.2 (0.5–2.5) | 11.5 (8.0–13.5) | 12.0 (8.0–19.5) |
| Short flagella (%) | 1.8 (1.5–2.0) | 43.3 (36.0–52.5) | 50.4 (47.0–55.0) |
| Coiled flagella (%) | 4.2 (2.0–5.5) | 32.0 (30.5–34.5) | 25.8 (20.5–33.0) |
| Angulation (%) | 1.7 (1.0–2.5) | 6.0 (4.0–8.5) | 5.5 (3.0–7.0) |
| Irregular caliber (%) | 0.3 (0–0.5) | 7.2 (4.5–8.0) | 6.3 (5.5–8.0) |
Values represent the mean (range).
Per single epididymis.
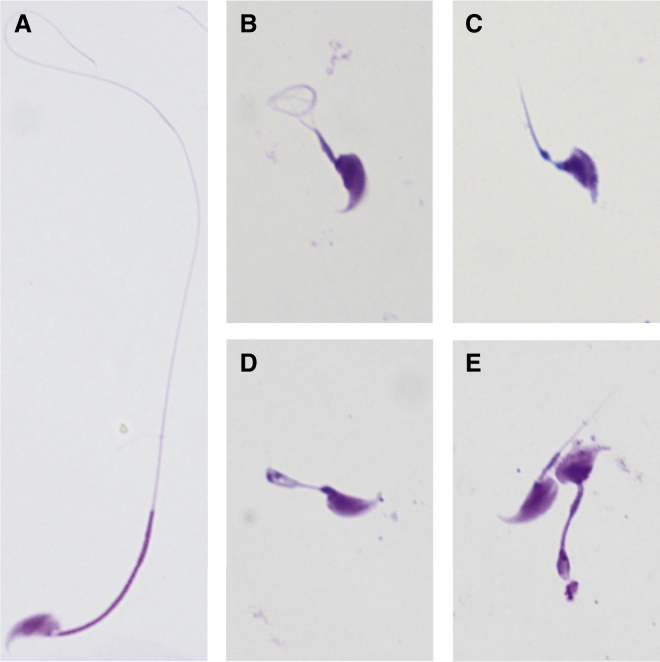
Figure 4.
Sperm Morphology in Cfap43- and Cfap44-Deficient Male Mice
(A) A spermatozoon with normal morphology from a wild-type male mouse.
(B–E) Almost all spermatozoa of Cfap43-deficient (B and C) and Cfap44-deficient (D and E) male mice had flagellar abnormalities. These flagella presented with short, coiled, or other distorted shapes, consistent with the clinical phenotypes in the subjects with MMAF.
We also investigated the epididymal and testicular tissues from the male mice deficient in Cfap43 and Cfap44. In seminiferous tubules, spermatogonia and spermatocytes had normal morphology. However, sperm flagella were distorted and contained excess cytoplasm in both Cfap43- and Cfap44-deficient male mice, whereas spermatozoa from wild-type male mice had normal elongated flagella (Figure S7). No apparent structural defects were observed in seminiferous tubules (Figure S7). In addition, cross sections of the cauda epididymides in Cfap43- and Cfap44-deficient male mice showed shedding of residual bodies, abnormal cells, and disorganized spermatozoa with flagellar malformations (Figure S7).
Discussion
Previously, only three genes (AKAP4, CCDC39, and DNAH1) and their autosomal-recessive inheritance had been formally identified to cause human MMAF, to which DNAH1 was the major genetic contributor.2, 3, 15, 16 However, the biallelic DNAH1 mutations identified in previous studies only accounted for 28%–44% of MMAF.3, 17, 18 These observations suggested the existence of unknown genes associated with MMAF. In this study, we employed both WES (for point mutations and indels) and aCGH assays (for deletions and other CNVs) in MMAF-affected individuals. Intriguingly, we identified mutations in CFAP43, CFAP44, and CFAP65 in 5 of 30 (17%) subjects with MMAF (Tables 1 and S2). The rare, potentially deleterious biallelic mutations in other genes in these five subjects are shown in Table S4, but none of these mutant genes have been associated with sperm flagellar formation or ciliopathies. Therefore, the MMAF in these subjects is preferentially explained by the biallelic mutations in CFAP43, CFAP44, and CFAP65.
Biallelic loss-of-function mutations in CFAP43, CFAP44, or CFAP65 are very rare in human populations. For example, homozygous loss-of-function mutations in these three genes are absent from the more than 100,000 individuals from gnomAD (Table S3), suggesting their pathogenicity.
Only homozygous missense mutations in CFAP43, CFAP44, and CFAP65 were reported in gnomAD. Among them, some were predicted to be potentially deleterious by both SIFT and PolyPhen-2 (Table S3). Interestingly, four of these homozygous missense mutations were too frequent (recurrent in more than ten individuals) to be considered disease causing in MMAF (Table S3).
Another explanation is that the deleterious mutations might not necessarily cause diseases in homozygous individuals. Previously, we identified a common TBX6 hypomorphic allele in which homozygosity did not result in disease in human populations.31 Rather, the compound inheritance of the hypomorphic allele plus a loss-of-function mutation led to congenital scoliosis.31 Interestingly, all five MMAF-affected subjects in Figures 1 and S3 had at least one loss-of-function mutation in CFAP43, CFAP44, or CFAP65, which is consistent with compound inheritance.31
CFAPs are conserved in many ciliated organisms. The orthologs in C. reinhardtii are the flagella-associated proteins (FAPs), whose expression is strongly induced during flagellar regeneration.39 The functions of most CFAPs and FAPs have not been characterized. Only several of these proteins have been associated with the function of cilia or flagella in humans. For example, CCDC39 (also known as FAP59) is required for the assembly of inner dynein arms and the dynein regulatory complex. Biallelic mutations in CCDC39 have been identified in cases of PCD. Some subjects with PCD also present with oligoasthenospermia and mild defects in sperm flagella.16
CFAP43 and CFAP44 were originally identified in centrioles of bovine sperm by mass spectrometry, suggesting their involvements in the formation of sperm flagella.40 Furthermore, CFAP43 was also identified by proteomic analysis in human spermatozoa.41 Both CFAP43 and CFAP44 contain WD repeat domains, which are highly enriched in the intraflagellar transport proteins of sperm flagella.42, 43 This evidence also supports the involvement of CFAP43 and CFAP44 in vertebrate spermatogenesis.
A biallelic mutation in CFAP65 was also identified to cause MMAF in this study (subject P019; Figure S3 and Table S5). CFAP65 was previously identified in centrioles of bovine sperm and in human spermatozoa by proteomic analyses.40, 41 CFAP65 contains a MSP (major sperm protein) domain that is present in sperm-specific proteins, and its mouse ortholog has been reported to be a putative ciliary protein.44 Furthermore, CFAP65 is expressed specifically in sexually mature male mice but is absent in juvenile mice.45 Although a knockout mouse model was not generated for CFAP65 in this study, evidence from previous studies strongly supports the roles of CFAP65 in spermatogenesis and flagellar development. It has been reported that the Rose-comb mutation in chickens can cause the partial loss of function of CFAP65 and lead to defective sperm motility in male homozygotes.46
In summary, our findings based on both human subjects and mouse models strongly suggest that biallelic mutations in CFAP43 and CFAP44 can cause MMAF and impair sperm motility. Experimental evidence also supports the involvement of CFAP65 in human MMAF. We suggest that other CFAP-encoding genes with uncharacterized functions could be considered candidate genes for sperm flagellar abnormalities and other related ciliopathies.47, 48
Acknowledgments
We would like to thank Drs. Yufang Zheng and Hexige Saiyin for their critical comments and technical support. This work was supported by the National Key Research and Development Program of China (2016YFC0905100 and 2016YFC1000600), Maternal and Child Health Research Project of Jiangsu Province (F201521), and National Natural Science Foundation of China (31625015, 31521003, and 31571297).
Published: May 25, 2017
Footnotes
Supplemental Data include seven figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.012.
Contributor Information
Shenmin Yang, Email: drim2004@126.com.
Feng Zhang, Email: zhangfeng@fudan.edu.cn.
Web Resources
ENCODE, https://www.encodeproject.org
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org
FANTOM, http://fantom.gsc.riken.jp
gnomAD, http://gnomad.broadinstitute.org
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2
SIFT, http://sift.jcvi.org
Supplemental Data
References
- 1.Boivin J., Bunting L., Collins J.A., Nygren K.G. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum. Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Coutton C., Escoffier J., Martinez G., Arnoult C., Ray P.F. Teratozoospermia: spotlight on the main genetic actors in the human. Hum. Reprod. Update. 2015;21:455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 3.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S.M., Li H.B., Wang J.X., Shi Y.C., Cheng H.B., Wang W., Li H., Hou J.Q., Wen D.G. Morphological characteristics and initial genetic study of multiple morphological anomalies of the flagella in China. Asian J. Androl. 2015;17:513–515. doi: 10.4103/1008-682X.146100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemes H.E., Brugo S., Zanchetti F., Carrere C., Lavieri J.C. Dysplasia of the fibrous sheath: an ultrastructural defect of human spermatozoa associated with sperm immotility and primary sterility. Fertil. Steril. 1987;48:664–669. doi: 10.1016/s0015-0282(16)59482-5. [DOI] [PubMed] [Google Scholar]
- 6.Chemes H.E., Alvarez Sedo C. Tales of the tail and sperm head aches: changing concepts on the prognostic significance of sperm pathologies affecting the head, neck and tail. Asian J. Androl. 2012;14:14–23. doi: 10.1038/aja.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chemes H.E. Phenotypes of sperm pathology: genetic and acquired forms in infertile men. J. Androl. 2000;21:799–808. [PubMed] [Google Scholar]
- 8.Knowles M.R., Daniels L.A., Davis S.D., Zariwala M.A., Leigh M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 10.Neesen J., Kirschner R., Ochs M., Schmiedl A., Habermann B., Mueller C., Holstein A.F., Nuesslein T., Adham I., Engel W. Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Hum. Mol. Genet. 2001;10:1117–1128. doi: 10.1093/hmg/10.11.1117. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Kostetskii I., Tang W., Haig-Ladewig L., Sapiro R., Wei Z., Patel A.M., Bennett J., Gerton G.L., Moss S.B. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol. Reprod. 2006;74:751–759. doi: 10.1095/biolreprod.105.049254. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Cochran D.A., Gargano M.D., King I., Samhat N.K., Burger B.P., Sabourin K.R., Hou Y., Awata J., Parry D.A. Regulation of flagellar motility by the conserved flagellar protein CG34110/Ccdc135/FAP50. Mol. Biol. Cell. 2011;22:976–987. doi: 10.1091/mbc.E10-04-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Mukherjee A., Wu J., Zhang L., Teves M.E., Li H., Nambiar S., Henderson S.C., Horwitz A.R., Strauss J.F., III Sperm Associated Antigen 6 (SPAG6) Regulates Fibroblast Cell Growth, Morphology, Migration and Ciliogenesis. Sci. Rep. 2015;5:16506. doi: 10.1038/srep16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teves M.E., Nagarkatti-Gude D.R., Zhang Z., Strauss J.F., 3rd Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton (Hoboken) 2016;73:3–22. doi: 10.1002/cm.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccetti B., Collodel G., Estenoz M., Manca D., Moretti E., Piomboni P. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum. Reprod. 2005;20:2790–2794. doi: 10.1093/humrep/dei126. [DOI] [PubMed] [Google Scholar]
- 16.Merveille A.C., Davis E.E., Becker-Heck A., Legendre M., Amirav I., Bataille G., Belmont J., Beydon N., Billen F., Clément A. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amiri-Yekta A., Coutton C., Kherraf Z.E., Karaouzène T., Le Tanno P., Sanati M.H., Sabbaghian M., Almadani N., Sadighi Gilani M.A., Hosseini S.H. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Jin H., Han F., Cui Y., Chen J., Yang C., Zhu P., Wang W., Jiao G., Wang W. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin. Genet. 2017;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]
- 19.Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 29.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F., Seeman P., Liu P., Weterman M.A., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arno G., Agrawal S.A., Eblimit A., Bellingham J., Xu M., Wang F., Chakarova C., Parfitt D.A., Lane A., Burgoyne T., UKIRDC Mutations in REEP6 Cause Autosomal-Recessive Retinitis Pigmentosa. Am. J. Hum. Genet. 2016;99:1305–1315. doi: 10.1016/j.ajhg.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lizio M., Harshbarger J., Abugessaisa I., Noguchi S., Kondo A., Severin J., Mungall C., Arenillas D., Mathelier A., Medvedeva Y.A. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017;45(D1):D737–D743. doi: 10.1093/nar/gkw995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pazour G.J., Agrin N., Leszyk J., Witman G.B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Firat-Karalar E.N., Sante J., Elliott S., Stearns T. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. J. Cell Sci. 2014;127:4128–4133. doi: 10.1242/jcs.157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jumeau F., Com E., Lane L., Duek P., Lagarrigue M., Lavigne R., Guillot L., Rondel K., Gateau A., Melaine N. Human Spermatozoa as a Model for Detecting Missing Proteins in the Context of the Chromosome-Centric Human Proteome Project. J. Proteome Res. 2015;14:3606–3620. doi: 10.1021/acs.jproteome.5b00170. [DOI] [PubMed] [Google Scholar]
- 42.Blacque O.E., Li C., Inglis P.N., Esmail M.A., Ou G., Mah A.K., Baillie D.L., Scholey J.M., Leroux M.R. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell. 2006;17:5053–5062. doi: 10.1091/mbc.E06-06-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Agustin J.T., Pazour G.J., Witman G.B. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol. Biol. Cell. 2015;26:4358–4372. doi: 10.1091/mbc.E15-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintock T.S., Glasser C.E., Bose S.C., Bergman D.A. Tissue expression patterns identify mouse cilia genes. Physiol. Genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- 45.Shima J.E., McLean D.J., McCarrey J.R., Griswold M.D. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 46.Imsland F., Feng C., Boije H., Bed’hom B., Fillon V., Dorshorst B., Rubin C.J., Liu R., Gao Y., Gu X. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012;8:e1002775. doi: 10.1371/journal.pgen.1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 48.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.