Abstract
Ichthyoses are a clinically and genetically heterogeneous group of genodermatoses associated with abnormal scaling of the skin over the whole body. Mutations in nine genes are known to cause non-syndromic forms of autosomal-recessive congenital ichthyosis (ARCI). However, not all genetic causes for ARCI have been discovered to date. Using whole-exome sequencing (WES) and multigene panel screening, we identified 6 ARCI-affected individuals from three unrelated families with mutations in Sulfotransferase family 2B member 1 (SULT2B1), showing their causative association with ARCI. Cytosolic sulfotransferases form a large family of enzymes that are involved in the synthesis and metabolism of several steroids in humans. We identified four distinct mutations including missense, nonsense, and splice site mutations. We demonstrated the loss of SULT2B1 expression at RNA and protein levels in keratinocytes from individuals with ARCI by functional analyses. Furthermore, we succeeded in reconstructing the morphologic skin alterations in a 3D organotypic tissue culture model with SULT2B1-deficient keratinocytes and fibroblasts. By thin layer chromatography (TLC) of extracts from these organotypic cultures, we could show the absence of cholesterol sulfate, the metabolite of SULT2B1, and an increased level of cholesterol, indicating a disturbed cholesterol metabolism of the skin upon loss-of-function mutation in SULT2B1. In conclusion, our study reveals an essential role for SULT2B1 in the proper development of healthy human skin. Mutation in SULT2B1 leads to an ARCI phenotype via increased proliferation of human keratinocytes, thickening of epithelial layers, and altered epidermal cholesterol metabolism.
Keywords: ichthyosis, autosomal-recessive congenital ichthyosis, exome sequencing, SULT2B1, 3D skin tissue culture model, cholesterol sulfate cycle, epidermis, epidermal proliferation, epidermal differentiation, skin permeability barrier
Introduction
Ichthyoses comprise a clinically and genetically heterogeneous group of hereditary cornification disorders characterized by differentiation defects in keratinocytes in the upper layers of epidermis, leading to a disturbed barrier function of the skin.1 Rare non-syndromic forms of ichthyosis present at birth are referred to as autosomal-recessive congenital ichthyoses (ARCI).2 ARCI are clinically subdivided into lamellar ichthyosis (LI [MIM: 242300]), congenital ichthyosiform erythroderma (CIE [MIM: 242100]), and the most severe, harlequin ichthyosis (HI [MIM: 242500]). Disease-causing mutations in nine different genes have been identified in the last two decades: TGM13, 4 (MIM: 190195), ALOX12B5 (MIM: 603741), ALOXE35 (MIM: ∗607206), NIPAL4/Ichthyin6 (MIM: 609383), CYP4F227 (MIM: 611495), ABCA128 (MIM: 607800), PNPLA19 (MIM: 612121), CERS310, 11 (MIM: 615276), and SDR9C712 (MIM: 609769). The genetic causes for more than 80% of individuals suffering from ARCI have been explained by mutations in these genes.13, 14 To date, for ∼18% of affected individuals, however, the underlying mutations remain unknown.
In this study, we performed whole-exome sequencing (WES) to identify the causative mutation in ARCI-affected families, followed by multi-gene panel screening to find additional individuals with mutations in the candidate gene. We clarified the underlying genetic cause in three unrelated families with six affected individuals by proving that mutations in Sulfotransferase family 2B, member 1 (SULT2B1 [MIM: 604125]) cause ARCI. SULT2B1 is a member of the large cytosolic sulfotransferase superfamily that is engaged in the synthesis and metabolism of steroids in humans.15 It further belongs to the SULT2 family of enzymes that are primarily involved in the sulfoconjugation of neutral steroids and sterols.16 Human SULT2B1 consists of two isoforms, SULT2B1a (GenBank: NP_004596.2) and SULT2B1b (GenBank: NP_814444.1), that differ only in 23 amino acids of their 5′ terminus17 resulting in divergent selectivity of substrates: SULT2B1b preferentially sulfonates cholesterol and oxysterols.15 The expression of SULT2B1b has been reported in tissues of human lung,18 breast, and the prostate.19 Paramount for this work, SULT2B1b is expressed in the human epidermis,19 whereas the related SULT2B1a has not been detected in skin.20 Accordingly, in this study the terms SULT2B1/SULT2B1 refers to the SULT2B1 transcript variant 2 (GenBank: NM_177973.1) and the SULT2B1 isoform b (GenBank: NP_814444.1), respectively, unless otherwise noted.
Our findings reveal a further player in epidermal cholesterol metabolism and emphasize the important role of SULT2B1 in the regulation of epidermal proliferation and differentiation and for constitution and maintenance of the barrier function in human skin.
Material and Methods
Sample Collection
We obtained blood samples from individuals with ARCI and their related family members in collaboration with physicians. The project was conducted according to the Declaration of Helsinki Principles. After receipt of written informed consents, DNA was extracted from whole blood according to standard procedures.
Genotyping and Sequencing
Genome-wide SNP genotyping was performed using a human SNP array (Illumina 370k Quad) with samples from family 1. Identification of causative mutations was performed by WES using the SureSelectHuman All Exon 50Mb Exome Enrichment Kit (Agilent) on an Illumina HiSeq instrument (Illumina). Screening for further affected families including families 2 and 3 was undertaken using a custom Agilent HaloPlex multi-gene panel. The sequencing was performed on an Illumina MiSeq sequencer using MiSeq reagent kit v2 (2 × 150 bp). After recalibration, realignments, and genotype calling with GATK tools,21 variants were annotated using ANNOVAR.22
Structural Visualization
The crystal structure of SULT2B1 (PDB: 1Q20) reported by Lee et al. was selected for visualization of the locations of residue substitutions.23 Residues were colored according to their conservation grade precalculated by the EPPIC software.24 The sequence entropy calculation is based on a multiple sequence alignment that was constructed for each chain in the query from homologs with at least 50% identity.
Keratinocyte and Fibroblast Cell Culture
Primary cultures of keratinocytes and fibroblasts were isolated according to standard protocols from skin biopsies taken from the thigh of individuals P5 and P6 of family three, both of whom carried the splice site mutation. Keratinocytes were cultured in defined, serum-free keratinocyte growth medium containing human keratinocyte growth supplement (SFM and Keratinocyte Supplements, Life Technologies). Fibroblasts were cultured in serum-free medium (FibroLife Serum-Free Cell Culture Medium, Cellsystems). For control experiments, we used the neonatal normal human epidermal keratinocyte cell line (NHEK-Neo, LONZA). Upon confluency, epidermal differentiation of keratinocytes was induced by maintaining the keratinocytes in medium supplemented with 1.1 mM Ca2+, 30 μM palmitic acid, and 15 μM linoleic acid.10, 25 Fatty acids were complexed to fatty acid-free BSA (both Sigma-Aldrich) in a ratio of 3:1. All cells were kept at 37°C in a humidified atmosphere with 5% CO2. Media were changed every second day.
3D Organotypic Tissue Culture Model
After isolation of both keratinocytes and fibroblasts from skin biopsies of individuals P5 and P6, cells (up to four passages) were seeded at final concentrations of 1 × 105 fibroblasts and 0.7 × 106 keratinocytes and cocultured for up to 14 days, as previously described.26, 27 To achieve equivalents that are as similar as possible to in vivo skin of diseased persons, the keratinocytes were cocultured on a dermal layer consisting of the fibroblasts from the affected individual and collagen Type I isolated from tendons of rat tails, serving as a scaffold for the epidermis. After 7 and 14 days of exposition to air (lifting), the models were harvested and prepared for downstream functional analyses.
Western Blotting Analysis
Whole cellular extracts of differentiated keratinocytes from individual P5 and the control cell line NHEK were collected at day 0, 4, 7, 14, and 21 after induction of keratinocyte differentiation. Western blot was performed as described in Radner et al.10 using a RIPA buffer (120 mM NaCl, 20 mM Tris HCl [pH 7.5], 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, and 1 mM EDTA containing protease inhibitor cocktails [Roche]), 20 min centrifugation at 14,000 rpm to remove cell debris, loading 7.5 μg protein per lane on 12% SDS-PAGE gels and subsequent blotting on Hybond-C extra nitrocellulose membranes (GE Healthcare Life Sciences). Due to the similar size of SULT2B1 and the loading control actin, stripping was required and performed after a mild stripping protocol (see Abcam entry in Web Resources) with the following minor difference: incubation time in stripping buffer was increased to 20 min. Primary and secondary antibodies are listed in Table S1.
Histological Analysis
Biopsies and organotypic models from individuals P5 and P6 were fixed in 4% formaldehyde, gradually dehydrated, and embedded in paraffin (Paraplast Plus, McCormick; formalin-fixed paraffin-embedded FFPE). Sections with a thickness of 7 μm were cut, gradually dewaxed, and incubated with hematoxylin (C16H14O6, Carl Roth) for 5 min and counterstained with 0.25% eosin Y (C20H6Br4Na2O5, Carl Roth) for 3 min. Sections were dehydrated and mounted in Roti-Histokitt medium (Carl Roth).
Immunofluorescence Analysis
Paraffin sections of biopsies were prepared as outlined above. 8 μm cryo sections of organotypic models were fixed according to Rosenberger et al.27 Double immunofluorescence staining was performed as described in Radner et al. with the following minor modifications.10 Antigen retrieval of paraffin sections was performed in a pressure cooker at pH 9.0 (Tris EDTA buffer). Additionally, an autofluorescence reducing kit (MaxBlock Kit, Dianova) was used according to the manufacturer’s instructions with an increased incubation time of reagent A to 20 min. Primary and secondary antibodies are listed in Table S1. Skin sections were nuclei-stained with 4’,6-diamidino-2-phenylindole (DAPI, 1:1,000, Sigma-Aldrich) for 5 min.
For immunocytofluorescence staining, 60,000 keratinocytes were cultured on each chamber of a slide (Nunc Lab-Tek II Chamber Slide System, Thermo Fisher Scientific). After 24 hr, cells were fixed for 15 min in 4% formaldehyde and permeabilized for 10 min with 0.5% Triton X-100 (Carl Roth). After 3 washes in PBS, blocking in PBS containing 5% BSA for 40 min followed. For further procedure, see immunofluorescence staining of paraffin sections above.
Electron Microscopy
Biopsies were fixed for at least 2 hr at room temperature in 3% glutaraldehyde solution in 0.1 M cacodylate buffer (pH 7.4), cut into pieces of ∼1 mm3, washed in buffer, postfixed for 1 hr at 4°C in 1% aqueous osmium tetroxide, rinsed in water, dehydrated through graded ethanol solutions, transferred into propylene oxide, and embedded in epoxy resin (glycidether 100). Ultrathin sections were cut with an ultramicrotome (Reichert Ultracut E). 60–80 nm ultrathin sections were treated with uranyl acetate and lead citrate.
Microscopy and Imaging
For histological analysis we used a Carl Zeiss Axioskop 40 light microscope equipped with an AxioCam MRc5 camera (software AxioVision Rel. 4.6). For immunofluorescence and immunocytofluorescence analysis we used a Carl Zeiss Axio Imager 2 microscope equipped with an AxioCam MRm camera (software Zen 2012). For ultrastructural analysis we used a JEM 1400 electron microscope equipped with a 2K TVIPS TemCam 216.
Reverse Transcription PCR
Keratinocytes from individuals P5, P6, and control cell line (NHEK) were collected before differentiation (day 0) and upon differentiation (day 14). Total RNA was extracted using the Roti-Quick kit (Carl Roth). The quality of RNA was analyzed by the NanoDrop spectrophotometer (PEQLAB) and 500 ng were reverse transcribed using ProtoScript II Reverse Transcriptase (NEB) and a 1:1 mix of oligo(dT)15 and random hexamer primers (Thermo Fisher Scientific) according to the manufacturer’s protocol. For the amplification of GAPDH and SULT2B1 transcripts, Taq Polymerase (QIAGEN) and primers listed in Table S2 were used. PCR products were measured by electrophoresis on a 2% agarose gel. The GAPDH transcript GenBank: NM_002046 and the SULT2B1 transcript variant 2 GenBank: NM_177973.1 from NCBI Nucleotide database were used as references.
Lipid Analysis
Total lipids were extracted from keratinocytes differentiated in monolayers and organotypic tissue culture models within 7 and 14 days. The extraction was carried out as previously described.10 Minor modifications included the extraction of polar lipids in methanol/2-propanol/acetic acid (66:33:1). The quantity of lipids for thin layer chromatography (TLC) was determined based on the protein concentrations. Lipids were normalized to corresponding 400 μg (polar lipids) or 100 μg (neutral lipids) of cellular protein. Cholesterol sulfate, cholesterol, sodium oleate, and mono-, di-, triglyceride-mix (all from Sigma-Aldrich) were used as standards at a concentration of 10 μg. Polar lipids were separated using chloroform/methanol/water/acetic acid (40:20:1:1 v/v/v/v), whereas neutral lipids were separated using hexan/diethyl ether/glacial acetic acid (70:29:1 v/v/v) as solvent system. Developed chromatograms were imaged by a scanner (CanoScan 4400F) and quantified applying the local background subtraction method using the Quantity One 1-D Analysis software 4.6.6 (Bio-Rad Laboratories). Statistical analysis was performed using Microsoft Excel 2010.
Results
Clinical Features of Affected Individuals
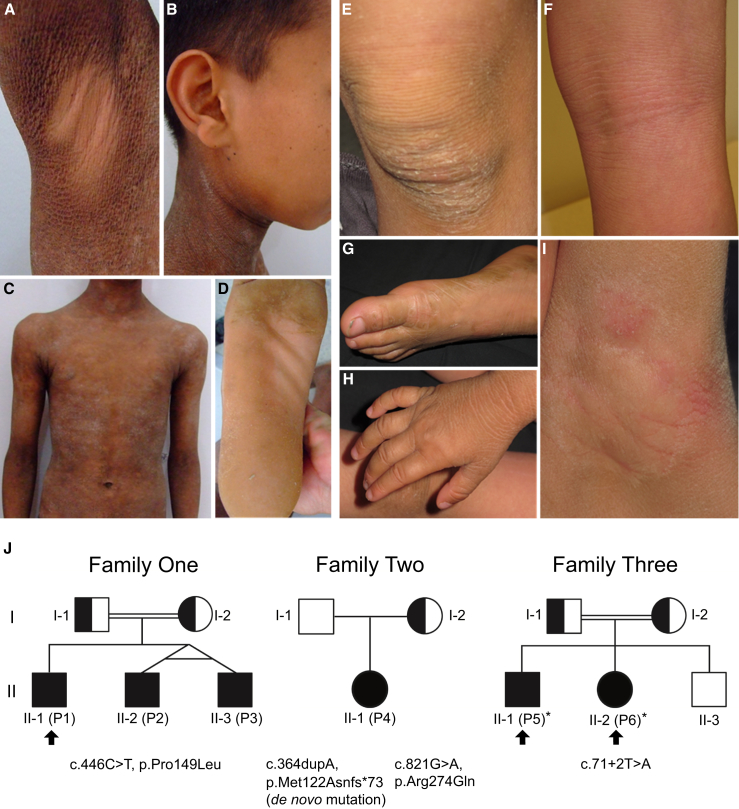
For genetic analysis, we examined three families with six individuals affected by ARCI (Figure 1). In family 1, all three affected individuals—P1 (II-1), P2 (II-2), and P3 (II-3)—were born as collodion babies. Individual P1 presented hyperkeratosis and generalized desquamation with large, dark scales typical of the lamellar form of ARCI (Figures 1A–1D). The only affected individual in the second family, P4 (II-1), was also born in a collodion membrane and presented hyperkeratosis and erythema. Individuals P5 (II-1) and P6 (II-2) of family 3 were not born as collodion babies and showed a generalized very dry, scaly skin with severe itching and erythema at birth. A thicker dry, scaly, greyish skin was particularly seen on the knees, elbows, and dorsal feet and hands, areas exposed to increased mechanical stress (Figures 1E–1I). The itching, and to some extent the scaling, progressively improved with age. No extra-cutaneous abnormalities have been reported and all affected individuals exclusively show the involvement of skin. Overall, the affected individuals of family 3 presented a milder ichthyosis phenotype compared to descendants of family 1. In most of these persons, mild or non-affected skin was observed in the axilla, face, popliteal fossa, and back (Figures 1A, 1B, and 1F). A similar pattern of ichthyosis was observed in individuals with X-linked ichthyosis (XLI [MIM: 308100]) caused by loss-of-function mutations in steroid sulfatase (STS [MIM: 300747]). This characteristic has not been observed in previously reported individuals with ARCI.
Figure 1.
Clinical Features, Pedigrees, and SULT2B1 Mutations of Individuals with ARCI
(A–D) At the age of 15 years, individual P1 (II-1, family 1) presents lamellar ichthyosis with generalized scaling and large brownish to gray scales. Hyperkeratotic plaques over the trunk (C) varied from mild to severe grade. A few areas such as the axilla (A), the face and ears (B), and the middle part of the soles (D) were not affected.
(E–I) Individuals P5 (II-1, family 3) at the age of 10 years and P6 (II-2, family 3) at the age of 6 years also show sparing of popliteal fossa (P5 shown in F) and axilla region (P6 shown in I) with a milder phenotype than individual P1. The axillary region appeared macerated. The back of feet (P5 shown in G) and hands (P6 shown in H) and knees (P6 shown in E) showed very dry skin and were more severely affected. Lichenification of the popliteal fossa in individual P5 shown in (F).
(J) Pedigrees of the three families and SULT2B1 mutations. In family 1, the homozygous missense mutation c.446C>T (p.Pro149Leu) was identified in individuals P1, P2, and P3, transmitted by consanguineous parents. In family 2, the only affected descendant, individual P4 (II-1), from non-consanguineous parents is compound heterozygous for the maternal inherited missense mutation c.821G>A (p.Arg274Gln) and a de novo insertion mutation with frameshift introducing a premature stop codon c.364dupA (p.Met122Asnfs∗73). Individuals P5 and P6 of family 3 are homozygous for the splice site mutation c.71+2T>A (p.Ser24Argfs∗42), which was transmitted by consanguineous parents. Clinical pictures from individuals P1, P5, and P6 are shown (arrows). Skin biopsies were obtained from individuals P5 and P6 (asterisks).
Identification of SULT2B1 Mutations in Three Families
By performing WES and genome-wide SNP-array based homozygosity mapping in the consanguineous Tunisian family (family 1), we identified a homozygous missense mutation, c.446C>T (p.Pro149Leu), in SULT2B1 located on chromosome 19 (Figure 2A). All three sons (individuals P1, P2, and P3) were homozygous for this missense mutation, transmitted by both parents (Figure 1J). In two additional families, mutations in SULT2B1 were subsequently identified by multi-gene panel screening.
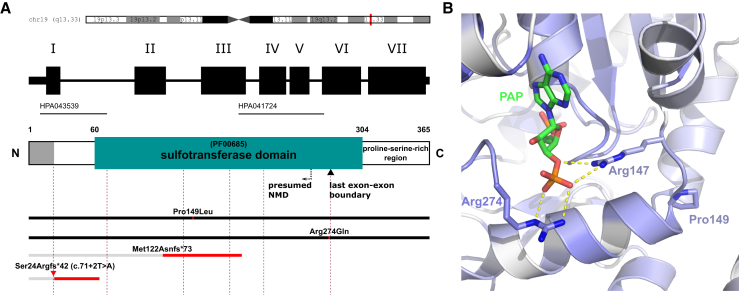
Figure 2.
Schematic Representation of SULT2B1 Structure, Positions of ARCI-Causing Mutations, and Localization of the Missense Mutations in the PAPS/PAP Binding Pocket of SULT2B1 in the Wild-Type Crystal Structure
(A) Localization of SULT2B1 on chromosome 19q13.33, exon/intron organization in SULT2B1, domain structures in SULT2B1, and positions of mutations. Dashed lines indicate exon/intron boundaries (rose), black bars fully expressed mutated proteins, and gray bars truncated proteins whose expression is assumed to be suppressed by NMD or other mechanisms. A proline-rich hydrophobic region (aa 305–365) is indicated at the C terminus of the protein. Near the C terminus, a black inverted triangle marks the position of the last exon-exon boundary. A dashed arrow labeled “NMD” (nonsense-mediated mRNA decay) indicates the ∼55 bp (∼18 aa) position upstream of the last exon-exon boundary. Assuming that the mutation c.71+2T>A (red triangle) leads to intron retention of the first intron, we have marked the hypothetical protein sequence leading to p.Ser24Argfs∗42. The epitopes of two SULT2B1 antibodies used in this study (HPA043539 and HPA041724, located at the N terminus and in the sulfotransferase domain, respectively) are indicated as black solid lines.
(B) The characteristic proline side chain at position 149 (Pro149) stabilizes a bending of the protein backbone, which places the positively charged arginine at position 147 (Arg147, blue) in the proper position for a strong polar interaction with the negatively charged 3′ phosphate group of PAPS/PAP (PAPS/PAP in green-blue, phosphate group in red-orange, polar interactions as yellow dashed lines). Arg274 (blue) interacts strongly with the same phosphate group of PAPS. The intensity of blue coloring of the protein backbone reflects the conservation grade of the amino acid residues. The darker the blue color of the backbone, the more conserved the amino acid residue. Amino acid residues no. 276–282 and 306–311 were hidden for visualization purposes.
In individual P4 from the second non-consanguineous Turkish family (family 2), we found a compound heterozygous mutation consisting of a missense mutation, c.821G>A (p.Arg274Gln), and an insertion mutation, c.364dupA (p.Met122Asnfs∗73), leading to a frameshift followed by a premature stop codon (Figure 2A). The missense mutation was transmitted by the mother whereas the insertion mutation appeared de novo (Figure 1J). Additionally, we found a paternally inherited in-frame deletion in exon 7 of individual P4, c.1054_1059delAGCCCC (p.Ser352_Pro353del, rs16989366), previously reported as a heterozygous variant in a healthy cohort with unknown consequences for SULT2B1 expression.28
In individuals P5 and P6, two siblings of a third consanguineous Kurdish family (family 3), we detected a homozygous splice site mutation, c.71+2T>A, which is predicted to affect the functionality of the splice donor site of intron 1 leading to intron retention and the hypothetical stop mutation p.Ser24Argfs∗42 (Figure 2A, red triangle).29 The parents were both carriers and the third sibling, a daughter, was not affected (Figure 1J). The consequences of the splice site mutation were the subject of further experiments in this study.
In our cohort of 577 families presenting with ARCI, approximately 0.52% of the individuals carry mutations in SULT2B1.
Disease-Associated Missense Mutations in SULT2B1 Are Localized in the Binding Pocket of PAPS/PAP
Both missense mutations—c.446C>T (p.Pro149Leu) and c.821G>A (p.Arg274Gln)—affect highly conserved residues24 in the binding pocket of PAPS/PAP (3′-phosphoadenosine 5′-phosphosulfate) as we highlighted in the crystal structure reported by Lee et al. (Figure 2B).23 Arg274 is positively charged and has been shown to directly bind the negatively charged 3′ phosphate group of PAPS/PAP.23 Disruption of this strong polar interaction by the missense mutation p.Arg274Gln would weaken this binding of the essential cosubstrate PAPS/PAP to the binding pocket. Pro149, in contrast, does not directly bind PAPS/PAP. However, it seems to contribute to the binding of PAPS/PAP by bending of the backbone with a phi torsion angle at −60°, typical for proline residues. Thereby, Pro149 properly positions the positively charged Arg147 for its strong polar interaction with the negatively charged 3′ phosphate group of PAPS/PAP. A mutated Pro149 very likely disrupts this binding by displacing Arg147, which inevitably will inhibit the sulfotransferase activity.
Therefore, we assume that both missense mutations contribute to disease etiology by weakening the interaction of SULT2B1 with its essential cofactor PAPS/PAP in a critical way. Moreover, the ExAC database lists a unique heterozygous allele for Arg274Gln, while Pro149Leu is not listed.30 Concordant with these findings, both missense mutations are predicted by MutationTaster and PolyPhen-2 as disease causing.31, 32
Mutations in SULT2B1 Cause Abnormal Skin Morphology with Orthokeratotic Hyperkeratosis
Histological analysis of a skin specimen from individual P5 revealed a pronounced hyperkeratosis with a compact orthokeratotic cornified layer as well as a prominent granular layer in the epidermis compared with control skin (Figures 3A and 3B). Minor acanthosis but no hints of epidermolytic hyperkeratosis were noticed. Interestingly, in the functionally altered epidermis numerous blood vessels were noticed, partly dilated and often accompanied by perivascular lymphocytic infiltrates frequently present in lower epidermal layers (Figure 3C). These signs indicate inflammatory processes taking place in the epidermis of individual P5.
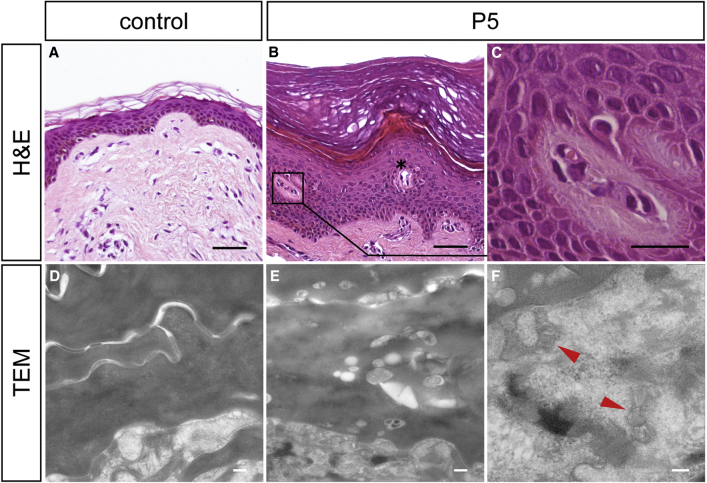
Figure 3.
Histological and Ultrastructural Analysis of Skin Biopsies
(A–C) Hematoxylin and eosin (H&E) staining of FFPE sections from (A) a healthy individual and (B, C) individual P5. Note the pronounced hyperkeratosis with a massive orthokeratotic cornified layer and a prominent granular layer in the epidermis of individual P5 (B). Blood vessels extended to epidermal layers (asterisk), frequently accompanied by (C) perivascular lymphocytic infiltrations with eosinophilic granulocytes appearing in the stratum spinosum and at the stratum spinosum-stratum granulosum junction.
(D–F) Transmission electron microscopy (TEM).
(D) Horny lamellae of healthy skin showing amorphous horny material of homogeneous electron density.
(E) Horny lamellae of individual P6 showing variable electron densities of horny material and numerous small vesicular inclusions, putatively representing irregularly processed lamellar body contents in the horny lamellae.
(F) Lamellated structures (arrowheads) within lamellar body profiles in skin of individual P6.
Scale bars represent 50 μm in (A) and (B), 10 μm in (C), 200 nm in (D) and (E), and 100 nm in (F).
Furthermore, we noticed abnormalities by ultrastructural analysis of the skin specimen from individual P6. The horny layer, which encompassed more than 30 layers of lamellae, showed a loose keratin pattern and some lipid droplets in upper cell layers and considerable amounts of small vacuolar inclusions and putatively irregularly processed lamellar body content in lower horny cell layers. In contrast, the horny layer of healthy skin was homogeneous (Figures 3D and 3E). However, within the granular layer of the functionally altered epidermis, tonofilaments, keratohyalin granules, and especially lamellar bodies exhibited normal morphology (Figure 3F).
SULT2B1 Localizes to the Stratum Granulosum-Stratum Corneum Junction in Healthy Human Skin
In order to study the precise localization of SULT2B1 in human skin, we performed immunofluorescence staining of healthy control skin. Co-staining of SULT2B1 with the terminal differentiation marker filaggrin revealed colocalization and therefore an expression of SULT2B1 at the stratum granulosum-stratum corneum junction, which is in agreement with data of the Human Protein Atlas (Figures S1A–S1C) and Higashi et al.33, 34 This finding points to a function in late keratinocyte differentiation and epidermal homeostasis. Further immunocytofluorescence studies by co-staining with the cytoskeletal marker tubulin revealed that SULT2B1 is present in the entire cytosol of differentiated keratinocytes (Figure S1D). The colocalization with tubulin may indicate a connection of SULT2B1 to the cytoskeleton.
The Splice Site Mutation c.71+2T>A Inhibits the Expression of SULT2B1
To study the effect of the splice site mutation c.71+2T>A on expression of SULT2B1, we performed RT-PCR (for primers see Table S2). Separation on agarose gel showed that SULT2B1 mRNA is weakly expressed in the control cell line before differentiation (day 0) and strongly expressed upon differentiation (day 14) (Figure 4A). In contrast, we did not observe any amplification product in the cell lines derived from individuals P5 and P6.
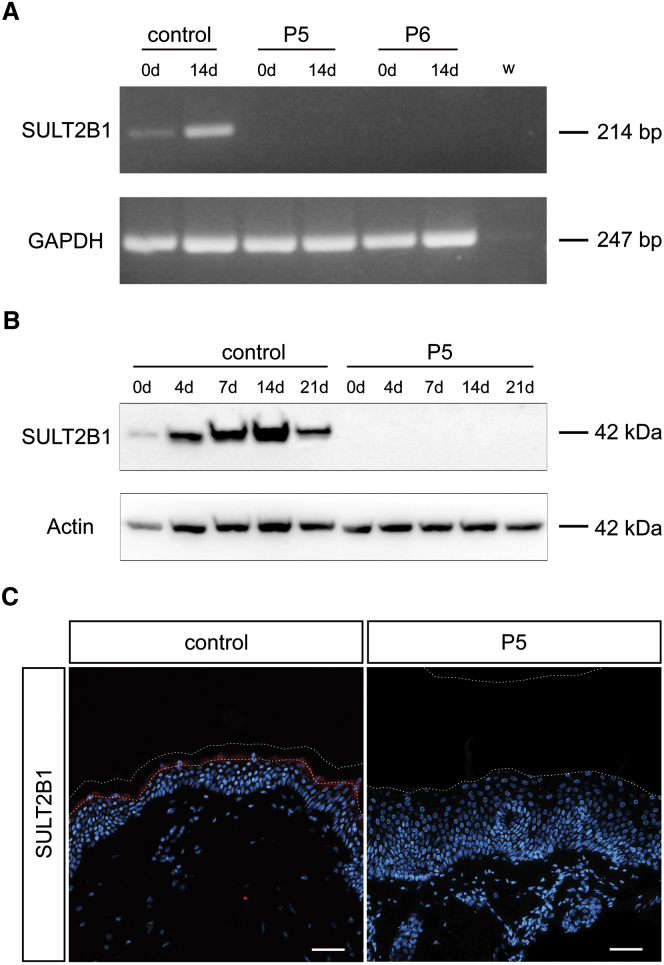
Figure 4.
SULT2B1 mRNA and Protein Expression in Affected Individuals Is Disturbed
(A) Absence of SULT2B1 expression at the mRNA level shown by RT-PCR on total RNA isolated from control cell line NHEK and keratinocytes from individuals P5 and P6 before differentiation (0d) and upon differentiation (14d). GAPDH served as positive control gene. Both samples of affected individuals did not result in a product for SULT2B1 whereas the control produced a weak signal at day 0 and an intense signal at day 14 of differentiation.
(B) Western blot analysis of SULT2B1 in control keratinocytes and those of individual P5 upon in vitro differentiation (day 0, 4, 7, 14, 21) using an antibody raised against the sulfotransferase domain of the protein. Actin served as loading control. In control keratinocytes SULT2B1 was detected at 42 kDa with increasing expression during the differentiation process until a maximum at day 14. In keratinocytes of individual P5, SULT2B1 was not detectable.
(C) Absence of SULT2B1 expression in the epidermis of individual P5 shown in vivo by immunofluorescence staining of FFPE sections. The healthy control epidermis shows strong expression of SULT2B1 in the granular layer (red). Counterstaining with DAPI (blue); dashed lines indicate the stratum granulosum-stratum corneum junction as well as the upper border of the stratum corneum. Abbreviations are as follows: w, water control; kDa, kilodalton; DAPI, 4’,6-diamidino-2-phenylindole. Scale bars represent 50 μm.
To examine the impact of this loss of SULT2B1 mRNA on protein expression, we performed western blot with keratinocytes from individual P5 and control keratinocytes upon in vitro differentiation (day 0, 4, 7, 14, 21) in conventional monolayers. Control blots showed SULT2B1-specific bands at a molecular weight of about 42 kDa with increasing amounts at higher differentiation levels (Figure 4B). At day 14 of differentiation, SULT2B1 expression reached a maximum and decreased at day 21. In contrast, the functionally altered keratinocytes did not show any signal, indicating that no functional protein is present in epidermis from individual P5.
Furthermore, we verified the absence of SULT2B1 expression in vivo via immunofluorescence staining of skin from individuals P5, P6, and control skin. SULT2B1 was not detectable in the individuals’ epidermis as shown by two distinct antibodies for SULT2B1 specific for two different epitopes of the protein listed in Figure 2A (Figures 4C and S2A). Moreover, these findings were confirmed on cellular level by immunocytofluorescence staining (Figure S2B). This allowed us to conclude that the splice site mutation prevents the correct expression and translation of SULT2B1.
Mutations in SULT2B1 Disturb the Epidermal Differentiation
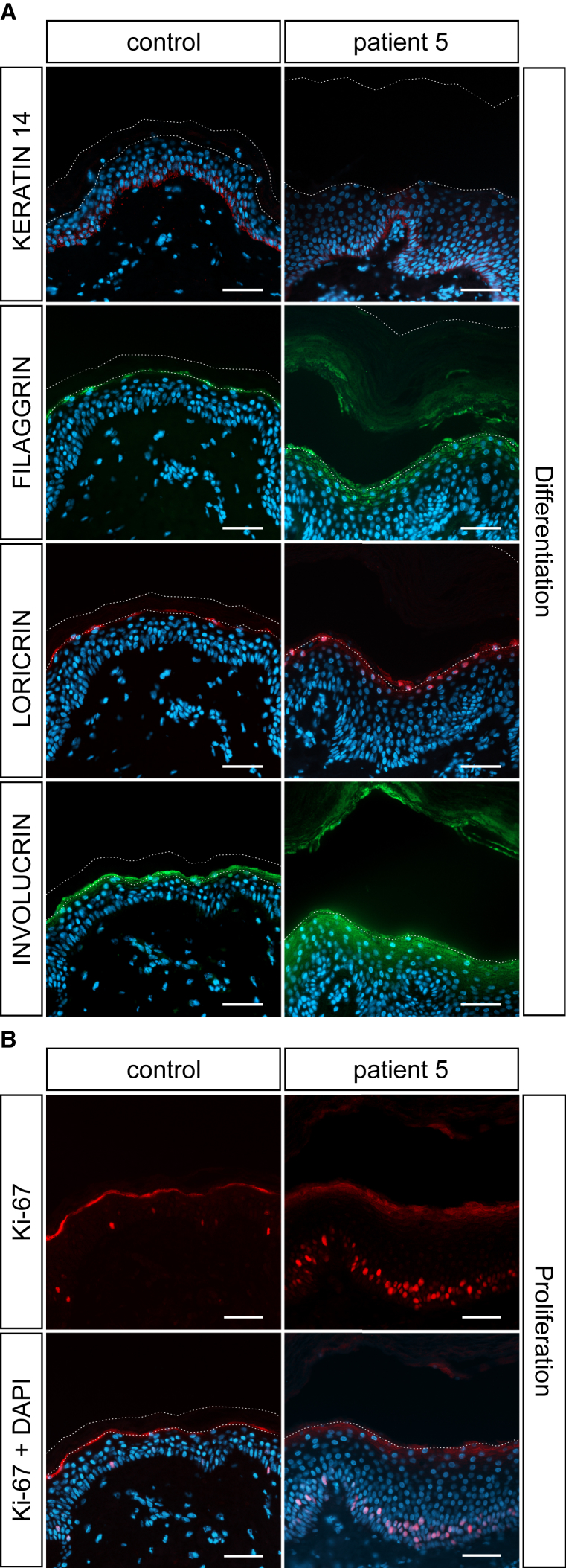
To study the effect of mutated SULT2B1 on epidermal differentiation, we performed immunofluorescence analysis with established markers for undifferentiated (keratin 14), early differentiated (involucrin), and terminal differentiated (filaggrin, loricrin) keratinocytes in skin sections from individual P5 and a healthy individual. Keratin 14 was mostly expressed in the basal layer (Figure 5A). No difference in the keratin 14 signal was observed between control and epidermis of individual P5. However, all differentiating layers in the SULT2B1-deficient epidermis were thickened and the expression pattern of the differentiation markers filaggrin, loricrin, and especially involucrin were considerably increased. Loricrin was restricted to the granular layer whereas filaggrin and involucrin were present even in the lower layers of the stratum corneum during differentiation. Since filaggrin, loricrin, and involucrin represent cornified envelope proteins, their abnormal expression points to an impaired skin barrier function. Thus, the loss of SULT2B1 also affects the proper expression of cornified envelope proteins such as loricrin, involucrin, and filaggrin and impairs the differentiation process in human skin.
Figure 5.
Disturbed Epidermal Proliferation and Differentiation upon SULT2B1 Depletion Shown by Immunofluorescence Staining with Antibodies Specific for Keratin 14, Filaggrin, Loricrin, Involucrin, and Ki-67
(A) FFPE sections from individual P5 show increased expression of terminal differentiation markers filaggrin, loricrin, and the early differentiation marker involucrin compared to control skin. Expression of the marker for undifferentiated keratinocytes, keratin 14, appears normal in the epidermis of individual P5. The stratum corneum in the individual’s skin is massively thickened, but often detached during staining procedure.
(B) Staining of FFPE sections with a nuclear marker for proliferation, Ki-67, indicates strongly enhanced proliferation in the epidermal basal layer of individual P5 in comparison to control skin. Counterstaining with DAPI (blue); dashed lines indicate the stratum granulosum-stratum corneum junction as well as the upper border of the stratum corneum.
Scale bars represent 50 μm.
Mutations in SULT2B1 Enhance Keratinocyte Proliferation
Due to the abnormal skin morphology and the thickening of the epidermal layers of individual P5 (Figure 3B), we determined whether the absence of SULT2B1 has an influence on the proliferation of keratinocytes. Immunofluorescence staining for Ki-67, a nuclear marker for cell proliferation, clearly displayed an increased number of proliferating keratinocytes in the basal layer of the SULT2B1-deficient epidermis of individual P5 compared to the control (Figure 5B).
A 3D In Vitro Skin Model Reproduces the Characteristics of SULT2B1-Deficient Skin
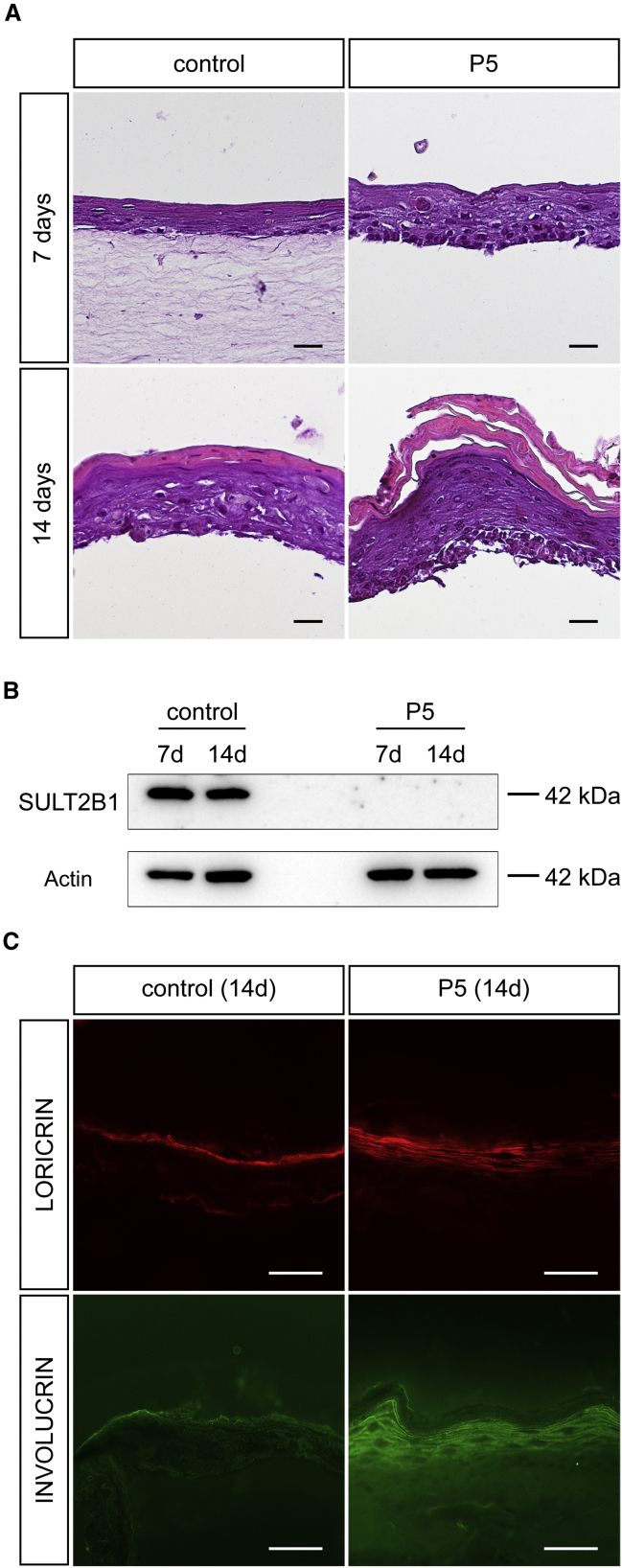
We reconstituted the abnormal skin morphology as observed in our affected individuals with SULT2B1 mutations in a 3D skin organotypic tissue culture model by cocultivating keratinocytes and fibroblasts of individual P5 on a collagen-based scaffold. Light microscopy revealed that 7 days after induction of differentiation, the organotypic models formed a multilayered stratified epithelium composed of layers that are representative for the stratum basale, stratum spinosum, and stratum granulosum. At this stage, the SULT2B1-deficient organotypic model developed a higher number of keratinocytes compared to the control (Figure 6A). Within 14 days, the keratinocytes completed their differentiation and reached the stratum corneum where they became corneocytes. The stratum corneum was significantly thickened in the organotypic model of individual P5 and presented massively accumulating cornified layers compared to the control. Western blot analysis of these models using an antibody targeting the sulfotransferase domain of SULT2B1 confirmed the absence of the protein in the individual’s model (Figure 6B). Because SULT2B1 is expressed in terminally differentiated keratinocytes (Figures 4B and 4C), its strong signal in the control organotypic model provides evidence that the process of differentiation was successful. By immunofluorescence analysis we verified the expanded expression of the differentiation markers loricrin and involucrin in the model of individual P5 (Figure 6C).
Figure 6.
Reconstitution of Affected Individual’s Skin Morphology in a 3D Organotypic Tissue Culture Model
(A) Hematoxylin and eosin staining of FFPE sections from the organotypic model of individual P5 reflects the thickening of the stratum corneum and the prominent granular layer compared to control 14 days after induction of differentiation. The dermal component was often detached during staining procedure (except the control at 7 days).
(B) Western blot analysis of SULT2B1 with organotypic models from control and mutated cell line confirms the absence of SULT2B1 in individual P5. Actin served as loading control.
(C) Immunofluorescence staining of cryo sections from organotypic cultures upon differentiation (14 days) verified increased expression of differentiation markers loricrin and involucrin in the model of individual P5.
Abbreviation: kDa, kilodalton. Scale bars represent 100 μm in (A) and 50 μm in (C).
The model reflected the in vivo morphological characteristics observed in the SULT2B1-deficient skin in Figure 3B and confirmed the absence of functional SULT2B1 in keratinocytes from individual P5 shown in Figure 4.
Mutations in SULT2B1 Cause Absence of Cholesterol Sulfate and Accumulation of Cholesterol
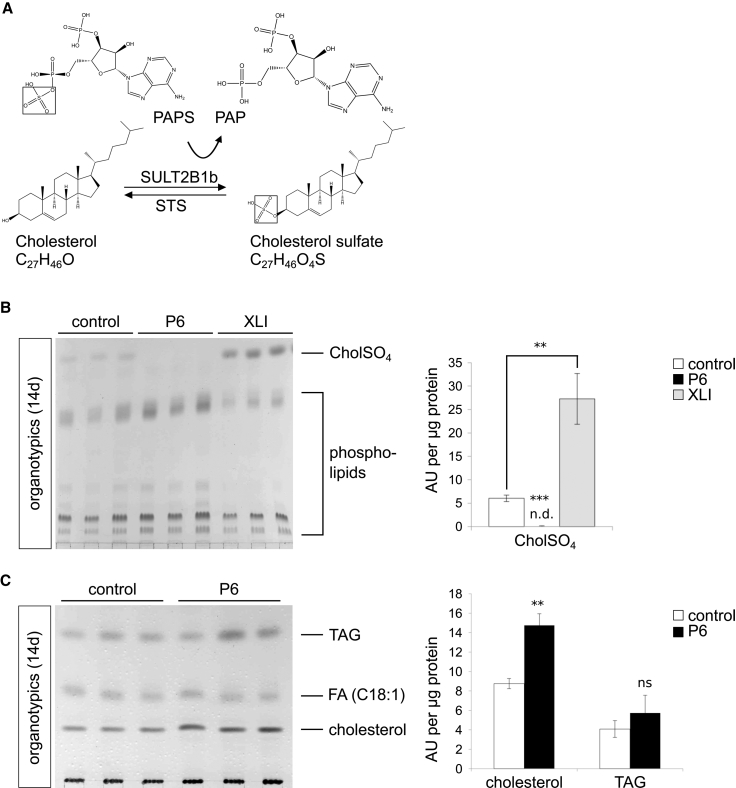
To gain further insights into the role of SULT2B1 in cholesterol metabolism and its involvement in epidermal lipid composition, we determined the concentration of neutral and polar lipids by TLC. SULT2B1 is known to convert cholesterol to cholesterol sulfate (drawn from Chem3D 15.1, PerkinElmer; Figure 7A).35 In this reaction PAPS serves as an essential cofactor providing the sulfate group (SO3−) for the sulfonation. Since an accumulation of cholesterol sulfate has been reported in red blood cells36 and stratum corneum37 of XLI-affected individuals, we determined the concentration of cholesterol sulfate in organotypic cultures of individual P6 and of a XLI-affected person with a not-yet-described hemizygous missense mutation—c.209C>A (p.Ala706Glu)—in STS (Figure 7B). We revealed a statistically significant increased cholesterol sulfate content in the organotypic extracts of the XLI-affected person (p < 0.01) compared to controls, while it was completely absent in the organotypic extracts of individual P6 (p < 0.001). As expected, the separation of neutral lipids revealed a statistically significant increase in the amount of cholesterol in organotypic extracts from individual P6 (Figure 7C; p < 0.01). Elevated cholesterol levels were already detectable 7 days after induction of differentiation compared to controls (Figure S3A; p < 0.01). We also measured a moderately increased triacylglyceride (TAG) content in the skin of individual P6 that was not statistically significant (p > 0.05). This enhanced cholesterol content based on organotypic models, however, could not be demonstrated for in vitro differentiated keratinocytes in conventional monolayers (Figure S3B; p > 0.05).
Figure 7.
Mutated SULT2B1 Causes Imbalance in Cholesterol Metabolism
(A) Enzymatic conversion of unconjugated cholesterol to cholesterol sulfate (drawn from Chem3D 15.1, PerkinElmer). SULT2B1 catalyzes the transfer of the 5′ sulfate from the donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to the hydroxyl group (OH) of cholesterol; consequently, cholesterol is converted to cholesterol sulfate by sulfonation. STS catalyzes the counter reaction converting cholesterol sulfate into cholesterol.
(B) TLC of polar lipids (left) and statistical analysis (right) of organotypic extracts from individual P6, the XLI-affected person, and control organotypic extracts upon differentiation (14 days) revealed a statistically significant increased amount of cholesterol sulfate in the XLI-affected person and its complete absence in individual P6. Lipids were normalized to corresponding 400 μg of cellular protein.
(C) TLC of neutral lipids (left) and statistical analysis (right) of extracts from individual P6 and control organotypic models upon differentiation (14 days) resulted in a statistically significant increased amount of cholesterol in individual P6. Lipids were normalized to corresponding 100 μg of cellular protein.
Data are presented as means + SD of triplicates and are representative for three independent experiments. Statistical significance was examined by unpaired two-tailed Student’s t test (∗∗p < 0.01; ∗∗∗p < 0.001; ns, p > 0.05). Abbreviations are as follows: CholSO4, cholesterol sulfate; TAG, triacylglycerol; FA, fatty acid (C18:1) Oleate cis-9-Octadecenoate; n.d., not detectable; ns, not significant.
Therefore, the absence of SULT2B1 leads to a loss of its reaction product, cholesterol sulfate. Our results further suggest that mutated SULT2B1 affects the cholesterol sulfate cycle but not TAG metabolism. Moreover, we highlighted reciprocal effects of accumulation and deficiency of either cholesterol or cholesterol sulfate as a consequence of a lack of enzymatic activity of either SULT2B1 or STS.
Discussion
The identification of mutations in SULT2B1 represents an important contribution to the analysis of cornification disorders by enlarging the group of genes in which defects are etiological for ARCI. Furthermore, it elucidates the role of the sulfotransferase SULT2B1 in the regulation of the human epidermal cholesterol metabolism and thereby in the maintenance of a functional cutaneous permeability barrier. Phenotypical and histological findings by electron and light microscopy were compatible with ARCI,3, 4, 5, 6, 7, 8, 9, 10, 11, 12 yet several body regions of our affected individuals, such as the popliteal fossa, axilla, back, and part of the soles (Figure 1), were characterized by very mild or non-affected skin, distinguishing them from previously reported individuals with ARCI. Interestingly, sparing of particular areas of skin has also been reported in XLI. Unaffected palms, soles, or popliteal fossae of persons suffering from XLI are similar to the clinical phenotype of our affected individuals.2 Therefore, this specific feature may represent a common characteristic of ichthyosis-affected individuals with affected steroid metabolism. A specific form of keratinopathic ichthyosis, superficial epidermolytic ichthyosis (SEI [MIM: 146800]), caused by mutations in KRT2 (MIM: 600194), also results in non-affected areas (e.g., the popliteal fossa) and is often accompanied by improvement with age resembling the symptoms of the individuals presented here (Figure 1F).38, 39 Similarly to SULT2B1 that interacts with cytoskeletal proteins via a proline/serine-rich C terminus, KRT2 is also involved in the cytoskeleton.40
We showed by structural and functional studies that the identified mutations considerably affect the function of SULT2B1. Structural visualization of the missense mutations c.446C>T (p.Pro149Leu) and c.821G>A (p.Arg274Gln) revealed that both residue substitutions very likely affect the binding of PAPS/PAP to SULT2B1. In contrast to Arg274, Pro149 is not directly involved in the interaction with PAPS/PAP, but is located in the terminal cap of the α-helix enclosing the pocket. The distinctive conformational rigidity of the cyclic structure of proline’s side chain might be necessary for the correct positioning of Arg147 toward the phosphate group of PAPS/PAP. Prolines located in the terminals of helices are often essential for the correct folding of proteins.41 A substitution with leucine might induce a misfolding of SULT2B1. The high conservation of all three residues (Arg274, Pro149, Arg147)24 further support the high relevance of the residues for the correct functioning of the enzyme (Figure 2B).
The inhibited splicing caused by the mutation c.71+2T>A is predicted in silico to result in a truncated SULT2B1 with a premature stop codon at amino acid position 65 downstream of the splice site. Since the premature stop codon occurs upstream of the theoretical border for nonsense-mediated decay (NMD),42 it is expected that SULT2B1 expression will be suppressed by NMD. To test this hypothesis, we performed RT-PCR and confirmed the absence of SULT2B1 mRNA in keratinocytes from individuals P5 and P6 for the respective region (Figure 4A). Thus we conclude that SULT2B1 is suppressed at the post-transcriptional level by mechanisms such as NMD. The absence of SULT2B1 on the protein level was further demonstrated in skin samples of individuals P5 and P6 via immunofluorescence staining. We confirmed the loss of SULT2B1 in the individuals’ skin (Figures 4C and S2) using two independent antibodies targeting the N terminus and the sulfotransferase domain of SULT2B1 (Figure 2A).
As reported for other ARCI genes, mutations in SULT2B1 also lead to an abnormal terminal differentiation process.9, 10 By immunofluorescence analysis of human skin and organotypic cultures, we detected an increased expression pattern of the differentiation markers filaggrin, loricrin, and involucrin as a result of a general thickening of all epidermal layers in the affected individual’s skin. Furthermore, cell proliferation was markedly increased in SULT2B1-deficient skin in contrast to control skin (Figure 5B). This reflects results of a previous study with prostate cancer cells in which SULT2B1 knockdown caused an increased cell proliferation due to altered 3beta-hydroxysteroid levels.43 We conclude that the enhanced proliferation at the initial stage may contribute to the thickening of the subsequent layers (stratum spinosum, stratum granulosum, and stratum corneum) of SULT2B1-deficient epidermis as shown in Figures 3B and 5A. Additionally, cholesterol sulfate, the product of the enzymatic reaction catalyzed by SULT2B1, has been reported to serve as a regulatory molecule in the signal transduction of differentiation in human skin,44 suggesting that its absence contributes to the thickening of epidermal layers observed in the skin of individual P6.
In vitro modeling of processes in the epidermis is conventionally performed by culturing keratinocytes in two-dimensional monolayers. In contrast, 3D organotypic skin models include the development of an extracellular matrix and thereby enable cell-to-cell-communication via paracrine signals and the formation of concentration gradients throughout the matrix.45 These conditions closely resemble the physiological situation of epidermal keratinocytes and lead to the in vitro formation of a multilayered, stratified epithelium that reflects key characteristics of in vivo skin. Thus, organotypic skin models provide large amounts of human skin equivalents for further functional analyses of mutation effects on keratinocyte differentiation and thereby overcome the limitation of the restricted amount of material available from affected individuals. Successful 3D skin models reconstructing ARCI phenotype have been previously described.27, 46, 47, 48, 49, 50 However, the presented organotypic skin models were constructed with both fibroblasts and keratinocytes of donor origin, providing an approach that is closer to the individual’s in vivo skin phenotype. Interestingly, we noted an accelerated differentiation process in organotypic models, in which the maximum of SULT2B1 expression is reached at day 7 compared to monolayers with a maximum at day 14 after induction of differentiation (Figures 4B and 6B).
The skin permeability barrier develops during cornification of living keratinocytes into the stratum corneum and implicates the cleavage of precursor lipids to ceramides, cholesterol, and non-esterified fatty acids.51 By lipid analysis via TLC, we noted only a 1.6-fold increased cholesterol content in the organotypic skin model with SULT2B1-deficient keratinocytes compared to wild-type (Figure 7C). It is possible that cholesterol levels were elevated only to a small degree due to a feedback mechanism on epidermal cholesterol synthesis. Furthermore, we revealed the total absence of cholesterol sulfate in the affected individual’s skin model (Figure 7B) and could show that SULT2B1 has a key role in cholesterol metabolism, in contrast to other ichthyosis-associated proteins such as CERS3,10 PNPLA1,52 and ABHD553 which impact epidermal ceramide synthesis. Thereby SULT2B1 is involved in the proper formation of a lipid-enriched extracellular matrix required for an effective cutaneous permeability barrier.
In XLI, the desulfation of cholesterol sulfate is prevented and thereby affects the steroid sulfate metabolism of the skin. As a result, cholesterol synthesis is deficient, whereas the cholesterol sulfate accumulates in the outer epidermis as we confirmed by TLC (Figure 7B).54 Considering the involvement of both proteins in the cholesterol sulfate cycle and the partial overlap of the clinical features, we speculate that STS and SULT2B1 expression might be interrelated. It is an interesting finding that two enzymes catalyzing reciprocal reactions both result in an ichthyosis phenotype due to loss-of-function mutations. Thus, more focus should be placed on the cholesterol sulfate cycle for future understanding of skin homeostasis.
Ultrastructural analysis of SULT2B1-deficient individuals by electron microscopy revealed abnormalities related to the epidermal lipid metabolism. Similar to the morphology of ARCI-affected individuals with ABCA12 mutations (especially in harlequin ichthyosis), horny lamellae do not have a homogeneous appearance, but contain many vesicular inclusions (Figure 3E). These inclusions may represent irregular released contents of lamellar bodies probably due to their disturbed processing. Yet the lamellar body profiles in granular cells of SULT2B1-deficient skin clearly show regular lamellated structures (Figure 3F). Since cholesterol is essential to form lamellar bodies55 and the inhibition of cholesterol is known to affect their formation,56 the accumulation of cholesterol may also disturb their proper formation and especially affect the processing of these organelles. This may eventually result in the irregular retention of some parts of LBs within cells during the transition from keratinocyte to corneocyte. It cannot be excluded that the moderately increased TAG content in skin of individual P6 has a causative role in the formation of the observed vesicular inclusions.
By histological analysis, we detected numerous perivascular lymphocytic infiltrates in the individual’s epidermis (Figures 3B and 3C) that may explain the severe itching and erythema reported for individuals P5 and P6. As 25HC3S (5-cholesten-3B, 25-diol 3-sulfate), a product of SULT2B1, has been reported to decrease nuclear NF-κB levels and proinflammatory cytokine expression,57 its decrease upon SULT2B1 mutation may cause the observed inflammation process in the individual’s skin. Therefore we assumed that SULT2B1 is also involved in the regulation of inflammatory responses.
After finding the underlying molecular cause of a skin disease, it is of particular interest to find a suitable therapeutic strategy to reduce scaling and improve the individual’s symptoms. Although no curative therapy exists for ichthyoses to date, there are a few approaches in terms of topical applications.46, 58, 59, 60 An example for a successful treatment of ichthyosis symptoms is a combined therapy with lovastatin (statin drug) and cholesterol for persons with congenital hemidysplasia with ichthyosiform erythroderma syndrome (CHILD [MIM: 308050]), an X-linked dominant cholesterol metabolic disorder caused by mutations in NSDHL (MIM: 300275).61 It is conceivable that treatment with cholesterol sulfate, the end product of the conversion catalyzed by SULT2B1, might enable the correct procession of lamellar bodies and consequently improve the phenotype of SULT2B1-deficient persons. However, finding the right dose of cholesterol sulfate would be critical, since an excess of cholesterol sulfate is also known to disturb epidermal differentiation and to cause abnormal desquamation.62, 63 In this regard, further studies to investigate the molecular mechanism of SULT2B1 especially for therapeutic purposes are required. For this type of functional study, the established 3D organotypic tissue culture model might provide a useful support.
In summary, our study highlights a key role of SULT2B1, a gene of the sulfotransferase family, in homeostasis and barrier function of the human skin by providing cholesterol sulfate, an important regulator of the keratinocyte differentiation. Mutations in SULT2B1 are shown to be associated with ARCI in humans. In this context we show that a loss-of-function mutation in SULT2B1 can lead to enhanced epidermal proliferation, disrupted differentiation processes, thickening of epidermal layers, and subsequently a spectrum of ARCI phenotypes.
Acknowledgments
The authors wish to express grateful thanks to Dr. Pavel Salavei and Dr. Susan Lauw for their technical support, to Prof. Dr. Stefan Günther for contributing the structural visualization, to Dr. Franz Radner for providing valuable advice for the performance of TLC, and to Prof. Dr. Dieter Metze for helpful comments about histological findings. We also wish to thank Dr. Susan Cure for critical reading of the manuscript, Dr. Sabine Rosenberger for helpful suggestions in performing 3D organotypic tissue culture models, Dr. Emmanuelle Bourrat and Dr. Anne-Marie Calza for providing clinical data, and Dr. Abderrahmen Masmoudi for cooperation. We are very grateful to all the individuals and probands who provided DNA samples and/or skin biopsy specimens. This study was supported by a grant from the German research foundation DFG (FI1767/3-1).
Published: June 1, 2017
Footnotes
Supplemental Data include three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.007.
Accession Numbers
The mutations reported in this paper are accessible in the ClinVar database under accession numbers SCV000576468 (c.446C>T), SCV000576469 (c.821G>A), SCV000576470 (c.364dupA), and SCV000576471 (c.71+2T>A).
Web Resources
Abcam, http://www.abcam.com/ps/pdf/protocols/stripping%20for%20reprobing.pdf
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Human Splicing Finder, v.2.4.1, http://www.umd.be/HSF3/HSF.html
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer3, http://bioinfo.ut.ee/primer3
Quantity One, http://www.bio-rad.com/de-de/product/quantity-one-1-d-analysis-software
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
The Human Protein Atlas, http://www.proteinatlas.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Oji V., Tadini G., Akiyama M., Blanchet Bardon C., Bodemer C., Bourrat E., Coudiere P., DiGiovanna J.J., Elias P., Fischer J. Revised nomenclature and classification of inherited ichthyoses: results of the First Ichthyosis Consensus Conference in Sorèze 2009. J. Am. Acad. Dermatol. 2010;63:607–641. doi: 10.1016/j.jaad.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 2.Traupe H., Fischer J., Oji V. Nonsyndromic types of ichthyoses - an update. J. Dtsch. Dermatol. Ges. 2014;12:109–121. doi: 10.1111/ddg.12229. [DOI] [PubMed] [Google Scholar]
- 3.Huber M., Rettler I., Bernasconi K., Frenk E., Lavrijsen S.P., Ponec M., Bon A., Lautenschlager S., Schorderet D.F., Hohl D. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 1995;267:525–528. doi: 10.1126/science.7824952. [DOI] [PubMed] [Google Scholar]
- 4.Russell L.J., DiGiovanna J.J., Rogers G.R., Steinert P.M., Hashem N., Compton J.G., Bale S.J. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat. Genet. 1995;9:279–283. doi: 10.1038/ng0395-279. [DOI] [PubMed] [Google Scholar]
- 5.Jobard F., Lefèvre C., Karaduman A., Blanchet-Bardon C., Emre S., Weissenbach J., Ozgüc M., Lathrop M., Prud’homme J.-F., Fischer J. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum. Mol. Genet. 2002;11:107–113. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 6.Lefèvre C., Bouadjar B., Karaduman A., Jobard F., Saker S., Ozguc M., Lathrop M., Prud’homme J.-F., Fischer J. Mutations in ichthyin a new gene on chromosome 5q33 in a new form of autosomal recessive congenital ichthyosis. Hum. Mol. Genet. 2004;13:2473–2482. doi: 10.1093/hmg/ddh263. [DOI] [PubMed] [Google Scholar]
- 7.Lefèvre C., Bouadjar B., Ferrand V., Tadini G., Mégarbané A., Lathrop M., Prud’homme J.-F., Fischer J. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum. Mol. Genet. 2006;15:767–776. doi: 10.1093/hmg/ddi491. [DOI] [PubMed] [Google Scholar]
- 8.Lefévre C., Audebert S., Jobard F., Bouadjar B., Lakhdar H., Boughdene-Stambouli O., Blanchet-Bardon C., Heilig R., Foglio M., Weissenbach J. Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum. Mol. Genet. 2003;12:2369–2378. doi: 10.1093/hmg/ddg235. [DOI] [PubMed] [Google Scholar]
- 9.Grall A., Guaguère E., Planchais S., Grond S., Bourrat E., Hausser I., Hitte C., Le Gallo M., Derbois C., Kim G.-J. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat. Genet. 2012;44:140–147. doi: 10.1038/ng.1056. [DOI] [PubMed] [Google Scholar]
- 10.Radner F.P.W., Marrakchi S., Kirchmeier P., Kim G.-J., Ribierre F., Kamoun B., Abid L., Leipoldt M., Turki H., Schempp W. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 2013;9:e1003536. doi: 10.1371/journal.pgen.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckl K.-M., Tidhar R., Thiele H., Oji V., Hausser I., Brodesser S., Preil M.-L., Onal-Akan A., Stock F., Müller D. Impaired epidermal ceramide synthesis causes autosomal recessive congenital ichthyosis and reveals the importance of ceramide acyl chain length. J. Invest. Dermatol. 2013;133:2202–2211. doi: 10.1038/jid.2013.153. [DOI] [PubMed] [Google Scholar]
- 12.Shigehara Y., Okuda S., Nemer G., Chedraoui A., Hayashi R., Bitar F., Nakai H., Abbas O., Daou L., Abe R. Mutations in SDR9C7 gene encoding an enzyme for vitamin A metabolism underlie autosomal recessive congenital ichthyosis. Hum. Mol. Genet. 2016;25:4484–4493. doi: 10.1093/hmg/ddw277. [DOI] [PubMed] [Google Scholar]
- 13.Fischer J. Autosomal recessive congenital ichthyosis. J. Invest. Dermatol. 2009;129:1319–1321. doi: 10.1038/jid.2009.57. [DOI] [PubMed] [Google Scholar]
- 14.Fischer J., Traupe H. Klinik und Genetik der Ichthyosen. Medgen. 2014;26:427–442. [Google Scholar]
- 15.Javitt N.B., Lee Y.C., Shimizu C., Fuda H., Strott C.A. Cholesterol and hydroxycholesterol sulfotransferases: identification, distinction from dehydroepiandrosterone sulfotransferase, and differential tissue expression. Endocrinology. 2001;142:2978–2984. doi: 10.1210/endo.142.7.8244. [DOI] [PubMed] [Google Scholar]
- 16.Yamazoe Y., Ozawa S., Nagata K., Gong D.W., Kato R. Characterization and expression of hepatic sulfotransferase involved in the metabolism of N-substituted aryl compounds. Environ. Health Perspect. 1994;102(Suppl 6):99–103. doi: 10.1289/ehp.94102s699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Her C., Wood T.C., Eichler E.E., Mohrenweiser H.W., Ramagli L.S., Siciliano M.J., Weinshilboum R.M. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 18.He D., Frost A.R., Falany C.N. Identification and immunohistochemical localization of Sulfotransferase 2B1b (SULT2B1b) in human lung. Biochim. Biophys. Acta. 2005;1724:119–126. doi: 10.1016/j.bbagen.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.He D., Meloche C.A., Dumas N.A., Frost A.R., Falany C.N. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochem. J. 2004;379:533–540. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elias P.M., Williams M.L., Choi E.-H., Feingold K.R. Role of cholesterol sulfate in epidermal structure and function: lessons from X-linked ichthyosis. Biochim. Biophys. Acta. 2014;1841:353–361. doi: 10.1016/j.bbalip.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K.A., Fuda H., Lee Y.C., Negishi M., Strott C.A., Pedersen L.C. Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3′-phosphoadenosine 5′-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2BG1 isoforms. J. Biol. Chem. 2003;278:44593–44599. doi: 10.1074/jbc.M308312200. [DOI] [PubMed] [Google Scholar]
- 24.Baskaran K., Duarte J.M., Biyani N., Bliven S., Capitani G. A PDB-wide, evolution-based assessment of protein-protein interfaces. BMC Struct. Biol. 2014;14:22. doi: 10.1186/s12900-014-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiden B., Gallala H., Doering T., Sandhoff K. Optimization of submerged keratinocyte cultures for the synthesis of barrier ceramides. Eur. J. Cell Biol. 2007;86:657–673. doi: 10.1016/j.ejcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Mildner M., Ballaun C., Stichenwirth M., Bauer R., Gmeiner R., Buchberger M., Mlitz V., Tschachler E. Gene silencing in a human organotypic skin model. Biochem. Biophys. Res. Commun. 2006;348:76–82. doi: 10.1016/j.bbrc.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberger S., Dick A., Latzko S., Hausser I., Stark H.-J., Rauh M., Schneider H., Krieg P. A mouse organotypic tissue culture model for autosomal recessive congenital ichthyosis. Br. J. Dermatol. 2014;171:1347–1357. doi: 10.1111/bjd.13308. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y., Moon I., Zlatkovic J., Salavaggione O.E., Thomae B.A., Eckloff B.W., Wieben E.D., Schaid D.J., Weinshilboum R.M. Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J. Pharmacol. Exp. Ther. 2007;322:529–540. doi: 10.1124/jpet.107.122895. [DOI] [PubMed] [Google Scholar]
- 29.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 32.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Higashi Y., Fuda H., Yanai H., Lee Y., Fukushige T., Kanzaki T., Strott C.A. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J. Invest. Dermatol. 2004;122:1207–1213. doi: 10.1111/j.0022-202X.2004.22416.x. [DOI] [PubMed] [Google Scholar]
- 35.Meloche C.A., Falany C.N. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J. Steroid Biochem. Mol. Biol. 2001;77:261–269. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 36.Bergner E.A., Shapiro L.J. Increased cholesterol sulfate in plasma and red blood cell membranes of steroid sulfatase deficient patients. J. Clin. Endocrinol. Metab. 1981;53:221–223. doi: 10.1210/jcem-53-1-220. [DOI] [PubMed] [Google Scholar]
- 37.Williams M.L., Elias P.M. Stratum corneum lipids in disorders of cornification: increased cholesterol sulfate content of stratum corneum in recessive x-linked ichthyosis. J. Clin. Invest. 1981;68:1404–1410. doi: 10.1172/JCI110391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traupe H., Kolde G., Hamm H., Happle R. Ichthyosis bullosa of Siemens: a unique type of epidermolytic hyperkeratosis. J. Am. Acad. Dermatol. 1986;14:1000–1005. doi: 10.1016/s0190-9622(86)70123-0. [DOI] [PubMed] [Google Scholar]
- 39.Hotz A., Oji V., Bourrat E., Jonca N., Mazereeuw-Hautier J., Betz R.C., Blume-Peytavi U., Stieler K., Morice-Picard F., Schönbuchner I. Expanding the clinical and genetic spectrum of KRT1, KRT2 and KRT10 mutations in keratinopathic ichthyosis. Acta Derm. Venereol. 2016;96:473–478. doi: 10.2340/00015555-2299. [DOI] [PubMed] [Google Scholar]
- 40.Kurogi K., Sakakibara Y., Kamemoto Y., Takahashi S., Yasuda S., Liu M.-C., Suiko M. Mouse cytosolic sulfotransferase SULT2B1b interacts with cytoskeletal proteins via a proline/serine-rich C-terminus. FEBS J. 2010;277:3804–3811. doi: 10.1111/j.1742-4658.2010.07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eyles S.J., Gierasch L.M. Multiple roles of prolyl residues in structure and folding. J. Mol. Biol. 2000;301:737–747. doi: 10.1006/jmbi.2000.4002. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z., Xin D., Wang P., Zhou L., Hu L., Kong X., Hurst L.D. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He D., Falany C.N. Inhibition of SULT2B1b expression alters effects of 3beta-hydroxysteroids on cell proliferation and steroid hormone receptor expression in human LNCaP prostate cancer cells. Prostate. 2007;67:1318–1329. doi: 10.1002/pros.20615. [DOI] [PubMed] [Google Scholar]
- 44.Strott C.A., Higashi Y. Cholesterol sulfate in human physiology: what’s it all about? J. Lipid Res. 2003;44:1268–1278. doi: 10.1194/jlr.R300005-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Maltman D.J., Przyborski S.A. Developments in three-dimensional cell culture technology aimed at improving the accuracy of in vitro analyses. Biochem. Soc. Trans. 2010;38:1072–1075. doi: 10.1042/BST0381072. [DOI] [PubMed] [Google Scholar]
- 46.Aufenvenne K., Larcher F., Hausser I., Duarte B., Oji V., Nikolenko H., Del Rio M., Dathe M., Traupe H. Topical enzyme-replacement therapy restores transglutaminase 1 activity and corrects architecture of transglutaminase-1-deficient skin grafts. Am. J. Hum. Genet. 2013;93:620–630. doi: 10.1016/j.ajhg.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckl K.-M., Alef T., Torres S., Hennies H.C. Full-thickness human skin models for congenital ichthyosis and related keratinization disorders. J. Invest. Dermatol. 2011;131:1938–1942. doi: 10.1038/jid.2011.126. [DOI] [PubMed] [Google Scholar]
- 48.O’Shaughnessy R.F.L., Choudhary I., Harper J.I. Interleukin-1 alpha blockade prevents hyperkeratosis in an in vitro model of lamellar ichthyosis. Hum. Mol. Genet. 2010;19:2594–2605. doi: 10.1093/hmg/ddq145. [DOI] [PubMed] [Google Scholar]
- 49.Thomas A.C., Tattersall D., Norgett E.E., O’Toole E.A., Kelsell D.P. Premature terminal differentiation and a reduction in specific proteases associated with loss of ABCA12 in Harlequin ichthyosis. Am. J. Pathol. 2009;174:970–978. doi: 10.2353/ajpath.2009.080860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youssef G., Ono M., Brown S.J., Kinsler V.A., Sebire N.J., Harper J.I., O’Shaughnessy R.F.L. Identifying a hyperkeratosis signature in autosomal recessive congenital ichthyosis: Mdm2 inhibition prevents hyperkeratosis in a rat ARCI model. J. Invest. Dermatol. 2014;134:858–861. doi: 10.1038/jid.2013.374. [DOI] [PubMed] [Google Scholar]
- 51.Wertz P.W. Epidermal lipids. Semin. Dermatol. 1992;11:106–113. [PubMed] [Google Scholar]
- 52.Grond S., Eichmann T.O., Dubrac S., Kolb D., Schmuth M., Fischer J., Crumrine D., Elias P.M., Haemmerle G., Zechner R. PNPLA1 deficiency in mice and humans leads to a defect in the synthesis of omega-O-acylceramides. J. Invest. Dermatol. 2017;137:394–402. doi: 10.1016/j.jid.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radner F.P.W., Streith I.E., Schoiswohl G., Schweiger M., Kumari M., Eichmann T.O., Rechberger G., Koefeler H.C., Eder S., Schauer S. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J. Biol. Chem. 2010;285:7300–7311. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Epstein E.H., Williams M.L., Elias P.M. The epidermal cholesterol sulfate cycle. J. Am. Acad. Dermatol. 1984;10:866–868. doi: 10.1016/s0190-9622(84)80144-9. [DOI] [PubMed] [Google Scholar]
- 55.Schurer N.Y., Elias P.M. The biochemistry and function of stratum corneum lipids. Adv. Lipid Res. 1991;24:27–56. doi: 10.1016/b978-0-12-024924-4.50006-7. [DOI] [PubMed] [Google Scholar]
- 56.Feingold K.R. Thematic review series: skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J. Lipid Res. 2007;48:2531–2546. doi: 10.1194/jlr.R700013-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Ren S., Ning Y. Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 2014;306:E123–E130. doi: 10.1152/ajpendo.00552.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerra L., Diociaiuti A., El Hachem M., Castiglia D., Zambruno G. Ichthyosis with confetti: clinics, molecular genetics and management. Orphanet J. Rare Dis. 2015;10:115. doi: 10.1186/s13023-015-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiritsi D., Schauer F., Wölfle U., Valari M., Bruckner-Tuderman L., Has C., Happle R. Targeting epidermal lipids for treatment of Mendelian disorders of cornification. Orphanet J. Rare Dis. 2014;9:33. doi: 10.1186/1750-1172-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sethuraman G., Marwaha R.K., Challa A., Yenamandra V.K., Ramakrishnan L., Thulkar S., Sharma V.K. Vitamin D: a new promising therapy for congenital ichthyosis. Pediatrics. 2016;137:137. doi: 10.1542/peds.2015-1313. [DOI] [PubMed] [Google Scholar]
- 61.Paller A.S., van Steensel M.A.M., Rodriguez-Martín M., Sorrell J., Heath C., Crumrine D., van Geel M., Cabrera A.N., Elias P.M. Pathogenesis-based therapy reverses cutaneous abnormalities in an inherited disorder of distal cholesterol metabolism. J. Invest. Dermatol. 2011;131:2242–2248. doi: 10.1038/jid.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abraham W., Wertz P.W., Landmann L., Downing D.T. Stratum corneum lipid liposomes: calcium-induced transformation into lamellar sheets. J. Invest. Dermatol. 1987;88:212–214. doi: 10.1111/1523-1747.ep12525375. [DOI] [PubMed] [Google Scholar]
- 63.Hatfield R.M., Fung L.W. A new model system for lipid interactions in stratum corneum vesicles: effects of lipid composition, calcium, and pH. Biochemistry. 1999;38:784–791. doi: 10.1021/bi981421k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.