Abstract
Yin and yang 1 (YY1) is a well-known zinc-finger transcription factor with crucial roles in normal development and malignancy. YY1 acts both as a repressor and as an activator of gene expression. We have identified 23 individuals with de novo mutations or deletions of YY1 and phenotypic features that define a syndrome of cognitive impairment, behavioral alterations, intrauterine growth restriction, feeding problems, and various congenital malformations. Our combined clinical and molecular data define “YY1 syndrome” as a haploinsufficiency syndrome. Through immunoprecipitation of YY1-bound chromatin from affected individuals’ cells with antibodies recognizing both ends of the protein, we show that YY1 deletions and missense mutations lead to a global loss of YY1 binding with a preferential retention at high-occupancy sites. Finally, we uncover a widespread loss of H3K27 acetylation in particular on the YY1-bound enhancers, underscoring a crucial role for YY1 in enhancer regulation. Collectively, these results define a clinical syndrome caused by haploinsufficiency of YY1 through dysregulation of key transcriptional regulators.
Keywords: YY1, haploinsufficiency, intellectual disability, chromatin, syndrome, transcription factor, neurodevelopment, epigenetics, H3K27Ac, enhancer
Introduction
The first study employing trio exome sequencing for the discovery of de novo mutations in individuals with intellectual disability (ID)1 reported a de novo mutation in YY1 (MIM: 600013), which was considered a prime candidate gene for ID given its known functions. YY1 encodes yin and yang 1 (YY1), a zinc-finger transcription factor (TF) that was originally identified to repress or activate the adeno-associated virus (AAV) P5 promoter in the absence or presence, respectively, of the adenovirus E1A oncoprotein,2 as well as repress an immunoglobulin enhancer and activate genes encoding ribosomal proteins.3, 4 The dual function inscribed in its name has been extended to a large number of genes and cell types and further articulated through an additional partitioning of YY1 activity between Polycomb-associated and -independent functions. YY1 is in fact the mammalian homolog of Drosophila pleiohomeotic (pho), one of the TFs that mediate recruitment of Polycomb group (PcG) proteins to target genes via Polycomb response elements (PREs). Although PREs have remained largely elusive in mammals, with a few exceptions,5 several observations have corroborated the functional interaction between YY1 and PcG proteins in selected cell types.5, 6, 7, 8 It has become clear, however, that a significant aspect of YY1 function is PcG independent through direct targeting by its four zinc fingers, which manifests mostly as positive regulation of gene expression both in mouse embryonic stem cells and in a variety of tumor cellular models.9, 10 The key interactors for YY1-mediated transcriptional activation include the INO80 chromatin remodeling complex,9, 11, 12 the p300/CBP histone acetyl transferase (HAT),13 and several other transcriptional co-activators reviewed elsewhere.14 Recently, compound-heterozygous and homozygous nonsense variants in YY1AP1 (MIM: 607860), a component of the INO80 chromatin remodeling complex, have been reported as the cause of Grange syndrome and a fibromuscular dysplasia-like vascular disease.15
The direct relevance of YY1 for neuronal development and function is suggested by several lines of convergent evidence.16 First, YY1 turned out to be haploinsufficient for mouse development, albeit at incomplete penetrance, such that a significant fraction of Yy1+/− embryos show exencephaly, pseudoventricles, and asymmetry of the developing brain.17 Moreover, in Schwann cells, YY1 was shown to mediate the neuregulin-dependent activation of the myelination gene expression program centered on the TF EGR2.18 In addition, YY1 has been shown to repress chromatin remodeler HCFC1, whose alterations lead to neurodevelopmental disorders.19, 20 Finally, the interaction with the PcG axis appears particularly relevant in light of the central role of this chromatin regulatory axis in neural development, as shown by us and others through the temporally highly specific neuronal phenotypes triggered by ablation of Polycomb repressive complex 2 (PRC2), which catalyzes methylation on lysine 27 of histone H3 (H3K27me), or of the demethylase JMJD3, which effects regulated H3K27me3 removal.21, 22, 23, 24, 25
Together, these considerations prompted us to search for YY1 mutations in additional individuals1 in order to establish a causal link between YY1 mutations and ID and to characterize the impact of YY1 mutations in terms of molecular alterations and pathogenic pathways. Here, we define YY1 dysfunction as a cause of ID by describing a cohort of ten individuals with de novo mutations in YY1 and 13 individuals with small de novo deletions that include YY1. A causal role for YY1 is further supported by the phenotypic overlap among individuals along with the functional equivalence of YY1 deletions and missense mutations. The genome-wide characterization of the molecular impact of YY1 haploinsufficiency reveals transcriptional and epigenetic dysregulation.
Material and Methods
Identification of Individuals with Mutations and Deletions of YY1
Upon the identification of the initial individual with a de novo mutation in YY1 (GenBank: NM_003403.4) by exome sequencing,1 an additional cohort of 500 individuals with unexplained ID was tested for mutations in YY1 by standard Sanger sequencing approaches on DNA extracted from peripheral blood. Primers are available upon request. DNA of the parents was tested for assessing the de novo occurrence of the identified mutations.
In addition to the two above-mentioned individuals, the remaining eight individuals with de novo mutations in YY1 were detected by exome sequencing in various cohorts of individuals with unexplained ID (Table 1), sequenced in different centers in Tokyo (n = 500), Zurich (n = 350), Oslo (n = 100), Houston (n = 5,500), Gaithersburg (n = 6,709), and Aliso Viejo (n = 1300), leading to a total cohort of 14,969 individuals with ID.26 Exome capture was performed with the SureSelect Human All Exon Kit V4 or V5, Agilent Clinical Research Exome Kit (Agilent Technologies), IDT xGen Exome Research Panel V1.0, or VCRome 2.1 (Roche NimbleGen).27 Sequencing was performed on a MiSeq, HiSeq 2000, or HiSeq 2500 (Illumina) or SOLID 5500XL (Life Technologies). Data annotation and analysis were performed with the Burrows-Wheeler Aligner for read alignment.28 Variant calling was performed with the Genome Analysis Toolkit,29 XomeDx, or in-house developed tools as described previously.30, 31
Table 1.
Individuals with YY1 Mutations
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | Individual 5 | Individual 6 | Individual 7 | Individual 8 | Individual 9 | Individual 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | ||||||||||
| cDNA changea | c.1138G>T | c.1097T>C | c.1096C>G | c.1030C>T | c.535A>T | c.1173delT | c.1174_1176del | c.385delG | c.1015A>C | c.958C>T |
| Protein changeb | p.Asp380Tyr | p.Leu366Pro | p.Leu366Val | p.Gln344∗ | p.Lys179∗ | p.Asn391Lysfs∗10 | p.Lys393del | p.Asp129Ilefs∗127 | p.Lys339Gln | p.His320Tyr |
| Chromosome positionc | g.100743830G>T | g.100743789T>C | g.100743788C>G | g.100742953C>T | g.100706116A>T | g.100743865del | g.100743869–110743871del | g.100239629delG | g.100742938A>C | g.100742881C>T |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo |
| PhyloPd | 6.18 | 5.13 | 2.14 | 6.10 | 0.12 | 1.01 | 4.51e | 2.87 | 4.97 | 6.26 |
| MutationTaster | damaging | damaging | damaging | NA | NA | NA | NA | NA | damaging | damaging |
| PolyPhen-2 | damaging | damaging | damaging | NA | NA | NA | NA | NA | damaging | damaging |
| Cohort size | 10 | 500 | 500 | 350 | 100 | 5,500f | 5,500f | 6,709f | 1,300 | 6,709f |
| Growth | ||||||||||
| Age | 2 years, 9 months | 15 years, 10 months | 5 years, 1 month | 39 years | 17 years, 6 months | 7 years, 10 months | 1 year, 3 months | 35 years | 9 years, 3 months | 1 year, 5 months |
| Gender | male | male | female | female | female | male | male | female | male | female |
| Birth weight in g (SD) | 2,010 (−2.5) | 2,220 (−3) | 2,290 (−1.6) | NA | 2,600 (−2) | 2,050 (−1.3) | 2,409 (−1.8) | 3.5 | 3.83 | 3,062 |
| Height in cm (SD) | 84 (−3) | 170 (−1) | 105.2 (−0.5) | 153.5 (−1.5) | 159 (+0.7) | 124 (0) | 77.2 (−0.8) | 154.4 (−2.5) | 125.5 (−1.5) | 79.2 (−0.8) |
| Weight in kg (SD) | NA | 47.5 (−1.5) | 12.5 (−2.1) | 51 (−1) | 65 (+0.7) | 24.6 (0) | 8.6 (−2.6) | 52.8 (+0.3) | 23 (−1.7) | 9.1 (−1.5) |
| HC in cm (SD) | 48.5 (−1) | 57 (+0.4) | 50.5 (+0.6) | 54 (−1.5) | 56 (+0.7) | 52.5 (0) | 47.3 (0) | 52.5 (−2) | 50 cm (−1.3) | 44.8 (−1.8) |
| Development | ||||||||||
| Motor development | sitting at 1 year; walking at 2 years, 9 months | walking at 2 years | sitting at 1 year; walking at 4 years, 8 months | mild delay | walking at 15 months | walking at 3 years | sitting at 1 year | walking at 15 months | walking at 6.5 years | sitting at 5 months; walking at 22 months |
| Speech development | first words at 2 years | first words at 2 years | mild delay | mild delay | first words at 2 years | delay, 2–3 word phrases | first words at 13 months | delay | non-verbal | first words 14 months |
| Intellectual disability | moderate | moderate | mild | mild | mild, learning difficulties | mild to moderate | mild (DQ 68) | special education | moderate to severe | NA |
| Neurological Features | ||||||||||
| Hypotonia | − | − | − | + | − | − | + | − | moderate | − |
| Behavioral abnormality | anxiety | ASD | NA or NK | schizoaffective disorder | ADHD | − | − | − | autism | − |
| Sleep disturbance | − | − | NA or NK | + | + | − | − | − | − | − |
| Abnormal movement | − | toe walking | NA or NK | tremor | progressive dystonia | waddling gait | − | progressive dystonia | − | − |
| Brain MRI | NA | normal | normal | frontal gliosis, enlarged SA spaces | normal | delayed myelination, cortical dysplasia, diffuse white-matter loss | minimal prominence of the right lateral ventricle | subcortical bifrontal white-matter foci | focal areas of encephalomalacia | normal |
| Facial Dysmorphisms | ||||||||||
| Facial asymmetry | + | + | + | + | − | − | mild | mild | + | − |
| Broad forehead | + | + | + | + | + | + | + | + | + | + |
| Simple posteriorly rotated ears | + | + | + | + | + | + | low-set | simple | NA or NK | protruding |
| Periorbital fullness | +/− | + | + | + | + | − | − | − | suborbital fullness | + |
| Downslant | − | + | + | + | − | + | − | − | + | − |
| Full nasal tip | + | + | + | + | + | − | + | − | + | + |
| Malar flattening | + | + | + | + | − | + | − | + | + | + |
| Indented upper lip | + | + | + | + | − | − | + | − | − | − |
| Thick lower lip | + | + | mild | + | + | + | − | − | + | + |
| Pointed chin | + | + | + | − | − | + | − | − | − | − |
| Other | telecanthus | − | NA or NK | epiblepharon, high palate | NA or NK | Pierre-Robin sequence with CP | − | NA or NK | micrognathia, ptosis, sparse eyebrows | dolichocephaly, hypotonic facies, frontal upsweep |
| Miscellaneous | ||||||||||
| Extremities | − | patella luxations, finger joint laxity | long fingers | finger joint laxity, Sydney line, sandal gap | hemihypotrophy distal left leg | laterally deviated halluces | − | NA or NK | distal arthrogryposis | − |
| Feeding problems | + | + | + (G-tube until 2 years, 5 months) | − | + | + (G-tube) | + (G-tube) | difficulty with chewing and swallowing | consistently underweight | + |
| Recurrent infections | − | + | − | − | − | − | − | − | − | − |
| Eye abnormalities | − | strabismus | − | strabismus | hypermetropia | − | strabismus | − | mild astigmatism | strabismus |
| Renal abnormalities | HN | HN, UPJ stenosis | − | − | − | − | − | NA | NA | |
| Cardiac abnormalities | − | − | NA or NK | − | − | Ebstein anomaly | PFO, small aorto-pulmonary collateral | NA or NK | − | − |
| Other | lacrimal duct stenosis | extensible skin, teeth abnormalities | neuroblastoma | none | esophageal atresia, febrile seizures, hypothyroidism, unilateral breast hypoplasia | none | bilateral cryptorchidism | hypothyroidism | craniosynostosis, GH deficiency, lacrimal duct hypoplasia, tooth abnormalities | hip clicking |
Abbreviations are as follows: +, present; −, absent; +/−, minor; ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; CP, cleft palate; DQ, developmental quotient; GH, growth hormone; G-tube, gastrostomy tube; HC, head circumference; HN, hydronephrosis; NA, not available; NK, not known; PFO, patent foramen ovale; SA, subarachnoid; TE, trachea-oesophageal; and UPJ, ureteropelvic junction.
GenBank: NM_003403.4.
GenBank: NP_003394.1.
UCSC Genome Browser build hg19.
PhyloP scores were calculated with UCSC track vertebrate Basewise Conservation (phyloP46wayAll, hg19).
This is the average score for the three deleted bases: 5.13, 5.13, and 3.27.
Individuals 6 and 7 were detected in the same cohort of 5,500 individuals. Individuals 8 and 10 were detected in the same cohort of 6,709 individuals.
The ten individuals with de novo mutations in YY1 were detected by exome sequencing in a total cohort of 14,969 individuals with ID. We calculated the probability of observing ten de novo mutations in YY1 in 14,469 individuals as described previously32 and corrected for the total number of tested genes (19,280, enrichment Agilent V5).
Individuals with deletions that included (part of) YY1 were identified from DECIPHER33 and our in-house database containing data from over 8,000 individuals with ID. Microarray analysis was performed with the 44K, 60K, or 400K Agilent array ’(Agilent Technologies) or 250K NspI SNP array (Affymetrix) according to the manufacturer’s protocol.
Detailed phenotype information of individuals with YY1 mutations and deletions was collected.
This study was approved by the institutional review board of the Radboud University Medical Center (Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen NL36191.091.11), by the University of Milan ethics committee, and by the Neurogenetics Research Program and South Australian Clinical Genetics Service of Women’s & Children’s Hospital (2361/3/2017). Written informed consent was obtained from all individuals.
LCL Culture
Lymphoblastoid cell lines (LCLs) were cultured in RPMI 1640, 15% fetal bovine serum, 1% HEPES, 1% L-glutamine, and 1% penicillin-streptomycin. All LCL samples were tested for mycoplasma.
Chromatin Immunoprecipitation and ChIP-Seq Analysis
Chromatin immunoprecipitation (ChIP) was performed as previously described34 with some modification. In brief, LCLs were centrifuged at 150 × g for 5 min, and an average of 150 × 106 LCLs were re-suspended in 1% formaldehyde in PBS for cross-linking. To stop the cross-linking reaction, glycine was added to the final concentration of 125 mM. Cells were re-suspended with ChIP SDS buffer (0.5% SDS, 5 mM EDTA, 100 mM NaCl, and 50 mM Tris-HCl [pH 8.1]). Pellets were collected by centrifuging at 400 × g for 30 min and re-suspended in immunoprecipitation (IP) buffer (0.5% SDS, 5 mM EDTA, 100 mM NaCl, and 50 mM Tris-HCl [pH 8.6] and 1.5% Triton X-100) To obtain a bulk of 300 bp DNA fragments, we sonicated cells with the Branson Digital Sonifier (Emerson Industrial Automation). Chromatin was quantified with the Bradford Protein Assay (Bio-Rad). To immunoprecipitate YY1, we incubated 1 mg of total proteins with YY1 antibody (sc-1703 or sc-281, Santa-Cruz Biotechnologies). For H3K27Ac ChIP, 100 μg of protein was immunoprecipitated with the antibody ab4729 (Abcam). H3K27me3 IPs were performed with 100 μg of proteins with the antibody C36B11 (Cell Signaling Technologies). Chromatin was incubated overnight with antibody at 4°C. G-Sepharose 4B (Thermo Fisher Scientific) beads were incubated with chromatin and antibody mix for 4 hr at 4°C and then washed with low- and high-salt buffers (1% Triton X-100, 0.1% SDS, 150 or 500 mM NaCl, 2 mM EDTA, and 20 mM Tris-HCl [pH 8]). De-crosslinking was performed at 65°C for 3 hr. DNA was collected with the QIAquick PCR Purification Kit (QIAGEN). Libraries were prepared as previously described35 with adaptations for the automated system Biomek FX.
ChIP sequencing (ChIP-seq) reads were trimmed for potential adaptor contamination with Scythe 0.981 (min 4 nucleotides) before being aligned to the hg38 genome (UCSC Genome Browser) with Bowtie 1.0 (-v 2 -m 1), and peaks were called by MACS 2.0.9 with default settings for YY1 and with the “broad” option for H3K27ac and H3K27me3. Peaks were defined as overlapping if they shared at least one nucleotide, and bound genes were defined as having a peak within 1 kb upstream of the transcription start site (TSS). For each target, all nucleotides falling under a peak in at least two IPs were then merged into non-overlapping windows in which reads were counted and used for quantitative analysis. In addition, for both histone modifications, an analysis was performed specifically on the regions bound by YY1 (with the addition of 500 bp on each side) and on active enhancers. In all cases and unless otherwise specified, the number of mapped reads was used as the library size, and edgeR v.3.12.1 (with exact test) was used for assessing the statistical significance of the changes at the specific windows. Because such an analysis rests on the assumption that the read-count distributions are globally similar, and the H3K27ac data showed signs of a global decrease, for the analysis of this dataset, we excluded regions called as having increased H3K27ac in the probands because these were likely to be artifacts of the normalization.
RNA Extraction and Quality Assessment
RNA was extracted with the RNeasy Micro Plus Kit (QIAGEN) according to the manufacturer’s protocol. Concentration and RNA quality were assessed by NanoDrop (NanoDrop Technologies). Prior RNA fragmentation for RNA sequencing (RNA-seq) library construction was assessed by the 2100 Bioanalyzer (Agilent Technologies). Only samples with an RNA integrity number > 9 were accepted for library preparation.
RNA-Seq Analysis and Differential Expression Analysis
RNA-seq libraries were prepared according to Illumina protocols (TruSeq Stranded Ribo-Zero depletion). Sequencing was performed with a HiSeq 2000. We performed paired-end 50 bp sequencing on independently cultured replicates of each line (see Table S1 for a summary of the samples and experiments). Only one of the two batches clustered by genotype, whereas the other showed high levels of variability partly attributable to broad variations in EP300 and a large amount of its targets. For this reason, and because the first batch represented the same cultures from which the IPs were performed, we first focused on this one. However, we then used the replicated datasets, along with the subset of HapMap LCLs, in a meta-analysis. Because the replicate samples were not fully independent (i.e., could not be considered additional individuals) yet were informative of the technical, stochastic, and epigenetic variability, we then focused on the high-confidence differentially expressed genes (DEGs) that were statistically significant both in the main dataset (with adjusted p value < 0.05) and in the meta-analysis (with adjusted p value < 0.1). We performed RNA-seq quantification directly from the reads with Salmon 6.1,36 which we have shown to be excellent in tracking relative gene expression differences,37 by using hg38 and the RefSeq annotation complementing the sequences of the ERCC spike-ins and Epstein-Barr virus. All differential expression analyses were performed with DESeq2 1.10.138 with the geometric normalization and the Wald test, considering only genes that had at least 20 reads in at least four samples. The merged analysis was performed with a variable accounting for the batch (i.e., ∼batch + condition). For the association with YY1 levels, “functional” YY1 levels were used; in other words, for the purpose of this analysis, the YY1 RNA levels of the two missense mutations were reduced so that their mean was comparable with that of the deletions, in line with the western blot (Figure 1C).
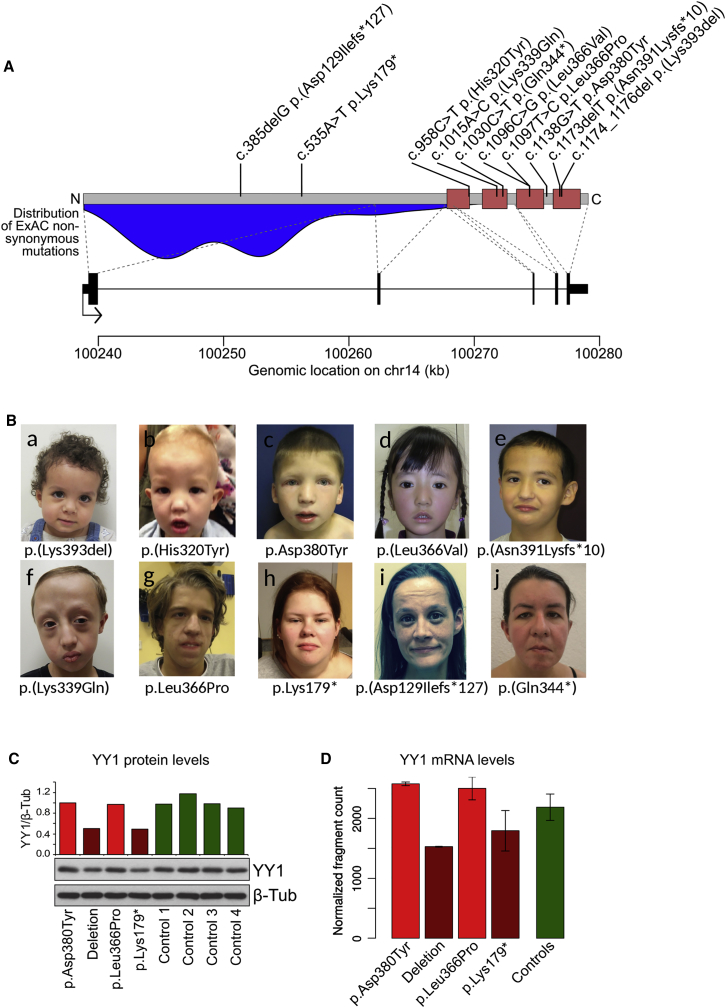
Figure 1.
Description of the Cohort
(A) YY1 locus, including the location of the mutations identified in this study (top) and the (smoothed) frequency of non-pathogenic (missense) mutations in the ExAC Browser (in blue, bottom).
(B) Frontal photographs sorted by age of individuals with YY1 mutations (a–j). Recurrent facial dysmorphism includes facial asymmetry, a broad forehead, fullness of the upper eyelids, and an upper lip shaped like a Gingko leaf. Each individual or his or her parents provided informed consent for the publication of these photographs.
(C and D) YY1 levels in affected individuals’ LCLs as measured by western blot (C; the y axis indicates the ratio to β-tubulin, normalized on the first sample) and RNA-seq (D). In (D), each bar represents the mean across two samples for the probands and nine samples for the control individuals; error bars indicate standard deviation.
Enrichment Analyses
Gene Ontology (GO) enrichment analyses were performed with the goseq R package,39 including correction for eventual RNA-seq transcript-length bias and exclusion of genes without annotation. Categories with at least 10 but no more than 1,000 genes were considered, and Fisher’s test was used. In the main figures, we filtered out the parent categories with significantly enriched offspring categories to maximize the specificity of the enrichments. The unfiltered enrichments are available in Table S4.
TF target enrichment analysis based on published ChIP-seq data was performed with the hypergeometric distribution. DNA motif enrichment analysis was performed with Homer v.4.8.40 Unless specified, the background used for enrichment analysis was the set of expressed and tested genes used for testing DEGs, and all genes were used otherwise.
Permutation Analysis on the HapMap LCLs
Using the RNA-seq profiles from the HapMap LCL collection,41, 42 we performed 300 random, sex-balanced differential expression analyses (DEAs) with three samples per group and identified a subset of genes with a higher tendency to be found differentially expressed in LCL lines derived from apparently healthy individuals (with an false-discovery rate [FDR] cutoff of <0.05 in at least 2% of the DEAs; see Table S2 for the detailed list).
Protein Extraction and Immunoblotting
For protein extraction, RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris [pH 8.0]) was used for lysating cells. A protease inhibitor cocktail (Sigma) was added during extraction. Cell lysates were sonicated with the Bioruptor Sonication System (UCD200) for three cycles of 30 s at high power with 30 s pauses and centrifuged at 13,000 × g for 15 min. The Bradford Protein Assay (Bio-Rad) was adopted for protein quantification according to the manufacturer’s protocol. For each sample, 20–40 μg of protein extract was loaded onto home-made 10% SDS-PAGE gels. Protein transfer were performed with a tension of 120 V for 1 hr on nitrocellulose membranes and blocked in 0.2% TBS-T (50 mM Tris [pH 7.5], 150 mM NaCl, and 0.2% Tween-20) and 5% milk prepared with 0.2% TBS-T. Primary and secondary antibodies were diluted in 5% milk and 0.2% TBS-T. Western blots were performed with the following antibodies and detected by peroxidase-based chemiluminescence (GE Healthcare Life Sciences): YY1 (sc-1703, 1:500; Sigma) and β-tubulin (ab6046, Abcam). Densitometry was performed with ImageJ software.
Structural Optimization of the C-Terminal Portion of YY1 Affected by De Novo Mutations
The 3D coordinates of the YY1 zinc domain structure were obtained from the Protein Data Bank (PDB: 1UBD 43). Hydrogens were added, and the structure was optimized with GROMACS 5.144 and force field Amber99-ILDN45 through 200 steepest descent minimization steps and 300 conjugate gradient minimization steps. The structures of probands 1–4 and 7 were built by Modeler 9.16.46 Surface accessibility was measured with GROMACS 5.1 with default parameters, whereby the protein was soaked in water explicit solvent with the TIP3P model.47, 48 Differences in binding energies resulting from point mutations were measured with Autodock Vina.49
External Datasets Used
The following published ChIP-seq datasets were used:
-
•
GEO: GSM935294 (p300 in GM12878)
-
•
GEO: GSM935611 (CTCF in GM12878; post-filtering q < 10 × 10−10)
-
•
GEO: GSM803391 (PAX5 in GM12878)
-
•
GEO: GSM935583 (CoREST in GM12878; post-filtering q < 10 × 10−5)
-
•
GEO: GSM935613 (ZNF143 in GM12878; post-filtering q < 10 × 10−10)
-
•
GEO: GSM733772 (H3K4me1 in GM12878)
-
•
GEO: GSM1003448 (HDAC1 in K562)
-
•
GEO: GSM1003574 (CBP in K562)
In all cases, ENCODE 2014 unified narrowPeak calls were downloaded and lifted over to hg38, eventually with the indicated post-filtering.
The list of chromatin interactions from GM12878 (called by the HiCUP Hi-C pipeline50) was downloaded from GEO: GSE63525 (file “GSE63525_GM12878_primary+replicate_HiCCUPS_looplist.txt.gz”) and lifted over to hg38. We considered a YY1 binding site to be distally in contact with a TSS if the YY1 peak overlapped an anchor of the loop and the second anchor overlapped the TSS.
Finally, the RNA-seq data from the HapMap LCLs were downloaded from the Sequence Read Archive (study ERP000101) and processed just like the RNA-seq data produced in the present study.
Results
Identification of Individuals with De Novo Mutations in YY1
Upon identification of the initial person with a de novo mutation in YY1,1 Sanger sequencing of YY1 in a cohort of 500 individuals with unexplained ID resulted in the identification of one additional person with a de novo mutation in YY1: c.1097T>C (p.Leu366Pro). In addition, different exome sequencing studies in individuals with idiopathic ID identified eight further individuals with de novo mutations in YY1 (Table 1). This resulted in a total of ten individuals with de novo mutations in YY1 (Figures 1A and 1B). The Bonferroni-adjusted probability of observing this or a greater number of de novo mutations in YY1 in our cohort was p = 2.8 × 10−6, calculated as previously described.32
The phenotype of these ten individuals consisted of cognitive impairment ranging from borderline to moderate ID. Other recurrently observed features included various behavioral problems, intrauterine growth restriction (IUGR), feeding problems, eye abnormalities, and movement abnormalities. In addition, a variety of congenital abnormalities, including esophageal atresia, cleft palate, craniosynostosis, hydronephrosis, and Ebstein anomaly, were observed in only one or two individuals each (Table 1). Overlapping craniofacial dysmorphisms included facial asymmetry with a broad forehead, fullness of the upper eyelids, and an upper-lip indentation shaped like a Gingko leaf (Figure 1B).
Additionally, we collected data on 13 individuals with deletions that encompassed YY1, among other genes (Table S3 and Figure S1). The deletions ranged in size from 75 kb to 13 Mb, and half of these also overlapped the UPD(14) gene cluster, hence resulting in a maternal or paternal UPD(14) phenotype (MIM: 608149). The phenotype of these 13 individuals overlapped the phenotype observed in individuals with point mutations in YY1, including developmental delay, ID, IUGR, feeding problems, and eye abnormalities in the majority of the individuals. Similar congenital abnormalities were observed between the two groups, albeit in a small percentage of individuals (Table S1).
Characterization of the YY1 Variants
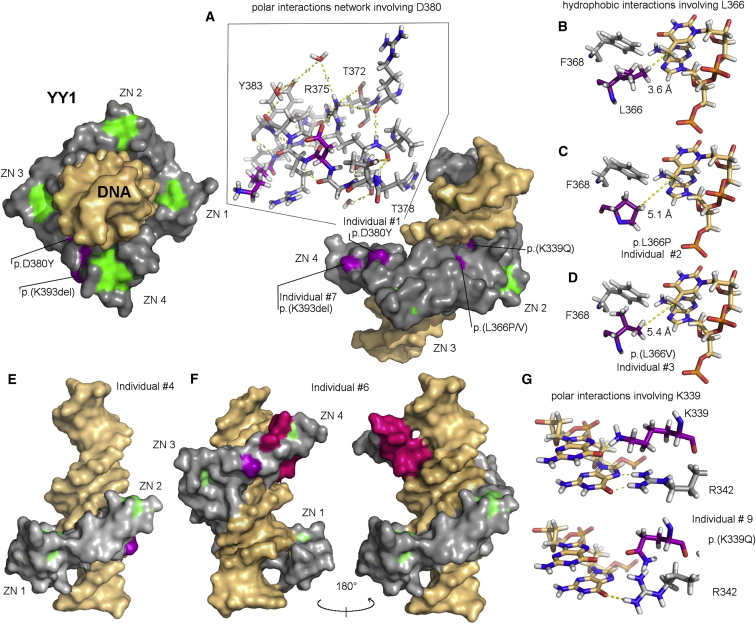
Of all the de novo mutations, six led to single amino acid changes in the zinc-finger domains of YY1 (Figure 1A): c.958C>T (p.His320Tyr), c.1015A>C (p.Lys339Gln), c.1097T>C (p.Leu366Pro), c.1096C>G (p.Leu366Val), c.1138G>T (p.Asp380Tyr), and c.1174_1176del (p.Lys393del). The affected amino acid residues are highly conserved, and at least two different programs predicted the introduced missense changes to damage protein function (Table 1).51, 52 Two mutations, nonsense c.1030C>T (p.Gln344∗) and frameshift c.1173delT (p.Asn391Lysfs∗10), were located in the (pen)ultimate exon and, as a result, are highly unlikely to trigger nonsense-mediated decay (NMD). Instead, these variants are predicted to give rise to a truncated YY1 without the last two zinc fingers. The two remaining mutations were the early frameshift c.385delG (p.Asp129Ilefs∗127) and nonsense c.535A>T (p.Lys179∗). Importantly, none of these mutations are present in dbSNP 139, our in-house database containing exome data of over 7,000 individuals, or the Exome Aggregation Consortium (ExAC) Browser. By mapping all known variation over the primary structure of YY1, we observed that all variants found in healthy individuals involve the unstructured amino-terminal portion of the protein, and none of them involve its zinc-finger domains (Figure 1A). We subsequently mapped the probands’ mutations over residues 295–408 of the 3D structure of YY1 (PDB: 1UBD) to gain insight into the mechanisms of action of these variants, which are predicted to result in loss of function (Figure 2). In brief, c.1138G>T (p.Asp380Tyr) is expected to affect a network of salt bridges and polar interactions that might be important for YY1 stability and for the fourth zinc finger to correctly bend toward DNA (Figure 2A). In addition, because residue 380 is on the solvent-accessible surface of the protein, the substitution of residue 380 with a tyrosine could disrupt the hydrogen bonds with surrounding residues, thus affecting the structure of the protein and its ability to interact with cofactors. Individuals 2 and 3 have different mutations (c.1097T>C and c.1096C>G, respectively) that both affect a leucine residue (Leu366) in the middle of the DNA recognition motif, constituting a hydrophobic core together with Phe368 and the seventh nucleotide of the target DNA motif (Figures 2B–2D). Thus, a substitution of Leu366 with proline or valine is likely to be detrimental because of the lower steric hindrance of these two amino acids. In fact, these mutations increase the surface-accessible area of residue Phe368 by ∼40%. Individual 4 has the nonsense mutation c.1030C>T (p.Gln344∗), which generates a protein missing the third and fourth zinc fingers (Figure 2E). Individuals 6 and 7 have mutations (c.1173delT [p.Asn391Lysfs∗10] and c.1174_1176del [p.Lys393del], respectively) that destroy the fourth zinc finger (Figure 2A and 2F). Individual 9 is affected by c.1015A>C (p.Lys339Gln), which disrupts the protein’s interaction with DNA. The loss in binding energy estimated by Autodock Vina is approximately 8.42 Kcal/mol.49 The mutation c.1015A>C (p.Lys339Gln) induces both bending of the Arg342 sidechain (partially preventing its interaction with a guanidine residue that is consecutive to the nucleosidic residues interacting with Leu366 and Phe368 in the YY1 binding motif) and impairment of its interaction with the next nucleotide in the sequence. Finally, we can expect c.958C>T (p.His320Tyr) in individual 10 to cause the loss of zinc coordination in the first zinc finger of the DNA binding domain and lead to a loss of DNA binding affinity on the resulting protein.
Figure 2.
YY1 Variants on the 3D Structure of the Protein’s DNA Binding Domain
The 3D structure of wild-type YY1’s C-terminal zinc-finger (ZN) domain is represented as a gray surface surrounding DNA, represented in gold. ZN coordination regions are reported in light green, and missense mutated amino acid residues are reported in violet.
(A) Residues involved in polar interactions with D380 are reported in gray sticks. Hydrogen bonds are represented as yellow dashes.
(B–D) Nucleotides interacting with residues L366 and F368 are reported in sticks. Distances between L366 (B), or its variants in individuals 2 (C) and 3 (D), and the closest nucleotides are reported with yellow dashes.
(E) Putative structure of the truncated form of YY1 in proband 4 is reported through depiction of its electrostatic surface.
(F) C-terminal portion of YY1 affected by proband 6 (variant in pink). The structure of the DNA-YY1 complex is reported from two opposite observation sides to highlight (1) its importance for ZN coordination in the fourth ZN and (2) its role in DNA binding.
(G) Nucleotides interacting with K339 and R342 are reported in sticks. Polar interactions are reported with yellow dashes.
A Panel of LCLs for the Functional Characterization of Different YY1 Mutations
To evaluate the functional impact of the different mutations on YY1 expression, we established LCLs from four individuals and compared them with LCLs from healthy control individuals. Specifically, we tested LCLs of two individuals with missense mutations (individuals 1 [c.1138G>T (p.Asp380Tyr)] and 2 [c.1097T>C (p.Leu366Pro)]), one with a nonsense mutation (individual 5 [c.535A>T (p.Lys179∗)]), and one with a complete deletion of the YY1 genomic region, including only two other genes (SLC25A29 [MIM: 615064] and SLC25A47 [MIM: 609911]) (individual 272547). Importantly, cDNA sequencing with and without cycloheximide treatment (Figure S2) indicated that the transcripts harboring the mutation causing a premature termination codon (individual 5) undergo NMD. This means that our panel effectively includes two missense mutations and two mutations leading to loss of protein levels from one YY1 allele. We confirmed YY1 mRNA expression and protein levels by RNA-seq and western blot (Figures 1C and 1D), which showed wild-type YY1 levels in LCLs from individuals 1 and 2 and 50% reduced levels in cells from individuals 5 and 272547. Thus, these findings allowed us to compare the molecular consequences of YY1 heterozygosity with respect to specific heterozygous missense mutations in the zinc-finger domains.
High-Resolution Profiling of YY1 Genome-wide Occupancy
To assess the impact of the different genetic lesions on YY1’s DNA-binding activity, we performed ChIP-seq in LCLs from control and affected individuals by using two different antibodies that recognize the N- and C-terminal regions of YY1. The two antibodies showed excellent agreement, including high overlap of the peak calling (Figure 3A and Figure S3) and a high Pearson correlation of fold enrichments over input in the union of enriched regions (>0.93). YY1 peaks showed a strong bias toward proximal promoters, such that over 77% of the peaks were less than 1 kb upstream of a TSS (Figure 3B). DNA motif enrichment analysis of these regions revealed two significant motifs that could be identified in a sizeable proportion of the peaks: a major one very similar to the canonical YY1 binding motif (MA0095.2) was identified in >50% of the peaks (p < 1 × 10−3106; ∼10-fold enrichment over background; Figure 3C), and a minor one similar to the ETS binding motifs, especially FLI1, was identified in >20% of the peaks (∼2-fold enrichment over background; Figure S4B). Virtually all YY1 bindings at promoters were co-occupied by ZNF143, and more than half of YY1-bound sites were shared among CoREST, PAX5, CTCF, and either HDAC1 or CBP (Figure S4A). The YY1-bound genes were enriched with genes regulating transcription, translation, and mRNA processing (Figure 3D and Table S4).
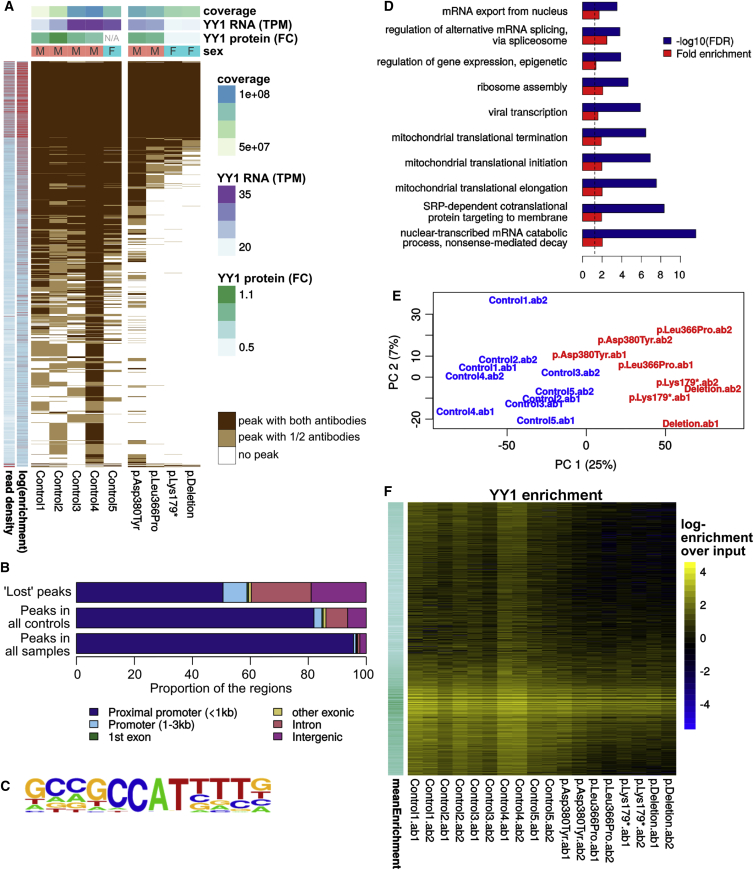
Figure 3.
ChIP-Seq for YY1 in LCLs Derived from Affected Individuals
(A) Overlap between the peaks detected in each sample and each antibody. Each horizontal line represents a peak, shown in white if it was undetected in a sample, in light brown if it was detected in only one of the two IPs, and in dark brown if it was detected with both antibodies. Above are shown the sex of each LCL line, the YY1 mRNA expression and protein levels, and the coverage of the corresponding ChIP-seq experiments. TPM stands for transcripts per million, and FC stands for fold change in relation to the control samples. In red on the left are shown the average log-transformed read density and log enrichment for each peak.
(B) Distribution of conserved and “lost” YY1 peaks across genomic features.
(C) Dominant YY1 DNA-binding motif identified.
(D) Top most-specific Gene Ontology (GO) enrichments of the YY1 target genes.
(E) Principal-component analysis of the log-transformed enrichments over the input across the union of YY1-enriched regions.
(F) YY1 enrichment over input across the different IPs (columns) and enriched regions (rows).
Missense Mutations and YY1 Hemizygosity Both Lead to a Global Loss of YY1 Binding
The LCL samples of the four affected individuals tested showed a marked decrease in the number of YY1 peaks, which was not accounted for by differences in coverage (p ∼ 4 × 10−4 by analysis of variance). Of note, the peaks that were lost or retained were consistent across samples irrespectively of genetic lesion (i.e., missense mutations, truncating mutations, or deletions), thus underscoring haploinsufficiency as the core mechanism underlying this condition. The high-occupancy peaks (defined by either high read density or high enrichment over the input) were preferentially retained (Figure 3A), as expected. “Lost” YY1 peaks, i.e., peaks found in at least seven of ten IPs from the control samples and in none of the probands (416 peaks), showed a similar prevalence of the major YY1 DNA-binding motif and were still enriched with promoter regions, but considerably less so than high-occupancy peaks detected (Figure 3B).
Although one proband sample showed a higher number of peaks than the other probands, this was mainly due to its higher coverage, which disappeared upon quantitative analysis (below). A very small number of peaks were called exclusively in proband IPs, particularly in the c.1138G>T (p.Asp380Tyr) sample. Although these regions did not have genome-wide significance in the control samples, all of them showed an enrichment over the input in most control samples (Figure S5). Moreover, the number of gained regions was smaller than the mean or median number of regions unique to any control sample, which strongly suggests that they were due to background genetic variability rather than to a gain of function of the mutant p.Asp380Tyr protein.
Because peak calling is coverage and threshold sensitive and because conserved peaks might still show quantitative differences in binding, we corroborated these results by looking at the read distribution over peaks. Independently of the normalization procedure, the IPs clearly clustered by genetic condition (Figure 3E), confirming the clear-cut impact of YY1 dosage on its genome-wide occupancy (Figure 3F and Figures S6–S8). Indeed, when the read distributions were linearly normalized on the total library size, proband samples showed a considerable decrease in the proportion of reads mapping to all YY1 binding sites (p < 0.00016, Mann-Whitney test; 95% confidence interval between 43% and 87%; see Figure S6). Because linear normalization of ChIP-seq data can be problematic,53 we confirmed this global decrease by using two additional normalization methods based on very different assumptions, both of which gave milder but statistically significant decreases in global YY1 binding (Figure S7). Differential-binding analysis based on library-size normalization showed a significant, 95% decrease in YY1-bound regions. We also confirmed, on the basis of read counts, the bias toward the conservation of highly enriched sites (Figure 3F and Figure S8), indicating that the facility of peak calling at those sites was not the only reason for their preferential retention upon the decrease in effective YY1 levels. We next investigated whether peaks more or less sensitive to YY1 dosage were associated with specific co-bindings. Compared with the 1,000 peaks showing the greatest reduction in the probands, the 1,000 YY1 peaks showing the lowest reduction were particularly enriched with bindings of cofactors such as CoREST (p ∼ 2 × 10−8), p300 (p ∼ 3 × 10−7), PAX5 (p ∼ 5 × 10−10), and HDAC1 (p ∼ 3 × 10−14, Chi-square test; see also Figure S9). In contrast, no significant difference was observed for bindings of CTCF and ZNF143, suggesting that YY1 binding is dependent on only a subset of its cofactors.
Finally, we did not detect any significant difference in the read distributions between the LCLs of individuals with YY1 deletions and those of individuals with missense mutations (excluding regions on chromosomes X and Y because of sex differences). This finding excludes that the missense mutations studied lead to aberrant recruitment of the protein to novel targets and support our hypothesis that the different mutations all lead to haploinsufficiency of YY1 function. As with every disorder caused by missense mutations, it remains possible that they could also contribute to phenotypic expressivity or variability through gains of function (not associated with DNA binding in this specific case).
YY1 Haploinsufficiency Leads to Differential Expression of Only a Minority of Its Targets
To assess the extent to which the observed reduction in YY1 binding results in transcriptional dysregulation of the target genes, we performed RNA-seq on the same set of LCL samples plus an additional control sample. YY1-associated changes did not dominate transcriptional variability, but the samples clustered by genotype according to the second principal component (Figure 4A). The first principal component correlated with no known biological or technical factor (sex, age, coverage, sequencing lane, RNA extraction efficiency, or proportion of ribosomal or Epstein-Barr virus RNA). Gene-set enrichment analysis on the whole expressed transcriptome revealed positive enrichments in several curated gene sets related to the cell cycle, transcription, and genome maintenance (Table S5). We identified 152 DEGs between proband and control LCLs (Figure 4B and Table S6), which were significantly enriched with genes related to chromatin silencing (p ∼ 9 × 10−10), aligning with the enrichment of chromatin remodelers among genes associated with autism spectrum disorder.54 The DEGs bound by YY1, along with the peaks detected in each sample, are shown in Figure 4C. Despite the massive decrease in YY1 DNA binding, only a minority (<1%) of YY1-bound genes turned out to be differentially expressed (and only 21% of the DEGs were bound by YY1).
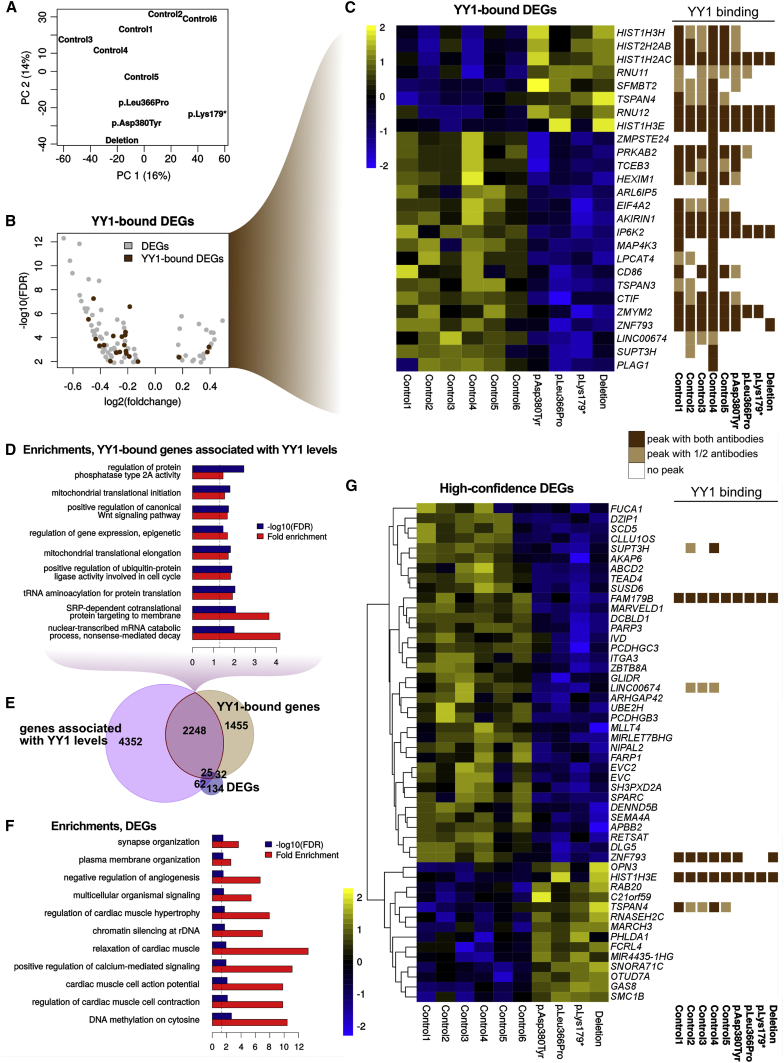
Figure 4.
Transcriptional Impact of the YY1 Mutation
(A) Principal-component analysis of the RNA-seq dataset.
(B) Volcano plot of the differentially expressed genes (DEGs) highlights YY1-bound genes.
(C) Expression of YY1-bound DEGs (left), along with the detected YY1 peaks at their TSS (right).
(D) Top most-specific GO enrichments of the YY1-bound genes that show a significant association with YY1 mRNA levels across HapMap LCLs.
(E) Overlap between the DEGs and YY1-bound and YY1-associated genes.
(F) Top most-specific GO enrichments of all DEGs.
(G) Expression of high-confidence DEGs (consistently dysregulated in the meta-analysis with the HapMap LCLs), along with eventual YY1 bindings at their TSS.
In (C) and (G), expression is shown as Z scores of log-transformed normalized counts, and YY1 binding is indicated in light brown if it was detected in only one of the two IPs and in dark brown if it was detected with both antibodies.
To improve the robustness of this analysis, we included transcriptomes from independently cultured replicates of the same samples and leveraged a panel of 73 transcriptional profiles from the HapMap collection of LCLs derived from apparently healthy individuals.42 We performed a large number of permutation analyses to identify genes often differentially expressed between healthy individuals (see Figures S10A and S10B and Material and Methods).55 Importantly, the HapMap dataset revealed important variations in chromatin regulators, including YY1 itself, as well as some of its key partners, such as EP300 and CREBBP, which explained a large proportion of the transcriptomic variance (Figure S8C). Although broad variation suggests that LCLs might not be the ideal cell type for studying these pathways, it also offers an opportunity to detect associated expression patterns. We therefore harnessed this variability to identify a large set of genes (6,687) that were significantly associated with YY1 levels (see Material and Methods) and that significantly overlapped YY1-bound genes (Figure 4E; see Figure 4D for the GO enrichments of the intersection). Finally, we performed a meta-analysis of the two RNA-seq datasets in this study (18 samples) and an equally sized subset of the HapMap LCLs by selecting the samples that had YY1 levels comparable with those of our control samples (see Material and Methods). On this basis, we identified both a lenient set of candidate DEGs (Table S6), whose GO enrichments are shown in Figure 4F, and a set of 50 high-confidence DEGs significant in both analyses (Figure 4G).
This analysis further corroborated the identified dysregulations and also confirmed the relatively small overlap with differential YY1 binding. On average, DEGs had a greater decrease in YY1 binding than genes not detected as having a statistically significant difference in expression (p ∼ 1 × 10−4), although the large overlap between the two distributions (see Figure S11A) suggests that other factors, such as the presence of binding partners, is likely to distinguish YY1-dosage-dependent, transcriptionally affected genes in any given cellular state. We therefore looked for enrichment among the DEGs for predicted targets of known TFs. After excluding very low (<2) enrichment scores, the only significant enrichment identified was for predicted targets of FLI1 (p ∼ 1 × 10−10, 13% of the DEGs and <3% of the expressed genes) on the basis of the MA0149.1 motif. It is interesting that, as mentioned earlier, a very similar motif was found to be enriched in the sequence surrounding the summit of YY1 peaks. However, neither FLI1 nor any of its known interactors were differentially expressed in the probands. In addition, on the basis of the published ChIP-seq data on related cell types, we detected a significant enrichment among DEGs for targets of ATF2 (p ∼ 2 × 10−97, ∼28% of the DEGs) and ATF3 (p ∼ 6 × 10−26, ∼20% of the DEGs). Of note, YY1 binds the TSS of both TFs, and although neither of them was detected as statistically significant in the differential expression analysis, ATF2 showed a significant correlation with YY1 mRNA levels across the whole cohort of LCLs from the meta-analysis (p ∼ 8 × 10−13; see Figure S11B). The DEGs were also enriched with targets of CEBPB (1.9-fold, FDR ∼ 0.001), CHD1 (1.5-fold, FDR ∼ 0.002), and EZH2 (1.9-fold, FDR ∼ 0.002), all of which showed statistically significant correlation with YY1 levels in the HapMap LCLs, pointing to potential mediators of the effects of YY1 haploinsufficiency.
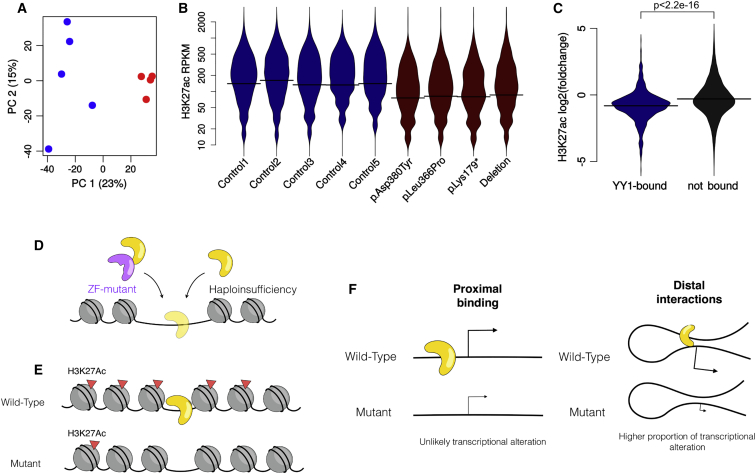
YY1 Haploinsufficiency Is Associated with Widespread Loss of H3K27 Acetylation
YY1 forms complexes with several important histone acetylases and de-acetylases, including HDAC1, HDAC2, p300, and CBP, whose antagonistic functions account at least in part for the bivalent role of YY1 in transcriptional repression and activation. Because most DEGs in YY1 probands were downregulated, we focused on the impact of YY1 haploinsufficiency on histone acetyltransferases by performing ChIP-seq for H3K27ac, selected as a marker of both active promoters and enhancers and hence allowing interrogation of both kinds of regulatory regions targeted by YY1. Consistently, we found that H3K27ac distribution clustered proband samples apart from control samples (Figure 5A and Figures S12A and S12B), confirming the hypothesis that the effect of the mutation is at least mostly mediated by the regulation of this modification. Linear normalization on library size suggested a global reduction of H3K27ac in the proband samples (Figures S12C and S12D), although the difference did not stand out with alternative normalization methods (Figure S13), and even though the difference in the number of peaks was statistically significant (p ∼ 0.012), it was relatively small in magnitude (∼10% fewer peaks). We therefore proceeded with the most conservative analysis, namely focusing on sites that were showing reduced H3K27ac even under the assumption of no difference in the global distribution (see Material and Methods). Although this analysis potentially underestimates the regions losing H3K27ac, it excludes the possibility that spurious regions will be identified as artifacts of normalization. Even with this most stringent method, 39% of the YY1-bound DEGs lost H3K27ac at their proximal promoter. Of note, however, 99% of the regions showing reduced H3K27ac in probands were on active enhancers (defined by enrichment in H3K4me1 and H3K27ac), the majority of which were not proximal to the TSS, suggesting an important component of distal regulation. Indeed, although the majority of YY1 bindings were located at the TSS (Figure 3B), these results were consistent with the involvement of YY1 in chromatin looping56, 57 and prompted us to test the possibility that YY1 binding at distal enhancers might have a stronger impact on gene expression. Indeed, of the YY1 binding sites that were not in the proximal promoter of a gene, 65% were on active enhancers, whereas fewer than 4% were on poised enhancers (defined by enrichment in H3K4me1 and H3K27me3). Proband samples showed a marked decreased in H3K27ac of YY1-bound active enhancers (Figures 5B and 5C and Figure S14) that was independent of read counts and normalization method (Figures S15A and S15B), and the fold changes in YY1 and H3K27ac enrichments at active enhancers were positively correlated (p < 2.2 × 10−16), further corroborating that the deacetylation was dependent on YY1 haploinsufficiency. Furthermore, after normalization on library size, 82% of YY1-bound enhancers showed a statistically significant decrease in H3K27ac, in contrast with merely 25% of the other active enhancers. However, only 15% of the enhancers overlapping the 500 YY1 peaks least affected by dosage showed a reduction in H3K27ac, underscoring a crucial cis role for YY1 in regulating enhancer activation. We therefore used the kilobase-pair-resolution Hi-C profiles of GM12878 LCLs58 to identify chromatin loops that would link distal regulatory sites to the TSS of putative target genes. We identified 545 genes that were distally bound by YY1 (Figures S16 and S17), of which 206 were also bound at the TSS. Although only a minority of these were differentially expressed, genes distally bound by YY1 were significantly enriched with DEGs (p ∼ 6 × 10−8 with respect to the whole genome and p ∼ 7 × 10−4 with respect to the expressed genes, hypergeometric test). Compared with genes proximally bound by YY1, distally bound genes were much more likely to be differentially expressed (p ∼ 1.5 × 10−5, chi-square test), indicating that despite its preferential enrichment at the TSS, YY1 affects its distal targets disproportionately more.
Figure 5.
YY1 Haploinsufficiency Results in Enhancer Dysregulation
(A) Principal-component analysis of the H3K27ac read-count distribution across all enriched regions segregates proband from control samples (probands in red and controls in blue).
(B) Distribution of H3K27ac read densities at YY1-bound enhancers shows a marked reduction in probands. RPKM stands for reads per kilobase pair per million reads mapped.
(C) Preferential loss of H3K27ac at YY1-bound versus non-YY1-bound enhancers. The p value is the result of a two-tailed t-test.
(D–F) Schematic representation of our key findings: YY1 haploinsufficiency and variants in the zinc-finger domain both result in a global loss of YY1 binding (D), associated with loss of H3K27ac (E); only a minority of differentially bound genes are differentially expressed, and the proportion is significantly higher for distal YY1 bindings (F).
YY1 Haploinsufficiency Leads to a Switch from Acetylation to Methylation
Loss of acetylation leaves the H3K27 substrate available for PRC2-mediated methylation, which is associated with repression. To investigate whether the YY1-dependent changes in H3K27ac were followed by Polycomb marking, we profiled H3K27me3 on the same set of samples. Although we observed no difference in the global amount of H3K27me3 between control and proband samples, the loss of H3K27ac was associated with an increase in H3K27me3 (p < 2 × 10−16; see Figure S18) across YY1-bound regions, particularly those enriched with H3K27ac and H3K27me3 in any sample. Although only a minority of these regions showed a statistically significant difference after correction for multiple testing, including cap binding complex dependent translation initiation factor (CTIF [MIM: 613178]), the increase in H3K27me3 is likely to underlie the predominant downregulation of DEGs in proband samples.
Discussion
We characterized pathogenic mutations in YY1 in ten individuals with overlapping phenotypes, whose molecular dissection defines a neurodevelopmental syndrome. Historically, YY1 was proposed as a candidate ID-associated gene in our original trio sequencing in 2010.1 Our combined clinical data indeed confirm ID as the core feature of the syndrome, which also includes variable expression of other comorbidities such as IUGR, feeding problems, behavioral problems, craniofacial dysmorphisms, and congenital anomalies. In addition, we showed relevant phenotypic overlaps with 13 more cases of deletions encompassing YY1. With the inherent limitations due to sample size and the phenotypic variability of a new syndrome, we observed no major differences between phenotypes associated with point mutations and those arising from deletions of YY1. Given that many of the deletions are large and encompass multiple genes, the phenotypic features in these persons might not be definitely attributed to YY1. As an example, half of the deletions also encompassed the UPD(14) gene cluster, whose rearrangements result in a maternal or paternal UPD(14) phenotype.59, 60 Because the majority of individuals with maternal UPD(14) have IUGR, the occurrence of this phenotype, for example, could be related to the YY1 deletion, to the deletion of the UPD(14) gene cluster, or to their combination. Interestingly, however, maternal UPD(14) is generally not associated with ID, so the concurrent deletion of YY1 could explain the ID that is observed in individuals with UPD(14)mat phenotypes. In order to further delineate the clinical spectrum associated with de novo mutations in YY1, we established a website to collect detailed clinical information on additional individuals who will be identified over the coming years (see Web Resources).
The overlapping phenotype of individuals with missense mutations, protein-truncating mutations, and deletions of YY1 indicates loss of function as the underlying mechanism. Importantly, neither YY1 loss-of-function mutations nor missense variations in YY1 zinc-finger domains have been identified in the ExAC Browser. Instead, all missense mutations in the probands map to zinc fingers, highlighting the pathogenic relevance of YY1 DNA binding. Indeed, our analyses of LCLs from control and affected individuals show that the missense mutations alter YY1 DNA binding as much as truncating mutations or deletions (as captured by two distinct antibodies recognizing each end of the protein), pointing to YY1 haploinsufficiency as the core pathogenic mechanism underlying this syndrome. This is also supported by the growth and neurological alterations in the Yy-1 heterozygous knockout mice17 and further corroborated by the observation that the phenotype of Yy-1 knockdown in mice is also dosage dependent.61
Our key findings on the molecular impact of the mutations are summarized in Figures 5D–5F. YY1 haploinsufficiency, regardless of whether it is due to deletions or missense mutations, leads to massive loss of genome-wide YY1 occupancy. The impact is larger on lower-occupancy YY1 binding sites and is accompanied by major changes in histone acetylation, in line with known interactions between YY1 and several histone acetyltransferases and deacetylases.62 Furthermore, our results show the specific loss of H3K27Ac at YY1-bound enhancers, independent of signal-to-noise ratio and normalization method, indicating an important and heretofore neglected role of YY1 in enhancer regulation and in line with recent evidence on enhancers as a key site of dysregulation for several ID syndromes, several of which are being reframed as enhanceropathies.62
Importantly, the targets differentially bound by YY1 include a number of genes causally involved in neurodevelopmental disorders and autism spectrum disorders, such as (1) GTF2I (MIM: 601679), encoding a TF of the genetic interval hemi-deleted or duplicated in, respectively, Williams-Beuren syndrome (MIM: 194050) and the reciprocal Williams-Beuren region duplication syndrome (MIM: 609757), for which converging lines of evidence indicate a key role in the cognitive-behavioral phenotypes of the two conditions;63, 64, 65, 66 (2) KANSL1 (MIM: 612452), whose deletion causes Koolen-de Vries syndrome (MIM: 610443), also involving ID;67, 68 (3) NRXN2 (MIM: 600566), which has been causally associated with autism69 and further corroborated by a recent mouse model;70, 71 (4) MED12 (MIM: 300188), mutations in which causes syndromic forms of ID; (5) NSD1 (MIM: 606681) mutations in which cause Sotos syndrome (MIM: 117550); and (6) ZBTB20, associated with Primrose syndrome (MIM: 259050), which encompasses ID and systemic abnormalities.72, 73, 74, 75, 76
Importantly, only a minority of the genes losing YY1 binding showed significant differential expression in LCLs, including additional genes already implicated in neurodevelopmental disorders, such as RNASEH2C (MIM: 610329) and NBEA (MIM: 604889).77 Thus, although the proportion of transcriptionally sensitive targets increased as YY1 binding decreased and was greater for enhancers than for TSS bindings, the overall delta between YY1-dosage-dependent occupancy changes and transcription suggest that although YY1 shows low rewiring78 and high conservation of binding sites across tissues, its transcriptional impact is highly cell-type specific and is most likely mediated by its co-factors. This offers a plausible explanation for the phenotypic variability of individuals, given that background genetic variation influencing the expression or binding of co-factors, which could be buffered in the context of normal YY1 dosage, could instead become clinically penetrant upon halving of YY1 dosage. Moreover, only a minority of the TSSs losing YY1 binding showed significant differential expression, an observation that might explain the comparatively moderate clinical phenotype for a transcription factor that has such a central role as YY1.
In conclusion, we have shown that both deletions and de novo point mutations affecting YY1 function cause a haploinsufficiency syndrome of ID with a broad range of growth and behavioral comorbidities.
Conflicts of Interest
The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered at the Baylor Miraca Genetics Laboratories. Z.P. is an employee of Ambry Genetics.
Acknowledgments
We are grateful to the individuals and their parents for their participation. We thank Lone Laulund (Department of Pediatrics, Odense University Hospital) for additional clinical information about deletion case 263711. We thank Giuseppe D’Agostino from the Testa lab for his help in creating the summary figure. This work was supported by the European Commission (GENCODYS grant 241995 under FP7 to A.T.V.-v.S. and B.B.A.d.V.), the European Research Council (grant DISEASEAVATARS 616441 to G.T.), the Telethon Foundation (grants GGP13231B and GGP14265 to G.T.), the Associazione Italiana per la Ricerca sul Cancro (investigator grant to G.T. and fellowship to M.G.), the Regione Lombardia (Ricerca Indipendente 2012 to G.T.), the Umberto Veronesi Foundation (P.-L.G.), the Italian Ministry of Health (ERANET-Neuron grant to P.-L.G. and Ricerca Corrente grant to G.T.), the Japan Agency for Medical Research and Development (research grant to K.K.), the Australian National Health and Medical Research Council (grant 1091593 to J.G.), and the Dutch Organization for Health Research and Development (ZON-MW grants 917-86-319 and 912-12-109 to B.B.A.d.V.).
Published: June 1, 2017
Footnotes
Supplemental Data include 18 figures, 6 tables, and author contributions and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.05.006.
Contributor Information
Giuseppe Testa, Email: giuseppe.testa@unimi.it.
Bert B.A. de Vries, Email: bert.devries@radboudumc.nl.
Accession Numbers
Raw ChIP-seq and RNA-seq data presented in this work are available in the Gene Expression Omnibus through accession number GEO: GSE98478.
Web Resources
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
YY1 individual website, http://humandiseasegenes.nl/yy1
Supplemental Data
References
- 1.Vissers L.E.L.M., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y., Seto E., Chang L.S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 3.Park K., Atchison M.L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc. Natl. Acad. Sci. USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hariharan N., Kelley D.E., Perry R.P. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc. Natl. Acad. Sci. USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satijn D.P., Hamer K.M., den Blaauwen J., Otte A.P. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V.E., Valente S., Mai A., Forcales S.V. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vella P., Barozzi I., Cuomo A., Bonaldi T., Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40:3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon S., Akopyan G., Garban H., Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y., Jin J., Yao T., Gottschalk A.J., Swanson S.K., Wu S., Shi Y., Washburn M.P., Florens L., Conaway R.C., Conaway J.W. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 12.Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H.H., Wang W., Nickoloff J.A. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.S., Galvin K.M., See R.H., Eckner R., Livingston D., Moran E., Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y., Lee J.S., Galvin K.M. Everything you have ever wanted to know about Yin Yang 1……. Biochim. Biophys. Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 15.Guo D.C., Duan X.-Y., Regalado E.S., Mellor-Crummey L., Kwartler C.S., Kim D., Lieberman K., de Vries B.B.A., Pfundt R., Schinzel A., University of Washington Center for Mendelian Genomics Loss-of-Function Mutations in YY1AP1 Lead to Grange Syndrome and a Fibromuscular Dysplasia-Like Vascular Disease. Am. J. Hum. Genet. 2017;100:21–30. doi: 10.1016/j.ajhg.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y., Casaccia-Bonnefil P. The Yin and Yang of YY1 in the nervous system. J. Neurochem. 2008;106:1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donohoe M.E., Zhang X., McGinnis L., Biggers J., Li E., Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y., Kim J.Y., Dupree J., Tewari A., Melendez-Vasquez C., Svaren J., Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat. Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L., Jolly L.A., Willis-Owen S., Gardner A., Kumar R., Douglas E., Shoubridge C., Wieczorek D., Tzschach A., Cohen M. A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 2012;91:694–702. doi: 10.1016/j.ajhg.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolly L.A., Nguyen L.S., Domingo D., Sun Y., Barry S., Hancarova M., Plevova P., Vlckova M., Havlovicova M., Kalscheuer V.M. HCFC1 loss-of-function mutations disrupt neuronal and neural progenitor cells of the developing brain. Hum. Mol. Genet. 2015;24:3335–3347. doi: 10.1093/hmg/ddv083. [DOI] [PubMed] [Google Scholar]
- 21.Burgold T., Spreafico F., De Santa F., Totaro M.G., Prosperini E., Natoli G., Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira J.D., Sansom S.N., Smith J., Dobenecker M.-W., Tarakhovsky A., Livesey F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testa G. The time of timing: how Polycomb proteins regulate neurogenesis. BioEssays. 2011;33:519–528. doi: 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- 24.Burgold T., Voituron N., Caganova M., Tripathi P.P., Menuet C., Tusi B.K., Spreafico F., Bévengut M., Gestreau C., Buontempo S. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2012;2:1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Park D.H., Hong S.J., Salinas R.D., Liu S.J., Sun S.W., Sgualdino J., Testa G., Matzuk M.M., Iwamori N., Lim D.A. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014;8:1290–1299. doi: 10.1016/j.celrep.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 32.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.-J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 33.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 36.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germain P.-L., Vitriolo A., Adamo A., Laise P., Das V., Testa G. RNAontheBENCH: computational and empirical resources for benchmarking RNAseq quantification and differential expression methods. Nucleic Acids Res. 2016;44:5054–5067. doi: 10.1093/nar/gkw448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houbaviy H.B., Usheva A., Shenk T., Burley S.K. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 45.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 47.Mahoney M.W., Jorgensen W.L. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8910. [Google Scholar]
- 48.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 49.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wingett S., Ewels P., Furlan-Magaril M., Nagano T., Schoenfelder S., Fraser P., Andrews S. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 53.Shao Z., Zhang Y., Yuan G.-C., Orkin S.H., Waxman D.J. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jabbi M., Chen Q., Turner N., Kohn P., White M., Kippenhan J.S., Dickinson D., Kolachana B., Mattay V., Weinberger D.R., Berman K.F. Variation in the Williams syndrome GTF2I gene and anxiety proneness interactively affect prefrontal cortical response to aversive stimuli. Transl. Psychiatry. 2015;5:e622. doi: 10.1038/tp.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medvedovic J., Ebert A., Tagoh H., Tamir I.M., Schwickert T.A., Novatchkova M., Sun Q., Huis In ’t Veld P.J., Guo C., Yoon H.S. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 2013;39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerasimova T., Guo C., Ghosh A., Qiu X., Montefiori L., Verma-Gaur J., Choi N.M., Feeney A.J., Sen R. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev. 2015;29:1683–1695. doi: 10.1101/gad.263871.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagami M., Kurosawa K., Miyazaki O., Ishino F., Matsuoka K., Ogata T. Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome) Eur. J. Hum. Genet. 2015;23:1488–1498. doi: 10.1038/ejhg.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Severi G., Bernardini L., Briuglia S., Bigoni S., Buldrini B., Magini P., Dentici M.L., Cordelli D.M., Arrigo T., Franzoni E. New patients with Temple syndrome caused by 14q32 deletion: Genotype-phenotype correlations and risk of thyroid cancer. Am. J. Med. Genet. A. 2016;170A:162–169. doi: 10.1002/ajmg.a.37346. [DOI] [PubMed] [Google Scholar]
- 61.Affar B., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith E., Shilatifard A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 63.Pober B.R. Williams-Beuren syndrome. N. Engl. J. Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 64.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somerville M.J., Mervis C.B., Young E.J., Seo E.-J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Pérez-Jurado L.A. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adamo A., Atashpaz S., Germain P.-L., Zanella M., D’Agostino G., Albertin V., Chenoweth J., Micale L., Fusco C., Unger C. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 67.Koolen D.A., Kramer J.M., Neveling K., Nillesen W.M., Moore-Barton H.L., Elmslie F.V., Toutain A., Amiel J., Malan V., Tsai A.C.-H. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat. Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 68.Zollino M., Orteschi D., Murdolo M., Lattante S., Battaglia D., Stefanini C., Mercuri E., Chiurazzi P., Neri G., Marangi G. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat. Genet. 2012;44:636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
- 69.Gauthier J., Siddiqui T.J., Huashan P., Yokomaku D., Hamdan F.F., Champagne N., Lapointe M., Spiegelman D., Noreau A., Lafrenière R.G. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dachtler J., Glasper J., Cohen R.N., Ivorra J.L., Swiffen D.J., Jackson A.J., Harte M.K., Rodgers R.J., Clapcote S.J. Deletion of α-neurexin II results in autism-related behaviors in mice. Transl. Psychiatry. 2014;4:e484. doi: 10.1038/tp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Born G., Grayton H.M., Langhorst H., Dudanova I., Rohlmann A., Woodward B.W., Collier D.A., Fernandes C., Missler M. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front. Synaptic Neurosci. 2015;7:3. doi: 10.3389/fnsyn.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vulto-van Silfhout A.T., de Vries B.B.A., van Bon B.W.M., Hoischen A., Ruiterkamp-Versteeg M., Gilissen C., Gao F., van Zwam M., Harteveld C.L., van Essen A.J. Mutations in MED12 cause X-linked Ohdo syndrome. Am. J. Hum. Genet. 2013;92:401–406. doi: 10.1016/j.ajhg.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Risheg H., Graham J.M., Jr., Clark R.D., Rogers R.C., Opitz J.M., Moeschler J.B., Peiffer A.P., May M., Joseph S.M., Jones J.R. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 74.Tarpey P., Parnau J., Blow M., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2004;75:318–324. doi: 10.1086/422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 76.Cordeddu V., Redeker B., Stellacci E., Jongejan A., Fragale A., Bradley T.E.J., Anselmi M., Ciolfi A., Cecchetti S., Muto V. Mutations in ZBTB20 cause Primrose syndrome. Nat. Genet. 2014;46:815–817. doi: 10.1038/ng.3035. [DOI] [PubMed] [Google Scholar]
- 77.Castermans D., Wilquet V., Parthoens E., Huysmans C., Steyaert J., Swinnen L., Fryns J.-P., Van de Ven W., Devriendt K. The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J. Med. Genet. 2003;40:352–356. doi: 10.1136/jmg.40.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.-K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.