Abstract
Populations maintain considerable segregating variation in the response to toxic, xenobiotic compounds. To identify variants associated with resistance to boric acid, a commonly-used household insecticide with a poorly understood mechanism of action, we assayed thousands of individuals from hundreds of strains. Using the Drosophila Synthetic Population Resource (DSPR), a multi-parental population (MPP) of inbred genotypes, we mapped six QTL to short genomic regions containing few protein-coding genes (3–188), allowing us to identify plausible candidate genes underlying resistance to boric acid toxicity. One interval contains multiple genes from the cytochrome P450 family, and we show that ubiquitous RNAi of one of these genes, Cyp9b2, markedly reduces resistance to the toxin. Resistance to boric acid is positively correlated with caffeine resistance. The two phenotypes additionally share a pair of QTL, potentially suggesting a degree of pleiotropy in the genetic control of resistance to these two distinct xenobiotics. Finally, we screened the Drosophila Genetic Reference Panel (DGRP) in an attempt to identify sequence variants within mapped QTL that are associated with boric acid resistance. The approach was largely unsuccessful, with only one QTL showing any associations at QTL-specific 20% False Discovery Rate (FDR) thresholds. Nonetheless, these associations point to a potential candidate gene that can be targeted in future validation efforts. Although the mapping data resulting from the two reference populations do not clearly overlap, our work provides a starting point for further genetic dissection of the processes underlying boric acid toxicity in insects.
Keywords: complex traits, MPP, RNAi, quantitative trait loci, xenobiotics, Multi-parent Advanced Generation Inter-Cross (MAGIC), multiparental populations
A major goal of quantitative genetics is to identify and characterize sequence variants contributing to the extensive interindividual variation observed for medically- and evolutionarily-relevant traits. Genetic mapping studies can implicate novel pathways, genes, and variants in the genetic control of disease risk and phenotypic variation. More broadly, similar investigations across multiple traits and systems have the potential to provide answers to fundamental questions about complex traits; for example, are causative alleles generally rare or at intermediate frequency, and do they segregate for a series of alleles that impact phenotype? Any general patterns uncovered can lead to predictions about the evolutionary forces acting on functional genetic variation, and aid in the development of novel, more powerful methods for the dissection of trait variation.
Essentially, any trait with a significant fraction of heritable variation is open to genetic dissection by linkage- or association-based mapping techniques. However, work in both model systems (e.g., Valdar et al. 2006b; Rat Genome Sequencing and Mapping Consortium et al. 2013) and in humans (e.g., Yang et al. 2010) indicates that, on average, causative loci confer only fairly subtle effects on phenotype. Thus, mapping studies must involve large numbers of individuals to find variants with realistic effect sizes, meaning that traits that can be assayed in a high-throughput fashion are more tractable. Toxic compounds, or xenobiotics, are easily delivered to flies by feeding, facilitating powerful mapping studies using hundreds of strains and tens of thousands of individuals (e.g., Najarro et al. 2015). Since xenobiotic resistance can also exhibit relatively high heritability in Drosophila melanogaster (Carrillo and Gibson 2002; Marriage et al. 2014; Najarro et al. 2015), genetic dissection of many such traits may help elucidate general properties of complex trait variation.
All animals face considerable pressure from exposure to xenobiotics in their diet and environment, from toxins produced by plants as protection against herbivory (Glendinning 2007; Mithofer and Boland 2012) to agricultural pesticides (Perry et al. 2011). As a result, animal genomes possess a diverse array of genes responsible for the metabolism and excretion of toxins. These genes, including cytochrome P450 monooxygenases (P450s), glutathione-S-transferases, UDP-glucuronosyltransferases, and ABC transporters, coordinate to form a sophisticated detoxification system (Xu et al. 2005; Li et al. 2007). These gene families are strong candidates to harbor segregating variation contributing to differences in xenobiotic resistance among individuals. Indeed, one of the best examples of xenobiotic metabolism in insects is the series of structural changes at the P450 gene Cyp6g1 that are largely responsible for resistance to the insecticide DDT in populations of D. melanogaster (Daborn et al. 2002; Chung et al. 2007; Schmidt et al. 2010).
Unbiased, genome-wide mapping studies for resistance to caffeine (Najarro et al. 2015) and nicotine (Marriage et al. 2014) have identified QTL collectively explaining significant fractions of trait heritability, and have implicated known detoxification genes in the genetic control of resistance. Here, we complement this work by dissecting the genetic basis of resistance to boric acid, a common household insecticide. Boric acid has been shown to be effective against various ant species (Klotz and Moss 1996; Klotz et al. 1996), and the larvae of both honeybees (da Silva Cruz et al. 2010) and wax moths (Hyrsl et al. 2007; Buyukguzel et al. 2013). Boric acid administration has morphological effects on the midgut and malpighian tubules in leaf-cutting ants (Sumida et al. 2010), leads to abnormalities in midgut cells in Argentine ants (Klotz et al. 2002) and honeybee larvae (da Silva Cruz et al. 2010), and promotes irregularities in both midgut tissue and the fat body in wax moths (Buyukguzel et al. 2013). Nonetheless, the physiological mechanisms responsible for the mortality associated with boric acid intake are not clear. Genetic dissection of variation in resistance in the D. melanogaster model system has the potential to provide insight into the genes involved in boric acid toxicity.
A powerful strategy to identify causative loci is to employ a series of recombinant genotypes derived from a small number of founding haplotypes. So-called MPPs have been developed for the major animal model systems, including the Collaborative Cross (Churchill et al. 2004; Threadgill and Churchill 2012) and the Diversity Outbred (DO, Churchill et al. 2012; Svenson et al. 2012) mouse populations, the NIH-HS rat heterogeneous stock (Rat Genome Sequencing and Mapping Consortium et al. 2013), and the DSPR (King et al. 2012b). These populations facilitate statistically powerful QTL mapping (Valdar et al. 2006a; King et al. 2012a; Gatti et al. 2014), are capable of resolving QTL to shorter intervals than is generally possible with an F2 mapping design due to the multiple generations of intercrossing typically employed, and as a result of being founded from multiple strains allow the characterization of allelic series at functional loci. In addition, stable reference populations such as the DSPR allow multiple traits, including “molecular phenotypes” such as chromatin accessibility and gene expression, to be measured on the same set of genotypes. In so doing, reference populations facilitate examinations of pleiotropy, as well as enabling characterization of the functional, molecular consequences of sequence-level variation.
In this study, we describe the genetic dissection of boric acid resistance in both the DSPR MPP and in a second reference mapping population, the DGRP (Mackay et al. 2012), a series of resequenced inbred strains allowing genome-wide association testing. We address three basic questions. First, can we identify QTL and uncover plausible candidate genes contributing to resistance to boric acid in flies? By working with D. melanogaster we can ultimately leverage the power of this model system to increase the depth of our mechanistic understanding of boric acid toxicity in insects. Second, can we find evidence for pleiotropic resistance loci? We previously used a nearly identical phenotyping assay to examine caffeine resistance in the DSPR (Najarro et al. 2015). Correlation between caffeine and boric acid resistance measured in the same set of genotypes, as well as any overlap in the QTL mapped, would imply some level of pleiotropy. Third, can we identify sequence variants contributing to trait variation by combining high-resolution QTL mapping with GWAS (genome-wide association study) data? GWAS have low power due to the strict correction for multiple tests that must be applied to avoid false positive associations, and one approach to mitigate this power deficit is to test associations only within those narrow intervals implicated by QTL mapping.
Materials and Methods
Phenotyping pipeline
Our measure of resistance to boric acid is taken as the lifespan (in hours) of mated females exposed to media supplemented with 1.5% boric acid (BH3O3, BP168; Fisher Scientific). This concentration was arrived at following a series of ad hoc experiments testing a number of D. melanogaster strains against a range of boric acid concentrations. We assayed resistance for all genotypes following an identical pipeline to that described previously for caffeine resistance (Najarro et al. 2015). Briefly, parental flies were permitted to lay eggs for up to 2 d, clearing when needed to maintain roughly similar egg density across experimental vials. After 8 d any adult progeny were cleared, and 2 d later female flies were collected under CO2 anesthesia. By this point all vials contained many flies of both sexes, so we assume that females were generally mated. Experimental females were kept in single-sex groups of ∼20 flies on regular media for 1 d to recover from anesthesia. Subsequently, test flies were aspirated individually, and without anesthesia into narrow polycarbonate tubes containing a small quantity of cornmeal-yeast-dextrose media supplemented with 1.5% boric acid. Media was made 24 hr prior to the start of the assay, and boric acid was added to the molten media at ∼50° to minimize any heat-induced degradation. Tubes were inserted into Drosophila Activity Monitoring System units (DAM2; TriKinetics), and the activity of each animal was recorded every minute for 6 d using the DAMSystem3 data collection software (TriKinetics). Fly death was indicated by a permanent cessation of activity.
All experimental individuals were reared and assayed at 25° and 50% relative humidity on a 12 hr light/12 hr dark cycle.
QTL mapping
We employed the DSPR MPP to find QTL contributing to boric acid resistance. The DSPR consists of two panels of Recombinant Inbred Lines (RILs) (pA and pB), each derived from a pair of replicate highly-recombinant synthetic populations (pA1 and pA2, and pB1 and pB2) initiated with a different set of eight inbred founder strains (pA RILs founded by strains A1–A7 and AB8, and pB RILs founded by strains B1–B7 and AB8). Detailed descriptions of RIL construction, founder resequencing, and RIL genotyping-by-sequencing are provided in King et al. (2012b). For this study, we screened through 835 pA and 855 pB RILs, measuring boric acid resistance for eight replicate individuals per RIL (Supplemental Material, File S1). The 1690 RILs were tested in 13 batches, with each RIL being tested in a single batch.
QTL were mapped by providing RIL means to the R package DSPRqtl (version 2.0-5), that depends on the data packages DSPRqtlDataA and DSPRqtlDataB (version 2.0-1) to provide the RIL genotypes. Packages are available at FlyRILs.org and GitHub (github.com/egking/DSPRqtl), and genotypes can be obtained via FlyRILs.org and Dryad (dx.doi.org/10.5061/dryad.r5v40). Methods for QTL mapping in the DSPR are thoroughly described in King et al. (2012a,b). Briefly, at each position in each RIL we assign an additive probability that the genetic material is derived from each of the eight possible founders, then regress mean RIL phenotype on these eight probabilities using the model where μ is the grand mean, Gi are the genotype probabilities, and βi are the additive genetic effects of each haplotype. The resulting F-statistic is then converted to a LOD score (Broman and Sen 2009). The pA and pB panels were analyzed separately, ignoring subpopulation since there was no significant difference in the average phenotype of RILs from each subpopulation (Figure 1; pA1 vs. pA2, Welch’s t-test, P = 0.34; pB1 vs. pB2, P = 0.21). Genome-wide 5% significance thresholds for calling QTL were estimated via 1000 permutations of the data (following Churchill and Doerge 1994), and we used 2-LOD support intervals to estimate the true positions of causative sites (King et al. 2012a).
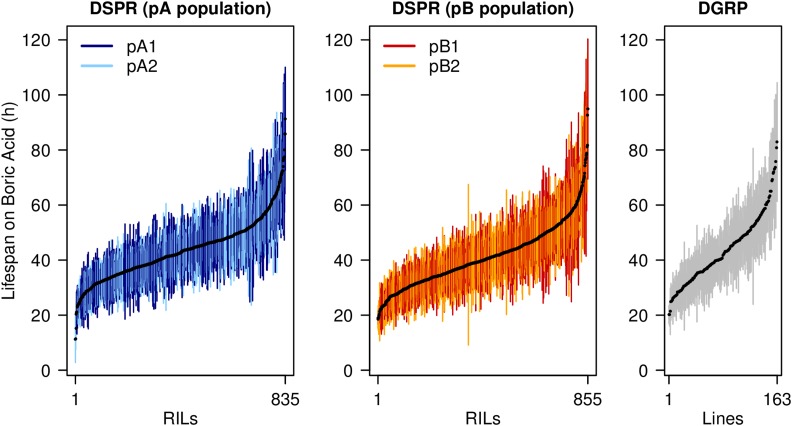
Figure 1.
Phenotype distributions in the reference populations. We measured lifespan on media supplemented with 1.5% boric acid for 1690 RILs from the DSPR and 163 inbred lines from the DGRP. We scored eight females from each DSPR RIL and 16 females from each DGRP strain. Each plot shows the mean (filled circles) and 1-SD (vertical lines) for each genotype. RILs derived from each of the DSPR subpopulations (pA1, pA2, pB1, pB2) are shown in different colors; No significant differences in RIL mean phenotype were found between subpopulations (Welch’s t-tests, P > 0.2). DGRP, Drosophila Genetic Reference Panel; DSPR, Drosophila Synthetic Population Resource; RILs, Recombinant Inbred Lines.
Association mapping
We employed strains from the DGRP (Mackay et al. 2012; Huang et al. 2014) to identify sites associated with variation in boric acid resistance. DGRP strains were purchased from the Bloomington Drosophila Stock Center (BDSC). Following the pipeline described above, we collected phenotypes for 16 replicate individuals from each of 163 inbred DGRP strains (File S1), assaying the strains in four batches. Strain means were subsequently submitted to the DGRP2 analytical engine (dgrp2.gnets.ncsu.edu) for GWAS analysis. Huang et al. (2014) provide thorough detail of the methodology employed. Briefly, the pipeline involves first adjusting line means to account for variation across lines in both Wolbachia infection status and the presence of large chromosomal inversions. Next, all common (minor allele frequency > 0.05) variants are tested for an association with phenotype using a linear mixed model that accounts for variation in relatedness among strains, y = Sb + Zu + e, where S is the design matrix for fixed SNP effect b, Z is the incidence matrix for the random polygenic effect u, and e is the residual (Huang et al. 2014). We sought to identify variants significantly associated with phenotype by using a Bonferroni correction for multiple tests, and by using a FDR (Benjamini and Hochberg 1995) as implemented by the p.adjust function in R (r-project.org).
Heritability of the phenotype
We calculated estimates of the broad-sense heritability of boric acid resistance for both panels of the DSPR and for the DGRP. Genetic and phenotypic variance components were calculated via the lme and VarCorr functions from the nlme R package (Pinheiro et al. 2011), using the linear mixed model Yij = μ + gi + εij, where Yij is the jth observation of the ith RIL, μ is the grand mean, gi is the random effect of RIL, and εij is the error term.
Functional testing of candidate genes via RNAi
We used the binary Gal4-UAS RNAi system to test a pair of candidate detoxification genes, Cyp6a2 and Cyp9b2, residing under DSPR QTL pB.Q1 for any effects on phenotype. We drove Gal4 in all cells at all timepoints using Act5C-GAL4 (from BDSC stock 25374). Transgenic RNAi Project (TRiP, Perkins et al. 2015) UAS-RNAi and co-isogenic control strains were obtained from the BDSC, specifically stock numbers 35786 (UAS-GFP control), 35788 (UAS-Luciferase control), 62996 (UAS-Cyp9b2-RNAi), and 64008 (UAS-Cyp6a2-RNAi). We generated RNAi knockdown/control genotypes by crossing five males from the Gal4 strain to 10 virgin females from each UAS strain, establishing four replicate cross vials per UAS. Sixteen Gal4-UAS females were tested from each vial, a total of 64 per genotype, following the phenotyping pipeline described above (see Phenotyping pipeline and File S1). In addition, a group of 20 Gal4-UAS females from each genotype was maintained on test media lacking boric acid for the duration of the assay. None of these Gal4-UAS animals died by the end of the exposure assay, indicating that RNAi-based knockdown of the two target genes does not lead to gross effects on viability in the absence of boric acid.
Xenobiotic metabolism genes in D. melanogaster
Five broad classes of genes are thought to be principally responsible for xenobiotic metabolism in eukaryotes. To determine which, if any, of these genes are present within QTL mapped in this study, we extracted from FlyBase (Attrill et al. 2016) all genes associated with the following Interpro IDs (Mitchell et al. 2015): cytochrome P450s (IPR001128), glutathione S-transferases (IPR004045, IPR004046, IPR010987), UDP-glucuronosyltransferases (IPR002213), esterases (IPR002018), and ABC transporters (IPR003439).
Data availability
All individual-level phenotypes are provided in File S1. Raw activity monitor datafiles, along with R scripts to generate individual- and strain-level phenotypes for the DSPR, have been uploaded as a Dryad data package (doi: 10.5061/dryad.3v55m). DSPR QTL mapping output, including positions and LOD scores, is provided in File S2. Lists of genes currently annotated within mapped DSPR QTL are provided in File S3. Positions and association statistics for all nominally significant (P < 0.05) variants tested in the DGRP GWAS are provided in File S4. DSPR RILs can be requested from FlyRILs.org.
Results and Discussion
Substantial trait variation and heritability
We measured the lifespan of mated female flies on 1.5% boric acid-supplemented media for 835 pA and 855 pB DSPR RILs, and 163 DGRP strains, testing 8 and 16 flies per genotype in the DSPR and DGRP, respectively (File S1). Figure 1 highlights the considerable among-genotype variation in boric acid resistance observed in all mapping panels. The three sets of genotypes exhibit similar average line phenotypes (41.3–43.6 hr), have comparable phenotypic ranges (DSPR pA, 11.3–91.3 hr; DSPR pB, 18.5–94.9 hr; and DGRP, 20.2–82.9 hr), and yield similar estimates of broad-sense heritability (DSPR pA = 47.0%; DSPR pB = 49.7%; and DGRP = 52.5%). Given that all replicate individuals tested for a single genotype were routinely derived from the same culture vial, and because every genotype was tested in just one experimental batch, some fraction of the variation we attribute to genetic factors is likely to result from environmental effects.
We previously measured adult, mated female resistance to 1% caffeine in the DSPR and DGRP using a phenotyping pipeline identical to that employed in the current study (Najarro et al. 2015). Using those strains assayed for both phenotypes we find strong and significant positive correlations between phenotypes in all three panels of lines (Figure 2): DSPR pA (n = 822, Pearson’s r = 0.37, P < 10−15), DSPR pB (n = 796, r = 0.37, P < 10−15), and DGRP (n = 154, r = 0.36, P < 10−5). This result suggests that some fraction of the genetic variation contributing to resistance might be shared by these traits.
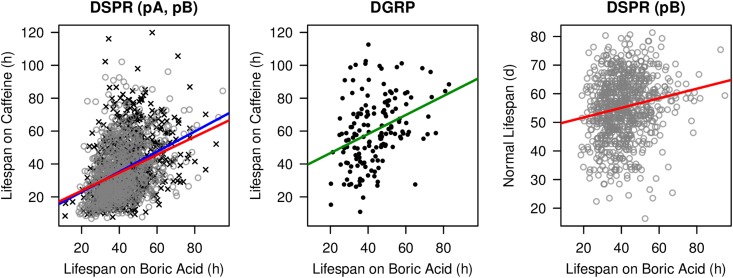
Figure 2.
Phenotypic correlations in the DSPR and DGRP. Boric acid resistance data are from the current study, resistance to caffeine is from Najarro et al. (2015), and lifespan data under no-drug conditions (available for DSPR pB only) is from Highfill et al. (2016). Each point represents the mean phenotype of a single line from the DSPR pA (black cross), DSPR pB (gray open circle), and DGRP (black filled circle) populations. Only those strains with data from all relevant phenotypes are shown (822 DSPR pA RILs, 796 DSPR pB RILs, and 154 DGRP lines). Correlations between phenotypes are shown with colored lines: DSPR pA (blue), DSPR pB (red), and DGRP (green). Boric acid and caffeine resistance are highly correlated in both the DSPR (r = 0.37, P < 10−15) and DGRP (r = 0.36, P < 10−5). Boric acid resistance shows a more modest relationship with normal lifespan in DSPR pB RILs (r = 0.17, P < 10−6). DGRP, Drosophila Genetic Reference Panel; DSPR, Drosophila Synthetic Population Resource; RILs, Recombinant Inbred Lines.
Highfill et al. (2016) measured the lifespan of mated females on regular, no-drug food in the DSPR pB panel. The correlation between lifespan and boric acid resistance is significant, although the magnitude of the relationship is relatively small (Figure 2, n = 796, r = 0.17, P < 10−6), and the phenotypic correlation between lifespan and caffeine resistance is low (n = 796, r = 0.07, P = 0.04). Calculating the correlation between the two measures of resistance while controlling for lifespan, using the pcor function in the R package ppcor (Kim 2015), reveals that the strong correlation between the resistance traits is maintained (n = 796, r = 0.36, P < 10−24). These results imply that variants contributing to overall longevity may generally not play a role in lifespan under drug-exposure conditions. Nonetheless, we note that the assay used to measure lifespan had little in common with that used to assay drug resistance, and these technical differences could lead to the underestimation of any correlation between traits.
Multiple modest-effect boric acid toxicity QTL
We mapped six, mapping panel-specific QTL in the DSPR (Figure 3, File S2, and Table 1). One QTL, pB.Q3, explains 7.9% of the phenotypic variation, while the remaining QTL each explain 4.5–5.0% of the variance. Assuming the QTL are independent and act additively, mapped QTL explain 9.1 and 21.9% of the variation in boric acid resistance in the pA and pB panels, respectively. Given that the power to map a 5% QTL in the DSPR with 800 RILs is 84% (King et al. 2012a), we reasoned that those genetic variants contributing to the remainder of the heritability are likely to be numerous and confer relatively small effects on phenotypic variance.
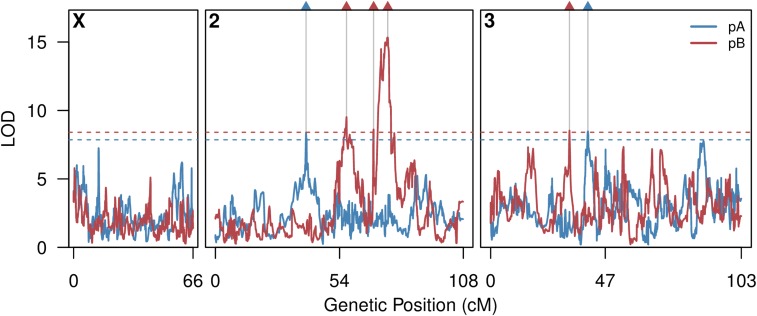
Figure 3.
Genome-wide scan for boric acid resistance QTL in the DSPR. Solid lines depict population-specific scans across the major D. melanogaster chromosomes. Horizontal dashed lines represent permutation-derived 5% critical thresholds (pA, 7.9; pB, 8.4). Autosomal centromeres are shown at positions 54 cM (chromosome 2) and 47 cM (chromosome 3). The locations of the six mapped QTL are highlighted along the top of the plot, with colors indicating population (left-to-right; pA.Q1, pB.Q1, pB.Q2, pB.Q3, pB.Q4, and pA.Q2). DSPR, Drosophila Synthetic Population Resource; QTL, quantitative trait loci.
Table 1. Boric acid QTL mapped in the DSPR.
| QTL Name | LOD Score | Chr | Interval (cM)a | Interval (Mb)a | Genesb | Var Explc |
|---|---|---|---|---|---|---|
| pA.Q1 | 8.4 | 2L | 39.1–39.7 | 10.07–10.21 | 14 (1) | 4.5 |
| pA.Q2 | 8.5 | 3L | 39.0–40.8 | 12.86–13.89 | 83 (42) | 4.6 |
| pB.Q1 | 9.5 | 2R | 55.7–57.3 | 2.66–3.85 | 188 (32) | 5.0 |
| pB.Q2d | 8.6 | 2R | 68.6–68.8 | 9.56–9.67 | 5 (1) | 4.5 |
| pB.Q3d | 15.3 | 2R | 73.2–75.5 | 11.00–11.57 | 63 (11) | 7.9 |
| pB.Q4 | 8.5 | 3L | 32.1–32.7 | 10.29–10.45 | 3 (8) | 4.5 |
QTL, quantitative trait locus; LOD, logarithm of the odds; Chr, chromosome; Var Expl, variation explained.
The 2-LOD support interval of the QTL. Physical positions (in Mb) are based on release 5 of the D. melanogaster reference genome.
The number of protein-coding genes within the 2-LOD support interval, and (in parentheses) the number of other genes (e.g., miRNA genes). Data were harvested from FlyBase (Attrill et al. 2016) after converting QTL positions from release 5 (reported by DSPRqtl) to release 6 coordinates.
The proportion of the phenotypic variance due to each QTL. This value comes directly from the linear model used for mapping (Broman and Sen 2009, page 77).
These QTL overlap with the positions of caffeine resistance QTL previously identified in the pB panel of the DSPR. pB.Q2 and pB.Q3 mapped in the current study overlap with Q4 and Q5, respectively, as reported in Table 1 from Najarro et al. (2015).
There is no overlap in the QTL identified in the pA and pB populations. Indeed, in regions harboring QTL in one panel, LOD curves in the other panel are not close to the genome-wide threshold, showing no signature of corresponding allelic effects (Figure 3). Similarly, Najarro et al. (2015) found that 7/10 caffeine resistance QTL were mapped in just one DSPR population. Absent power concerns, and given there is similar trait heritability in both DSPR panels (above), these results imply there is heterogeneity in the trait genetic architecture between DSPR mapping panels. This is perhaps unsurprising since panels were initiated from small, largely nonoverlapping sets of founding lines (just one founder is shared between populations). Causative alleles present in one panel may be absent in the other, or the alleles may be present in both but at very different frequencies, affecting mapping power, and/or the alleles may have background-specific effects. Without identifying the precise causative sites it will be difficult to ascertain why the DSPR panels reveal different sets of QTL.
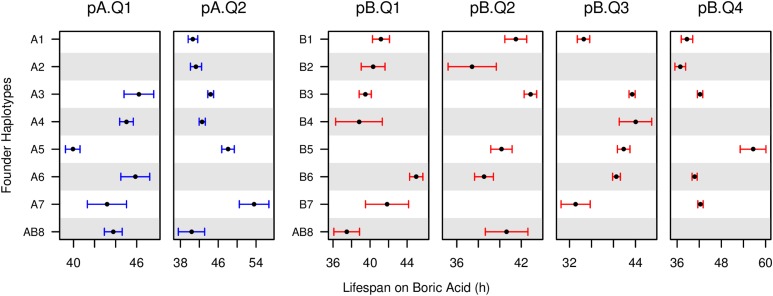
A feature of MPP approaches is that one can estimate the phenotypic effects of each founder haplotype at mapped QTL. The estimated effects at the six boric acid QTL do not clearly suggest that the QTL are each the result of a single biallelic variant (Figure 4). This observation is consistent with data from previous studies in the DSPR for a number of traits (Kislukhin et al. 2013; King et al. 2014a,b) and has been seen in studies using other MPPs, such as the mouse DO population (Gu et al. 2016), the rat NIH-HS population (Rat Genome Sequencing and Mapping Consortium et al. 2013), and in maize (Giraud et al. 2014). The strain effects shown in Figure 4 could result from the action of multiple, tightly-linked QTL in each region, or from allelic heterogeneity, where multiple functional alleles segregate at single causative genes, as is routinely observed in human Mendelian disease genes. The strain effects will be valuable for elucidating the causative loci in the future. For example, one might expect to see a correlation between the founder mean phenotypes at a QTL, and the level of expression of a causative gene in a relevant tissue among founders.
Figure 4.
Founder haplotype means at mapped boric acid resistance QTL. RILs can harbor any of the eight possible founder haplotypes at each position. Averaging the phenotypes of RILs carrying each founder haplotype describes their contribution to a QTL. Founder haplotype means (filled circles) and 1-SE (whiskers) are calculated using only RILs for which we confidently assign a founder genotype at the position (probability > 0.95). Means are plotted only when based on at least 10 such observations; Certain haplotypes can be nearly absent at particular loci due to drift and/or selection occurring during the maintenance of the synthetic populations that led to the DSPR (King et al. 2012b). DSPR, Drosophila Synthetic Population Resource; QTL, quantitative trait loci; RILs, Recombinant Inbred Lines.
Three of the QTL (pA.Q1, pB.Q2, and pB.Q4) are mapped to small genetic (0.27–0.61 cM) and physical (110–140 kb) intervals, and harbor 3–14 protein-coding genes (File S3 and Table 1). The interval for pA.Q1 harbors the gene Sur (Sulfonylurea receptor, FBgn0028675), which encodes an ABC transporter. This locus represents a strong candidate to carry functional variation impacting resistance to boric acid, since transporters are required to excrete the products of xenobiotic metabolism from cells (Xu et al. 2005). Notably, proper expression of Sur has been shown to be required for heart function (Akasaka et al. 2006), and heart-specific RNAi knockdown of Sur leads to reduced survival following injection with the flock house RNA virus (Croker et al. 2007). One of five protein-coding genes implicated by the pB.Q2 interval is fas (faint sausage, FBgn0000633), which when mutated alters the morphology of malpighian tubules (Jack and Myette 1999; Liu et al. 1999), structures critical for xenobiotic metabolism (Dow and Davies 2006; Yang et al. 2007). QTL pB.Q4 harbors a pair of ionotropic receptors, Ir67b (FBgn0036083) and Ir67c (FBgn0052058), a gene family involved in chemosensation (Benton et al. 2009; Stewart et al. 2015) but lacking any clear link to xenobiotic detoxification. However, the phenotyping assay we employed does not involve a check on the amount of media ingested by test animals. Our screen could therefore result in QTL that contribute to the propensity to ingest a toxic substance, and variation in chemosensory genes would be strong candidates to underlie such QTL.
The remaining three QTL (pA.Q2, pB.Q1, and pB.Q3) are more broadly mapped, implicating 63–188 protein-coding genes (File S3 and Table 1), making it more difficult to hypothesize plausible candidates for follow-up, functional testing. However, a search for members of those gene families known to be involved in xenobiotic metabolism (see Materials and Methods) revealed four P450s among the 188 protein-coding genes residing within the pB.Q1 interval; Cyp6a2 (FBgn0000473), Cyp6u1 (FBgn0033121), Cyp9b1 (FBgn0015038), and Cyp9b2 (FBgn0015039). Expression of Cyp6a2 has been positively associated with resistance to the insecticide DDT (Wan et al. 2014), but other than being members of the P450 family no specific evidence links the other three genes to roles in xenobiotic resistance (Attrill et al. 2016). Chung et al. (2009) used in situ hybridization against the majority of D. melanogaster P450s, finding expression of Cyp6a2 and Cyp9b2 in the third instar larval midgut and malpighian tubules. Cyp6u1 was expressed in gonadal tissue rather than in the gut, while no expression of Cyp9b1 was detected. Thus, Cyp6a2 and Cyp9b2 represent the strongest candidates within pB.Q1 to contribute to boric acid resistance.
Cyp9b2 RNAi reduces boric acid resistance
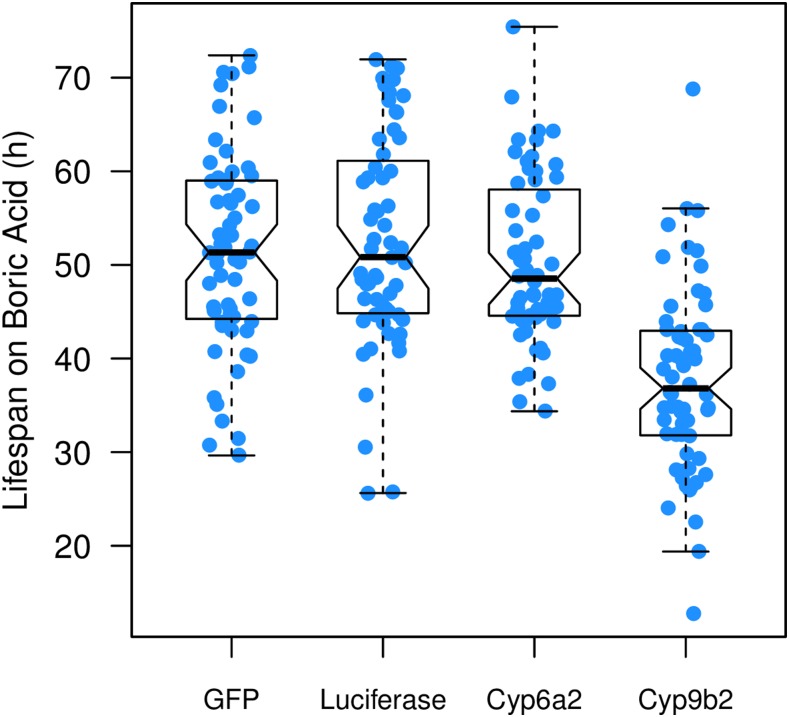
To test for any functional effects of genes Cyp6a2 and Cyp9b2 on boric acid resistance we employed RNAi, knocking down each gene ubiquitously, and comparing to control genotypes expressing the markers GFP or luciferase (Figure 5). Knockdown of Cyp6a2 yielded no change in phenotype compared to controls (Welch’s t-tests, P > 0.2), whereas reduction in Cyp9b2 expression markedly lowered resistance to boric acid (P < 10−11), suggesting that Cyp9b2 is functionally important for resistance.
Figure 5.
RNAi knockdown of candidate cytochrome P450 genes. We expressed Gal4 in all cells at all timepoints under the control of the Actin 5C promoter, driving GFP and luciferase as controls, and knocking down expression of the genes Cyp6a2 and Cyp9b2 (both residing under DSPR QTL pB.Q1). Raw phenotypes (59–64 animals per genotype) are presented as blue filled circles, along with a standard boxplot (generated using the R boxplot command). The Cyp9b2 knockdown genotype shows a significant reduction in resistance compared to all other genotypes (Welch’s t-tests, P < 10−11). All other genotype pairs are not significantly different (P > 0.2). DSPR, Drosophila Synthetic Population Resource; GFP, green fluorescent protein; QTL, quantitative trait loci; RNAi, RNA interference.
Clearly, experimental knockdown of gene expression in all cells and at all timepoints may not reflect the effect of any natural Cyp9b2 allele segregating within the DSPR, and positive RNAi knockdown is not strong evidence for the existence of such allelic variation. Preadult knockdown of Cyp9b2 could additionally negatively impact adult fitness, indirectly resulting in reduced lifespan during drug exposure. Furthermore, RNAi can lead to nonspecific effects, affecting the expression of genes other than the target (Ma et al. 2006), potentially falsely implicating a given gene in the control of a phenotype. Notwithstanding these caveats, Cyp9b2 represents a good target for future analyses seeking to identify causative variants impacting boric acid resistance. We note that we cannot rule out the possibility that genes other than Cyp9b2 could additionally be responsible for the QTL effect observed, particularly since pB.Q1 is relatively broad (Table 1).
Overlap among xenobiotic resistance QTL in the DSPR
Two of the six boric acid QTL, pB.Q2 and pB.Q3, overlap with QTL contributing to variation in caffeine resistance that we previously mapped in the DSPR pB panel (Table 1 in Najarro et al. 2015). Given the relatively short intervals implicated by mapped QTL, the nearly identical phenotyping pipelines employed in each study, and the phenotypic correlations observed between the two traits (Figure 2), it is conceivable that resistance to both drugs is associated with the same loci in these intervals. A correlation between the strain effects estimated at each overlapping pair of QTL would provide additional support for this contention. However, while the correlations are positive and high, since only 6/8 founders are present at these locations (see Figure 4), they are not significant (Pearson’s r = 0.53–0.70, P > 0.12 in each case).
Assuming the causative variants are pleiotropic, since boric acid QTL pB.Q2 resides completely within caffeine resistance QTL Q4, this would reduce the number of protein-coding genes implicated in this latter QTL from 26 (see Najarro et al. 2015) to 5 (File S3 and Table 1). The intervals for boric acid QTL pB.Q3 and caffeine QTL Q5 also overlap, and if the QTL have the same genetic cause the size of the region containing any pleiotropic variant(s) would be reduced from 570 kb (boric acid pB.Q3, Table 1) and 650 kb (caffeine QTL Q5) to 460 kb, a region containing 47 protein-coding genes.
Simple overlap of QTL does not provide unambiguous evidence for pleiotropy, particularly when each QTL implicates multiple genes as we see here. Overlap may be purely stochastic, with different, very closely-linked loci within the overlapping region contributing independent effects to the different phenotypes. Functional tests of plausible candidate genes within overlapping QTL regions can directly address whether some of the genetic control of resistance to boric acid and caffeine is shared.
GWAS for boric acid resistance loci
MPP mapping designs have considerable power to detect relatively short regions of the genome containing alleles influencing complex trait variation (Valdar et al. 2006a; King et al. 2012a; Gatti et al. 2014). This power derives from the fact that in an MPP one is testing the effect of a local haplotype on phenotype, in effect integrating over any and all causative variants in that region in a single test. A caveat with this approach is that mapping resolution is necessarily diminished relative to a population-based association study. In such a study LD (linkage disequilibrium) likely extends a relatively short distance, and any associated variant is likely to be the causative site, or at worst be very close to the causative site (Risch and Merikangas 1996). In the DSPR, within fragments of the genome not subject to crossing over, many variants are in perfect LD, and all tag the exact same haplotype(s). This phenomenon can be seen in QTL-specific association scans. In these analyses, we use the variants identified in the founders together with the estimated mosaic haplotype structure of the RILs (King et al. 2012b) to infer the alleles present in each RIL, and carry out a series of single marker association tests. Figure S1 shows that many SNPs within each QTL interval are strongly associated with phenotype, and while these tests might allow variants to be ranked, they do not confidently point to highly likely candidate causative sites.
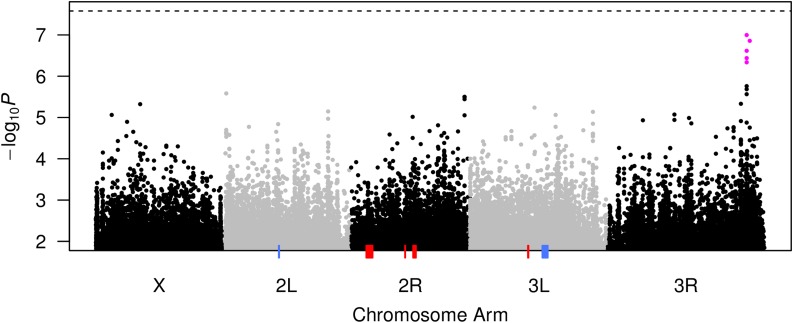
To attempt to resolve nucleotide sites contributing to variation in boric acid resistance we turned to the DGRP (Mackay et al. 2012; Huang et al. 2014), a series of inbred strains of D. melanogaster derived from a single collection site, and subjected to short-read sequencing. The distribution of phenotypes across the 163 strains assayed, and the heritability of the phenotype in the DGRP is comparable to that seen in the DSPR (Figure 1). Following GWAS analysis, no variant survives a conservative genome-wide 5% Bonferroni multiple testing threshold (Figure 6, horizontal dashed line).
Figure 6.
Manhattan plot of DGRP-based boric acid resistance GWAS. Association testing was carried out at 1.9 million common variants, accounting for variation across strains in relatedness, Wolbachia infection status, and chromosomal inversion genotype. The strength of each association is presented as a −log10(P-value) with variants on the major chromosome arms delineated by alternating black/gray symbols. The set of five associations toward the right end of chromosome 3R (magenta symbols) survive a genome-wide 20% FDR threshold. The horizontal dashed line at the top of the plot represents a 5% genome-wide Bonferroni correction for multiple tests (P = 2.6 × 10−8). The blue/red colored rectangles along the x-axis represent the physical positions of the six QTL mapped in the DSPR (left-to-right; pA.Q1, pB.Q1, pB.Q2, pB.Q3, pB.Q4, and pA.Q2). DGRP, Drosophila Genetic Reference Panel; DSPR, Drosophila Synthetic Population Resource; FDR, False Discovery Rate; GWAS, genome-wide association study; QTL, quantitative trait loci.
Identifying no or very few variants surviving a strict Bonferroni threshold is common in GWAS studies employing the DGRP (e.g., Arya et al. 2015; Dembeck et al. 2015; Carbone et al. 2016; Akhund-Zade et al. 2017), and such analyses commonly report interesting associations as those surviving an arbitrary threshold of P < 10−5. In the present study, this threshold corresponds to an FDR of 79% (File S4), likely providing limited value in elucidating causative genes/variants. Instead, we explored several more stringent FDR thresholds, identifying five variants toward the end of chromosome 3R that survive a genome-wide 20% FDR threshold (Figure 6, magenta symbols; File S4). These five are the only variants that survive a genome-wide 50% FDR threshold, and none are present within those QTL intervals implicated by the DSPR (Figure 6).
One of the 20% FDR variants (3R:24708824 in release 5 of the D. melanogaster reference genome) is present within an intron of gene CG33203 (FBgn0053203), for which there is limited information on FlyBase (Attrill et al. 2016). Approximately 5–7 kb downstream is a cluster of three variants (3R:24714040, 3R:24716025, and 3R:24716095) residing within Doa (Darkener of apricot, FBgn0265998). This locus encodes a serine/threonine protein kinase, and structural mutations in the gene increase resistance to paraquat-induced oxidative stress (James et al. 2009). These four variants are present within a 7 kb section of the genome, show relatively high levels of LD (r2 = 0.55–0.88), and thus may all be proxies for a single causative variant. The fifth associated variant is > 500 kb further downstream (3R: 25288112), shows limited LD with the four upstream variants (r2 = 0.07–0.12), and is present within the Ptp99A gene (FBgn0004369). Ubiquitous RNAi of this protein tyrosine phosphatase has been shown to exhibit increased levels of tolerance to ethanol and benzyl alcohol (Ghezzi et al. 2013).
Validation of DGRP “hits” (meaning sites with association statistics significant at P < 10−5) via RNAi of genes containing those hits, or analysis of mutations in such genes, is frequently quite successful. This suggests that the sets of genes containing sites yielding small P-values in the GWAS may be enriched for those with true effects on phenotype. For instance, four recent DGRP studies showed that 64–88% of genes containing hits that were functionally tested showed a significant effect on phenotype (Arya et al. 2015; Carbone et al. 2016; Vonesch et al. 2016; Akhund-Zade et al. 2017). Notably, Vonesch et al. (2016) additionally showed that the rate of functional validation for GWAS hits was significantly higher than that for a set of random genes, again suggesting that despite the modest level of statistical support for DGRP-based GWAS hits, as a class they appear to be enriched for true positives. Based on these results, future functional testing targeted at Doa and Ptp99A may help to confirm any effects of these loci on boric acid resistance.
Limited overlap between loci implicated by the DSPR and DGRP
A key question is whether there is any signature of associated variants in the DGRP within DSPR-derived QTL intervals, since these would help to implicate nucleotide sites in the control of trait variation. We extracted association tests carried out at variants within each QTL region, and explored various QTL-specific multiple testing thresholds. No variants survive QTL-specific 5% Bonferroni or 10% FDR thresholds. At 20% FDR, 5/6 QTL still show no associated variants, but 4/4415 variants tested within pB.Q4 do survive. All four of these sites are within 90 bases of each other (3L:10341876–10341966) in CR46006 (FBgn0267668), one of the 11 genes residing within the pB.Q4 interval. No information is currently available regarding the function of this nonprotein-coding gene (Attrill et al. 2016), but given the precision with which pB.Q4 is mapped in the DSPR (Table 1), and the presence of these suggestive associations in the DGRP, future testing of the effects of this gene on boric acid resistance are warranted.
Ultimately, the level of overlap between the DSPR and DGRP is disappointingly low, consistent with our previous work on caffeine resistance (Najarro et al. 2015). Combining results over the two panels does not strongly improve the ability to identify causative nucleotide sites underlying trait variation. Part of the difficulty is certainly the low power in the DGRP (Turner et al. 2013; Long et al. 2014). With 163 lines, the power to identify a common (50% minor allele frequency) variant contributing 5% to the total phenotypic variation, which broadly corresponds to the estimated effects of our mapped QTL (Table 1), is ∼6% at a genome-wide 5% Bonferroni threshold (based on testing close to 1.9 million sites). QTL intervals contain 2615–18754 tested variants, and at a more liberal threshold of P < 10−5 appropriate for a QTL-centric association study, power remains at a relatively low ∼39%. Power will be lower still if DSPR QTL are multiallelic, as implied by Figure 4 and by a range of MPP studies indicating that the loci contributing to complex traits are commonly multiallelic (Kislukhin et al. 2013; Rat Genome Sequencing and Mapping Consortium et al. 2013; Giraud et al. 2014; King et al. 2014a,b; Gu et al. 2016). Multiallelic QTL are the result of the action of many, likely rare alleles, with independent small effects on phenotype that will be very difficult to detect individually using a traditional GWAS framework (Pritchard 2001; Thornton et al. 2013).
A further concern with the GWAS approach is that one can only detect the effects of variants that are directly genotyped, or those that are in strong LD with a genotyped variant. Any other variants are effectively invisible to the approach. This is in contrast to MPP-based mapping, where methods test for the effects of short haplotypes on phenotype and are agnostic as to the molecular nature of the causative variants. In this respect, DGRP genotypes are currently based on short-read sequencing data, which is likely to miss a range of structural variants (Pendleton et al. 2015). Copy number variants and transposable element insertions have been previously implicated in xenobiotic resistance (e.g., Daborn et al. 2002; Chung et al. 2007; Schmidt et al. 2010). So, to the extent that this class of variants are important contributors to boric acid and caffeine resistance, are absent from the DGRP variant catalog, and are not “tagged” by genotyped sites due to the very low LD in the DGRP (Mackay et al. 2012), and in D. melanogaster more generally (e.g., Macdonald et al. 2005), the ability to find resistance loci in the DGRP may be undermined.
Finally, despite the similar trait distributions (Figure 1), the genetic architecture of variation in boric acid resistance may show heterogeneity between the DGRP and DSPR populations. The DSPR is constructed from a worldwide collection of founding strains that lack P transposable elements, and that have been maintained for decades in labs since their collection (King et al. 2012b), whereas the DGRP is constructed from lines harboring P-elements derived more recently from a single location in North America (Mackay et al. 2012). We might expect such populations to differ in the genetic control of trait variation. Indeed, hundreds of thousands of SNPs are unique to one of the two mapping populations (Najarro et al. 2015; King and Long 2017), so any phenotypic effects they confer cannot be replicated across mapping populations. Even for those variants that are shared between the DSPR and DGRP, allele frequencies are not strongly correlated between the panels (King and Long 2017). Thus, even if a causative variant is present in both the DSPR and DGRP, different allele frequencies could markedly impact the power to find it in each panel. For instance, GWAS methods are highly underpowered to detect very rare causative alleles (Pritchard 2001), so a causative variant that is truly rare in the population but was captured in one of the DSPR founders and led to a mapped QTL would be hard to replicate in the DGRP. Finally, shared variants may be background-dependent, conferring detectable effects on phenotype only in one of the two mapping panels. Epistasis as an explanation for the failure to replicate variants contributing to complex trait variation across mapping populations has been proposed previously (Huang et al. 2012), although other phenomena, such as mapping power and Beavis effects (King and Long 2017), are likely important contributors to such lack of validation across mapping panels.
There is an array of biological and technical factors that could lead to differences between the population-based DGRP GWAS approach and the MPP-based DSPR linkage mapping approach. However, a principal difference is that the strains from which the populations are derived are distinct. Deriving recombinant MPP mapping populations from subsets of the DGRP strains can control for the difference in mapping methodology, may allow a more effective contrast between the DGRP and DSPR genotypes, and permit an exploration of the heterogeneity in trait architecture across populations.
Conclusions
In this study, we mapped several loci that impact resistance to the xenobiotic boric acid, resolving these modest-effect QTL to regions implicating relatively small numbers of genes. We highlight plausible candidate genes for some mapped QTL that can be targets of future studies to elucidate the mechanisms of boric acid toxicity in insects. Perhaps surprisingly, only two of the QTL implicate members of gene families known to be responsible for metabolism of xenobiotics: one QTL contains an ABC transporter (Sur) and a second contains multiple P450s. We confirm that one of these P450s, Cyp9b2, affects boric acid resistance using RNAi knockdown, although we recognize that while RNAi is a useful and efficient tool to implicate a gene in the control of a phenotype, a positive result is a weak standard of evidence that the gene harbors variants that contribute to natural trait variation. It is possible that those QTL lacking recognized detoxification genes are generated by variants that lead to morphological or behavioral differences across strains that impact xenobiotic resistance indirectly, rather than by directly affecting boric acid metabolism. A benefit of large, inbred reference panels is that one can pursue more detailed phenotypic characterization of phenotypically-extreme strains, collect genomics data at a range of levels (e.g., transcriptomics and metabolomics), use a systems genetics approach to generate a more granular picture of phenotypic variation, and help identify the variants that produce it.
Our attempt to marry DSPR-based QTL mapping with a DGRP-based association study to identify putative causative variants within QTL was largely unsuccessful, as it was in our previous effort (Najarro et al. 2015). While the approach has appeal—broadly map a QTL in the DSPR and fine map that QTL to the nucleotide level with the DGRP—its success is predicated both on the genetic architecture of a trait being fairly consistent across mapping panels, and on having power in both panels to detect genes segregating for alleles with fairly subtle effects on phenotype that may be at relatively low frequency. At the level of both SNP allelic variation and transposable elements, the DGRP and DSPR show modest overlap (Cridland et al. 2013; King and Long 2017), suggesting genetic architecture for a trait may often not be the same over panels. In addition, straightforward power calculations (Long et al. 2014) demonstrate that QTL-centric association scans employing 200 DGRP strains will typically be insufficient to identify intermediate (or lower) frequency alleles with small effects on phenotype. As shown by a wealth of human genetics work, much larger and therefore more powerful association studies employing thousands of genotypes are necessary to pinpoint causative loci. And if the causative variants, their allele frequencies, or their effects on phenotype are not recapitulated in different populations, fully elucidating trait architecture will entail mapping in a number of separate populations.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041418/-/DC1.
Acknowledgments
We thank Carla Burford, Kristen Cloud-Richardson, Theresa Melhem, and Brittny Smith for help acquiring phenotypic data, and Rob Unckless for helpful comments on an earlier version of this manuscript. We thank the TRiP [Transgenic RNAi Project, National Institutes of Health (NIH) R01-GM084947] for generating the transgenic RNA interference (RNAi) fly stocks used in this study. Drosophila Genetic Reference Panel and RNAi stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40-OD018537). This project was supported by NIH grant R01-OD010974 to S.J.M. and Anthony D. Long (University of California, Irvine).
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Akasaka T., Klinedinst S., Ocorr K., Bustamante E. L., Kim S. K., et al. , 2006. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc. Natl. Acad. Sci. USA 103: 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhund-Zade J., Bergland A. O., Crowe S. O., Unckless R. L., 2017. The genetic basis of natural variation in Drosophila (Diptera: Drosophilidae) virgin egg retention. J. Insect Sci. 17: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G. H., Magwire M. M., Huang W., Serrano-Negron Y. L., Mackay T. F., et al. , 2015. The genetic basis for variation in olfactory behavior in Drosophila melanogaster. Chem. Senses 40: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., et al. , 2016. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44: D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B., 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer Dordrecht, New York. [Google Scholar]
- Buyukguzel E., Buyukguzel K., Snela M., Erdem M., Radtke K., et al. , 2013. Effect of boric acid on antioxidant enzyme activity, lipid peroxidation, and ultrastructure of midgut and fat body of Galleria mellonella. Cell Biol. Toxicol. 29: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M. A., Yamamoto A., Huang W., Lyman R. A., Meadors T. B., et al. , 2016. Genetic architecture of natural variation in visual senescence in Drosophila. Proc. Natl. Acad. Sci. USA 113: E6620–E6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo R., Gibson G., 2002. Unusual genetic architecture of natural variation affecting drug resistance in Drosophila melanogaster. Genet. Res. 80: 205–213. [DOI] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., Ffrench-Constant R. H., et al. , 2007. Cis-regulatory elements in the accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Sztal T., Pasricha S., Sridhar M., Batterham P., et al. , 2009. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc. Natl. Acad. Sci. USA 106: 5731–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Gatti D. M., Munger S. C., Svenson K. L., 2012. The diversity outbred mouse population. Mamm. Genome 23: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Macdonald S. J., Long A. D., Thornton K. R., 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol. Biol. Evol. 30: 2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker B., Crozat K., Berger M., Xia Y., Sovath S., et al. , 2007. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nat. Genet. 39: 1453–1460. [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Yen J. L., Bogwitz M. R., Le Goff G., Feil E., et al. , 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- da Silva Cruz A., da Silva-Zacarin E. C., Bueno O. C., Malaspina O., 2010. Morphological alterations induced by boric acid and fipronil in the midgut of worker honeybee (Apis mellifera L.) larvae: morphological alterations in the midgut of A. mellifera. Cell Biol. Toxicol. 26: 165–176. [DOI] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Magwire M. M., Lawrence F., Lyman R. F., et al. , 2015. Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genet. 11: e1005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow J. A., Davies S. A., 2006. The Malpighian tubule: rapid insights from post-genomic biology. J. Insect Physiol. 52: 365–378. [DOI] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 4: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A., Krishnan H. R., Lew L., Prado F. J., III, Ong D. S., et al. , 2013. Alcohol-induced histone acetylation reveals a gene network involved in alcohol tolerance. PLoS Genet. 9: e1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud H., Lehermeier C., Bauer E., Falque M., Segura V., et al. , 2014. Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the flint and dent heterotic groups of maize. Genetics 198: 1717–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning J. I., 2007. How do predators cope with chemically defended foods? Biol. Bull. 213: 252–266. [DOI] [PubMed] [Google Scholar]
- Gu T., Gatti D. M., Srivastava A., Snyder E. M., Raghupathy N., et al. , 2016. Genetic architectures of quantitative variation in RNA editing pathways. Genetics 202: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill C. A., Reeves G. A., Macdonald S. J., 2016. Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population. BMC Genet. 17: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Richards S., Carbone M. A., Zhu D., Anholt R. R., et al. , 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA 109: 15553–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrsl P., Buyukguzel E., Buyukguzel K., 2007. The effects of boric acid-induced oxidative stress on antioxidant enzymes and survivorship in Galleria mellonella. Arch. Insect Biochem. Physiol. 66: 23–31. [DOI] [PubMed] [Google Scholar]
- Jack J., Myette G., 1999. Mutations that alter the morphology of the malpighian tubules in Drosophila. Dev. Genes Evol. 209: 546–554. [DOI] [PubMed] [Google Scholar]
- James B. P., Staatz W. D., Wilkinson S. T., Meuillet E., Powis G., 2009. Superoxide dismutase is regulated by LAMMER kinase in Drosophila and human cells. Free Radic. Biol. Med. 46: 821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., 2015. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22: 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Long A. D., 2017. The Beavis effect in next-generation mapping panels in Drosophila melanogaster. G3 (Bethesda) 7: 1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Macdonald S. J., Long A. D., 2012a Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics 191: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012b Genetic dissection of a model complex trait using the Drosophila synthetic population resource. Genome Res. 22: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Kislukhin G., Walters K. N., Long A. D., 2014a Using Drosophila melanogaster to identify chemotherapy toxicity genes. Genetics 198: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Sanderson B. J., McNeil C. L., Long A. D., Macdonald S. J., 2014b Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 10: e1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislukhin G., King E. G., Walters K. N., Macdonald S. J., Long A. D., 2013. The genetic architecture of methotrexate toxicity is similar in Drosophila melanogaster and humans. G3 3: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz J. H., Moss J. I., 1996. Oral toxicity of a boric acid - sucrose water bait to Florida carpenter ants (Hymenoptera: Formicidae). J. Entomol. Sci. 31: 9–12. [Google Scholar]
- Klotz J. H., Oi D. H., Vail K. M., Williams D. F., 1996. Laboratory evaluation of a boric acid liquid bait on colonies of Tapinoma melanocephalum Argentine ants and Pharaoh ants (Hymenoptera: Formicidae). J. Econ. Entomol. 89: 673–677. [Google Scholar]
- Klotz J. H., Amrhein C., McDaniel S., Rust M. K., Reierson D. A., 2002. Assimilation and toxicity of boron in the argentine ant (Hymenoptera: Formicidae). J. Entomol. Sci. 37: 193–199. [Google Scholar]
- Li X., Schuler M. A., Berenbaum M. R., 2007. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52: 231–253. [DOI] [PubMed] [Google Scholar]
- Liu X., Kiss I., Lengyel J. A., 1999. Identification of genes controlling malpighian tubule and other epithelial morphogenesis in Drosophila melanogaster. Genetics 151: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Macdonald S. J., King E. G., 2014. Dissecting complex traits using the Drosophila synthetic population resource. Trends Genet. 30: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L., Beachy P. A., 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363. [DOI] [PubMed] [Google Scholar]
- Macdonald S. J., Pastinen T., Long A. D., 2005. The effect of polymorphisms in the Enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics 171: 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriage T. N., King E. G., Long A. D., Macdonald S. J., 2014. Fine-mapping nicotine resistance loci in Drosophila using a multiparent advanced generation inter-cross population. Genetics 198: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., et al. , 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43: D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithofer A., Boland W., 2012. Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63: 431–450. [DOI] [PubMed] [Google Scholar]
- Najarro M. A., Hackett J. L., Smith B. R., Highfill C. A., King E. G., et al. , 2015. Identifying loci contributing to natural variation in xenobiotic resistance in Drosophila. PLoS Genet. 11: e1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton M., Sebra R., Pang A. W., Ummat A., Franzen O., et al. , 2015. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., et al. , 2015. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry T., Batterham P., Daborn P. J., 2011. The biology of insecticidal activity and resistance. Insect Biochem. Mol. Biol. 41: 411–422. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J., D. Bates, S. DebRoy, D. Sarkar, and R Core Team, 2011 nlme: linear and nonlinear mixed effects models. R package version 3.1–101. Available at: https://CRAN.R-project.org/package=nlme. Accessed: January 19, 2016.
- Pritchard J. K., 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing and Mapping Consortium. Baud A., Hermsen R., Guryev V., Stridh P., et al. , 2013. Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat. Genet. 45: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., Merikangas K., 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Koh T. W., Ghosh A. C., Carlson J. R., 2015. Candidate ionotropic taste receptors in the Drosophila larva. Proc. Natl. Acad. Sci. USA 112: 4195–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida S., Silva-Zacarin E. C., Decio P., Malaspina O., Bueno F. C., et al. , 2010. Toxicological and histopathological effects of boric acid on Atta sexdens rubropilosa (Hymenoptera: Formicidae) workers. J. Econ. Entomol. 103: 676–690. [DOI] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the mouse diversity outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. R., Foran A. J., Long A. D., 2013. Properties and modeling of GWAS when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS Genet. 9: e1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A., 2012. Ten years of the collaborative cross. G3 2: 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Miller P. M., Cochrane V. A., 2013. Combining genome-wide methods to investigate the genetic complexity of courtship song variation in Drosophila melanogaster. Mol. Biol. Evol. 30: 2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R., 2006a Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006b Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887. [DOI] [PubMed] [Google Scholar]
- Vonesch S. C., Lamparter D., Mackay T. F., Bergmann S., Hafen E., 2016. Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12: e1005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H., Liu Y., Li M., Zhu S., Li X., et al. , 2014. Nrf2/Maf-binding-site-containing functional Cyp6a2 allele is associated with DDT resistance in Drosophila melanogaster. Pest Manag. Sci. 70: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Xu C., Li C. Y., Kong A. N., 2005. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 28: 249–268. [DOI] [PubMed] [Google Scholar]
- Yang J., McCart C., Woods D. J., Terhzaz S., Greenwood K. G., et al. , 2007. A Drosophila systems approach to xenobiotic metabolism. Physiol. Genomics 30: 223–231. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All individual-level phenotypes are provided in File S1. Raw activity monitor datafiles, along with R scripts to generate individual- and strain-level phenotypes for the DSPR, have been uploaded as a Dryad data package (doi: 10.5061/dryad.3v55m). DSPR QTL mapping output, including positions and LOD scores, is provided in File S2. Lists of genes currently annotated within mapped DSPR QTL are provided in File S3. Positions and association statistics for all nominally significant (P < 0.05) variants tested in the DGRP GWAS are provided in File S4. DSPR RILs can be requested from FlyRILs.org.