Abstract
The oligoadenylate-synthetase (Oas) gene locus provides innate immune resistance to virus infection. In mouse models, variation in the Oas1b gene influences host susceptibility to flavivirus infection. However, the impact of Oas variation on overall innate immune programming and global gene expression among tissues and in different genetic backgrounds has not been defined. We examined how Oas1b acts in spleen and brain tissue to limit West Nile virus (WNV) susceptibility and disease across a range of genetic backgrounds. The laboratory founder strains of the mouse Collaborative Cross (CC) (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/HlLtJ) all encode a truncated, defective Oas1b, whereas the three wild-derived inbred founder strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ) encode a full-length OAS1B protein. We assessed disease profiles and transcriptional signatures of F1 hybrids derived from these founder strains. F1 hybrids included wild-type Oas1b (F/F), homozygous null Oas1b (N/N), and heterozygous offspring of both parental combinations (F/N and N/F). These mice were challenged with WNV, and brain and spleen samples were harvested for global gene expression analysis. We found that the Oas1b haplotype played a role in WNV susceptibility and disease metrics, but the presence of a functional Oas1b allele in heterozygous offspring did not absolutely predict protection against disease. Our results indicate that Oas1b status as wild-type or truncated, and overall Oas1b gene dosage, link with novel innate immune gene signatures that impact specific biological pathways for the control of flavivirus infection and immunity through both Oas1b-dependent and independent processes.
Keywords: Oas, flavivirus, viral infection, innate immunity, multiparental populations, Multi-parent Advanced Generation Inter-Cross (MAGIC), MPP
WNV is a mosquito transmitted flavivirus that emerged from Africa and is now endemic within Asia, middle Eastern Europe, Australia, and the Americas (Courtney et al. 2012). WNV is an enveloped virus carrying a genome of a positive sense, single-stranded RNA of roughly 11,000 nucleotides in length. While only 20% of individuals infected with WNV develop symptoms (www.cdc.gov/westnile), symptomatic individuals can develop clinical illness ranging from West Nile fever to encephalitis or meningitis linked with severe inflammation in the brain and spinal cord, and leading to death (Graham et al. 2016). WNV is among a group of emerging flaviviruses including dengue virus, Zika Virus, and Japanese encephalitis virus that are global health concerns. Understanding their molecular pathogenesis is a major step toward a therapeutic or adjunctive therapy.
During viral infection, the infected cell senses viral replication products, including viral nucleic acid, as foreign, nonself macromolecules through the actions of pathogen recognition receptors (PRRs), including RIG-I-like receptors (RLRs), Toll-like receptors (TLRs), and other PRRs (Suthar et al. 2013). In particular, flaviviruses including WNV are sensed by the RLRs through binding of viral RNA (Loo and Gale 2008, 2011; Errett et al. 2013). RNA binding induces RLR activation and interaction with downstream signaling proteins that activate transcription factors IRF3, IRF7, and NF-κB to drive innate immune gene expression and the production of type 1 and III interferons (IFNs). IFN is secreted by the infected cell to signal through the IFN receptors on both the infected cell and on bystander cells, to drive the expression of hundreds of IFN-stimulated genes (ISGs) across the local tissue (Loo and Gale 2011). Innate immune genes and ISGs have antiviral and immune modulatory activity to limit viral replication and spread such that their induction and function are essential for the control of WNV infection and immunity (Daffis et al. 2009; Suthar et al. 2010; Lazear et al. 2013; Lazear and Diamond 2015).
The Oas genes including Oas1b are ISGs whose expression is induced by IFN in most cell types (Elkhateeb et al. 2016, Choi et al. 2015). In humans, the Oas family contains Oas1, Oas2, Oas3, and Oas-like (OasL) genes. Oas2 and Oas3 genes have high human–mouse sequence similarity with a 1:1 orthogonal copy. The mouse Oas gene cluster is located on chromosome 5 and includes Oas1, Oas2, Oas3, and OasL genes. The Oas1 gene has eight orthogonal copies (Oas1a, Oas1b, Oas1c, Oas1d, Oas1e, Oas1f, Oas1g, and Oas1h) in mouse compared to one copy (Oas1) in humans on chromosome 12. These eight mouse Oas1 genes are the result of gene duplications, rearrangements, and other evolutionary processes [Choi et al. 2015, for more details of such genome regions in the CC, see Morgan et al. (2017)]. Most members of the Oas gene family encode 2′-5′ oligoadenylate (2-5A) enzymatic activity that catalyzes adenosine into 2′-5′-linked oligonucleotides referred to as oligoadenylates or 2-5A (Kristiansen et al. 2011). 2-5A serves as a ligand to bind and activate ribonuclease L (RNaseL), a latent endoribonuclease. When activated during viral infection, RNaseL serves to suppress viral replication by nucleolytic targeting viral and host RNAs. RNaseL products of RNA degradation can also serve as activator ligands of the RLRs, thus stimulating and amplifying further rounds of innate immune signaling (Malathi et al. 2007, Siddiqui et al. 2012, Drappier et al. 2015). Among the Oas family members, Oas1b lacks 2-5A activity, revealing that the antiviral function of Oas1b is unique and independent of RNaseL activation for controlling WNV infection (Scherbik et al. 2006; Courtney et al. 2012; Elbahesh et al. 2011). Oas1b is found truncated in classical inbred mouse strains due the presence of a stop codon/nonsense mutation within the mRNA (Elbahesh et al. 2011). When otherwise expressed as a full-length protein, OAS1b provides functional antiviral activity against WNV (Elbahesh et al. 2011). Recent studies found that OAS1b has a C-terminus domain targeting the endoplasmic reticulum with binding partner protein ABCF3 assisting in antiviral activity (Courtney et al. 2012). The same study showed that full-length (nontruncated) Oas1b inhibited OAS1a activity and lowered 2-5A levels in a dose-dependent manner in vivo. Polymorphisms in Oas1b are also thought to impact the overall host immune response to virus infection, but how Oas1b operates in immune regulation is not defined (Bigham et al. 2011).
In vivo studies assessing Oas1b function during WNV infection and immunity have been largely conducted with traditional inbred mouse strains, thus limiting analyses of the natural genetic variation of Oas1b found within populations. Genetic variation of immune genes plays an important role in infection outcome and contributes to host susceptibility and resistance (Ferris et al. 2013). As noted above, most wild derived inbred mouse strains produce a full-length Oas1b protein, whereas most if not all inbred mouse strains, including the strain of the reference genome, C57BL/6J, have a premature stop codon in Oas1b that links with susceptibility to WNV infection (Elbahesh et al. 2011; Mashimo et al. 2002, see Figure 6, Supplemental Material, Figure S1, and Table 1 and Table 2). To capture the genetic diversity of Oas1b for functional studies of WNV susceptibility, we evaluated its expression and linkage with WNV infection outcome and innate immune transcriptional signatures in the CC mouse population (Iraqi et al. 2012).
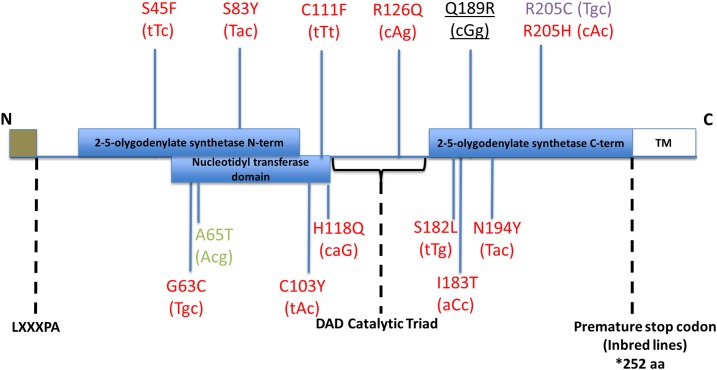
Figure 6.
Oas1b protein. SNPs that impact aa sequence identified over annotated (Uniprot) functional domains. Changes in aa are coded relative to the reference sequence, which are identical for the five classical inbred founder strains, and changes are assigned to strains (PWK/PhJ = red; WSB/EiJ = purple; WSB/EiJ and Cast/EiJ = green; and PWK/PhJ, WSB/EiJ, and CAST/EiJ = underlined black). aa, amino acid; DAD (Motif); SNP, single nucleotide polymorphism; TM, transmembrane domain.
Table 1. QTL Loci.
| Innate Immune Activation QTL | ||
|---|---|---|
| Chromosome and Region | Phenotype | Candidate Genes |
| Chr 5: 120–123 Mb | Weight loss (D12) | Oas1b* (see Table S1 for a complete list) |
The genomic region identified in chromosome 5 where Oas1b was found. The QTL ranged between 2 and 3 Mb and includes Oas1b. QTL, quantitative trait locus; Chr, chromosome; D12, day 12 postinfection.
Table 2. Table of F1s screened for transcriptomics analysis.
| Cross | OASlb Status | Phenotype | Oaslb Origin | Dam | Sire |
|---|---|---|---|---|---|
| CC017×CC004 | F/F | Asymptomatic | CAST/WSB | Functional | Functional |
| CC019×CC004 | F/F | Asymptomatic | CAST/WSB | Functional | Functional |
| CC009×CC040 | N/N | Symptomatic | N/A | Null | Null |
| CC006×CC007 | N/N | Symptomatic | N/A | Null | Null |
| CC055×CC028 | N/F | Symptomatic | PWK | Null | Functional |
| CC003×CC062 | F/N | Asymptomatic | CAST/WSB | Functional | Null |
| CC030×CC061 | F/N | Asymptomatic | CAST/WSB | Functional | Null |
The F1s used in analysis along with their Oas1b status, and Oas1b founder origin. Gene expression changes were assessed against mock-infected congenic control mice for each cross. F/F, functional Oas1b alleles from Dam and Sire; N/N, homozygous null Oas1b; N/A, not applicable; N/F, heterozygous RIX lines where only the Sire has a functional Oas1b allele; F/N, heterozygous RIX lines where only the Dam has a functional Oas1b allele.
The CC is a MPP, which was generated to improve systems genetic research using the mouse as a model organism. The CC resource is composed of ∼70 independently bred, octo-parental recombinant inbred mouse strains (Srivastava et al. 2017), out of several hundred lines started (Shorter et al. 2017). Each CC line inherits haplotypes from three wild-derived (CAST/EiJ, PWK/PhJ, and WSB/EiJ) and five classical (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShILtJ, and NZO/HiLtJ) inbred strains contributing both functional and nonfunctional OAS1b proteins into this MPP. Here, we use F1 crosses between CC strains, generated to understand complex and diverse phenotypic responses to virus infection (Rasmussen et al. 2014; Graham et al. 2016). The CC mice are an excellent model of WNV infection because their intron and exon structures in the Oas gene family are conserved (Choi et al. 2015). However, utilization of F1s allows sequences to differ between alleles at a locus, causing allelic heterozygosity between the wild-derived and classical inbred Oas1b sequences in this panel. Within this panel of F1s, we can generate classes of F1s based on their Oas1b alleles: functional Oas1b alleles from Dam and Sire (F/F), heterozygous RIX lines where only the Dam has a functional Oas1b allele (F/N), only the Sire (N/F), or neither parent has functioning Oas1b alleles (N/N). In previous work, we used the CC model to evaluate host responses to WNV and observed phenotypic variation that recapitulates the diversity in WNV susceptibility and outcomes observed in human populations (Graham et al. 2015, Green et al. 2016a, Green et al. 2016b).
In this study, we performed a coupled analysis of in vivo WNV infection, viral replication, host transcriptomics, and bioinformatics analyses to expand this genetic screen to identify regulatory loci (Table 1) and determine underlying transcriptional responses contributing to infection outcome. We screened 90 different F1 hybrids between CC strains infected with WNV to identify quantitative trait loci (QTL) contributing to disease phenotypes (Figure 1). To investigate the host response across F1s of different Oas1b haplotypes, transcriptomics was performed on the spleens of WNV-infected mice at 2, 4, 7, and 12 d postinfection. This transcriptional profiling revealed innate immune regulatory networks associated with the Oas1b haplotype. Our observations show that the CC is an important biomedical model for studies of human disease, and leveraging newly developed genomic resources in this model can improve our understanding of host responses controlling WNV infection and immunity.
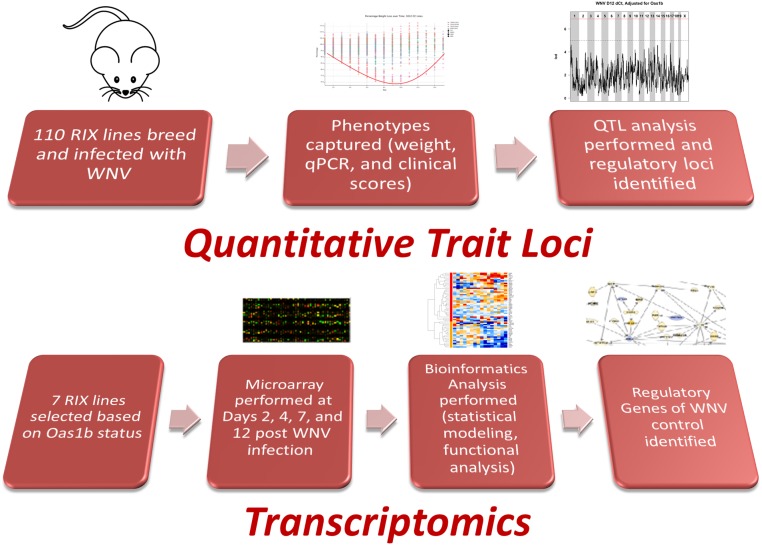
Figure 1.
Analysis workflow. The analysis steps for generating innate immune regulatory networks and pathways influenced by Oas1b. qPCR, quantitative polymerase chain reaction; QTL, quantitative trait loci; RIX, recombinant inbred intercross; WNV, West Nile virus.
Materials and Methods
Mice and infection
F1s were bred at the University of North Carolina at Chapel Hill under specific-pathogen-free (SPF) conditions. The 6- to 8-wk-old male mice were transferred to the University of Washington and housed directly in a biosafety level 2 (BSL-2) laboratory within an SPF barrier facility. After a resting period, age- and sex-matched 8- to 10-wk-old mice were subcutaneously inoculated in the rear footpad with 100 PFU WNV TX-2002-HC (WN-TX). Mice were monitored daily for morbidity (percentage of initial weight loss) and clinical disease scores. Mice were then housed under BSL-3 conditions throughout the experiments, and tissues were processed under BSL-3 conditions. All animal experiments were approved by the University of Washington Institutional Animal Care and Use Committee. The Office of Laboratory Animal Welfare of the National Institutes of Health (NIH) has approved the University of Washington (A3464-01), and this study was carried out in strict compliance with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Virus
WN-TX was propagated using previously described methods (Graham et al. 2016; Green et al. 2016). Viral stocks were generated using supernatants collected from infected Vero cell lines and stored at 80°.
QTL analysis
To identify polymorphic host loci impacting phenotypic differences, we used a modified approach of the DOQTL bioconductor package (version 1.6) in R. The additive haplotype model was used in our analyses and in previous studies (Gatti et al. 2014). Under this model the regression equation is defined as:
where Yi indicates the phenotype for animal i, x is the indicator of covariate (k) for animal (i), and α is the effect of covariate (k). dij(h) is assigned to allelic dosage of the founder (h) at locus j. βh is a coefficient of the genetic effects in each founder. The second Yi adjusts for kinship and εi for residual error in the animal (i). In order to generate additive allelic dosage probabilities for each F1, we generated eight-allele probabilities for each F1 with half of the probability for each allele coming from the inbred dam RI, and half coming from the sire RI, based on the MegaMUGA most recent common ancestor (MRCA) probabilities (http://csbio.unc.edu/CCstatus/index.py?run=FounderProbs) (Srivastava et al. 2017). The X-chromosome of these F1 males presents a special case, where the eight-allele probabilities are inherited fully from the dam RI. QTL genome scans were performed by regression on the day 12 weight change on genotype probabilities for each of the eight founder strains using R (see Supplemental Material for executable code). A random-effect term was included in the model to account for kinship among animals. A LOD score for each marker was calculated from the likelihood ratio comparing the regression model described above to a regression model without the founder genotype probabilities. The statistical significance of LOD scores was determined via a permutation test. A threshold of p < 0.05 was used to select significant associations.
DOQTL genotype probability files were generated with each animal having a unique genome probability entry. In this way, even animals from the same RIX would have individual (albeit identical) genome probability entries. This approach allowed us to assess both within- as well as between-strain phenotypic differences.
An adjusted p-value for each marker was not included because a permutation test was used to determine statistical significance of the LOD scores. The supplemental code provided contains hard coded significance thresholds to match the original analysis from the figures. Since there is some randomness to the permutation test, the threshold may display slightly different values if repeated.
Identification of Oas1b allele status
We utilized haplotype probability reconstructions [based on a hidden Markov model, described in Iraqi et al. (2012) and Srivastava et al. (2017)] for each CC strain to identify which founder strain haplotype was present at the Oas1b locus. Previous work (Graham et al. 2015) had confirmed that Oas1b alleles from the CAST/Eij, PWK/PhJ, and WSB/EiJ strains had read-through codons relative to the reference genome (Keane et al. 2011), and this read-through variant is associated with protection from WNV infection. F1s were identified as Oas1b N/N if each parent had an Oas1b haplotype without the read-through variant (A/J, C57BL/6J, 129s1/SvImJ, NOD/ShILtJ, or NZO/HILtJ); Oas1b F/F if each parent had an Oas1b haplotype with the read-through variant (CAST/EiJ, PWK/PhJ, or WSB/EiJ); and Oas1b heterozygous if only one parent had the read-through variant.
Oas1b sequence reconstruction
We integrated high confidence nonsynonymous SNPs from the Sanger mouse genomes project (www.sanger.ac.uk) (Iraqi et al. 2012; Oreper et al. 2017; Srivastava et al. 2017) of the eight CC founder strains with the annotated Oas1b exon sequence (Figure S1).
Clinical scoring: CC F1s and their disease definitions
The clinical scoring system used to evaluate WNV-infected mice was as follows: 0, healthy mouse (baseline); 1, ruffled fur, lethargy, hunched posture, no paresis, and normal gait; 2, altered gait and limited movement in one hind limb; 3, lack of movement and paresis in one or both hind limbs; and 4, moribund.
RNA extraction and quantitative PCR (qPCR) of WNV
Spleen and brain tissue were removed from mock- or WNV-infected mice. Half of the brain was homogenized immediately after harvest in 1 × PBS at 5500 RPM for 20 sec using a Precellys 24 machine and then centrifuged. Brain supernatant was added to TRI reagent (Ambion). Spleen tissues were stored in RNAlater (Ambion) and later homogenized in TRI reagent (Ambion). Total RNA was extracted using the Ribopure RNA Purification Kit (Ambion), with the addition of bromochloropropane (Acros Organics). RNA was converted to cDNA using the iScript Select cDNA Synthesis Kit (Bio-Rad). Using SYBR Green (Applied Biosystems) RT-PCR, WNV was quantified relative to GAPDH by probing cDNA with WNV-specific probes. qPCR results were recorded as fold change over mock-infected mice using the R statistical programming language (version 3.1). Results were loaded into Spotfire (http://spotfire.tibco.com, version 7.5.0.86) to produce box plots. Relative quantification was performed for WNV detection through qPCR. The forward and reverse primer used were: Primer WNV 1160F, 5′-TCA GCG ATC TCT CCA CCA AAG-3′ and Primer WNV 1229R, 5′-GGG TCA GCA CGT TTG TCA TTG-3′.
Correlation matrix
Phenotypic results (clinical score, weight loss, and qPCR in spleen and brain) were loaded into R. Phenotypes were averaged by their genetic background (F1), tissue type, and time point. The Pearson correlation function (cor) was performed using R’s stats package and scores were displayed using the ggplot2 bioconductor package. For a complete listing of package information, data, and commands, please refer to the data reproducibility document in Github under correlation analysis.
Determining outcome: symptomatic and asymptomatic
To quantify disease outcome, we used weight loss and clinical scoring to segregate the F1s into two broad pathogenic phenotype categories: asymptomatic or symptomatic. Three animals were assessed for each F1 and outcome was based on at least one mouse out of three meeting weight loss or clinical score criteria at any time point. Symptomatic were defined as having weight loss > 10% of original preinfection weight, clinical score > 1, and/or death, whereas asymptomatic was defined as having weight loss < 10% of original preinfection weight, clinical score of 0 or 1, and no death.
Affymetrix target preparation and microarray hybridization
RNA spleen samples were prepared for whole-transcriptome expression analysis using the WT PLUS Reagent Kit following the manufacturer’s recommended protocol (Affymetrix, Inc.). Next, 100 ng total RNA was used to prepare the hybridization-ready targets. Individual sense-strand DNA targets were randomized and hybridized to Mouse Gene 2.1 ST 96-Array Plates (Affymetrix, Inc.) using the GeneTitan Multi-Channel Instrument for hybridization, staining, and washing of arrays, as well as for scanning. Quality control (QC) metrics for hybridization, labeling and sample quality were generated using the Affymetrix Expression Console (version 1.3.187) software. All samples passed QC criteria.
CC-probe masking
To ensure that all Affymetrix probes were identified across all CC lines, previously described masking techniques were applied based on the CC founder data (Graham et al. 2016; Rasmussen et al. 2014). To mask out inconsistent Affymetrix probes, we used the oligomask R package designed for CC mouse data (Bottomly et al. 2014). Oligomask uses VCF files and the oligo package to filter probes prior to normalization and statistical analysis.
Transcriptomic analysis
Samples were screened for QC and outlier detection using the Affymetrix expression console using boxplots, as well as multi-dimensional scaling analysis and interarray correlation plots using the R statistical programming language (https://www.r-project.org, version 3.1), Bioconductor (version 2.13), and packages oligo (1.32.0), oligomask (https://github.com/dbottomly/oligoMask), limma (3.28.4), and corresponding dependencies. Background correction, normalization, probe masking, and probe summarization were performed with RMA (Robust Multichip Average) within the oligomask package. The sva package (3.20) in R was used to correct for plate batch effects. Differential expression (DE) analysis was performed using the limma bioconductor package (see statistical analysis under supplemental wiki in Github). A statistical cutoff of > 1.5-fold change over mock with a Benjamini–Hochberg-adjusted p-value < 0.05 was applied. The union of all DE genes for at least one time point in at least one F1 was first obtained and then filtered for immune genes. Immune genes were selected as genes identified in immune-related canonical pathways from Ingenuity pathway analysis (IPA). This list of immune-related genes was used for the global heatmap (Figure 4). The ABC (ATP binding cassette) and oxysterol-binding proteins were added to the screen to observe their correlation with other known Oas1b binding partners (Courtney et al. 2012). The heatmap was created using the heatmap.2 function in gplots (3.0.1) in R and bioconductor. A data report containing executable code can be found in Github under transcriptional analysis. Data were deposited in the GEO repository (access number GSE91003).
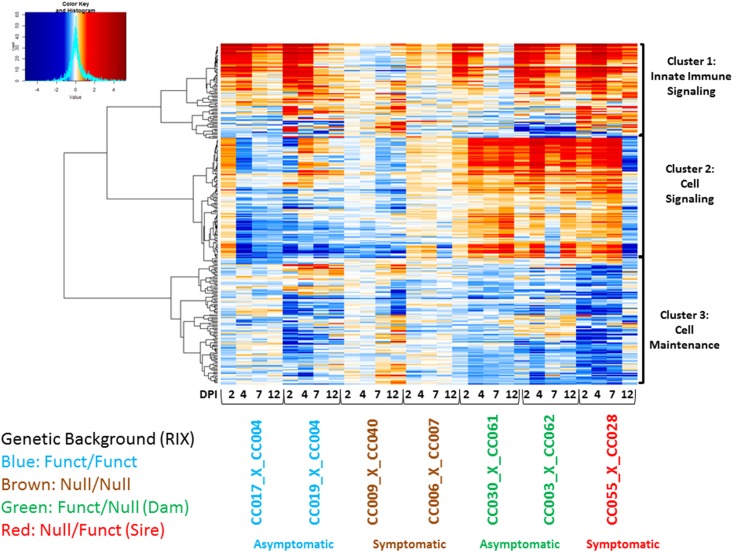
Figure 4.
Heatmap of immune disease modules. Coexpression in spleen at days 2, 4, 7, and 12 postinfection. The heatmap represents differential expression of 450 genes across seven F1s with different Oas1b functionality (F/F, N/N, F/N, and N/N). Expression is shown as log2(FC) WNV infected relative to mock. Red marks genes that are upregulated, blue for downregulated, and white represents no log2(FC). The y-axis shows genes that coregulate based on their directionality and magnitude. The x-axis shows the days postinfection, haplotype, and disease outcome according to the legend. Listed on the right as cluster 1: innate immunity, cluster 2: Cell maintenance, and cluster 3: cell signaling. log2(FC), log2 fold change; RIX, recombinant inbred intercross; WNV, West Nile virus.
Mock correlation
Due to infection and sample collection scheduling, most mock samples were collected at either 2 or 12 d postinfection per F1. To ensure that this had no impact on normalized transcriptional activity, probe-masked expression data were compared in mocks for two separate F1s, CC(041x012)F1 and CC(004x011)F1, at days 12 and 28 postmock-infection (data not shown). Pearson correlation in R gave scores (0.9688 and 0.9687) that confirmed high correlation in the expression between mocks of the same F1s at different time points.
Coexpression heatmap
Coexpression was performed only on genes that were determined to be statistically significant from the differential expression analysis (threshold: log2 fold change ≥ 0.58 and FDR ≤ 0.05) in at least one comparison and considered immune-related (see transcriptional analysis). Pearson correlations were run on the union of log2FC using the WGCNA and heatmap.2 bioconductor packages in R (Loraine et al. 2015; Gentleman et al. 2004; Smyth 2004)
Pathway and regulator analysis
A list of statistically significant, differentially expressed genes (threshold of significance of a > 1.5-fold change over mock with a Benjamini–Hochberg-adjusted p-value ≤ 0.05) were uploaded into IPA for core analysis to identify enrichment of biological pathways. The tools produce a list of known biological functions with an enrichment score (−log p-value) determining how significant those genes are to each function, and an activation z-score that tells the proposed activation of that pathway (activated or inhibited).
The z-score is based on knowledge of expression changes (and functions) in the Ingenuity knowledge base (Krämer et al. 2014). After the core analysis was performed in IPA, it was exported into Spotfire (version 7.5.0.86). Plots were created conservatively based on the following criteria: each biological pathway needed a −log p-value ≥ 1.30 (corresponds to p-value ≤ 0.05), two or more genes in each pathway, and had to produce an activation z-score. For the regulator effects network, the F/F, CC(019x002)F1 was analyzed for expression changes at 2 d postinfection and the top scoring network (antiviral response, 63% known regulators) was observed. Network generation was performed within IPA.
Innate immune networks
Genes detected in linear modeling and correlation analyses were loaded into IPA and networks were selected based on enrichment for immune-related pathways. The F/F network was based on the top two highest scoring networks (immunological disease and inflammatory response with network scores 36 and 33). The F/N network was based on immune and inflammatory functions (network score 35). The N/N network was built on functions in cell-to-cell signaling (network score 27). Protein–protein interactions from the Ingenuity knowledge base were used to connect ABCF3 and ORPL1 with the transcriptional data. This was done using the network build and connect tools in IPA. Network data were then exported into cytoscape (version 3.1.1) to make custom network figures.
Data availability
A complete description of the data and methods can be found on the manuscript’s Github page (https://github.com/greener98103/oas1b/wiki) and in the supplemental file titled “Green_Oas1b_data_policy_documentation.” This file includes information on strains/lines, variant files from Sanger, and marker files [MUGA platform (Morgan et al. 2015)]. The data document also contains detailed descriptions, file locations, and links to executable code for QTL, transcriptional, and correlation analyses. Gene expression data are in GEO with the accession number GSE91003. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
A QTL identified in chromosome 5 found in day 12 weight loss
We conducted QTL mapping across our panel of 90 F1s (Table 1), and identified an approximate 2.5 Mb region spanning from 120–122.5 Mb on chromosome 5 (here termed the host response to WNV, region 1, or Hrw1) (Figure 1A). HrW1 impacted day 12 weight loss (Figure 2, A and C). The Hrw1 aggregate peak contained 70 genes and microRNAs (excluding predicted genes). The genes under this locus include all the murine Oas genes as well as genes associated with biological pathways including actin cytoskeleton signaling, CD28 signaling in T helper cells, and integrin signaling (Table S1 and Table S2). The QTL peak sits over Oas1b. Based on previous mouse studies (Suthar et al. 2013; Mashimo et al. 2002; Ferguson et al. 2008; Kajaste-Rudnitski et al. 2006), Oas1b is a known driver of disease phenotypes. To determine which of the eight strains in the CC were influencing the loci, we next looked at the founder effects driving this QTL.
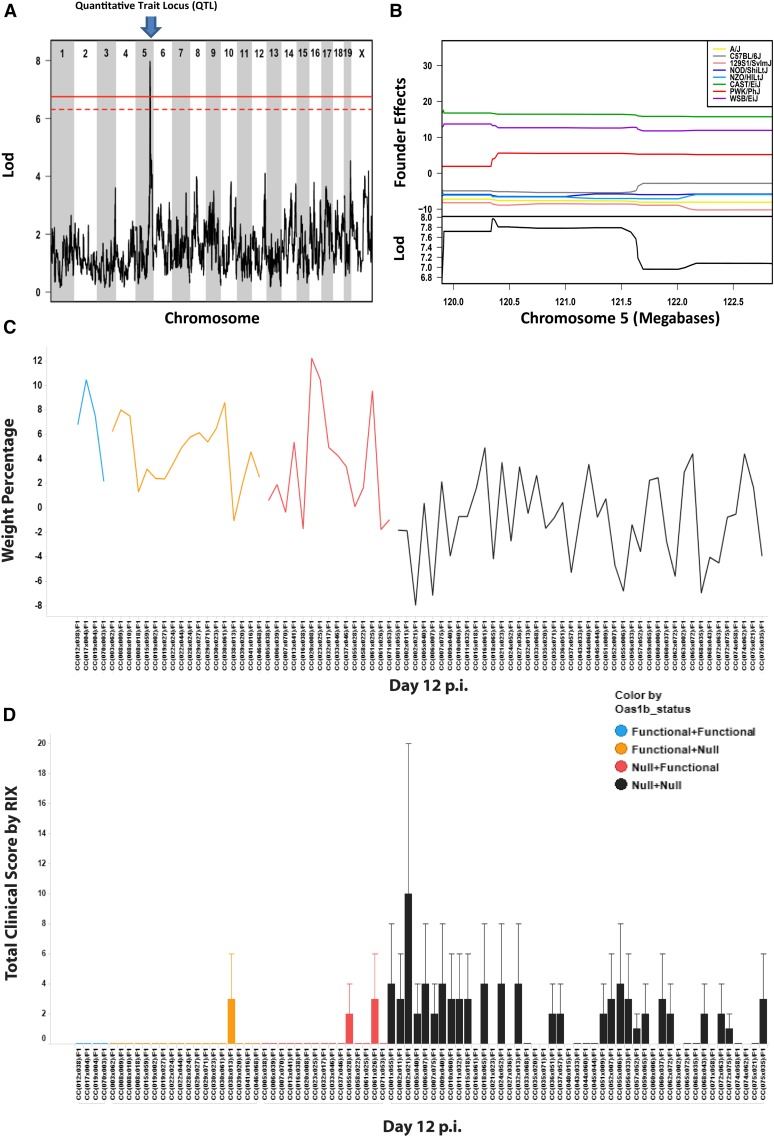
Figure 2.
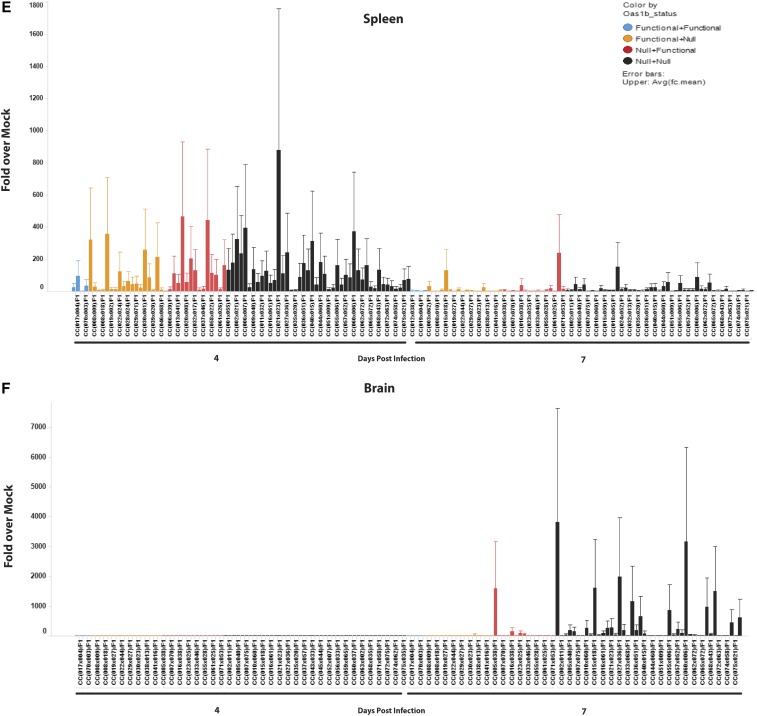
WNV QTL. (A) Genome scan of 90 F1s from day 12 weight loss with a significant QTL peak on chromosome 5. The top row indicates chromosome number and the y-axis is the LOD score. The LOD is the degree of concordance for a genetic marker for a phenotype (weight loss at day 12). The solid red line is p-value = 0.05 and the dotted line is p = 0.1. (B) The upper image is the contribution of each founder in the QTL (founder effects), where the population average is zero and each colored line represents a different founder’s predicted effect of one allelic copy on an individual’s phenotype (weight percentage at day 12). The lower image shows the LOD score in the lower y-axis. The x-axis shows the locus in megabases at chromosome 5. Each colored line represents a different founder’s predicted effect on phenotype. The positive alleles on the top show founder lines contributing to greater than the population mean, and the negative values contributing to less than the population mean. (C) Weight change at day 12 p.i. The y-axis represents weight percentage (gain or loss) across all RIXs. The x-axis shows the F1 identity and the color indicates Oas1b haplotype. Blue = F/F, orange = F/N, red = N/F, and black =N/N. (D) Total clinical score per F1 at day 12 p.i. Clinical score was determined as follows: 0, healthy mouse (baseline); 1, ruffled fur, lethargy, hunched posture, no paresis, and normal gait; 2, altered gait and limited movement in one hind limb; 3, lack of movement and paresis in one or both hind limbs. qPCR of WNV virus in the spleen (E) and brain (F) at days 4 and 7 p.i. The y-axis shows fold change over mock and the x-axis shows each F1 background and their days p.i. Each color represents Oas1b haplotype. Blue = F/F, Orange = F/N, Red = N/F, Black = N/N. LOD, log odds ratio; p.i., postinfection; qPCR, quantitative polymerase chain reaction; QTL, quantitative trait loci; RIX, recombinant inbred intercross; WNV, West Nile virus.
Loci founder effects
As the CC is derived from eight founder strains, QTL can contrast any combinations of these founder haplotypes. Figure 2B shows the estimated phenotypic effect of each founder strain haplotype at the locus on day 12 weight. Haplotype effects are determined as the deviation from the population mean due to one copy of that founder allele at the locus. In this way, we can see that the wild-derived inbred strains CAST/EiJ, PWK/PhJ, and WBS/EiJ ameliorate weight loss (higher value = less weight lost), whereas the five classic inbred strain alleles show enhanced weight loss (lower effects). These results are consistent with a previously reported analysis of Oas1b (Courtney et al. 2012) and the Sanger mouse genomes project (Keane et al. 2011), which show that classical strains have an early stop codon, whereas wild-derived founders encode a fully functional protein. These different founder contributions yield a range of complex phenotypes seen in the F1s. Previous CC studies showed that F1s heterozygous for Oas1b did not guarantee protection (Graham et al. 2016) and there were likely other immune factors beyond Oas1b driving WNV infection outcome. To get a global view of WNV pathogenesis in the CC, we studied disease responses by haplotype.
Contribution of Oas1b in preventing viral growth and neuroinvasion
Guided by the observed allele effects, and the presence of the truncated stop codon in Oas1b, we binned our F1s into four classes: F/F (any F1 containing CAST/EiJ, PWK/PhJ, and/or WSB/EiJ alleles), N/N (any F1 containing A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, and/or NZO/HILtJ), and the two reciprocal heterozygous classes F/N (a maternally inherited functional allele and a paternally inherited defective allele) and N/F (a maternally inherited defective allele and a paternally inherited functional allele). We measured a variety of WNV disease response phenotypes and assessed their differences between these classes. F/F and heterozygous F1s displayed similar fluctuations in weight at day 12 postinfection (Figure 2C) compared to N/N F1s. Of the heterozygous F1s, N/F showed a slightly higher distribution in weight at day 12 compared to F/N haplotype mice. Clinical scores were also evaluated at day 12 (Figure 2D). Clinical score was calculated by animal’s appearance and physical function characteristics (ruffled fur, lethargy, hunched posture, and no paresis) and movement (see Materials and Methods). The only mice that developed overt clinical scores associated with WNV disease were from the N/N Oas1b F1s.
WNV is a neuroinvasive virus that first replicates in the spleen and then progresses to the central nervous system in a manner controlled by the innate immune response (Suthar et al. 2013; Elbahesh et al. 2011). To assess viral growth and neuroinvasion, we measured the levels of WNV virus in the spleen and brain using qPCR. An inability to effectively control WNV growth in the periphery (such as the spleen) facilitates viral production and neuroinvasion (Suthar et al. 2013; Green et al. 2016a; Fredericksen 2014). qPCR results showed that F1s without functioning Oas1b alleles displayed an increased viral load in the spleen at day 4 (Figure 2E). By day 7, this lack of viral control resulted in WNV neuroinvasion characterized by virus in the brain (Figure 2F). There were lower levels of virus in heterozygous F1s in both the spleen at day 4 and in the brain by day 7. The accumulation of virus in N/N was statistically significant in the brain compared to heterozygous F1s (p-value = 0.0005) across all time points (2, 4, 7, and 12 d postinfection). Viral accumulation was not statistically significant in the spleen, but we observed a correlation between viral qPCR and clinical score in the N/N and heterozygous F1s (see Figure S2 and Github).
Distribution of infection response
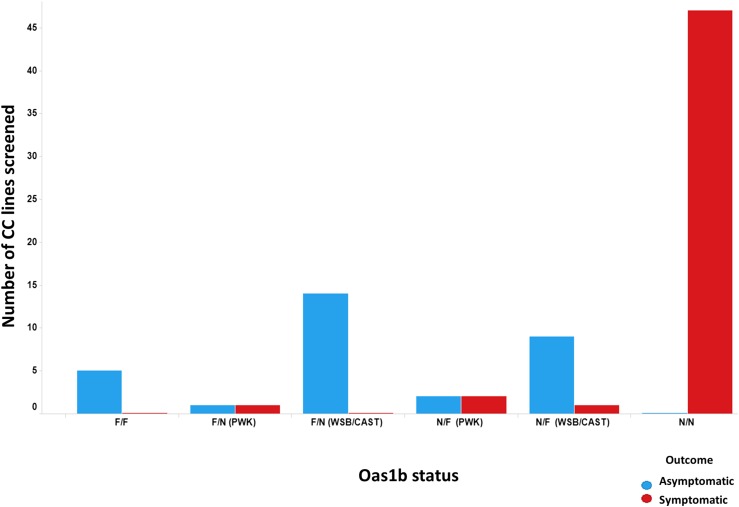
From the 90 F1s used for QTL mapping based on weight loss, we evaluated 83 F1s because they had clinical metrics of susceptibility to WNV infection. Responses of each line were captured and an outcome was determined as asymptomatic or symptomatic. For a line to be considered symptomatic, infected animals had to exhibit a clinical score > 1 or a > 10% loss in weight since the time of infection (see Materials and Methods). We evaluated infection outcome based on haplotype (Figure 3). Out of the 83 F1s screened, 47 of them (56%) were symptomatic and did not contain full-length Oas1b alleles. Only five F1s lines were asymptomatic with F/F alleles (6%), and zero lines with F/F alleles were symptomatic. Likewise, no F1s with N/N alleles were determined to be asymptomatic. F/F Oas1b alleles provided complete protection against WNV clinical disease. Of the asymptomatic heterozygous F1s, (F/N and N/F), 16 were F/N (19%) compared to 14 N/F (17%). We only observed four symptomatic N/F F1s (5%) compared to one asymptomatic F/N F1 (1%). We also observed a relationship between viral load and disease phenotypes (see Figure S2).
Figure 3.
Disease outcome under Oas1b functionality. The number of F1s summarized by disease outcome. Outcome was determined as symptomatic (loss of 10% or greater body weight or succumbed to WNV infection) or asymptomatic (fought off infection and never lost 10% weight). Each group of mice is binned according to their Oas1b status. The heterozygous F1s are shown by their disease outcome and their allelic founder contribution. F/N(PWK) means the F1 contains at least one PWK allele (Null/PWK or PWK/Null). F/N(WSB/CAST) means the F1 contains at least one WSB/CAST allele. The colors indicate each’s F1’s outcome (blue = asymptomatic and red = symptomatic). CC, Collaborative Cross; WNV, West Nile virus.
To identify differences between the heterozygous lines, we looked at allelic contributions among the eight founder strains. There was a subtle difference in disease outcome among the heterozygous lines but this was likely driven by lines containing an Oas1b allele from the WSB/EiJ or CAST/EiJ founder strains. F1s that contained the Oas1b allele from the PWK/PhJ founder strain appeared to only provide moderate protection compared to either the WSB/EiJ or CAST/EiJ alleles (see Materials and Methods). These observations support previous studies implying a major role for Oas1b in protection against WNV disease in mice, although this is the first time a comparison has been made between heterozygous F1s and their Oas1b’s founder effects.
To reveal innate immune signatures driving WNV susceptibility, we performed a multi-tier bioinformatics analysis (see Figure 1) involving genome-wide microarray analysis on a set of F1s with different Oas1b haplotypes (F/F, F/N, N/F, and N/N), focusing on revealing the profile of immune-related genes in the spleen at days 2, 4, 7, and 12 post-WNV infection as compared to mock-infected congenic control mice (Table 2). We centered our transcriptional profiling assessment on the spleen to better understand peripheral immune programming and possible linkage with the control of viral neuroinvasion.
Coregulatory patterns between Oas1b haplotypes
We performed a transcriptional correlation analysis using the union of statistically significant genes across F1s with different haplotypes (Figure 4). Since the genetic backgrounds among F1s vary and those without Oas1b produced a large amount of differentially expressed genes (especially in later time points due to increased illness), we focused on statistically significant immune-related genes. Correlation analysis identified three distinct gene clusters. Functional analysis was performed to summarize the clusters into known biological functions. Genes within cluster 1 are largely involved in innate immunity, genes within cluster 2 include those involved in cellular signaling, and cluster 3 genes include a role in cell maintenance (Figure 4).
Genes in cluster 1 (innate immune signaling) include several molecules involved in RLR signaling, IFN responses, and death receptor signaling. The coregulated gene module contained ISGs, Myd88, Oas2, Oas3, Stat2, and Tlr7. Cluster 1 was also enriched for IFN regulatory factors (Irf3, Irf7, and Irf9). Expression of genes in this cluster appeared to be activated across all F1s, although not all met statistical significance. Within the F/F F1s, activation of this cluster was highly induced only until day 4. This profile is different from the heterozygous F1s, especially the symptomatic N/F F1 CC(055x028)F1, which remained induced through day 12. The Oas genes also appear in cluster 1. These heatmap and qPCR results display an early termination of innate immune signaling in the asymptomatic lines that could be influencing disease outcome. Expression changes in the second cluster (cell signaling) are primarily influenced by the three heterozygous F1s. The genes in this module are associated with the cell cycle, DNA replication, DNA repair, growth, and proliferation. This cluster/gene module showed little variation across heterozygous F1s and was sparsely suppressed in the F/F F1s. Expression was inverted at day 12 between the asymptomatic and symptomatic F1s (F/N and N/F, see Table 2), suggesting that this subcluster links with viral infection and differential WNV disease.
The expression changes found in CC(055x028)F1 between days 7 and 12 postinfection correspond with viral load. At day 12 postinfection, we found an increase in viral load in the brain as measured by qPCR (Figure S6). The gene expression changes between days 7 and 12 postinfection could mark viral spread and subsequent neuroinvasion.
The coexpressed genes found in the third cluster (cell maintenance) are involved in cell/tissue morphology and leukocyte extravasation (diapedesis). Genes in this biological pathway are suppressed in expression relative to mock-infected controls, implying that leukocyte migration is less active during infection in CC(009x040)F1 (N/N) and CC(055x028)F1 (F/N). Inhibition of leukocyte migration (and supporting chemokines) to infection sites could destabilize protection against WNV in these F1s.
To validate select innate immune gene expression, we performed qPCR on two virus-inducible genes: tetratricopeptide repeats 1 (Ifit1) and IFNβ (Ifnβ1) using the F1s shown in our heatmap and in Table 2. Ifit1 and Ifnβ1 are IRF3 target genes induced upon WNV infection; Ifit1 is also an ISG and is highly expressed in response to type 1 or type 3 IFNs. Not surprisingly, Ifit1 induction was highest at day 2 postinfection, indicative of IRF3 activation and the onset of IFN signaling (Figure S3 and Figure S4). All F1s appeared elevated for Ifit1 but it was highest in F/F CC(019x004)F1. Ifit1 decreased by day 4 post-WNV infection, indicating that viral replication was being controlled. The Ifit1 elevation in N/N F1s at day 2 post-WNV infection indicated induction of IFN production. Ifnβ1 appeared low across most of the F1s in the spleen except the one N/N F1 at day 2 post-WNV infection. This IFN response was also seen in the brain in the N/N along with virus at day 7 and suggests that the F/F F1 was protective from the neuroinvasion (Figure S4). In summary, the gene expression profile of both F/F and N/N Oas1b haplotypes revealed expressed IFN signaling at day 2, but this alone did not link with protection in the N/N line.
Transcriptional analysis of genes within the Hrw1 locus
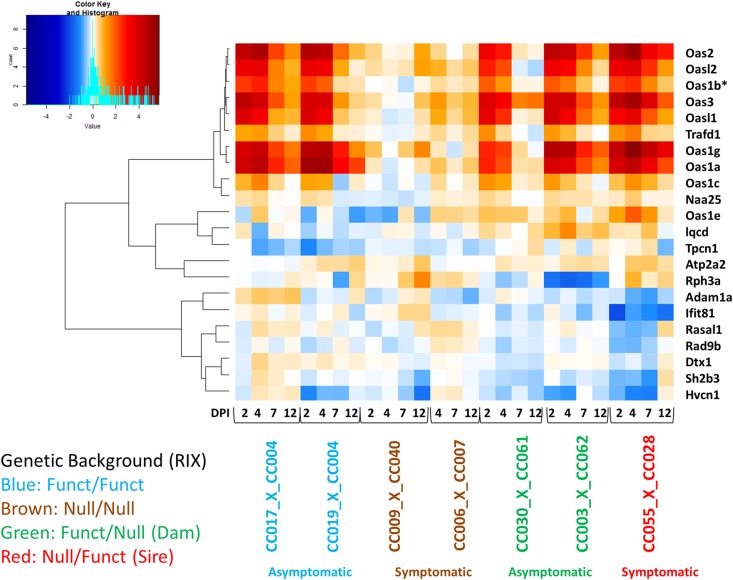
To determine if expression differences in the Oas family members and other genes within the Hrw1 locus on chromosome 5 (Figure 5) linked with WNV infection outcome, we applied a targeted transcriptional analysis within the locus. The genes that flank the Oas cluster (Dtx1 and Rph3a) were identified as expressed compared to mock-infection in our heatmap. There are several genes whose expression was reduced (downregulated) independently of Oas signatures in the heterozygous F1s. Interestingly, Rasal1 appeared downregulated in CC(055x028)F1, which was previously observed with elevated viral load in the spleen and brain (Scherbik et al. 2006), and this finding was confirmed with qPCR in the spleen at day 4 postinfection (see Materials and Methods). Rasal1 is an inhibitory regulator of the Ras-cyclic AMP pathway and is involved in dendrite formation (observed in melanocytes), intercellular signal transduction, and GTPase activity, and could be implicated in WNV disease. We also evaluated the expression of other Oas gene family members and found that the F1s with the largest induction of Oas genes were CC(017x004)F1 (F/F), CC(019x004)F1 (F/F), and CC(055x028)F1 (N/N). Oas1b is included in Figure 5 and was induced by WNV infection to be differentially regulated compared to mock, but was not always statistically different across infection time points after multiple statistical comparisons. Oasl1 (Oas-like gene 1), Oas3, and Oas1a were the most highly induced (upregulated) Oas gene family members across the panel. Oas1a and Oas1g, which are influential in viral detection and the innate immune response, were the next highest in expression. Oasl1 has been shown to regulate IRF7 production (Lee et al. 2013). Oas3 was found upregulated in each F1 we screened using qPCR and transcriptomics in at least one time point except for N/N CC(090x040)F1, which had low gene expression levels. The importance of Oas3 in virus response is corroborated by a previous study that found that OAS3 displayed a higher affinity for dsRNA than OAS1 or OAS2, which indicated its contributive role in the antiviral response (Li et al. 2016). Importantly, none of the significant Oas genes were suppressed or “downregulated” in our analyses. We validated our findings with qPCR in Oas target genes Oas1g, Oas2, and Oas3. Thus, our analysis of Oas gene family members is supportive of Oas genes playing a significant role in antiviral immunity.
Figure 5.
Heatmap of QTL genes. Coexpression in spleen at days 2, 4, 7, and 12 postinfection. The heatmap represents differential expression of 21 genes found under the QTL across seven F1s with different Oas1b haplotype (F/F, N/N, F/N, and N/N). Expression is shown as log2(FC) WNV infected relative to mock. Red marks genes that are upregulated, blue for downregulated, and white represents no log2(FC). The y-axis shows genes that coregulate based on their directionality and magnitude. The x-axis shows the days postinfection, haplotype, and disease outcome according to the legend. log2(FC), log2 fold change; QTL, quantitative trait loci; RIX, recombinant inbred intercross; WNV, West Nile virus.
Oas1b haplotype distinguishes gene expression modules
We conducted pathway analysis to assess the biological differences of significant genes in F1s of differential Oas1b haplotype. As shown in Figure 6, the Sanger sequencing revealed several polymorphisms within functional domains of the full-length Oas1b protein among alleles encoded in the PWK/PhJ, WSB/EiJ, and CAST/EiJ outbred parental CC strains compared to the C57BL6/J reference strain, while all of the classical inbred founder strains encoded a stop codon within the OAS1b C-terminal domain. We first focused on the presence and absence of full-length Oas1b to reveal differences in gene module expression between the two groups.
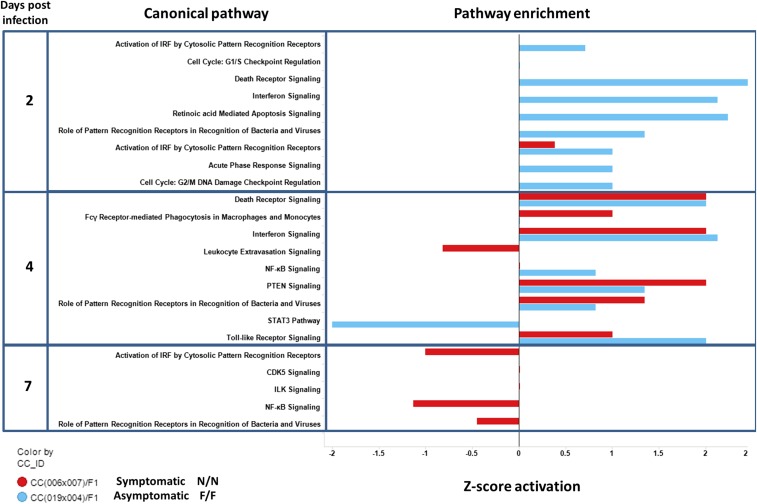
The immune response at day 2 postinfection (Figure 7) was impacted by Oas1b status. Immune pathways in F/F F1s were predominantly activated between days 2 and 4 postinfection. Aside from low level viral recognition, the N/N CC(006x007)F1 did not show significant activation in immune pathways until day 4 postinfection. By day 4 postinfection, several pathways were expressed in parallel, including pathways involving death receptor signaling, IFN response, toll like receptors, and viral recognition receptors, thus, marking the onset of the innate immune response. In contrast, STAT3 signaling was downregulated by day 4 postinfection in the F/F asymptomatic F1s, reflecting differential STAT3 signaling activity between the two groups. By day 7 postinfection, the N/N F1 profile marked a reduced transcriptional response within innate immune signaling pathways including NF-κb, and pattern recognition receptor signaling, possibly reflecting the progression of WNV infection out of the spleen onward to the CNS.
Figure 7.
Pathway analysis in the presence (F/F, Functional + Functional Oas1b) and absence (N/N, Non-functional + Non-functional Oas1b) of functioning Oas1b. A bar plot quantifying enrichment of biological pathways across F1s and time points (2, 4, and 7 days) postinfection. The y-axis indicates time point and biological pathway. The x-axis shows the activation z-score for a pathway. The z-score is based on literature findings and determines the magnitude of gene regulation for a pathway. Values to the left indicate that pathway is inhibited and values to the right indicate that pathway is activated. The colors in the bar plot mark the different F1 backgrounds [blue = CC(009x040)F1 and red = CC(006x007)F1].
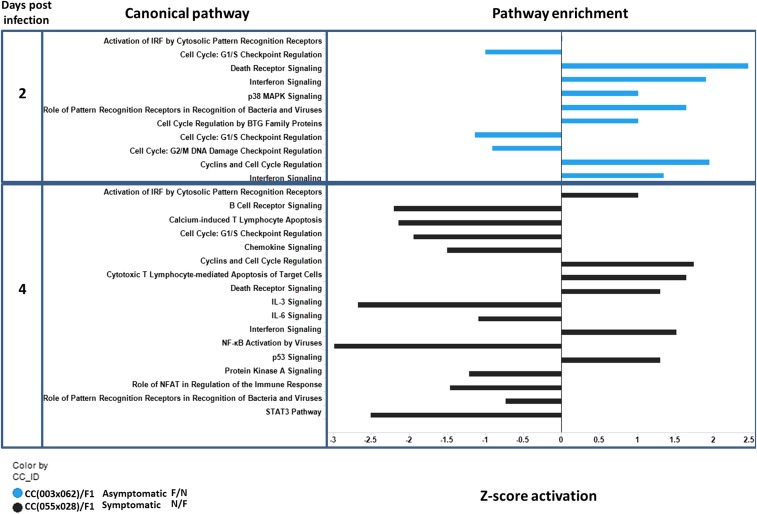
To assess gene expression pathways among Oas1b heterozygous F1s, we concentrated on the day 2 and 4 postinfection time points (Figure 8). These pathway analyses compared the downstream effects of Oas1b between symptomatic CC(055x028)F1, whose Oas1b allele is derived from PWK/PhJ, vs. asymptomatic F/N CC(003x062)F1, whose Oas1b is from WSB/EiJ. We observed differences in G1/S checkpoint cell cycle pathways between CC(055x028)F1 and CC(003x062)F1. The pathway was downregulated in the asymptomatic line, but had very weak induction in the symptomatic line, possibly marking differential pathology among the lines. At day 2 postinfection, N/F CC(055x028)F1 had reduced expression of p53 signaling wherein the other heterozygous lines showed differential p53 module expression and cell signaling response modules at day 4 post-WNV infection. We observed more extensive gene module regulation in the Oas1b heterozygous mice overall.
Figure 8.
Pathway analysis in the heterozygous Oas1b F1s at day 2 and 4 post-WNV infection. A bar plot similar to day 4 showing enriched biological pathways. The y-axis indicates the haplotype, time point, and biological pathway. The x-axis shows the activation z-score for a pathway. Values to the left indicate that pathway is inhibited. Values to the right indicate that pathway is activated. The colors in the bar plot mark the different F1 backgrounds [blue = CC(003x062)F1 and black = CC(055x028)F1]. CC_ID, Collaborative Cross identifier.
Innate immune regulatory networks
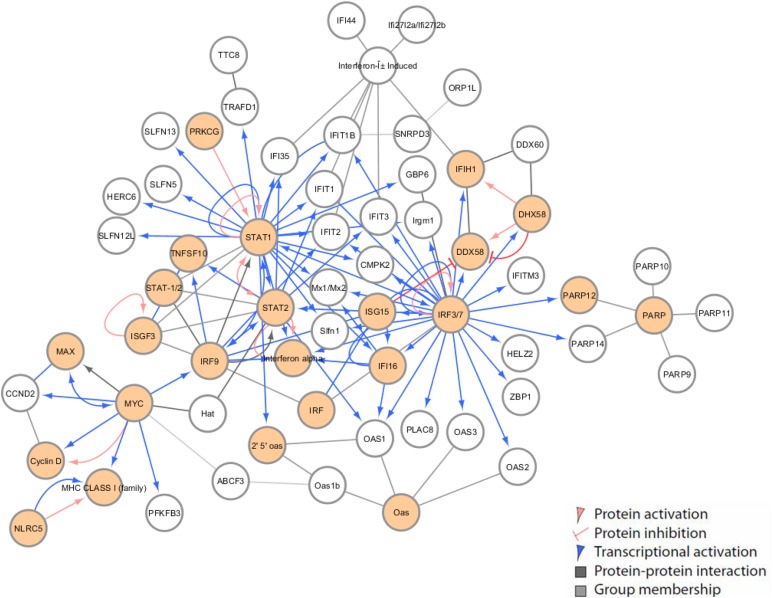
Based on correlation and functional analysis, we proceeded to construct novel innate immune regulatory networks to discern the unique host response associated with Oas1b allelic differences. Using transcriptional data from F1s of different Oas1b haplotypes, we identified three immune regulatory networks during WNV infection. Figure 9 represents the Oas1b allele from both parents (F/F), Figure 10 shows the network from Oas1b heterozygous (F/N), and Figure 11 shows the network from Oas1b homozygous nonfunctioning (N/N) mice. The F/F innate (day 2 postinfection) immune network (Figure 9) is comprised of Irf3, Irf7, Stat1, STAT2, Myc, and the poly(ADP-ribose) polymerase family of proteins (PARP) response networks, and includes the Oas gene cluster. Irfs and Stats were the major hubs launching virus-responsive genes and ISG networks, along with the induction of the Oas pathway. IRF3/7 also connects with several pattern recognition receptors including DDX58 (the RLRs), IFIH1, and DDX60. PARP genes contribute to death receptor signaling and apoptosis while Mx1/Mx2 are ISGs involved in antiviral innate immunity against several viruses (Shaffer et al. 2001). Antigen presentation is clearly activated, as indicated through network connections among the MHC class I family of genes. ABCF3 is connected by MYC (myelocytomatosis) and performs various roles in the cell, and has been identified as an Oas1b-binding protein of WNV control (Courtney et al. 2012). IFIT1B and SNRPD3 (small nuclear ribonucleoprotein) connect to ORP1L, another Oas1b-binding protein. Thus, network analysis of F/F Oas1b mice reveals innate immune and antigen processing induction, OAS protein expression, and network interaction of known Oas1b-binding proteins in response to WNV associated with homozygous full-length Oas1b genotype.
Figure 9.
Innate immune regulation network [Oas1b allele (F/F)]. Innate immune regulatory network identified in F1s with F/F Oas1b alleles. Each node represents a gene with different colored lines showing types of connectivity. Filled-in nodes are target genes from the dataset.
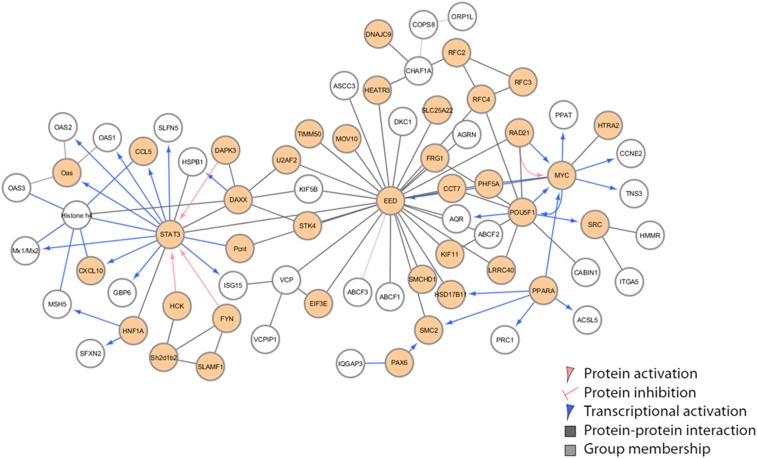
Figure 10.
Innate immune regulation network [Oas1b allele (F/N)]. Innate immune regulatory network identified in F1s with F/N Oas1b alleles. Each node represents a gene with different colored lines showing types of connectivity. Filled-in nodes are target genes from the dataset.
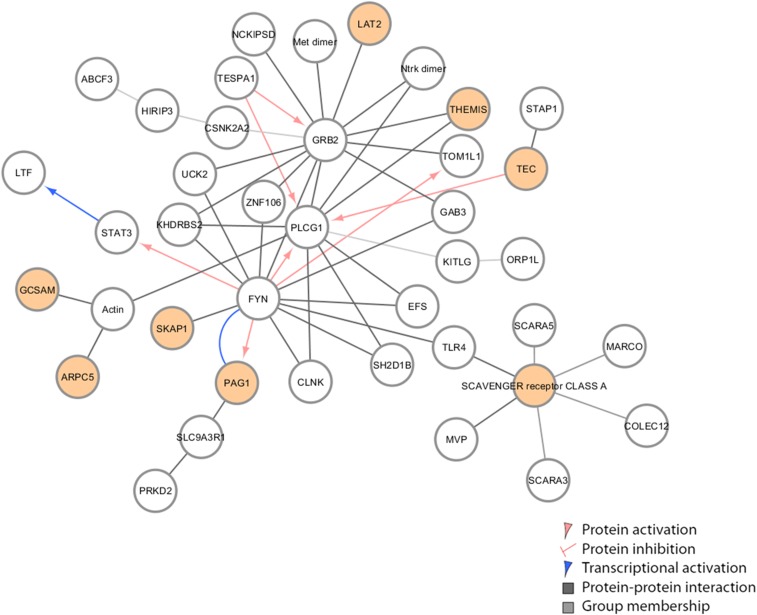
Figure 11.
Innate immune regulation network [Oas1b allele (N/N)]. Innate immune regulatory network identified in F1s with N/N Oas1b alleles. Each node represents a gene with different colored lines showing types of connectivity. Filled-in nodes are target genes from the dataset.
The innate (day 2 postinfection) heterozygous network (F/N) in Figure 10 shared genetic signatures with F/F and included STAT3 as one of the network hubs. Additional hubs included EED (embryonic ectoderm development), MYC, and POU5F1 (POU class 5 homeo box 1), major cell signaling pathways (Neri et al. 2012; Shaw and Martin 2009). Along with STAT3, the heterozygous network also connects the OAS pathway genes (1–3 including Oas1b). Unique to this network is the incorporation of DAXX (death domain associated protein) and Mov10 (RISC complex RNA helicase). DAXX is involved in death receptor signaling and targets infected cells for apoptosis (Engelhardt et al. 2001; Li et al. 2000; Perlman et al. 2001). Mov10 is an IFN-inducible gene recently shown to have antiviral activity against RNA viruses like Sendai and VSV, but not seen before in WNV. This network again supports previous work (Courtney et al. 2012) where ABC proteins are binding partners to Oas1b, as noted above. In our network, we also identified connections with ABCF1 and ABCF2.
The N/N network, derived from later time points (days 7 and 12 postinfection), are involved in cell-to-cell signaling interactions and RNA post-translational modifications. There are several connector hubs in this network driven by genes PLCG1, GRB2, and FYN. PLCG1 is involved in CD28 signaling in T helper cells and apoptosis. GRB2 is a growth factor receptor signaling-adaptor protein and is involved in a variety of cell signaling programs (Jang et al. 2009). Interestingly, ABCF3 is connected to GRPB2 through protein interactions with HIRIP3 (HIRA-interacting protein 3) and CSNK2A2 (casein kinase 2 α 2). It is believed that HIRIP3 functions as part of a multi-protein complex with roles in chromatin and histone metabolism. CSNK2A2 is a kinase, and has roles in apoptosis and G2/M phase cell cycling. ORP1L is connected to GRB2 through interactions with growth factor KITLG (Kit ligand). KITLG has several cellular roles including differentiation and apoptosis. The N/N network shares a STAT3 hub with the F/N network but its interaction partner is the FYN proto-oncogene (Src family tyrosine kinase family), which is involved in ATP binding as well as CD4 and CD8 receptor binding. Toll-like receptor 4 (TLR4), a well-documented immune signaling receptor, is downregulated. Taken together, these results link Oas1b haplotype with differential gene networks and WNV infection outcome.
Identifying regulatory factors in host innate immune signaling
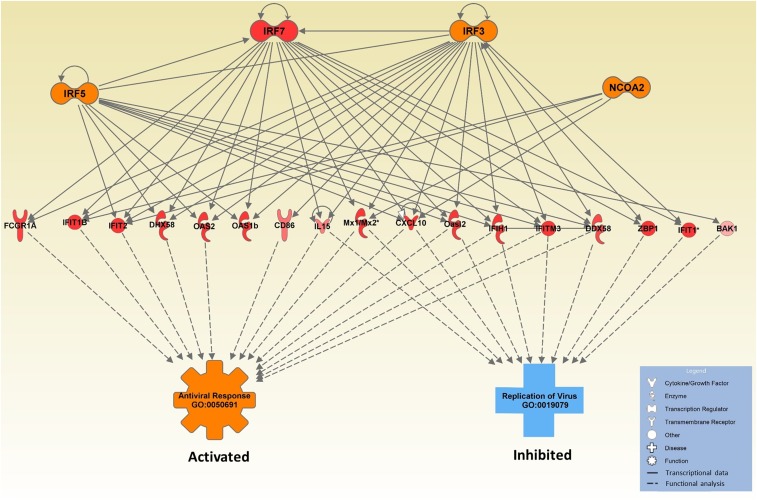
Our results show that F1s containing functional Oas1b alleles from both parents do not succumb to WNV infection. To determine the regulators of transcriptional responses in these Oas1b F/F F1s, we performed a predicted regulator analysis to localize target genes and find additional transcriptional regulators necessary for protection. Our regulator network (Figure 12) revealed that gene expression upregulation occurred predominantly in ISGs at day 2 postinfection. The network analysis determined activation of four transcriptional regulators (IRF3, IRF5, IRF7, and NCOA2). Three of these—IRF3, IRF5, and NCOA2—have been functionally linked to innate immune regulation or WNV infection in previous studies (Lazear et al. 2013; Flammer et al. 2010). The regulators and their candidate genes shared two distinct operations: antiviral response and inhibition of viral replication.
Figure 12.
Infection regulator network F/F. A regulatory network generated from differentially expressed genes in CC(019x004)F1 at day 2 postinfection. The top nodes are transcriptional regulators. The red regulator was observed in the expression data and the orange nodes are predicted based on the target genes in our data set. The row of red-colored nodes indicate target genes that are upregulated. The lower orange and blue nodes refer to functional Gene Ontology (GO) terms. Antiviral response (GO:0050691) is predicted to be activated and replication of virus (GO:0019079) is predicted to be inhibited.
Among the target genes in the network were Oas members, Mx1, and various chemokines. Additionally, BAK1 (BCL antagonist/killer 1) and ZBP1 (Z-DNA binding protein 1) are present in the network and play roles in cell cycle regulation and proapoptotic activity; each are putative IRF-target genes. FCGR1A FC γ receptor is also linked to this network and is known to function in antigen presentation and removal of infected cells via apoptosis. NCOA2 is an Oas1b F/F network gene and is a transcriptional coregulatory protein with multiple nuclear interacting domains allowing DNA to become more accessible for transcription. Oas1 and 2 expression is in part mediated by IRFs and NCOA2; studies using mouse bone-derived macrophages lacking NCOA2 expression showed a decrease in ISG expression (Flammer et al. 2010). Thus, WNV infection in the context of the F/F Oas1b haplotype engages specific IRF, IFN, and virus-responsive transcriptional networks that mediate protection from neuroinvasion and disease.
Discussion
Infectious diseases cause a wide range of responses across genetically diverse populations. The use of genetically diverse MPP within the infectious disease community has been increasing with an effort to characterize (Iraqi et al. 2012; Ferris et al. 2013; Rasmussen et al. 2014; Graham et al. 2015, 2016) and genetically define the range of disease responses under host genetic control during viral infections (Boon et al. 2009; Ferris et al. 2013; Gralinski et al. 2015, 2017). Here, we confirmed a role for variants at the Oas1b locus in driving major disease outcomes following WNV infection. By leveraging prior gene sequence information of the Oas1b gene, we subclassified our F1s into four diplotype classes at the Oas1b locus. We found significant within-class variation in disease outcomes in these F1s, utilized transcriptional analysis to identify key host genes (and pathways) involved in allelic differences, and obtained a comprehensive view of the innate immune response associated with Oas1b haplotype variation.
Genetic variation impacts disease
The Oas1b gene has been shown (Bigham et al. 2011; Courtney et al. 2012) to play a major role in controlling WNV disease by limiting viral replication (Kajaste-Rudnitski et al. 2006) within the C3H mouse strain (Perelygin et al. 2002). We identified a major-effect QTL over the Oas1b locus within a large population of F1s, and this QTL showed allele effects consistent with the previously described sequence variation within Oas1b. Importantly, this variant appears to segregate between wild-derived inbred strains and classical inbred strains derived from small population fancy mice (Yang et al. 2011). While our study and previous ones cannot definitively answer the question of whether the mutation arose in natural populations or in laboratory mice, it highlights the utility of assessing a broader range of genetic variants (both at targeted loci and genome-wide) in dissecting disease responses.
The presence of genes of major effect within genetic reference or other mapping populations can overshadow effects of other genetic variants cocirculating within these populations. As seen in our study and others (Ferris et al. 2013), it is possible to detect, transcriptionally characterize, and genetically map variants modifying disease responses in the presence of a major gene. As many pathogens and other diseases are impacted by large-effect genes (Ferris and Heise 2014), care in the design of experiments and their resultant analysis within the context of reference populations must be given. As we have shown here, studies further characterizing between-class responses at a molecular level can improve insight into disease mechanisms. Alternatively, studies such as F2 crosses or molecular investigations of strains within an allele class can identify specific variants modifying disease and will help to expand our knowledge of naturally polymorphic networks driving aberrant disease responses.
Oas1b has been shown to act independently from the RNase L pathway to mediate protection against WNV infection. OAS1b lacks OAS activity, and hence does not produce 2-5A that otherwise binds and activates RNaseL (Brinton and Perelygin 2003; Perelygin et al. 2002). In addition to ABC and ORP1L protein binding by OAS1b, our network analysis also observed additional factors implicated in Oas haplotype control of WNV infection including DAXX (mediated apoptosis), Mov10 (antiviral), GRB2 (cell signaling and leukocyte migration), and Trafd1. Trafd1 has not previously been associated with the Oas family but appears within our QTL heatmap (Figure 5). Trafd1 is an immune regulator known to be activated by STAT1 in mouse bone-derived macrophages (Mashima et al. 2005), and possibly contributes to innate immune protection mediated by this Oas1b network.
Evolutionary studies have observed that there is a balancing selection occurring at the Oas1 gene locus across the human and nonhuman primate species. The allelic drift between primates produced variations in amino acids associated with RNA binding and this contributed to the susceptibility to flaviviruses (Ferguson et al. 2008). By performing a large genome scan across our diverse mouse backgrounds, we confirmed a QTL (containing the Oas genes and others) correlated with WNV infection. We chose here to observe the transcriptional differences influenced by Oas1b using the CC and allelic effects to determine the differential outcome (Figure S1). Our transcriptional analysis showed that there is still considerable strain-specific variation within the F1 backgrounds with functional Oas1b. While true disentangling of the relationship between genetic variants within these transcriptional networks and Oas1b allelic variants (e.g., via genome editing Oas1b loci within CC strains) is beyond the scope of this study, our results strongly suggest that the anti-WNV effect of Oas1b leads to differential activation of molecular programs. Further work to understand these programs, the role of genetic variants within its members, and the causal role of Oas1b, will help to elucidate the mechanisms of WNV disease and point to potential therapeutic responses to this disease.
We observed for the first time the role of parental effects of Oas1b and their differential host transcriptomics. Among the heterozygous F1s, we noted a slightly higher survival rate in F1s where Oas1b was functional in the mother (F/N). We also observed three immune regulatory networks influenced by WNV infection and the Oas1b haplotype. Our analysis found a surprising relationship with cell cycle checkpoint signaling, diapedesis, and Oas1b haplotype. We found that the Mov10 gene was included in this network of gene activation in F1s with heterozygous F/N alleles. Interestingly, Mov10 has been shown previously to function in an IRF3-dependent manner independent of the RIG-I pathway (Cuevas et al. 2016), implicating possible RIG-I-independent signaling within the Oas1b network to drive gene expression within this module.
We were also able to assemble an innate immune network linking functional Oas1b to proteins including IRFs, specific ISGs, ubiquitin protein ligases, and PARP genes. The F/F and some F/N F1s displayed earlier viral recognition, which appeared in transcriptional pathway analysis and qPCR through Ifit1 and Ifnβ1 expression. In symptomatic F1s lacking functional Oas1b (N/N), transcriptomics profiling indicated a lack of innate immune induction and cell migration that included monocyte migration and macrophage development. These observations indicate that, without activation of key processes such as innate immune signaling, diapedesis, and IFN responses, the host is unable to develop a strong defense and thus succumbs to infection in a manner linked to loss of Oas1b function. For example, the symptomatic N/F CC(055x028)F1 was unable to control viral replication in the spleen and developed neuroinvasive infection by day 12 (Figure S4). Lastly, our regulatory network (Figure 12) identified nuclear receptor coactivator 2 (NCOA2) as a novel transcriptional regulator producing an antiviral effect linked with Oas1b.
Understanding why some people are susceptible to viral infection while others are resistant is both a genetics and a genomics challenge that can be studied through genetics reference populations and infection models like the CC. Our study identifies new features of gene regulation linked with differential Oas1b genotypes within varied genetic backgrounds. This work also affirms the CC as a valuable tool for revealing the genetics of immune regulation and WNV control. CC mouse population is therefore an optimal model of virus infection amenable to defining genetic traits of disease outcome.
Supplementary Material
Supplemental material is available online at http://www.g3journal.org/content/7/6/1665.supplemental.
Acknowledgments
Thanks to Amy Green for proofreading, and to Darla Miller and Ginger Shaw for their support with mouse resources. Supported by National Institutes of Health grants U19AI100625, R01AI104002, and U19AI083019. The funders had no role in the design, data acquisition, analysis, or preparation of the manuscript.
Footnotes
Communicating editor: A. Long
Literature Cited
- Bigham A. W., Buckingham K. J., Husain S., Emond M. J., Bofferding K. M., et al. , 2011. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One 6(9): e24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon A. C., deBeauchamp J., Hollmann A., Luke J., Kotb M., et al. , 2009. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 83(20): 10417–10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D., Wilmot B., McWeeney S. K. 2014. Oligomask: a framework for assessing and removing the effect of genetic variants on microarray probes. R J. 6(1): 159–163. [Google Scholar]
- Brinton M. A., Perelygin A. A., 2003. Genetic resistance to flaviviruses. Adv. Virus Res. 60: 43–85. [DOI] [PubMed] [Google Scholar]
- Cartmell T., Poole S., Turnbull A. V., Rothwell N. J., Luheshi G. N., 2000. Circulating interleukin-6 mediates the febrile response to localised inflammation in rats. J. Physiol. 526(Pt. 3): 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi U. Y., Kang J. S., Hwang Y. S., Kim Y. J., 2015. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp. Mol. Med. 47: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney S. C., Di H., Stockman B. M., Liu H., Scherbik S. V., et al. , 2012. Identification of novel host cell binding partners of Oas1b, the protein conferring resistance to flavivirus-induced disease in mice. J. Virol. 86: 7953–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas R. A., Ghosh A., Wallerath C., Hornung V., Coyne C. B., et al. , 2016. MOV10 provides antiviral activity against RNA viruses by enhancing RIG-I-MAVS-independent IFN induction. J. Immunol. 9: 3877–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Suthar M. S., Gale M., Jr, Diamond M. S., 2009. Measure and countermeasure: type I IFN (IFN-alpha/beta) antiviral response against West Nile virus. J. Innate Immun. 5: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drappier M., Michiels T., 2015. Inhibition of the OAS/RNase L pathway by viruses. Curr. Opin. Virol. 15: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahesh H., Jha B. K., Silverman R. H., Scherbik S. V., Brinton M. A., 2011. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2–5A synthesis in intact cells. Virology 409(2): 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhateeb E., Tag-El-Din-Hassan H. T., Sasaki N., Torigoe D., Morimatsu M., et al. , 2016. The role of mouse 2′,5′-oligoadenylate synthetase 1 paralogs. Infect. Genet. Evol. 45: 393–401. [DOI] [PubMed] [Google Scholar]
- Engelhardt O. G., Ullrich E., Kochs G., Haller O., 2001. Interferon induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp. Cell Res. 271: 286–295. [DOI] [PubMed] [Google Scholar]
- Errett J. S., Suthar M. S., McMillan A., Diamond M. S., Gale M., Jr, 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J. Virol. 87(21): 11416–11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson W., Dvora S., Gallo J., Orth A., Boissinot S., 2008. Long-term balancing selection at the west nile virus resistance gene, Oas1b, maintains transspecific polymorphisms in the house mouse. Mol. Biol. Evol. 8: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M. T., Heise M., 2014. Quantitative genetics in the study of virus-induced disease. Adv. Virus Res. 88: 193–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M. T., Aylor D. L., Bottomly D., Whitmore A. C., Aicher L. D., et al. , 2013. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flammer J. R., Dobrovolna J., Kennedy M. A., Chinenov Y., Glass C. K., et al. , 2010. The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol. Cell. Biol. 19: 4564–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen B. L., 2014. The neuroimmune response to West Nile virus. J. Neurovirol. 2: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 4(9): 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. B., Thomas S., Swarts J., McMillan A. A., Ferris M. T., et al. , 2015. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio 6(3): e00493–e00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. B., Swarts J. L., Wilkins C., Thomas S., Green R., et al. , 2016. A mouse model of chronic West Nile virus disease. PLoS Pathog. 12(11): e1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L. E., Ferris M. T., Aylor D. L., Whitmore A. C., Green R., et al. , 2015. Genome wide identification of SARS-CoV susceptibility loci using the collaborative cross. PLoS Genet. 11(10): e1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L., Menachery V., Morgan A., Totura A., Beall A., et al. , 2017. Allelic variation in mouse Ticam2 contributes to SARS-CoV pathogenesis. G3 (Bethesda) 7: 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Wilkins C., Thomas S., Sekine A., Ireton R. C., et al. , 2016a Identifying protective host gene expression signatures within the spleen during West Nile virus infection in the collaborative cross model. Genom. Data 10: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Wilkins C., Thomas S., Sekine A., Ireton R. C., et al. , 2016b Transcriptional profiles of WNV neurovirulence in a genetically diverse collaborative cross population. Genom. Data 10: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi F.A., Mahajne M., Salaymah Y., Sandovski H., Tayem H., et al. Collaborative Cross Consortium , 2012. The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190(2): 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I. K., Zhang J., Gu H., 2009. Grb2, a simple adapter with complex roles in lymphocyte development, function, and signaling. Immunol. Rev. 232(1): 150–159. [DOI] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A., Mashimo T., Frenkiel M. P., Guénet J. L., Lucas M., et al. , 2006. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 281(8): 4624–4637. [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477(7364): 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Tugendreich S., 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen H., Gad H. H., Eskildsen-Larsen S., Despres P., Hartmann R., 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 1: 41–47. [DOI] [PubMed] [Google Scholar]
- Lazear H. M., Diamond M. S., 2015. New insights into innate immune restriction of West Nile virus infection. Curr. Opin. Virol. 11: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H. M., Lancaster A., Wilkins C., Suthar M. S., Huang A., et al. , 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 1: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Kim B., Oh G. T., Kim Y. J., 2013. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol. 4: 346–355. [DOI] [PubMed] [Google Scholar]
- Li H., Leo C., Zhu J., Wu X., O’Neil J., et al. , 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20: 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Banerjee S., Wang Y., Goldstein S. A., et al. , 2016. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. USA. Feb 23(8): 2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y. M., Gale M., Jr, 2008. Unveiling viral enablers. Nat. Biotechnol. 10: 1093–1094. [DOI] [PubMed] [Google Scholar]
- Loo Y. M., Gale M., Jr, 2011. Immune signaling by RIG-I-like receptors. Immunity 34(5): 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine A. E., Blakley I. C., Jagadeesan S., Harper J., Miller G., et al. , 2015. Analysis and visualization of RNA-Seq expression data using RStudio, bioconductor, and integrated genome browser. Methods Mol. Biol. 1284: 481–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., Dong B., Gale M., Jr., Silverman R. H., 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 448: 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T., Lucas M., Simon-Chazottes D., Frenkiel M. P., Montagutelli X., et al. , 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. USA 99(17): 11311–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima R., Saeki K., Aki D., Minoda Y., Takaki H., et al. , 2005. FLN29, a novel interferon- and LPS-inducible gene acting as a negative regulator of toll-like receptor signaling. J. Biol. Chem. 280(50): 41289–41297. [DOI] [PubMed] [Google Scholar]
- Morgan A. P., Fu C. P., Kao C. Y., Welsh C. E., et al. , 2015. The Mouse Universal Genotyping Array: From Substrains to Subspecies. G3 (Bethesda). 2015 12: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Gatti D. M., Keane T. M., Galante R. J., Pack A. I., et al. , 2017. Structural variation shapes the landscape of recombination in mouse. Genetics 206: 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F., Zippo A., Krepelova A., Cherubini A., Rocchigiani M., et al. , 2012. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol. Cell. Biol. 32(4): 840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreper D. G., Cai Y., Tarantino L. M., Pardo-Manuel de Villena F., Valdar W., 2017. Inbred strain variant database (ISVDB): a repository for probabilistically informed sequence differences among the collaborative cross strains and their founders. G3 (Bethesda) 7: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin A. A., Scherbik S. V., Zhulin I. B., Stockman B. M., Li Y., et al. , 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA 99(14): 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman R., Schiemann W. P., Brooks M. W., Lodish H. F., Weinberg R. A., 2001. TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat. Cell Biol. 3: 708–714. [DOI] [PubMed] [Google Scholar]
- Rasmussen A. L., Okumura A., Ferris M. T., Green R., Feldmann F., et al. , 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346(6212): 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbik S. V., Paranjape J. M., Stockman B. M., Silverman R. H., Brinton M. A., 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80(6): 2987–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A. L., Rosenwald A., Hurt E. M., Giltnane J. M., Lam L. T., et al. , 2001. Signatures of the immune response. Immunity. 15: 375–385. [DOI] [PubMed] [Google Scholar]
- Shaw T., Martin P., 2009. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 8: 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. R., Odet F., Aylor D. L., Pan W., Kao C.-Y., et al. , 2017. Male infertility is responsible for nearly half of the extinction observed in the Collaborative Cross. Genetics 206: 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui M. A., Malathi K., 2012. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 287: 43651–43664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K., 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Morgan A. P., Najarian M., Sarsani V. K., Sigmon J. S., et al. , 2017. The genomes of the collaborative cross. Genetics 206: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M. S., Ma D. Y., Thomas S., Lund J. M., Zhang N., et al. , 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 6(2): e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M. S., Diamond M. S., Gale M., Jr, 2013. West Nile virus infection and immunity. Nat. Rev. Microbiol. 2: 115–128. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43(7): 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A complete description of the data and methods can be found on the manuscript’s Github page (https://github.com/greener98103/oas1b/wiki) and in the supplemental file titled “Green_Oas1b_data_policy_documentation.” This file includes information on strains/lines, variant files from Sanger, and marker files [MUGA platform (Morgan et al. 2015)]. The data document also contains detailed descriptions, file locations, and links to executable code for QTL, transcriptional, and correlation analyses. Gene expression data are in GEO with the accession number GSE91003. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.