Abstract
Multi-parental populations are promising tools for identifying quantitative disease resistance loci. Stem rot caused by Ganoderma boninense is a major threat to palm oil production, with yield losses of up to 80% prompting premature replantation of palms. There is evidence of genetic resistance sources, but the genetic architecture of Ganoderma resistance has not yet been investigated. This study aimed to identify Ganoderma resistance loci using an oil palm multi-parental population derived from nine major founders of ongoing breeding programs. A total of 1200 palm trees of the multi-parental population was planted in plots naturally infected by Ganoderma, and their health status was assessed biannually over 25 yr. The data were treated as survival data, and modeled using the Cox regression model, including a spatial effect to take the spatial component in the spread of Ganoderma into account. Based on the genotypes of 757 palm trees out of the 1200 planted, and on pedigree information, resistance loci were identified using a random effect with identity-by-descent kinship matrices as covariance matrices in the Cox model. Four Ganoderma resistance loci were identified, two controlling the occurrence of the first Ganoderma symptoms, and two the death of palm trees, while favorable haplotypes were identified among a major gene pool for ongoing breeding programs. This study implemented an efficient and flexible QTL mapping approach, and generated unique valuable information for the selection of oil palm varieties resistant to Ganoderma disease.
Keywords: plant disease, multiparental population, quantitative disease resistance loci, survival analysis, identity-by-descent, MPP
Plant diseases cause high yield losses for almost all crops worldwide, thus constituting a major food security constraint in a context of high population growth and shrinking cropland (Oerke and Dehne 2004). The genetics of plant resistance to pathogens is thus an active research area to support the selection of resistant crop varieties in the framework of integrated pest management programs geared toward limiting the use of chemical pesticides. R-genes controlling qualitative disease resistance have been identified in plants mainly by using biparental populations derived from crosses between resistant and susceptible parents (Gururani et al. 2012). Despite the efficiency of R-genes in the selection of resistant varieties, the sustainability of resistance could be improved by taking quantitative resistance loci into account (Poland et al. 2009), as shown empirically in studies on oilseed rape (Brun et al. 2010) and pepper (Quenouille et al. 2014). Broadening the genetic diversity screened using association panels is a way to identify such QTL (Wei et al. 2006; Adeyanju et al. 2015). Based on improved genetic and statistical properties (Verhoeven et al. 2005), multi-parental populations (MPPs) are increasingly being developed in many organisms (see the MPP collection http://www.genetics.org/content/multi-parental_populations), and their use for mapping quantitative resistance loci is promising (Kump et al. 2011; Benson et al. 2015; Cockram et al. 2015; Bajgain et al. 2016). Studies carried out in different contexts are needed to explore alternative experimental designs, appropriate statistical analyses, and to assess the potential of MPPs to gain insight into the genetic architecture of plant disease resistance.
Oil palm (Elaeis guineensis Jacq.) is a perennial allogamous species of African origin. It is currently the world’s leading fat resource, accounting for >30% of global vegetable oil production (USDA 2015), with a constantly increasing demand (FAOSTAT, http://www.fao.org/faostat/). Southeast Asia represents almost 90% of global production, but oil palm cultivation in this area is highly hampered by stem rot diseases, i.e., basal (BSR) or upper (USR). BSR and USR are caused by Ganoderma boninense, a soil hemi-biotroph fungal pathogen that is also found in Africa and South America, where the disease is currently developing. Ganoderma disease is economically problematic when affecting ∼10–20% of palm trees, while 30–70% could be lost by the end of a typical 25-yr planting cycle, leading to substantial yield losses, premature replantation, and a waste of cropland (Durand-Gasselin et al. 2005; Cooper et al. 2011). The identification of genetic resistance in breeding material, based on field observations (de Franqueville et al. 2001) or screening tests with seedling inoculation (Idris et al. 2004; Breton et al. 2006), has led to the selection of partly resistant varieties. Information on the genetic architecture and molecular nature of Ganoderma resistance could help shorten the long oil palm breeding cycle (up to 20 yr), and enable its multi-criteria improvement using a marker-assisted selection strategy. All studies on Ganoderma disease to date have been based on seedling inoculation at the nursery stage: transcriptomic (Tee et al. 2013; Ho et al. 2016), proteomic (Al-Obaidi et al. 2014), or metabolomic (Nusaibah et al. 2016) approaches have identified genes, proteins, and pathways affected by Ganoderma infection, while an analysis based on 58 simple sequence repeat markers found alleles associated with Ganoderma symptoms in resistant vs. susceptible families (Hama-Ali et al. 2015). Field experiments are needed to assess broader genetic diversity in the agronomic context, but QTL analyses on oil palm crosses in the field are often not sufficiently effective because of population size and allelic segregation limitations (Jeennor and Volkaert 2014; Lee et al. 2014; Pootakham et al. 2015). Recent alternatives, based on data from breeding programs involving multiple connected families, have yielded promising results on agronomic traits (Billotte et al. 2010; Tisné et al. 2015).
In this study, an oil palm multi-parental population (Eg9PP) derived from nine founders of an ongoing breeding program was used to map Ganoderma disease resistance loci. Natural Ganoderma infection was monitored in the field, and the infection status of the 1200 Eg9PP individuals were recorded over 25 yr to generate a unique dataset. An innovative statistical modeling approach was implemented to map genetic loci affecting the survival of Eg9PP individuals, by combining a Cox regression model used for survival time QTL mapping in animal (Durrant et al. 2011; Gonen et al. 2015) or plant (Luo et al. 2013; Jiang et al. 2014) species, with an identity-by-descent (IBD)-based variance component approach (George et al. 2000) adapted to MPP design and with spatial effects modeling in order to control the confounding effects of Ganoderma epidemics. As Eg9PP parents are from genetic origins that are widely used in oil palm breeding programs, loci identified in this study provided highly relevant information to guide efficient marker-assisted selection for Ganoderma resistance.
Materials and Methods
Genetic material
The oil palm (Elaeis guineensis Jacq.) multi-parental population (Eg9PP) was developed from 14 full-sib families generated by crossing five outbred parents of heterotic group A with four outbred parents of heterotic group B in an incomplete factorial design (Figure 1 and see Supplemental Material, Table S1). The group A parents belonged to the “Deli” population derived from four individuals planted in 1848 in Indonesia, and the group B parents belonged to “La Mé” (Cote d’Ivoire) or “Yangambi” (Democratic Republic of the Congo) populations (Corley and Tinker 2003). The Eg9PP parents are major founders of the PalmElit breeding program, a CIRAD subsidiary and leading oil palm breeding company (www.palmelit.com). This breeding program is shared and conducted with partners, i.e., PT Socfin Indonesia (Indonesia) and INRAB (Benin). The 1200 Eg9PP individuals were planted in 1986 on the SOCFINDO estate (Medan, Indonesia), according to a randomized complete block design (RCBD) with at least five replications per family, each replication consisting of a plot with 15 full-sib individuals (see Figure S1). Despite the highly favorable agro-climatic conditions for oil palm growing, this location is hampered by the presence of G. boninense—the causal agent of basal stem rot disease that naturally infected Eg9PP.
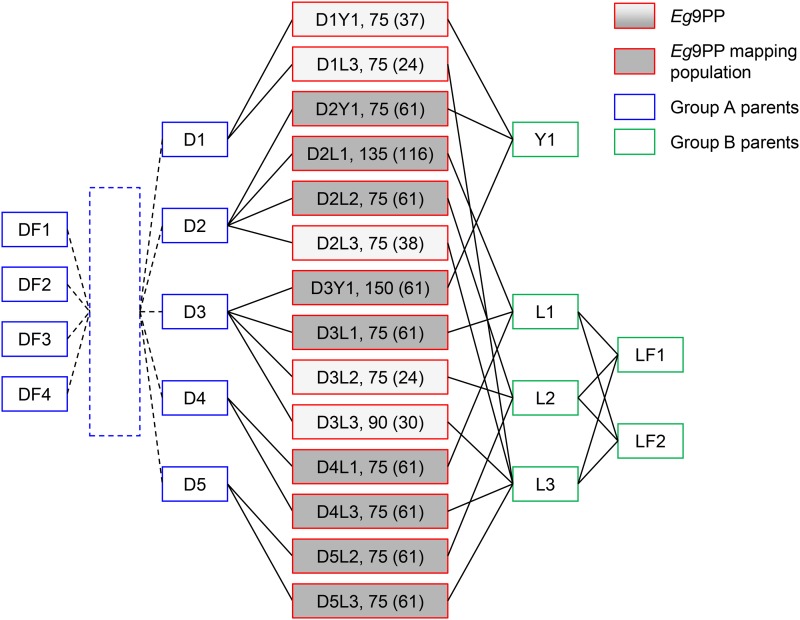
Figure 1.
Pedigree of the Eg9PP oil palm multi-parental population. Eg9PP (red boxes, n = 1200) consisted of 14 full-sib families obtained by crossing parents from heterotic group A (blue boxes) and B (green boxes). The five parents from group A (D1–D5) derived from four founders (DF1–DF4) by partially known successive crosses and self-pollination (dotted lines). The number of individuals per family are indicated at the right side of the family name, with the number of sampled individuals in parenthesis (n = 757). Shaded boxes represent the fully genotyped families that constituted the mapping population (n = 604).
Pedigree information was available for the La Mé parents, while the pedigree of the Deli parents was only known for a few (one or two) generations before the Eg9PP parents. The relatedness of the five Deli parents was estimated using pedigree reconstruction with MOLCOANC 3.0 software, as described in Cros et al. (2014), by using a larger Deli palm population (Tisné et al. 2015).
Phenotype acquisition
The G. boninense infection status was recorded biannually on the 1200 Eg9PP individuals over 25 yr, from the first year after planting (June 1987) to the uprooting of the genetic trial (June 2012). Symptoms were scored blindly on the basis of a six level scale, set up in the 1980s, and used since that time in the SOCFINDO plantations, with zero being a healthy palm tree and six a dead and fallen palm tree with the presence of Ganoderma fruiting bodies on the palm trunk (see Figure S2 and Table S2). A seventh level represented palm trees absent at the visit time, or dead for reasons other than Ganoderma disease. Raw data were then curated, and the occurrence of two events was recorded with the time associated: the first Ganoderma symptom appearance (T1S, first observation of score 2–6), and the death of the palm tree due to Ganoderma disease (TD, first observation of score 6). Events of interest corresponded to T1S and TD, and the associated times were considered as survival times, i.e., times from the start of the experiment to the time the event occurred (see Table S3 for phenotypic data). When the events were observed, the associated times were called exact or uncensored times, otherwise they were called censored observations.
Molecular data and genetic map construction
A total of 757 individuals out of the 1200 from Eg9PP were sampled before uprooting the genetic trial because of premature death or illegitimacy (Figure 1). In a previous study, 116 individuals from the reference family D2L1 were genotyped with 390 single sequence repeat (SSR) markers, and a first reference high density linkage map grouping 255 SSRs and 688 amplified fragments length polymorphisms (AFLP) markers was published (Billotte et al. 2005). The SSR markers, plus 26 SSR additional markers previously mapped in a pedigree-based linkage analysis that was recently published (Cochard et al. 2015), were used to genotype 604 individuals from nine Eg9PP families, including D2L1, which constituted the mapping population (Figure 1). Based on genotyping of the Eg9PP parents, and the genetic position of SSR markers, different SSR marker subsets were used depending on the family, to optimize the genome coverage and marker information level (see Table S4 and Table S5 for individual genotypes). A QTL region identified in the linkage analysis of the mapping population (n = 604) was investigated in further detail by genotyping all of the 757 sampled individuals from the 14 Eg9PP families with all of the SSR markers mapped in the region plus two markers (see Table S4). DNA extraction, microsatellite amplification, and analyses were performed according to the protocol described in Cochard et al. (2015), and raw genotype data are available in Table S5.
The Eg9PP genetic map was constructed with CRI-MAP version 2.507 (Green et al. 1990). This program allows simultaneous analysis of multiple families and performs multipoint linkage analysis. The nine families of the Eg9PP mapping population were treated as unrelated families, and all individuals were considered as being of undetermined sex. Markers were grouped in separate input files based on the linkage group defined in the previously published genetic map (Billotte et al. 2005; Cochard et al. 2015). Markers were ordered in each linkage group using several runs of the BUILD command on the most informative markers, then the remaining markers were placed in the framework obtained using the ALL command. The FLIPS command was used to check alternative marker orders at the different linkage group construction stages, using different values up to four, depending on the number of markers and length of the linkage group analyzed. The FIXED option was finally used to calculate the Kosambi genetic distances between markers (see Figure S3 and Table S4). The genetic map obtained was drawn using MapChart software (Voorrips 2002).
We attempted to infer a physical position for each marker mapped using BLASTn (Zhang et al. 2000). We used forward and reverse primer sequences for each marker as query sequences against the oil palm genome sequence as the target database (Singh et al. 2013). We filtered the result hits of BLASTn with a minimum e-value threshold of 0.5. The marker position was assigned easily when forward and reverse primers were found in the same genome location. In case of multiple hits on different chromosomes for the same primer pair, the marker position was assigned based on the hit with the greatest e-value. Inferred physical positions of markers are available in Table S4.
Based on the marker physical positions, the physical positions of 210 predicted R-genes in the oil palm genome (Singh et al. 2013; Chan et al. 2017) were projected on the Eg9PP genetic map in order to identify colocalizations with Ganoderma resistance QTL (See Table S8).
Linkage mapping
The linkage mapping of Ganoderma disease resistance was performed using a combination of survival data analyses and the two-step variance component approach described in George et al. (2000). The first step involved computation of IBD kinship matrices over a grid of genetic positions. The second step consisted of QTL presence tests over the grid of genetic positions by comparison of various models including random effect terms.
IBD matrix computation:
The variance component approach requires IBD kinship matrices as variance-covariance matrices for testing QTL effects at each genetic position. IBD analyses using SimWalk2 software (Sobel et al. 2001) were conducted to estimate the IBD matrices formed by empirical kinship coefficients for each pair of Eg9PP individuals and their parents. Simwalk2 implements a Bayesian framework using a Markov chain Monte Carlo (MCMC) algorithm to select the most likely genetic descent graph depicting inheritance patterns within pedigrees (Sobel and Lange 1996). Empirical kinship coefficients were first calculated between the 604 fully genotyped Eg9PP individuals and their parents at every marker position and over a 3 cM interval grid, leading to 1006 evaluation points for QTL presence testing (available at https://doi.org/10.6084/m9.figshare.4780489.v1).
Statistical modeling:
The Cox regression model (Cox 1972) is a popular choice for analyzing the relationship between the survival outcome and explanatory covariates X. This semiparametric model has the following hazard function:
| (1) |
where is a unknown vector of parameters reflecting the effects of covariates on survival, is the time to the event or censoring, and denotes the baseline hazard function. Note that no particular shape was assumed for the baseline function.
In this study, the analysis of data based on natural field infection required taking into account the spatial component of the propagation of Ganoderma disease. The spatial (plot) information was included to model (1), as well as genetic (family and individual) information to avoid the genetic variance being absorbed by spatial variance due to the experimental design. One way of considering this information is to extend the model (1) to a mixed effect Cox model by adding one random effect with a covariance matrix based on pedigree, and one with a spatially structured covariance matrix based on the geographic coordinates of the plot. Vaupel et al. (1979) were the first to introduce random effects (called frailties) in the survival model in order to group time-to-event data within clusters.
Model (2) is then defined as follows:
| (2) |
with corresponding to the number of individuals, is the design matrix relating the observations to the vector of family fixed effects, the design matrix relating the survival outcome to the vector of spatial random effects at the plot level, and is the design matrix relating the survival outcome to the additive genetic random effects with m the number of parents.
Concerning the spatial random effects, a conditionally auto-regressive (CAR) structure (Cressie 1993) was used in order to model dependence among observations in neighboring plots (Banerjee et al. 2003). Where denotes the neighborhood of plot including the plots which share a common boundary, the distribution of is a multivariate Normal distribution defined by:
where α is the spatial autocorrelation parameter, the adjacency matrix (i.e., if s and s′ are neighbors, and otherwise), is the spatial dispersion parameter, and D is a diagonal matrix with diagonal elements equal to i.e., the number of neighbors for the plot s. The additive genetic random effects also follow a multivariate Normal distribution:
with being the kinship matrix based on pedigree information, and the additive genetic variance.
To save computation time in the QTL mapping step, the model (2) was used for predicting spatial random effects associated with each plot, and the spatial effects predictions were included as fixed effects in the following QTL models (see Table S7 for model (2) results). The model considered as null model for QTL mapping was then defined by:
| (3) |
where is the design matrix relating the survival outcome for genotyped individuals to fixed effects (family and spatial effects). The null model defined in (3) was compared to the model (4) such that:
| (4) |
with being the design matrix relating the survival outcome for genotyped individuals to the QTL random effect at the pth position on the genome. The QTL random effect at the pth position followed a multivariate normal distribution, with a variance-covariance matrix equal to the IBD kinship matrix at the pth position then being
A genome scan was performed to test the QTL presence by computing the log-likelihood ratio to compare models (3) and (4) at each p position. Best linear unbiased predictions (BLUPs) of QTL effect were computed for each Eg9PP individual using model (4) at the retained genetic positions (see next paragraph).
Log-likelihood ratio test:
The log-likelihood ratio test (LRT) was used to test whether frailty (QTL) variance was null at each p position. With a non-negative restriction on the variance component, the LRT is one-sided and has null value on the boundary of parameter space, so the classical likelihood ratio asymptotic chi-squared distribution theory is no longer valid. The distribution of LRT is then unknown under the null hypothesis H0, so an empirical approach by permutations (Churchill and Doerge 1994) and a theoretical approach were used to estimate significance levels. In the permutation approach, different LRT significance levels were calculated for each putative QTL based on 500 chromosome-wide permutations obtained by shuffling the identity of individuals in the data to break genotype-phenotype associations. For each permutation, the maximum LRT value obtained was recorded to construct the distribution of the maximum LRT value under the null hypothesis (see Figure S4A). The empirical type I error rates associated with each LRT distribution were calculated so as to estimate the significance level of each LRT peak detected (see Figure S4B). The theoretical approach was based on different works in shared frailty model framework (Duchateau et al. 2002; Vu and Knuiman 2002; Claeskens et al. 2008) that assessed the limiting distribution of LRT. The resulting distribution is the asymptotic 50:50 mixture of the and the chi-squared distributions of 0 and 1 degree of freedom, respectively, that was used to calculate p-values associated to each QTL detected.
Haplotype analysis:
QTL mapping identified a strong QTL in the top of linkage group 1, and this region was analyzed in further detail. Based on the genotyping of all the 757 sampled individuals from the 14 Eg9PP families with seven SSR markers covering 20 Mb (see Table S4), the kinship coefficients were calculated between the 757 Eg9PP individuals and used to perform QTL mapping with this extended dataset in order to determine precise QTL location in the candidate region. Then, haplotype analysis was performed in the candidate region using SimWalk2 to identify the funder allele segregation in the Eg9PP individuals. Funder allele effects were then estimated including two fixed effects in model (3)—one for each heterotic group—in order to identify favorable alleles among the Eg9PP parents at the QTL peak closest marker.
All analyses were performed with R software version 3.2.3 (R Development Core Team 2012) and the coxme package (Therneau and Therneau 2015), and R scripts, with example data files and documentation, are available at the github repository https://github.com/DenisMarie/Eg9PP_Ganoderma. The genome scan results are available in Table S6.
Data availability
Eg9PP was uprooted in 2012, but Eg9PP families can be reproduced by crossing individuals derived from the self-fertilized progenies of Eg9PP parents. Requests for such genetic material should be addressed to PalmElit (www.palmelit.com). Raw genotype data are available in Table S5 and IBD kinship matrices at https://doi.org/10.6084/m9.figshare.4780489.v1. Phenotyping of disease events are available in Table S3. All R scripts, data file and documentation are available at https://github.com/DenisMarie/Eg9PP_Ganoderma.
Results
Variations in Ganoderma resistance in Eg9PP
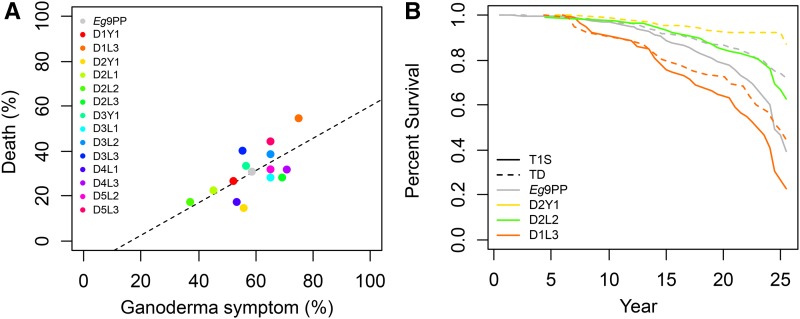
Ganoderma disease resistance was assessed in 1200 Eg9PP individuals by biannually recording their infection status over 25 yr. At the end of the experiment, 58.5% of the individuals exhibited Ganoderma symptoms over this period, and 30.5% died due to Ganoderma disease (Figure 2A and see Table S7). These percentages ranged from 74.7 and 54.7% for infected and dead individuals, respectively, for the worst family D1L3, to 37.3 and 14.7% for the best families D2L2 and D2Y1 (Figure 2A and see Table S7). The percentages of individuals per family infected and dead at the uprooting of the trial were significantly correlated (r = 0.66, Figure 1A), but families with a similar percentage of infected individuals exhibited a substantially different percentage of death (e.g., D3L3 and D2Y1, Figure 2A). The times of the first Ganoderma symptom appearance (T1S) and death due to Ganoderma (TD) were modeled using Cox regression including spatial and family effects, and both effects were found to be highly significant (P < 0.001). Risks per family ranged from 0.32 to 0.98 for T1S, corresponding to 3.1-fold lower risk for the best family (D2L2) compared to the worst family (D1L3) (Figure 1B and see Table S7). For TD, risks per family ranged from 0.2 to 0.96, corresponding to 4.9-fold lower risk for the best family (D2Y1) compared to the worst family (D1L3) (Figure 1B and see Table S7). In addition to this interfamily variance, large confidence intervals for risk per family indicated other sources of variance that could be captured by QTL effects in families (see Table S7). Kaplan-Meier estimates of survival showed that the economically significant threshold of 20% affected palm trees was reached 15 yr after planting for the worst Eg9PP family as compared to 24 yr for the best (Figure 1B).
Figure 2.
Ganoderma disease epidemics in the Eg9PP oil palm multi-parental population. (A) Relationship between the percentages of affected and dead individuals per family at the trial uprooting. Colors corresponding to families are indicated in the legend at the top left, with gray circles indicating the Eg9PP value (n = 1200). Dotted lines represent the linear regression between the percentages of affected and dead individuals (slope = 0.72, Pearson correlation coefficient r = 0.66, p-value = 0.009). (B) Survival curves for Eg9PP and Eg9PP families exhibiting maximum (D1L3) and minimum (D2Y1, D2L2) percentages of affected and dead individuals at the trial uprooting (color legend at bottom left). Survival estimates are plotted for two events, the time of the first Ganoderma symptom observation (T1S, solid lines), and death due to Ganoderma infection (TD, dotted lines).
Linkage mapping of Ganoderma resistance loci
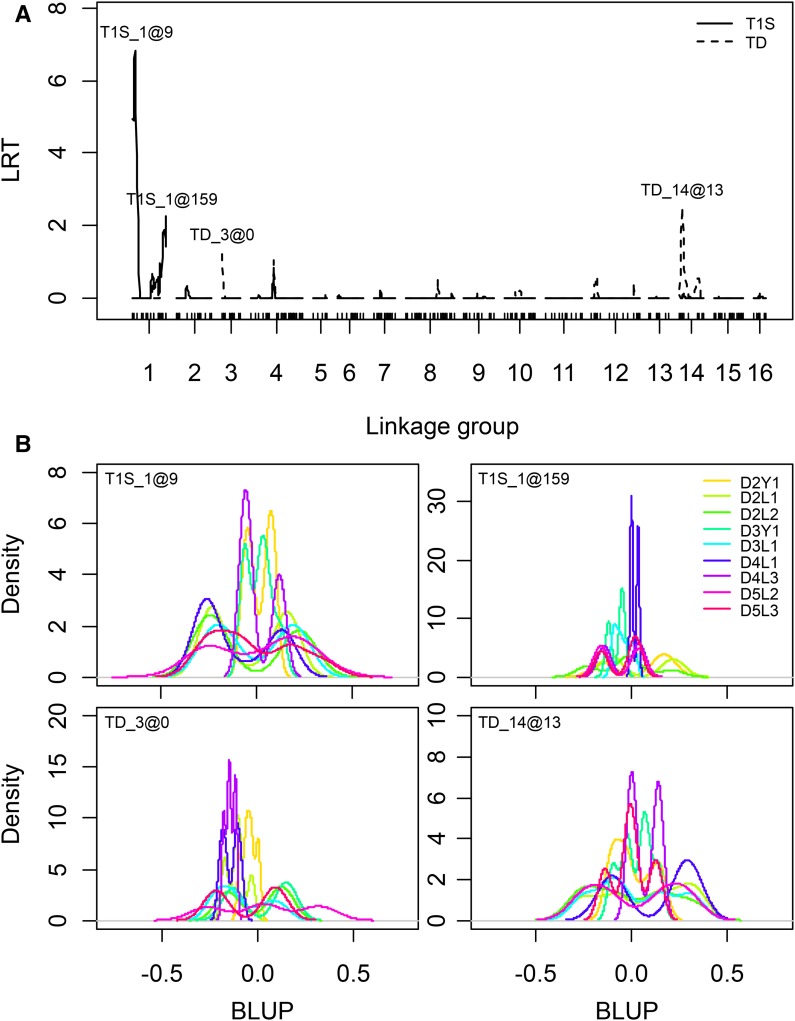
QTL analysis in the Eg9PP mapping population (n = 604) identified two regions associated with the time of the first symptom observation (T1S), and two with the time of death due to Ganoderma infection (TD), with no overlap between them (Figure 3A). The inclusion of spatial effects in the base model did not change the QTL results for T1S despite the reduced overall LRT level (see Figure S5). On the contrary, three LRT peaks for TD were lost by adding the spatial effect (middle of LG1 and LG4, bottom of LG14), but a peak at the top of LG3 was increased (see Figure S5). The strongest QTL was found for T1S at the top of LG1, with an LRT of 6.83 at a significance level (α) of 0.01 (T1S_1@9, Table 1). The variance associated with T1S_1@9 was 0.16, corresponding to a risk = 1.49-fold above the norm for ∼15% of the individuals, with a similar fraction of individuals having lower risk. The distribution of individual BLUP indicated the greatest risk segregation in families from La Mé parents L1, L2, and L3 (Figure 3B). The second QTL for T1S (T1S_1@159) was detected at the bottom on LG1, with a risk of 1.30 (α = 0.29), with risk segregation found mainly in families from parent D2 (Figure 3B and Table 1). Both QTL detected for TD, TD_3@0, and TD_14@13 at the top of LG3 and LG14, respectively, had high risks (1.45) but low significance levels (α = 0.38 and 0.19, respectively), with a risk BLUP distribution indicating the greatest QTL segregation in families from parents D3 and D5 for TD_3@0, and L1 and L2 for TD_14@13 (Figure 3B and Table 1).
Figure 3.
Genetic determinism of Ganoderma resistance in the Eg9PP oil palm multi-parental population. (A) LRT testing the presence of a QTL for the time of the first Ganoderma symptom observation (T1S, solid lines) and death due to Ganoderma infection (TD, dotted lines) in the Eg9PP mapping population (n = 604). Dashes on the x-axis represent markers on the genetic map. Labels above LRT peaks (Trait_Linkage group@position in cM) indicate QTL with an LRT below a type I error rate of 0.5. (B) BLUP distributions for random QTL effects (QTL names at top left of each panel) for the Eg9PP families of the mapping population (n = 604, color legend at top right).
Table 1. QTL associated with Ganoderma resistance in the Eg9PP oil palm multi-parental population.
| Traits | QTLa | LRTb | αc | p-valued | Variance | Closest Marker | Chromosomee | Physical Positionf |
|---|---|---|---|---|---|---|---|---|
| T1S | ||||||||
| T1S_1@9 | 6.83 | 0.01 | 0.004 | 0.16 | mEgCIR3803 | 3 | 58.17 | |
| T1S_1@159 | 2.24 | 0.29 | 0.067 | 0.07 | mEgCIR0332 | 3 | — | |
| TD | ||||||||
| TD_3@0 | 1.23 | 0.38 | 0.134 | 0.14 | mEgCIR3698 | 8 | 30.15 | |
| TD_14@13 | 2.50 | 0.19 | 0.057 | 0.14 | mEgCIR2427 | 11 | 23.86 |
QTL name indicates the trait, the number of linkage group and the peak position on the genetic map in cM.
LRT Log-likelihood ratio test.
Empirical type I error rate determined by permutation tests.
p-values computed from a 50:50 mixture of the and the
Chromosome number according to Singh et al. (2013).
Physical position of the closest marker in Mb.
Haplotype analysis of the major Ganoderma resistance QTL
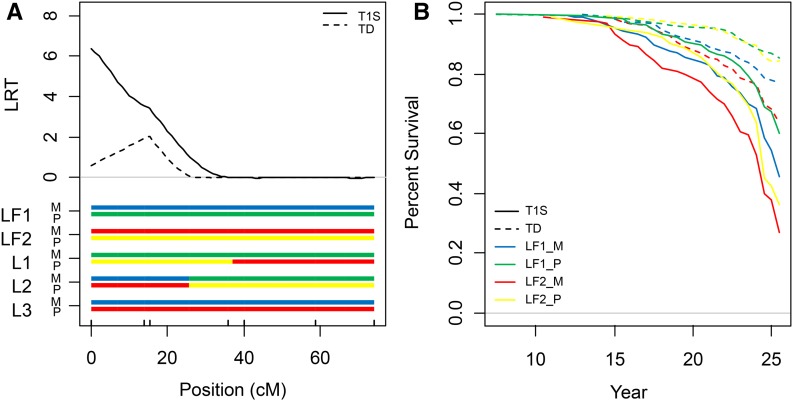
T1S_1@9, the major QTL for Ganoderma resistance, was investigated in further detail by genotyping all sampled Eg9PP individuals (n = 757) with the five SSR markers in the region (LG1, 0–44.5 cM), by adding two additional SSR markers and performing IBD and haplotype analyses. IBD-QTL mapping on the augmented dataset confirmed the presence of a QTL for T1S on LG1, but the LRT peak position shifted to 0 cM on the mEgCIR1713 marker (Figure 4A). While no QTL were found for TD in this region, a LRT peak was detected with the augmented dataset close to the mEgCIR3803 marker (Figure 4A). Cox regression models including two fixed effects for haplotypes at the closest markers in each heterotic group indicated significant effects in group A (p-value < 0.01) and group B (p-value < 0.001) for T1S as well as for TD (p-values < 0.01 and < 0.001 for groups A and B, respectively). The four La Mé founder IBD alleles were present in this region in the three La Mé Eg9PP parents, with two recombination events in the T1S_1@9 region in founder meiosis (Figure 4A). Marked differences in estimated risk were observed for individuals carrying alternative La Mé founder haplotypes: individual with an LF1 paternal allele (LF1_P, Figure 4B) had a 2.1-fold lower risk of being infected by Ganoderma and 2.07-fold lower risk of dying due to Ganoderma compared to the worst allele (LF2_M). The survival estimates showed that the 20% affected palm tree threshold was reached 19.5 yr after planting for individuals carrying the LF2_M allele, as compared to >23.5 yr for those carrying the LF1_P allele (Figure 4B). Moreover, this haplotype analysis allowed us to identify favorable alleles within the Yangambi and Deli genetic backgrounds associated with contrasted risks of Ganoderma infection and death (see Figure S6).
Figure 4.
Haplotype analysis of Ganoderma resistance QTL T1S_1@9. (A) LRT testing the presence of a QTL at the top of linkage group 1 for the time of the first Ganoderma symptom observation (T1S, solid lines) and death due to Ganoderma infection (TD, dotted lines) in the Eg9PP sampled individuals (n = 757). Dashes on the x-axis represent markers on the genetic map. La Mé founder (LF1, LF2), and parent (L1 to L3) maternal (M) and paternal (P) IBD alleles are represented by different colors below the LRT lines. (B) Survival curves for the Eg9PP sampled individuals (n = 757) according to the IBD allele they inherited from their La Mé parent. Survival estimates are plotted for two events, i.e., the time of the first Ganoderma symptom observation (T1S, solid lines), and death due to Ganoderma infection (TD, dotted lines).
Discussion
This study was based on data from monitoring natural field infection of an oil palm multi-parental population over 25 yr. This unique dataset provided an opportunity to map Ganoderma resistance loci in natural conditions according to an economic and efficient in silico approach, i.e., based on data from breeding programs (Parisseaux and Bernardo 2004). Four Ganoderma resistance loci were identified by combining different statistical methods, including survival analysis, infection spatial modeling, and a variance component approach for QTL mapping.
Disease resistance loci mapping using the Cox regression model
The QTL mapping approach was developed in the framework of normally distributed quantitative traits. In this study, Ganoderma infection data were treated as survival traits, with censoring because of unobserved events. Such data are commonly found in human or animal studies in which most of the statistical methods were developed. Zhao (2005) proposed a statistical approach using a mixed-effects Cox model to study familial effects in alcohol dependence. In salmon, disease resistance loci were mapped in complex populations using a variance component approach on binary survival data (Gonen et al. 2015). In plants, especially annual ones, survival analyzes are seldom performed because of the experimental potential, but methods have been developed to map QTL associated with flowering time in biparental rice (Luo et al. 2013) and barley (Jiang et al. 2014) populations. As suggested in Zhao (2005), QTL mapping was performed in this study using IBD kinship matrices, calculated with Simwalk2 (Sobel et al. 2001), as covariance matrices of the QTL random effect. This approach is flexible in terms of population design, and was found to be efficient in a former study on agronomic quantitative traits in a complex oil palm pedigree using a linear mixed effect model (Tisné et al. 2015). Modeling of a spatial effect to account for the variance associated with the natural inoculation of the palm trees was found to be highly significant, and changed the time of death mapping results. However, it is unclear whether it improved the mapping results without additional information, e.g., from simulation studies. Further developments could improve the mapping method. Modeling QTL effects including two variance components, one for each heterotic group, would enable independent testing of the QTL presence in each genetic background and assessing the importance of nonadditive effects, i.e., dominance or epistasis. A multi-QTL approach would also be worthwhile as the multiple QTL methods proved their superiority in estimating QTL effects and position (Jansen 1993). This could be achieved using a variable selection approach in the Bayesian framework, as developed for quantitative traits (Bink et al. 2008).
The approach described in this study—flexible, efficient, with further possible extensions—could be interesting for studying disease resistance in plants, especially for perennials, which have limited experimental potential.
Genetic architecture of Ganoderma disease resistance
Plant disease resistance is commonly partitioned between R-gene mediated qualitative or complete resistance, and quantitative or incomplete resistance controlled by many quantitative resistance loci (Poland et al. 2009). In the present study, no QTL were found that completely prevented Ganoderma infection or death, which is consistent with field observations that could not highlight such complete resistance (Durand-Gasselin et al. 2005). Colocalizations of Ganoderma resistance QTL with oil palm predicted R-genes were observed (see Table S8), but further work would be needed to validate these genes as candidate. Moreover, R-gene mediated complete resistance was observed almost exclusively with biotrophic pathogens, so it is unlikely that such resistance occurs in oil palm, with Ganoderma being an hemi-biotrophic pathogen (Ho and Tan 2015). However, marked delayed effects on the time of the first symptom observation (T1S) and the time of death (TD) were observed among families (threefold to fivefold, respectively), with high variance associated with QTL noted for both T1S and TD. The different QTL patterns observed for T1S and TD, without any colocalizations noted, except at the top of LG1, was a striking result. This is consistent with the correlation at the family level between T1S and TD being looser than expected, indicating variation in the survival time when infected by Ganoderma. This could still be linked to the two stages of infection reported for Ganoderma, i.e., biotrophic then necrotrophic, as it is known that both induce different immune responses that could lead to different genetic architectures for T1S and TD. Poland et al. (2009) put forward six hypotheses on the mechanisms underlying quantitative resistance loci, with the first being the effect of pleiotropic genes regulating morphological and developmental phenotypes. Mapping of yield and morphology traits in Eg9PP (unpublished results) revealed colocalization of TD QTL with fruit weight and composition (TD_3@0) and stem height (TD_14@13), indicating possible pleiotropic effects as a basis of quantitative resistance to Ganoderma.
This is the first study that identified the genetic determinism of Ganoderma resistance in the field, so the detected QTL need further validation in independent experiments, accompanied by a fine mapping approach to identify the underlying genes. Despite a marker density suitable for QTL mapping (one marker every 8 cM on average), the analysis of Eg9PP would benefit from denser marker sets, e.g., SNP obtained by genotyping-by-sequencing (Pootakham et al. 2015) or high density genotyping array (Kwong et al. 2016) that were recently developed in oil palm. Haplotype analysis of the T1S_1@9 region exhibited many recombination events (105 in 1.9 Mb), indicating that densifying Eg9PP genotyping in the candidate regions could lead to candidate gene identification. Eg9PP individuals were uprooted but leaf samples are still available, and acquisition of a denser genotyping is in progress. Although field experiments are certainly more relevant, nursery tests with inoculation are interesting to accumulate genetic and omic knowledge on Ganoderma resistance. Unfortunately, the only nursery-based genetic study to date (Hama-Ali et al. 2015) showed no overlap between field resistant QTL and SSR markers associated with Ganoderma disease incidence in the nursery despite some common markers and genetic backgrounds (Deli × Yangambi). The scope of the study, i.e., involving 79 individuals from one tolerant and two susceptible families, genotyped with 58 SSRs, made it difficult to conclude on the transferability of the QTL results from the nursery to the field, and further research is needed to map resistance QTL based on nursery inoculated populations.
Contribution to Ganoderma resistance loci in oil palm breeding programs
The multi-parental population design allowed us to identify quantitative resistant loci among an extended genetic diversity, and to test their effects in various genetic backgrounds, which should enhance the transferability of results and the sustainability of the selected resistances. The most widely used oil palm breeding scheme was developed in the 1950s from a reciprocal recurrent selection (RRS) scheme to take advantage of heterotic effects on bunch production observed between group A and B genetic backgrounds, represented by Deli and La Mé/Yangambi parents, respectively, in the present study (Gascon and de Berchoux 1964). Despite substantial differences in Ganoderma resistance levels among the three genetic origins assessed, with Deli being highly susceptible in comparison to Yangambi and La Mé material (Durand-Gasselin et al. 2005), allelic segregation was found in the Deli background for the major QTL T1S_1@9 (D1 and D5 parents), QTL T1S_1@159 (D2 parent), and QTL TD_3@0 (D3 and D5 parents). Hence, RRS would be efficient for both heterotic groups by selecting quantitative resistant alleles at different loci within genetic origins and combining them in A × B producer hybrids.
Localization of Ganoderma resistance loci will facilitate multi-criteria improvement of oil palm, with the possibility of combining known favorable genomic regions, e.g., for disease and yield related traits, while avoiding unintended unfavorable selection on the others. This is currently feasible since an increasing number of studies have reported QTL for agronomic traits such as bunch production and sex ratio (Ukoskit et al. 2014; Tisné et al. 2015), mesocarp oil content (Teh et al. 2016), fatty acid composition (Montoya et al. 2014), stem height (Lee et al. 2014), and lipase activity (Morcillo et al. 2013). The selection of varieties resistant to a combination of diseases will be essential for this multi-criteria improvement, as Ganoderma is found throughout the oil palm cultivation area, combined with other devastating diseases like the Fusarium wilt in Africa, and bud rot in South America. Hence, the global genetic architecture of disease resistance in oil palm needs to be investigated, especially potential trade-off effects related to different pathogens, as shown for other species (Poland et al. 2009). The multi-parental population presented in this study, and the implemented QTL mapping approach, provide powerful tools for investigating such global genetic architecture of disease resistance in oil palm.
Supplementary Material
Supplemental material is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041764/-/DC1.
Acknowledgments
We thank N. Marseillac and A. Berger for partly genotyping the population and N. Billotte for providing former genotyping results and collected samples. We acknowledge SOCFIN Indonesia (Indonesia) for planning and carrying out the field trials with CIRAD (France), and authorizing use of the phenotypic data for this study. This research was partly funded by a grant from PalmElit SAS.
Footnotes
Communicating editor: J. Holland
Literature Cited
- Adeyanju A., Little C., Yu J., Tesso T., 2015. Genome-wide association study on resistance to stalk rot diseases in grain sorghum. G3 5: 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Obaidi J. R., Mohd-Yusuf Y., Razali N., Jayapalan J. J., Tey C.-C., et al. , 2014. Identification of proteins of altered abundance in oil palm infected with Ganoderma boninense. Int. J. Mol. Sci. 15: 5175–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgain P., Rouse M. N., Tsilo T. J., Macharia G. K., Bhavani S., et al. , 2016. Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PLoS One 11: e0155760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Wall M. M., Carlin B. P., 2003. Frailty modeling for spatially correlated survival data, with application to infant mortality in Minnesota. Biostatistics 4: 123–142. [DOI] [PubMed] [Google Scholar]
- Benson J. M., Poland J. A., Benson B. M., Stromberg E. L., Nelson R. J., 2015. Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PLoS Genet. 11: e1005045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billotte N., Marseillac N., Risterucci A.-M., Adon B., Brottier P., et al. , 2005. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 110: 754–765. [DOI] [PubMed] [Google Scholar]
- Billotte N., Jourjon M. F., Marseillac N., Berger A., Flori A., et al. , 2010. QTL detection by multi-parent linkage mapping in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 120: 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bink M., Boer M. P., Ter Braak C. J. F., Jansen J., Voorrips R. E., et al. , 2008. Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 161: 85–96. [Google Scholar]
- Breton F., Hasan Y., Hariadi S., Lubis Z., De Franqueville H., 2006. Characterization of parameters for the development of an early screening test for basal stem rot tolerance in oil palm progenies. J. Oil Palm Res. 2006: 24–36. [Google Scholar]
- Brun H., Chèvre A.-M., Fitt B. D., Powers S., Besnard A.-L., et al. , 2010. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 185: 285–299. [DOI] [PubMed] [Google Scholar]
- Chan K. L., Tatarinova T., Rozana R., Nadzirah A., Azizi N., et al. , 2017. Evidence-based gene models for structural and functional annotations of the oil palm genome. arXiv:1702.07246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeskens G., Nguti R., Janssen P., 2008. One-sided tests in shared frailty models. Test 17: 69–82. [Google Scholar]
- Cochard B., Carrasco-Lacombe C., Pomiès V., Dufayard J.-F., Suryana E., et al. , 2015. Pedigree-based linkage map in two genetic groups of oil palm. Tree Genet. Genomes 11: 1–12. [Google Scholar]
- Cockram J., Scuderi A., Barber T., Furuki E., Gardner K. A., et al. , 2015. Fine-mapping the wheat Snn1 locus conferring sensitivity to the Parastagonospora nodorum necrotrophic effector SnTox1 using an eight founder multiparent advanced generation inter-cross population. G3 5: 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R. M., Flood J., Rees R. W., 2011. Ganoderma boninense in oil palm plantations: current thinking on epidemiology, resistance and pathology. Planter 87: 515–526. [Google Scholar]
- Corley R. H. V., Tinker P. B., 2003. Selection and Breeding. Oil Palm Blackwell Sci. Ltd., Oxford. [Google Scholar]
- Cox D. R., 1972. Models and life-tables regression. J. R. Stat. Soc. B 34: 187–220. [Google Scholar]
- Cressie N., 1993. Statistics for Spatial Data: Wiley Series in Probability and Statistics Wiley-Intersci. New York. [Google Scholar]
- Cros D., Sánchez L., Cochard B., Samper P., Denis M., et al. , 2014. Estimation of genealogical coancestry in plant species using a pedigree reconstruction algorithm and application to an oil palm breeding population. Theor. Appl. Genet. 127: 981–994. [DOI] [PubMed] [Google Scholar]
- Duchateau L., Janssen P., Lindsey P., Legrand C., Nguti R., et al. , 2002. The shared frailty model and the power for heterogeneity tests in multicenter trials. Comput. Stat. Data Anal. 40: 603–620. [Google Scholar]
- Durand-Gasselin T., Asmady H., Flori A., Jacquemard J. C., Hayun Z., et al. , 2005. Possible sources of genetic resistance in oil palm (Elaeis guineensis Jacq.) to basal stem rot caused by Ganoderma boninense—prospects for future breeding. Mycopathologia 159: 93–100. [DOI] [PubMed] [Google Scholar]
- Durrant C., Tayem H., Yalcin B., Cleak J., Goodstadt L., et al. , 2011. Collaborative cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 21: 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Franqueville H., Asmady H., Jacquemard J. C., Hayun Z., Durand-Gasselin T., 2001. Indications on sources of oil palm (Elaeis guineensis Jacq.) genetic resistance and susceptibility to Ganoderma sp., the cause of basal stem rot, pp. 420–431 in Cutting-edge Technologies for Sustained Competitiveness: Proceedings of the 2001 PIPOC International Palm Oil Congress, Agriculture Conference, 20–22 August, 2001. Malaysian Palm Oil Board (MPOB), Kuala Lumpur, Malaysia. [Google Scholar]
- Gascon J. P., de Berchoux C. H. 1964. Caractéristiques de la production d’Elaeis guineensis (Jacq.) de diverses origines et leurs croisements. Appl. À Sélection Palmier À Huile. Oléagineux 19: 75–84. [Google Scholar]
- George A. W., Visscher P. M., Haley C. S., 2000. Mapping quantitative trait loci in complex pedigrees: a two-step variance component approach. Genetics 156: 2081–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Baranski M., Thorland I., Norris A., Grove H., et al. , 2015. Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar). Heredity 115: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., Falls K., Crooks S., 1990. Cri-Map CRI-MAP Doc. [Google Scholar]
- Gururani M. A., Venkatesh J., Upadhyaya C. P., Nookaraju A., Pandey S. K., et al. , 2012. Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78: 51–65. [Google Scholar]
- Hama-Ali E. O., Panandam J. M., Tan S. G., Alwee S. S. R. S., Sheong T. J., et al. , 2015. Association between basal stem rot disease and simple sequence repeat markers in oil palm, Elaeis guineensis Jacq. Euphytica 202: 199–206. [Google Scholar]
- Ho C.-L., Tan Y.-C., 2015. Molecular defense response of oil palm to Ganoderma infection. Phytochemistry 114: 160–169. [DOI] [PubMed] [Google Scholar]
- Ho C.-L., Tan Y.-C., Yeoh K.-A., Ghazali A.-K., Yee W.-Y., et al. , 2016. De novo transcriptome analyses of host-fungal interactions in oil palm (Elaeis guineensis Jacq.). BMC Genomics 17: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris A., Kushairi A., Ismail S., Ariffin D., 2004. Selection for partial resistance in oil palm progenies to Ganoderma basal stem rot. J. Oil Palm Res. 16: 12–18. [Google Scholar]
- Jansen R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeennor S., Volkaert H., 2014. Mapping of quantitative trait loci (QTLs) for oil yield using SSRs and gene-based markers in African oil palm (Elaeis guineensis Jacq.). Tree Genet. Genomes 10: 1–14. [Google Scholar]
- Jiang D., Wang H., Li J., Wu Y., Fang M., et al. , 2014. Cox regression model for dissecting genetic architecture of survival time. Genomics 104: 472–476. [DOI] [PubMed] [Google Scholar]
- Kump K. L., Bradbury P. J., Wisser R. J., Buckler E. S., Belcher A. R., et al. , 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. [DOI] [PubMed] [Google Scholar]
- Kwong Q. B., Teh C. K., Ong A. L., Heng H. Y., Lee H. L., et al. , 2016. Development and validation of a high-density SNP genotyping array for African oil palm. Mol. Plant 9: 1132–1141. [DOI] [PubMed] [Google Scholar]
- Lee Y. W., Gould B. A., Stinchcombe J. R., 2014. Identifying the genes underlying quantitative traits: a rationale for the QTN programme. AoB Plants 6: pii: plu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Piao Z., Zhou X., Yang T., Yang R., 2013. An optimal parametric proportional hazards model for mapping heading time loci in rice. Euphytica 192: 37–44. [Google Scholar]
- Montoya C., Cochard B., Flori A., Cros D., Lopes R., et al. , 2014. Genetic architecture of palm oil fatty acid composition in cultivated oil palm (Elaeis guineensis Jacq.) compared to its wild relative E. oleifera (HBK) cortés. PLoS One 9: e95412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo F., Cros D., Billotte N., Ngando-Ebongue G.-F., Domonhédo H., et al. , 2013. Improving palm oil quality through identification and mapping of the lipase gene causing oil deterioration. Nat. Commun. 4: 2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusaibah S. A., Akmar A. S. N., Idris A. S., Sariah M., Pauzi Z. M., 2016. Involvement of metabolites in early defense mechanism of oil palm (Elaeis guineensis Jacq.) against Ganoderma disease. Plant Physiol. Biochem. 109: 156–165. [DOI] [PubMed] [Google Scholar]
- Oerke E.-C., Dehne H.-W., 2004. Safeguarding production—losses in major crops and the role of crop protection. Crop Prot. 23: 275–285. [Google Scholar]
- Parisseaux B., Bernardo R., 2004. In silico mapping of quantitative trait loci in maize. Theor. Appl. Genet. 109: 508–514. [DOI] [PubMed] [Google Scholar]
- Poland J. A., Balint-Kurti P. J., Wisser R. J., Pratt R. C., Nelson R. J., 2009. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14: 21–29. [DOI] [PubMed] [Google Scholar]
- Pootakham W., Jomchai N., Ruang-areerate P., Shearman J. R., Sonthirod C., et al. , 2015. Genome-wide SNP discovery and identification of QTL associated with agronomic traits in oil palm using genotyping-by-sequencing (GBS). Genomics 105: 288–295. [DOI] [PubMed] [Google Scholar]
- Quenouille J., Paulhiac E., Moury B., Palloix A., 2014. Quantitative trait loci from the host genetic background modulate the durability of a resistance gene: a rational basis for sustainable resistance breeding in plants. Heredity 112: 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Singh R., Ong-Abdullah M., Low E.-T. L., Manaf M. A. A., Rosli R., et al. , 2013. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E., Lange K., 1996. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58: 1323. [PMC free article] [PubMed] [Google Scholar]
- Sobel E., Sengul H., Weeks D. E., 2001. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum. Hered. 52: 121–131. [DOI] [PubMed] [Google Scholar]
- Tee S.-S., Tan Y.-C., Abdullah F., Ong-Abdullah M., Ho C.-L., 2013. Transcriptome of oil palm (Elaeis guineensis Jacq.) roots treated with Ganoderma boninense. Tree Genet. Genomes 9: 377–386. [Google Scholar]
- Teh C.-K., Ong A.-L., Kwong Q.-B., Apparow S., Chew F.-T., et al. , 2016. Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci. Rep. 6: 19075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. M., Therneau M. T. M., 2015 Package “coxme” Mixed Effects Cox Models R Package Version 2. https://CRAN.R-project.org/package=coxme.
- Tisné S., Denis M., Cros D., Pomiès V., Riou V., et al. , 2015. Mixed model approach for IBD-based QTL mapping in a complex oil palm pedigree. BMC Genomics 16: 798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukoskit K., Chanroj V., Bhusudsawang G., Pipatchartlearnwong K., Tangphatsornruang S., et al. , 2014. Oil palm (Elaeis guineensis Jacq.) linkage map, and quantitative trait locus analysis for sex ratio and related traits. Mol. Breed. 33: 415–424. [Google Scholar]
- USDA, 2015 Oilseeds: world markets and trade. http://www.thecropsite.com/reports/?id=4943.
- Vaupel J. W., Manton K. G., Stallard E., 1979. The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography 16: 439–454. [PubMed] [Google Scholar]
- Verhoeven K. J. F., Jannink J. L., McIntyre L. M., 2005. Using mating designs to uncover QTL and the genetic architecture of complex traits. Heredity 96: 139–149. [DOI] [PubMed] [Google Scholar]
- Voorrips R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Vu H. T., Knuiman M. W., 2002. Estimation in semiparametric marginal shared gamma frailty models. Aust. N. Z. J. Stat. 44: 489–501. [Google Scholar]
- Wei X., Jackson P. A., McIntyre C. L., Aitken K. S., Croft B., 2006. Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor. Appl. Genet. 114: 155–164. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203–214. [DOI] [PubMed] [Google Scholar]
- Zhao J. H., 2005. Mixed-effects Cox models of alcohol dependence in extended families. BMC Genet. 6: S127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Eg9PP was uprooted in 2012, but Eg9PP families can be reproduced by crossing individuals derived from the self-fertilized progenies of Eg9PP parents. Requests for such genetic material should be addressed to PalmElit (www.palmelit.com). Raw genotype data are available in Table S5 and IBD kinship matrices at https://doi.org/10.6084/m9.figshare.4780489.v1. Phenotyping of disease events are available in Table S3. All R scripts, data file and documentation are available at https://github.com/DenisMarie/Eg9PP_Ganoderma.