Abstract
Cdk1 (Cdc28 in yeast) is a cyclin-dependent kinase (CDK) essential for cell cycle progression and cell division in normal cells. However, CDK activity also underpins proliferation of tumor cells, making it a relevant study subject. While numerous targets and processes regulated by Cdc28 have been identified, the exact functions of Cdc28 are only partially understood. To further explore the functions of Cdc28, we systematically overexpressed ∼4800 genes in wild-type (WT) cells and in cells with artificially reduced Cdc28 activity. This screen identified 366 genes that, when overexpressed, specifically compromised cell viability under conditions of reduced Cdc28 activity. Consistent with the crucial functions of Cdc28 in cell cycle regulation and chromosome metabolism, most of these genes have functions in the cell cycle, DNA replication, and transcription. However, a substantial number of genes control processes not directly associated with the cell cycle, indicating that Cdc28 may also regulate these processes. Finally, because the dataset was enriched for direct Cdc28 targets, the results from this screen will aid in identifying novel targets and process regulated by Cdc28.
Keywords: Cdc28, Cdk1, cell cycle, synthetic lethality, Mutant Screen Report
CDKs drive the cell cycle in eukaryotic cells. A single CDK, Cdc28, is necessary and sufficient for cell cycle regulation in the budding yeast Saccharomyces cerevisiae (Mendenhall and Hodge 1998; Enserink and Kolodner 2010), although many of its functions are supported by the nonessential CDK Pho85, and there exists substantial cross talk between these kinases in the regulation of cell cycle-related processes (Huang et al. 2007). Cdc28 is activated by its cyclin partners, which are differentially expressed throughout the cell cycle. Cyclin–Cdc28 complexes coordinate the cell cycle by phosphorylating specific proteins involved in DNA replication and repair, telomere homeostasis, cell growth and morphogenesis, lipid synthesis, formation of the mitotic spindle, and transcriptional programs (Enserink and Kolodner 2010). Cdc28 is a proline-directed kinase that preferentially phosphorylates the consensus sequence S/T-P-x-K/R (where x is any amino acid), although it also phosphorylates the minimal consensus sequence S/T-P (Moreno and Nurse 1990).
Aberrant CDK activity underlies uncontrolled proliferation of tumor cells (Hunter and Pines 1994), which is why it is important to study its functions. However, while Cdc28 is one of the best studied kinases with a well-described repertoire of substrates (Enserink and Kolodner 2010), its exact molecular functions are not fully understood. An important technological improvement was the development of the engineered cdc28-as1 allele (Bishop et al. 2000), which encodes a form of Cdc28 in which the gatekeeper phenylalanine at position 88 is replaced with a much smaller glycine residue. This creates an enlarged ATP binding pocket that can accommodate the bulky, nonhydrolyzable ATP analog 1-NM-PP1 (Bishop et al. 2000). Treatment of cdc28-as1 mutants with the cell-permeable 1-NM-PP1 results in specific inhibition of Cdc28 (Bishop et al. 2000). The effect of 1-NM-PP1 on cell cycle progression of cdc28-as1 mutants is dose-dependent; at lower doses (≤500 nM) it causes a delay or arrest with 2N DNA content and large hyperpolarized buds, whereas higher doses (5000 nM) arrest cell cultures as a mix of unbudded G1 cells and large-budded M phase cells. However, it should be noted that the gene product of cdc28-as1, Cdc28-as1, has a 10-fold reduction in ATP-binding affinity and a sixfold reduction in maximum ATP turnover rate (Bishop et al. 2000). As a consequence, the cdc28-as1 mutant is hypomorphic and has a 20% longer doubling time in liquid culture (Bishop et al. 2000), although this increased doubling time is not obvious in spot assays (Zimmermann et al. 2011).
Despite its hypomorphic nature, the cdc28-as1 allele is a powerful tool that has been used in multiple studies to identify novel functions and targets of Cdc28; for instance, it has been used to identify Cdc28-dependent phosphorylations in vivo using a SILAC-based phosphoproteomics approach (Holt et al. 2009). Another interesting feature of Cdc28-as1 is that it can use N6-(benzyl) ATP as an ATP source, which cannot be used by other kinases due its bulkiness. One drawback of N6-(benzyl) ATP is that it is not cell permeable, restricting its use to in vitro experiments. Nevertheless, a screen for proteins that are directly phosphorylated by Cdc28 in whole-cell extracts identified ∼200 Cdc28 substrates, several of which were confirmed to be phosphorylated in vivo in a Cdc28-dependent manner (Ubersax et al. 2003).
Significant insight into the function and regulation of Cdc28 has also come from classic dosage suppression screens, in which genes were identified that complement the slow growth phenotype of temperature-sensitive cdc28 alleles. For example, these screens identified several cyclins as well as Cks1, which is a subunit of the Cdc28 holoenzyme (Hadwiger et al. 1989a,b; Surana et al. 1991). Such classic genetic screens can be relatively laborious, often involving the organization of suppressors in complementation groups followed by mapping and identification of the genes responsible for suppression of the mutant phenotype. More recently, automated high-throughput genetic screens have been developed to systematically interrogate the genetic landscape of cells. One commonly used method is synthetic genetic array (SGA; Tong et al. 2001). Most SGA screens use synthetic lethality as a readout. The concept of synthetic lethality was first developed in Drosophila to describe the observation that the combination of two mutations results in a significantly worse fitness defect than either single mutation alone (Dobzhansky 1946). Genes with synthetic lethal interactions often function in parallel nonessential pathways that control an essential cellular process (Dixon et al. 2009). Therefore, synthetic lethality can be used to identify new regulators of specific cellular processes, or assign new functions to genes (Enserink 2012). For instance, we recently reported an SGA screen in which we used low doses of 1-NM-PP1 that partially reduced the growth of cdc28-as1 single mutants, and screened for deletion mutants that aggravated the growth defect of the cdc28-as1 mutant. This screen identified 107 genes that are essential for maintaining cell viability under conditions of reduced Cdc28 activity, and revealed several cellular processes that were not previously known to affect the cell cycle (Zimmermann et al. 2011). Follow-up studies revealed novel functions of Cdc28; for instance, we have shown that Cdc28 has a function in stimulating the transcription of housekeeping genes, which is important for cell homeostasis (Chymkowitch et al. 2012).
Another high-throughput method to identify novel regulators of cellular processes is synthetic dosage lethality (SDL) screening, which is based on the idea that increasing the levels of a protein results in a significantly worse fitness defect in a mutant strain than in an otherwise WT strain (Kroll et al. 1996). Because it induces specific regulatory imbalances in pathways that regulate critical cellular pathways, systematic gene overexpression has revealed novel regulatory pathways and phenotypes (Kroll et al. 1996; Measday et al. 2005; Sopko et al. 2006). For example, recently, SDL screening was applied to interrogate the yeast kinome using 92 kinases as a query, revealing potential novel functions for many kinases (Sharifpoor et al. 2012). SDL datasets tend to be enriched for kinase substrates, enabling the identification of key downstream components of cell signaling pathways (Sopko et al. 2006; Sharifpoor et al. 2012). Thus, SDL screening is a useful tool for the identification of novel kinase functions and kinase substrates, and in the present study we performed an SDL screen to probe the functions of Cdc28.
Materials and Methods
Yeast strains
The WT strain (BY4741) and the isogenic cdc28-as1 query strain (JEY4250) have been published previously (Brachmann et al. 1998; Zimmermann et al. 2011).
Genetic screen
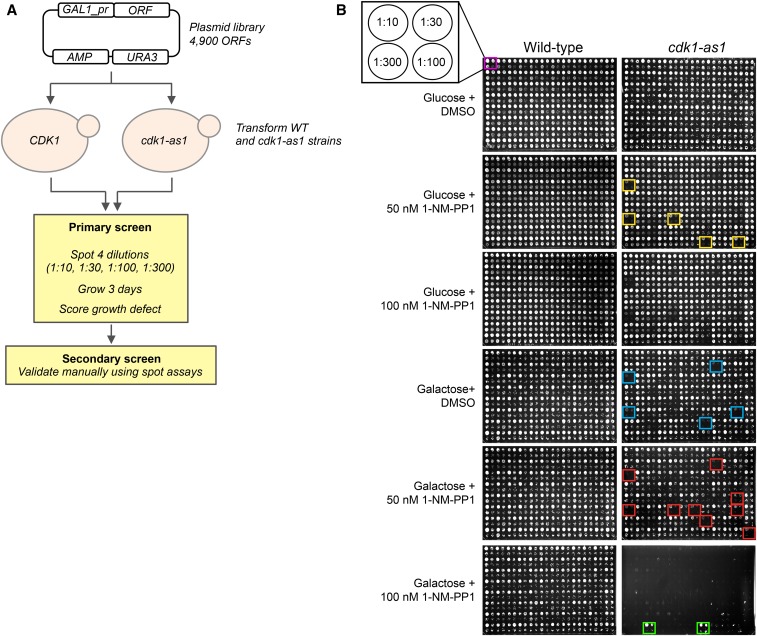
Strains BY4741 and JEY4250 were systematically transformed with the arrayed yeast movable ORF library (Gelperin et al. 2005), and grown overnight in liquid dropout media lacking uracil (to select for the plasmid library) and supplemented with 2% glucose. Overnight cultures were then pinned onto synthetic dropout plates lacking uracil and containing either 2% glucose or 2% galactose, and supplemented with either DMSO, 50 nM 1-NM-PP1, or 100 nM 1-NM-PP1. Plates were then incubated at 30° for 2–3 d and photographed. Phenotypes that were scored were either aggravation of the growth defect of the cdc28-as1 mutant with respect to the WT control, or relatively improved growth compared with a cdc28-as1 mutant overexpressing YEL074W, which is a dubious ORF with no known genetic interactions (overexpression of YEL074W does not affect growth of either WT cells or cdc28-as1 mutants). The entire screen was repeated once using essentially the same set-up, although we spotted four different dilutions (1:10, 1:30, 1:100, and 1:300) of overnight cultures onto the selection plates instead of pinning WT cells and cdc28-as1 mutants at one single concentration.
STRING analysis
STRING analysis (Szklarczyk et al. 2015) was carried out at highest confidence (0.900) using the query proteins only and using default settings, with the following exceptions: Active interaction sources: “Experiments,” “Databases.” Network edges: “Evidence.”
Gene Ontology (GO) analysis and visualization
GO analysis and visualization of the GO network was performed using Metascape (http://metascape.org; Tripathi et al. 2015) using parameters specific for S. cerevisiae. Additional analysis of the overrepresented biological processes of the SDL network was performed using GO Slim mapper at the SGD database using the term “Yeast GO-Slim: Process,” selecting all terms present in the database.
Comparison with other datasets
High- and low-throughput genetic interaction data (synthetic lethality, negative and positive genetic interactions, dosage rescue, and dosage lethality) were obtained from BioGRID (Chatr-Aryamontri et al. 2017). Physical interactions with the Cdc28 holoenzyme (Cln1-3, Clb1-6, Cdc28, and Cks1; both high- and low-throughput data) were also derived from BioGRID. Because physical interaction data from high-throughput studies can consist of up to 50% false positives (von Mering et al. 2002; Patil and Nakamura 2005), we only included physical interactions that were observed at least twice per Cdc28 holoenzyme subunit. Data regarding in vitro phosphorylation by Cdc28 was obtained from Ubersax et al. (2003) (185 proteins; cut-off for inclusion: P score ≥ 2) and Ptacek et al. (2005) (67 proteins). In vivo Cdc28-dependent phosphorylation data were compiled from high- and low-throughput data derived from PhosphoGRID (Sadowski et al. 2013) and Phosphopep (Bodenmiller and Aebersold 2011; Bodenmiller et al. 2010), from Holt et al. (2009) (cut-off for inclusion: log2 ≤ 1), and from publications that appeared to be absent from PhosphoGRID and Phosphopep (Han et al. 2005; Liu et al. 2011; Tao et al. 2011; Yaglom et al. 1995; Zhu et al. 2010). The list of known Cdc28 substrates was based on our previous compilation of Cdc28 targets (Enserink and Kolodner 2010), which was updated with information from recent literature.
RNA extraction, reverse transcription, and qPCR
Log phase cdc28-as1 cells were treated with 500 nM of 1-NM-PP1 in YPD. After 4 hr of treatment, cell cycle arrest in M phase was confirmed by flow cytometry (see below). CLN3 expression was induced by switching cells to galactose-containing medium for 4 hr. Then, RNA was isolated using the RNeasy Mini Kit (QIAGEN) and equal amounts of RNA were reverse transcribed using the QuantiTect Reverse Transcription Kit (QIAGEN). qPCRs were performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) and the StepOnePlus real-time PCR system (Applied Biosystems).
Primer sequences
Primer sequences used are as follows: CLN2: 5′-TATCCCAGGATAGTGATGCCACTG-3′ and 5′-TCTAAGTAAGTCGTACTGCCACGC-3′; CLN3: 5′-CAAAGAGCGCTACGGTTTCATCTG-3′ and 5′-TGGAGAGGATGAAGATGAGGTTGG-3′; CLB6: 5′-CATCACTTGCCTGTTCATTGCCTG-3′ and 5′-AGCTCAGCCTTCCTAATTCCTTCG-3′; and ACT1: 5′-TGAGGAGCACCCTTGCTTGT-3′ and 5′-TCTTCTCACGGTTGGATTTGG-3′.
Cell cycle analysis by flow cytometry
cdc28-as1 mutant cells were treated as described under RNA extraction, reverse transcription, and qPCR before being processed for cell cycle analysis by flow cytometry as previously described (Enserink et al. 2009). Briefly, cells were fixed in 70% EtOH (vol/vol), centrifuged, and resuspended in 50 mM sodium citrate + 0.2 µg/µl of RNAse A. After 2 hr at 37°, cells were centrifuged and resuspended in 50 mM sodium citrate + 5 mg/ml of pepsin. After 30 min at 37°, cells were centrifuged and resuspended in 50 mM sodium citrate + 2 µg/ml of propidium iodine and analyzed by flow cytometry using a BD FACSCalibur instrument. Cell cycle profiles were analyzed with FlowJo software.
Quantification of spindle alignment and morphology
To determine the effect of the overexpression of different genes on mitotic spindle alignment and morphology, WT (JEY5235) and cdc28-as1 (JEY5239) strains expressing the Tub1-GFP fusion protein were transformed with the plasmids harboring YEL074W (control), CIN8, FIN1, KAR9, and SPC97. Cultures were grown to log phase in synthetic dropout media without uracil containing 2% glucose, followed by incubation in dropout medium containing 2% galactose for 4 hr, after which cells were imaged in a Zeiss Axioplan2 microscope coupled to a Zeiss AxioCam HRC camera. To estimate the mitotic spindle orientation, the angle between the bud axis and the spindle axis was measured for each large-budded cell and the spindle was counted as misaligned when this angle was bigger than 45°. The percentage of aberrant mitotic spindles was measured by comparison with the spindle morphology of WT cells. All image processing and quantification was performed in ImageJ.
Quantification of nuclear localization of Whi5-GFP
Whi5-GFP-expressing cells were grown overnight in dropout medium lacking uracil, centrifuged, and resuspended in YPD. Cells were then arrested in M phase using 15 µg/ml nocodazole for 2.5 hr and M phase arrest was confirmed by light microscopy, by staining a small sample of the culture with DAPI followed by fluorescence microscopy, and for a few samples also by flow cytometry. Cells were then washed and incubated in nocodazole-containing YP medium supplemented with 2% galactose to induce expression for 3 hr, after which cells were imaged with a Zeiss Axioplan2 microscope coupled to a Zeiss AxioCam HRC camera. The relative amount of nuclear Whi5-GFP was determined by determining the increase of the intensity of the nuclear Whi5-GFP signal over the Whi5-GFP intensity of the cytoplasm.
Venn diagrams
Venn diagrams were calculated and visualized using http://bioinformatics.psb.ugent.be/webtools/Venn/.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Mapping the SDL network and comparison with previous genetic screens
We systematically transformed a WT strain and a cdc28-as1 mutant strain (Bishop et al. 2000; Zimmermann et al. 2011) with the yeast movable ORF library, which is an arrayed plasmid library consisting of ∼4900 ORFs under control of the galactose-inducible GAL1 promoter (Gelperin et al. 2005). We screened for genes that, when overexpressed, either specifically inhibited the growth of cdc28-as1 mutant cells (see Figure 1A and Materials and Methods) or that (at least partially) complemented the growth defect of cdc28-as1 mutant cells. We found that overexpression of 366 genes in total resulted in reduced cell viability of the cdc28-as1 mutant (Figure 1B, red boxes and Supplemental Material, Table S1) and that 20 genes may suppress the growth defect of this mutant (Figure 1B, green boxes). Most of the 366 SDL genes already caused significant growth defects under noninducing conditions (e.g., see Figure 1B, yellow boxes), which is likely due to leakage from the GAL1 promoter. Furthermore, several SDL genes caused growth defects in the absence of 1-NM-PP-1 (e.g., blue boxes in Figure 1B), which is probably caused by the fact that the cdc28-as1 allele is hypomorphic (Bishop et al. 2000). This indicates that just a slight increase in the expression of these genes is sufficient to perturb cell homeostasis in cells with reduced Cdc28 activity.
Figure 1.
Set-up of the screen. (A) Schematic overview of the screen. (B) Example of the data. Red boxes indicate genes that aggravate the cdc28-as1 phenotype, whereas green boxes indicate genes that ameliorate the slow-growth defect of cdc28-as1 mutants on 1-NM-PP1. Yellow boxes indicate genes that induce the SDL phenotype even under noninducing conditions, whereas blue boxes indicate genes that induce the SDL phenotype even in the absence of 1-NM-PP1. DMSO, dimethyl sulfoxide; ORF, open reading frame; SDL, synthetic dosage lethality; WT, wild-type.
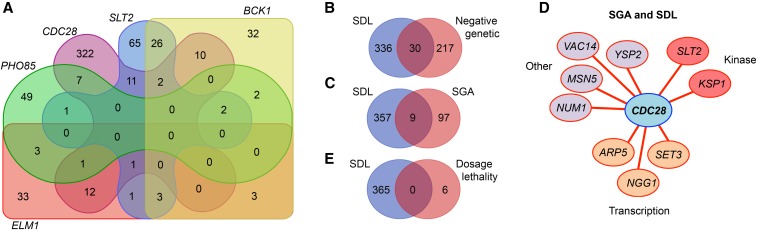
Notably, the SDL network of CDC28 is substantially larger than previously identified kinase SDL networks [366 unique SDL interactions for CDC28 compared to 65 for PHO85 (Sharifpoor et al. 2012)]. This may reflect the importance of Cdc28 in cell cycle control, cell morphogenesis, and cell homeostasis, and is consistent with the previous finding that kinases that coordinate cell cycle progression with cell polarity tend to exhibit “hub” SDL profiles (Sharifpoor et al. 2012). Perhaps surprisingly, there appeared to be limited overlap between these kinase SDL hubs (Figure 2A). The reason for the lack of overlap is not clear, but it could reflect the unique cellular functions of these kinases. For example, several of the hub kinases have important functions in response to cellular stress, such as Slt2, which promotes cell survival in response to cell wall stress. In contrast, Cdc28 is important for the regulation of multiple cellular processes under normal growth conditions. An alternative, technical explanation for the relative lack of overlap between these kinase hubs could be that different GAL overexpression systems were used with different epitope tags (such as N-terminal GST compared to the C-terminal tag used here).
Figure 2.
Comparison of the SDL dataset with other genome-wide screens. (A) Overlap between the “hub” SDL networks of CDC28 and the kinases SLT2, BCK1, ELM1, and PHO85. Data were obtained from Sharifpoor et al. (2012). (B) Overlap between the SDL screen and previously identified genes with a negative genetic relationship with CDC28. Data were obtained from BioGRID. (C and D) Overlap between the SDL screen and the SGA screen described in Zimmermann et al. (2011). (E) Overlap between the SDL screen and previously identified genes that cause dosage lethality of cdc28 mutants (data from BioGRID). SDL, synthetic dosage lethality; SGA, synthetic genetic array.
Limited overlap between the SDL network and previously identified negative genetic interactions with CDC28
To gain further understanding of the genetic network of CDC28, we compared our dataset to genes previously shown to negatively genetically interact with CDC28. It has been observed that SGA and SDL screens tend to uncover nonoverlapping genetic interactions, reflecting the distinct properties associated with loss-of-function mutation (SGA) and potential gain-of-function (gene overexpression; SDL) (Measday et al. 2005). Indeed, there exists <10% overlap (30 genes) between the SDL dataset and all previously reported negative genetic interactions that were identified with various hypomorphic cdc28 alleles (Figure 2B and Table S2; see Materials and Methods). However, these cdc28 alleles have different cell cycle defects (Reed and Wittenberg 1990; Reed 1980; Hadwiger et al. 1989b; Surana et al. 1991). Therefore, to be able to better compare the SDL data with the negative genetic interaction network of CDC28, we focused on data from our own previously published SGA screen in which we used the same cdc28-as1 query mutant as in the current SDL screen (Zimmermann et al. 2011). This comparison identified nine genes that were present in both the SDL and the SGA datasets, i.e., ARP5, KSP1, MSN5, NGG1, NUM1, SET3, SLT2, VAC14, and YSP2 (Figure 2, C and D and Table S3). The fact that these genes display both a synthetic lethal as well as a dosage lethal interaction with CDC28 shows that cells are very sensitive to changes in the activity of these genes when Cdc28 activity has been compromised, suggesting that these genes and Cdc28 have similar cellular functions. Supporting this interpretation, three out of the nine genes are involved in the regulation of basal transcription (i.e., ARP5, NGG1, and SET3), and we have recently shown that Cdc28 promotes basal transcription of housekeeping genes to maintain cell homeostasis (Chymkowitch et al. 2012).
Genes that aggravate the cdc28-as1 slow growth phenotype
According to information retrieved from the SGD database, only six genes have previously been shown to induce dosage lethality in cdc28 mutants, i.e., CLN1 (Tobe et al. 2009), ELM1 (Sharifpoor et al. 2012), FCP1 (Chymkowitch et al. 2012), MPS1 (Rudner et al. 2000), RTR1 (Chymkowitch et al. 2012), and SSU72 (Chymkowitch et al. 2012). Surprisingly, the SDL screen did not identify any of these six genes (Figure 2E), even though three of these interactions were identified previously by our own laboratory using the same cdc28-as1 mutant [i.e., RTR1, SSU72, and FCP1 (Chymkowitch et al. 2012)]. This finding reflects the previous observation that SDL screens can result in a false negative rate as high as 50% (Sharifpoor et al. 2012). There may be multiple explanations for this high false negative rate. For instance, not all ORFs are present in the SDL library, such as e.g., YKL048C/ELM1. Furthermore, we have not fully sequenced the plasmids harboring these ORFs and it is possible that at least some of them either do not contain the correct ORF, or they may contain loss-of-function mutations resulting from the PCR step during library construction. Furthermore, overexpression of highly toxic genes may result in rapid selection of cells that have acquired mutations in either their genetic background or in the plasmid, resulting in bypass of the SDL phenotype. It is also possible that the activity of the overexpressed proteins is impaired by the relatively large C-terminal ProtA-HA-HIS6 tag, thus suppressing the SDL phenotype. Keeping in mind these drawbacks, which are inherent to high-throughput screens, we believe that the SDL dataset provides useful information about the function of Cdc28.
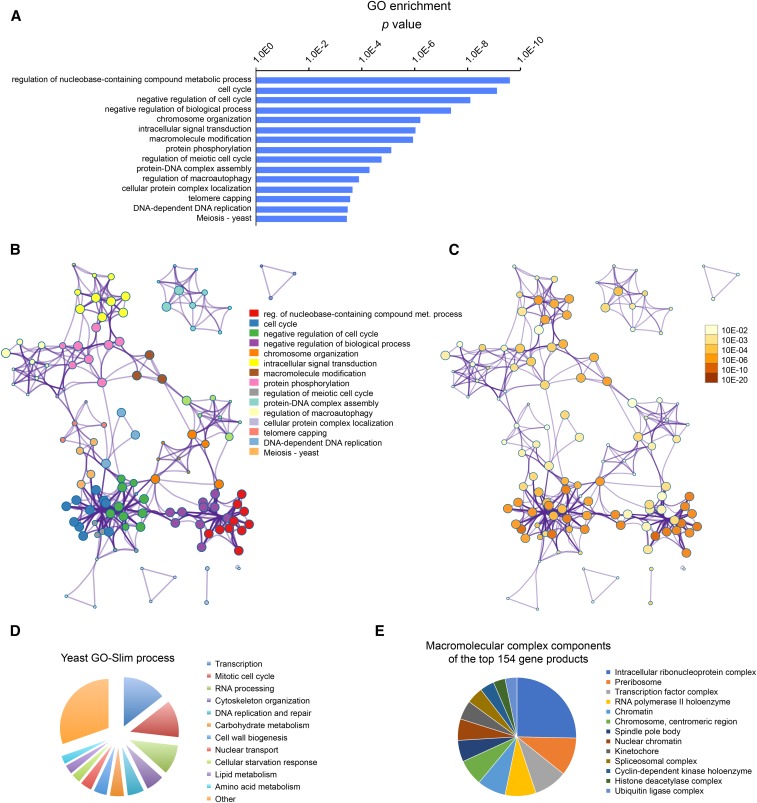
To better understand the SDL network, we analyzed the functions of the genes in the SDL dataset. GO analysis revealed overrepresentation of genes important for the cell cycle, nucleic acid metabolism, and transcription (Figure 3, A–E), as might be expected based on the important functions of Cdc28 in these processes (Enserink and Kolodner 2010). STRING analysis of the relationships between the proteins encoded by these genes yielded an interaction network centered on Cdc28 (Figure S1, A and B), which was highly enriched for factors involved in cell cycle control (1.58E−08; Figure S1C). Thus, as previously reported (Sharifpoor et al. 2012), SDL screening is a powerful tool for functional characterization of kinases, and while we cannot discuss all the SDL genes in detail, we will highlight some of the major overrepresented groups below.
Figure 3.
Analysis of the data reveals overrepresentation of genes involved in the cell cycle, DNA metabolism, and transcription. (A) GO Biological Process analysis of the genes that aggravate the cdc28-as1 phenotype (negative SDL interactions). GO analysis was performed using Metascape. (B) Network plot of the relationships among GO terms. Nodes represent enriched terms colored by its cluster ID. (C) The same network as presented in (B) but showing P values for the nodes. (D) Analysis of the negative SDL interactors using the GO Slim mapper tool of the SGD. (E) Analysis of the macromolecular complex components of the top 154 genes in the SDL network. Only those GO terms are shown for which at least five terms genes were identified. GO, gene ontology; ID, identifier; SDL, synthetic dosage lethality; SGD, Saccharomyces Genome Database.
Cell cycle regulators
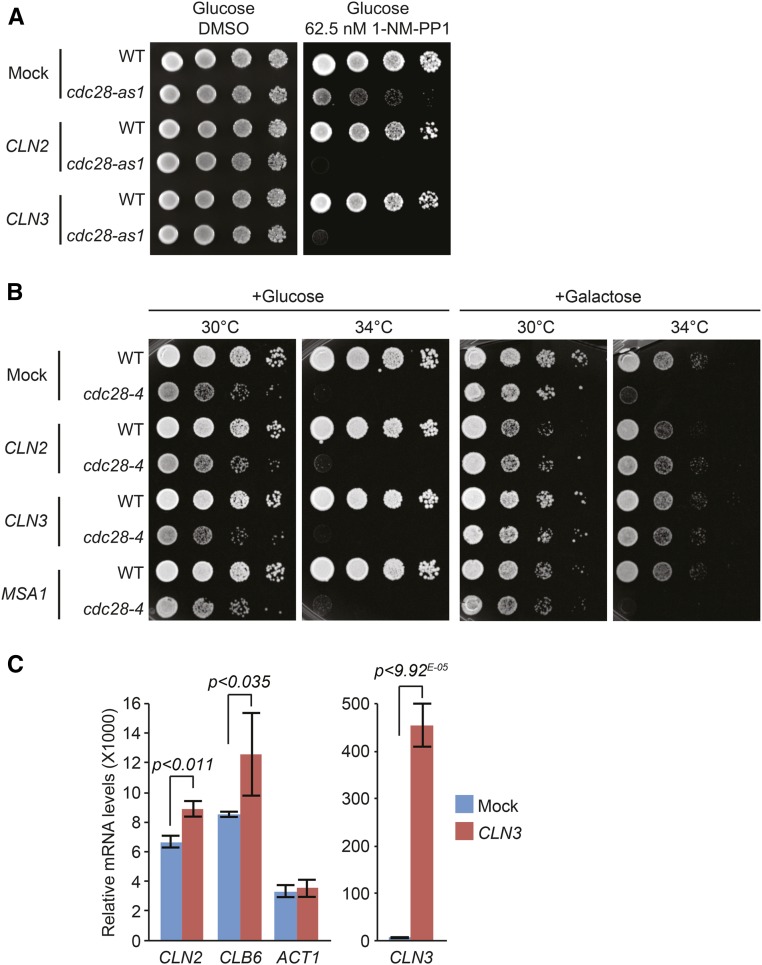
CLN2 and CLN3: we found that increased expression of multiple cell cycle genes resulted in impaired survival of cdc28-as1 mutants. In particular, cdc28-as1 mutants were very sensitive to overexpression of CLN2 and CLN3, which caused SDL in the absence of galactose (i.e., noninducing conditions; Figure 4A), indicating that even a minor increase in the levels of these cyclins (due to leakage of the GAL1 promoter) is toxic to these cells. This was surprising because previous studies had identified cyclins as high dosage suppressors of temperature-sensitive cdc28 alleles; for instance, CLN1 and CLN2 suppress cdc28-4, cdc28-9, and cdc28-13, whereas B-type cyclins suppress cdc28-1N mutants (Hadwiger et al. 1989b; Surana et al. 1991). This incongruity between the different cdc28 mutants could be due to the fact that the cdc28-as1 allele confers a different cell cycle defect than the aforementioned temperature-sensitive cdc28 alleles; for instance, cdc28-4 mutant cells primarily have a defect early in the cell cycle, whereas cdc28-as1 mutants mainly have a M phase defect (Reed 1980; Hadwiger et al. 1989b; Surana et al. 1991; Bishop et al. 2000). Consistent with this idea, we found that overexpression of CLN2 and CLN3 in the cdc28-4 mutant suppressed the temperature-sensitive growth defect, whereas overexpression of the control ORFs YEL074W and MSA1 had no effect (Figure 4B).
Figure 4.
The effect of overexpression of cyclins. (A) Overexpression of CLN2 and CLN3 causes SDL in cdc28-as1 mutants even under noninducing conditions. Cultures of cells transformed with plasmids containing the ORFs YEL074W (“Mock”), CLN2, or CLN3 were spotted on glucose-containing SC-URA plates supplemented with either DMSO or 1-NM-PP1, followed by incubation at 30° until colonies appeared. (B) Overexpression of CLN2 and CLN3 suppresses the growth defect of cdc28-4 mutants. Cultures of cells transformed with plasmids containing the control ORF YEL074W (“Mock”), CLN2, CLN3, or MSA1 were spotted on YPD or on YP-galactose and incubated at either 30° or 34° until colonies appeared. (C) cdc28-as1 strains transformed with plasmids harboring either the control ORF YEL074W (Mock) or CLN3 were grown to log phase and treated with 500 nM 1-NM-PP1, after which transcription was induced with galactose as described in the Materials and Methods. Subsequently, mRNA levels of CLN2, CLB6, ACT1, and CLN3 were determined by RT-qPCR. DMSO, dimethyl sulfoxide; ORF, open reading frame; RT-qPCR, quantitative reverse transcription polymerase chain reaction; SC-URA, synthetic complete-uracil; SDL, synthetic dosage lethality; WT, wild-type; YP, yeast extract peptone; YPD, YP and dextrose.
One explanation for the toxicity of overexpressed cyclins in cdc28-as1 mutant cells could be that these cells spend much more time in the M phase of the cell cycle when exposed to sublethal doses of 1-NM-PP1 (Zimmermann et al. 2011), and that overexpression of CLN2 or CLN3 in this phase of the cell cycle titrates away B-type cyclins and inappropriately triggers activation of the G1/S phase program. We tested this by overexpressing CLN3 in the cdc28-as1 mutant strain treated with 500 nM 1-NM-PP1. This dose of drug has been reported to strongly delay cells in early M phase (Bishop et al. 2000), which we confirmed by flow cytometry (Figure S2A). We then analyzed activation of the G1 transcriptional program by monitoring the transcription of CLN2 and CLB6, which are part of the G1 cluster of cell cycle-regulated genes (Spellman et al. 1998). Interestingly, overexpression of CLN3 in these M phase-arrested mutants resulted in a modest but significant increase in CLN2 and CLB6 mRNA levels, whereas expression of the actin-encoding gene ACT1 remained unchanged (Figure 4C). In contrast, overexpression of CLN3 in M phase-arrested cdc28-4 mutants did not result in increased expression of CLN2 and CLB6 (Figure S2B). This indicates that overexpression of CLN3 can to a certain degree activate the G1/S phase transcriptional program specifically in cdc28-as1 cells. This supports our hypothesis that ectopic expression of CLN3 in M phase-arrested cdc28-as1 mutants may inappropriately activate cellular processes normally associated with early stages of the cell cycle, thus contributing to the SDL phenotype. In addition, overexpressed Cln3 may further exacerbate the SDL phenotype by outcompeting the binding of Clb1-4 to Cdc28, resulting in reduced Clb-Cdc28 activity.
We identified several other cell cycle regulators in the SDL screen, which we will only discuss briefly.
SIC1: Sic1 is an inhibitor of Clb5/6–Cdc28 complexes (Schwob et al. 1994), and phosphorylation of Sic1 by Cdc28 promotes cell cycle entry. Depending on the cell cycle stage, SIC1 overexpression can result in cell cycle arrest (Nugroho and Mendenhall 1994), but it can also prematurely induce the formation of prereplicative complexes at origins of replication, causing some cells to rereplicate their DNA (Dahmann et al. 1995). Overexpression of SIC1 may be toxic to the hypomorphic cdc28-as1 mutant by preventing efficient cell cycle progression, by interfering with the timing of DNA replication, or both.
CDC20: Cdc20 is an activator of the anaphase promoting complex (Visintin et al. 1997). Mitotic Cdc28 activity is required to activate Cdc20, and cdc28 mutants have difficulty leaving mitosis (Rudner and Murray 2000). Overexpression of Cdc20 under conditions of reduced Cdc28 activity likely interferes with regulation of mitosis, potentially inducing premature entry or exit from mitosis leading to genome instability.
DNA replication
CDC6, ORC2, SLD2, POL12, POL32, RFA1, TOF1, and TOP2: Sld2, Orc2, and Cdc6 are important for initiation of DNA replication (Kamimura et al. 1998) (Bell and Stillman 1992; Hartwell 1976; Park and Sternglanz 1999); Rfa1 is a component of the heterotrimeric replication protein A complex, which is essential for DNA replication (Brill and Stillman 1991); and Tof1 is a component of the DNA replication complex and is particularly important for recovery from DNA replication stress (Park and Sternglanz 1999; Katou et al. 2003). Pol12 is a component of the DNA polymerase α-primase complex which is required for initiation of DNA replication (Foiani et al. 1994), whereas Pol32 is a component of DNA polymerase δ and is involved in efficient DNA replication (Gerik et al. 1998). Top2 is the catalytic subunit of topoisomerase II (Holm et al. 1985), and plays a major role in maintaining genome stability by alleviating DNA supercoiling and catenation that arise as a consequence of DNA replication. Changes in the expression levels of RFA1, ORC2, CDC6, SLD2, POL32, TOF1, and TOP1 may either perturb the stoichiometry of the factors involved in DNA replication, disturb the timing of DNA replication, or affect DNA supercoiling. For instance, overexpression of fission yeast CDC6 is sufficient to trigger DNA replication (Nishitani and Nurse 1995). We and others have previously shown that minor changes in the timing and efficiency of DNA replication can have severe consequences for the viability of cells with compromised Cdc28 activity (Tanaka and Diffley 2002; Enserink et al. 2009; Lengronne and Schwob 2002). For instance, Cdc28 activity is required for the repair of DNA double-strand breaks (Ira et al. 2004; Aylon et al. 2004), and even a slight reduction in Cdc28 activity can result in mitotic catastrophe and loss of cell viability in response to DNA damage emanating from problems with DNA replication (Enserink et al. 2009). However, there may exist additional mechanisms by which overexpression of these genes causes SDL of cdc28-as1 mutants; overexpression of CDC6 from the GAL1 promoter is known to cause an M phase delay (Bueno and Russell 1992), and because Cdc28 activity is required for entry into M phase (Pines and Hunter 1990) it is possible that a combination of high Cdc6 levels with reduced Cdc28 activity results in M phase arrest.
Chromosome cohesion
SCC1/MCD1: Chromosome cohesion occurs concomitantly with DNA replication (Uhlmann and Nasmyth 1998). The α-kleisin subunit Mcd1 is involved in recruiting the cohesion complex to chromosomes (Guacci et al. 1997; Michaelis et al. 1997). DNA replication-induced chromosome cohesion is limited to S phase, and this is regulated by Cdc28. Specifically, Cdc28 inhibits chromosome cohesion after S phase (Lyons and Morgan 2011). Thus, overexpression of cohesion factors in a cell with reduced Cdc28 activity may result in establishment of cohesion outside of S phase, leading to chromosome separation defects in anaphase.
Chromosome condensation
SMC4: After DNA replication, chromosomes are compacted by the Smc2/4 condensin complex. Chromosome condensation is regulated by Cdc28 (Enserink and Kolodner 2010), and Smc4 is activated by Cdc28 in this process (Robellet et al. 2015). In fact, Smc4 is highly sensitive to Cdc28 activity, such that initiation of chromosome condensation is induced at levels of Cdc28 activity that are too low to activate other mitotic processes (Robellet et al. 2015). Therefore, one can envision that overexpression of SMC4 in a cdc28-as1 mutant results in high levels of unphosphorylated Smc4, such that the cell fails to reach the critical level of Smc4 phosphorylation required for inducing chromosome condensation.
Mitotic spindle, kinetochore, and spindle pole body
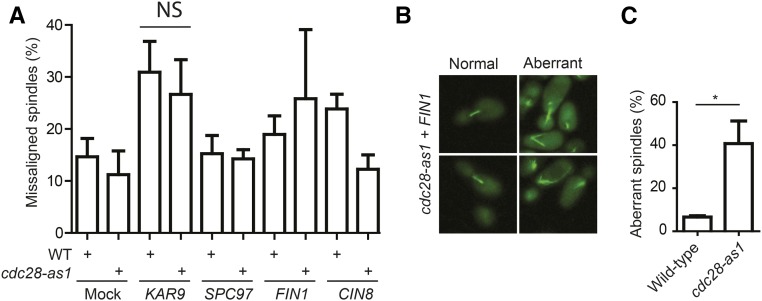
CIN8, CSE4, SGO1, FIN1, KAR9, MAD3, SLK19, MPS2, SPC29, and SPC97: Cin8 is a kinesin motor protein involved in assembly of the mitotic spindle (Saunders and Hoyt 1992); Cse4 is a histone H3-like protein that is a component of the kinetochore (Stoler et al. 1995), whereas Sgo1 is a kinetochore protein that senses tension on the mitotic spindle (Indjeian et al. 2005); Fin1 is an intermediate filament protein that forms filaments between spindle pole bodies, which is important for mitotic spindle stability (van Hemert et al. 2002); Kar9 is involved in spindle positioning (Miller and Rose 1998); and Mad3 is a component of the mitotic spindle checkpoint. Formation of the mitotic spindle is strictly regulated by Cdc28 (Enserink and Kolodner 2010). Therefore, overexpression of these proteins under conditions of reduced Cdc28 activity could perturb spindle assembly, potentially resulting in chromosome missegregation and cell death. To test this, we overexpressed KAR9, SPC97, FIN1, and CIN8, and monitored mitotic spindle assembly and alignment in strains expressing Tub1-GFP (Figure 5A and Figure S2C). Consistent with the function of Kar9 in spindle positioning under normal conditions (Miller and Rose 1998), we found that overexpression of KAR9 resulted in an increase in the number of cells with misaligned spindles, although there was no difference between WT and cdc28-as1 cells (Figure 5A). Furthermore, and consistent with the previous observation that overexpression of KAR9 induces migration of the nucleus into the bud (Korinek et al. 2000), we found that KAR9 overexpression resulted in increased translocation of the entire mitotic spindle into the bud (Figure S2, D and E). Together, these data indicate that the SDL phenotype of KAR9 overexpression may be caused by increased misalignment or mislocalization of the mitotic spindle, but we cannot exclude the possibility that Cdc28 activity is also required for the survival of cells with such misaligned/mislocalized spindles.
Figure 5.
Overexpression of FIN1 in cdc28-as1 mutants induces mitotic spindle aberrancies. (A) No obvious differences in spindle misalignment between WT cells and cdc28-as1 mutants after overexpression of KAR9, SPC97, FIN1, or CIN8. TUB1-GFP-expressing WT cells and cdc28-as1 mutants were transformed with plasmids containing the indicated ORFs (Mock: YEL074W), after which cells were treated with 1-NM-PP1 and overexpression was induced by galactose as described in Materials and Methods. Spindle alignment in at least 100 M phase cells was imaged and quantified using fluorescence microscopy. (B) Overexpression of FIN1 in the cdc28-as1 mutant background results in aberrant spindle assembly. Cells were treated and imaged as in (A). At least 300 cells were analyzed per treatment and genotype. (C) Quantification of the data shown in (B). Error bars indicate SD. *P < 0.05. NS, not significant; WT, wild-type.
While spindle alignment did not appear to be affected in response to overexpression of SPC97, FIN1, and CIN8, we did observe that overexpression of FIN1 resulted in substantial malformation of the mitotic spindle in ∼40% of the cdc28-as1 cells, and these spindles often had unusually long astral microtubules (Figure 5, B and C). This phenotype is remarkably similar to that of cells overexpressing fin1-5A, which encodes a form of Fin1 that can no longer be phosphorylated by Cdc28 (Woodbury and Morgan 2007). Therefore, the SDL phenotype is likely caused by accumulation of unphosphorylated Fin1. Currently, we do not know what causes the SDL phenotype imparted by overexpression of SPC97 and CIN8, which did not cause obvious spindle defects in either the WT cells or in cdc28-as1 mutants (Figure 5A and Figure S2C). It is possible that overexpression of these genes results in phenotypes too subtle to be detected in our assays. For instance, overexpression of CIN8 has been shown to induce premature spindle elongation in WT cells, but it was only clearly detectable when cells were maintained in prolonged hydroxyurea-induced S phase arrest (Saunders et al. 1997).
Cell morphogenesis
BEM1, BUD2, and BUD4: Cell cycle entry is accompanied by the assembly of a new bud (Schwann 1837), which is driven by cell polarity factors and which strongly depends on the cell cycle (Howell and Lew 2012). Bem1 is an adaptor protein that, together with the small GTPase Bud1, is important for bud site selection and bud growth (Bender and Pringle 1991). Bud2 is a GTPase-activating protein (GAP) for Bud1 (Benton et al. 1993; Cvrckova and Nasmyth 1993), whereas the anillin-like protein Bud4 is important for formation of the septin ring (Eluere et al. 2012), which is an essential aspect for bud assembly. Bud growth is dependent on Cdc28 activity (Hartwell et al. 1973; McCusker et al. 2007; Howell and Lew 2012), Bem1 and Bud4 are direct Cdc28 targets (Han et al. 2005; Eluere et al. 2012), and it is likely that altered levels of Bem1, Bud2, and Bud4 perturb bud growth under conditions of reduced Cdc28 activity. Bem1 may also affect cell cycle progression, since BEM1 overexpression has previously been shown to enhance G1 arrest (Lyons et al. 1996) in response to α factor. Because α factor inhibits Cdc28 activity (Tyers and Futcher 1993), it is conceivable that overexpression of BEM1 in the hypomorphic cdc28-as1 mutant background results in G1 phase arrest.
Transcription
A very large number of genes that induce dosage lethality in the cdc28-as1 mutant are involved in transcription. This is not surprising, given the fact that Cdc28 is important for activation of transcription at several stages (Chymkowitch and Enserink 2013). For instance, Cdc28 regulates a compendium of cell cycle-specific transcription factors that play an important role in executing the various stages of the cell cycle (Enserink and Kolodner 2010; Wittenberg and Reed 2005). Cdc28 also directly controls the basal transcription machinery at a subset of genes mainly involved in housekeeping and cell homeostasis (Chymkowitch et al. 2012; Chymkowitch and Enserink 2013). Consistently, analysis by GO Slim Mapper revealed that nearly all of the SDL-inducing genes involved in the process of transcription are part of complexes that regulate basal transcription, such as TFIID, INO80, Mediator, and the HDAC and HAT complexes, as well as the RNA polymerase II holoenzyme (Table S4). It is likely that overexpression of these genes alters the stoichiometry of these complexes, thereby interfering with efficient transcription. Consistently, we have previously shown that transcription of cell cycle genes (such as cyclins) is strongly affected by even very modest changes in transcription efficiency (Zimmermann et al. 2011), which is likely to have severe consequences for the cell cycle progression and viability of 1-NM-PP1-treated cdc28-as1 cells.
Genes that may alleviate the cdc28-as1 slow growth phenotype
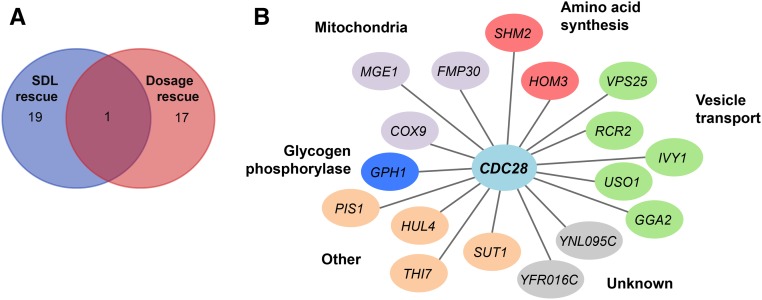
We identified 20 potential suppressor genes that may partially compensate the growth defect of the cdc28-as1 mutant (Table S1). We compared these genes to previously identified dosage suppressors of cdc28 alleles (Figure 6A) and found that, apart from CDC28 itself, there was no overlap between the SDL screen and these previous dosage rescue studies, which as discussed above mainly identified cyclins. GO analysis did not reveal any overrepresented gene functions of the genes that alleviate the cdc28-as1 growth defect. However, we did notice that five of these genes (GGA2, VPS25, RCR2, IVY1, and USO1) have functions in vesicle transport (Nakajima et al. 1991; Zhdankina et al. 2001; Robinson et al. 1988; Kota et al. 2007; Lazar et al. 2002) (Figure 6B), indicating that one consequence of chronically reduced Cdc28 activity could be defective vesicle transport, which might be overcome by overexpression of these vesicle transport genes. Indeed, Cdc28 has been shown to be important for membrane trafficking (McCusker et al. 2012).
Figure 6.
Genes that may promote dosage rescue of cdc28-as1 mutant cells. (A) Comparison of previously identified genes that mediate dosage suppression of cdc28 alleles with the ORFs that mediate dosage suppression identified by the synthetic dosage lethality (SDL) screen. The only ORF in common between these datasets was YBR160W, which encodes Cdc28. (B) Putative dosage suppressors identified by the SDL screen organized by their cellular functions.
In addition to genes involved in vesicle transport, we identified genes involved in amino acid synthesis. We have previously shown that deletion of one of these genes, HOM3 (which encodes aspartokinase), results in synthetic lethality with the cdc28-as1 mutation (Zimmermann et al. 2011). Thus, loss of HOM3 aggravates the growth defect of cdc28-as1 mutants whereas its overexpression suppresses this defect, suggesting that HOM3 may have an important function in the cell cycle. Indeed, we previously found that HOM3 may support survival of the cdc28-as1 mutant cells by promoting the synthesis of dNTPs (Zimmermann et al. 2011).
Another gene worth mentioning is GPH1. Gph1 functions as glycogen phosphorylase, which mobilizes glycogen to increase glucose-1-phosphate. Intriguingly, Gph1 was recently found to be a direct Cdc28 substrate and phosphorylation of Gph1 may coordinate carbohydrate metabolism with cell division (Zhao et al. 2016), providing a potential explanation for why its overexpression suppresses the cdc28-as1 phenotype. Interestingly, in addition to Gph1, the SDL screen identified several Cdc28 targets (see below), although all of them (except Gph1) induced a dosage lethality phenotype.
Overrepresentation of Cdc28 targets
It was previously reported that SDL screens can identify novel functional relationships and pathways regulated by kinases, and SDL screens are enriched for kinase substrates (Sharifpoor et al. 2012; Sopko et al. 2006). Interestingly, our SDL screen was also enriched for direct targets of Cdc28 (Tab2 in Table S1). We identified 29 of the 100 currently known Cdc28 targets (known Cdk1 targets are defined here as proteins shown in low-throughput experiments to be phosphorylated in a Cdk1-dependent manner in vivo; we previously compiled a list of targets (Enserink and Kolodner 2010) to which we added more recently identified Cdk1 substrates). However, given that 20 known Cdc28 targets were not present in the library and that at least three clones were incorrect, we identified 37% of the currently known Cdc28 substrates that could theoretically be identified by the SDL screen.
To gain further insight in potential Cdc28 targets, we compared the SDL dataset to previously published datasets. We compiled data obtained with phosphoproteomics, physical interactions, and in vitro kinase assays (see Materials and Methods). Figure S3A shows a Venn diagram that combines all these datasets, including a list of known Cdc28 targets (see Table S5 for ORF identities). This comparison indicates that only a minority of direct Cdc28 substrates has been found to actually physically interact with the Cdc28 holoenzyme (Figure S3B and Table S6), suggesting that phosphorylation by Cdc28 involves transient interactions of the Cdc28 holoenzyme with its substrates that elude detection by high-throughput proteomic approaches.
To simplify the interpretation of the data, we removed the physical interaction dataset from the Venn diagram (Figure 7A and Table S7), as well as the dataset containing known Cdc28 targets (Figure 7B and Table S8). This revealed that most (201 out of 229) proteins that have previously been found to be phosphorylated in vivo in a Cdc28-dependent manner also cause dosage lethality of cdc28-as1 mutants, suggesting a functional relationship between these proteins and Cdc28. However, it is unlikely that many of these proteins are direct Cdc28 targets, because most of them do not appear to be phosphorylated by Cdc28 in vitro (Figure 7B and Table S8). Furthermore, even though the phosphorylation of these proteins depends on Cdc28 activity, a large number of these phosphorylations occur on non-Cdc28 consensus sites [see Datasets S1 and S2 in Holt et al. (2009)]. While Cdc28 has been shown to be able to phosphorylate substrates on nonconsensus sites (Harvey et al. 2005; McCusker et al. 2007), this appears to be relatively rare (Suzuki et al. 2015). Therefore, it is more likely that Cdc28-dependent phosphorylation of proteins on nonconsensus sites in the SDL dataset depends on cell cycle stage rather than on Cdc28 activity per se. Alternatively, Cdc28 may indirectly control their phosphorylation by regulating kinases/phosphatases that in turn regulate phosphorylation of these proteins. It should also be mentioned that high-throughput phosphoproteomics studies that used the hypomorphic cdc28-as1 allele may have missed poor substrates whose phosphorylation is very sensitive to Cdk1 activity; as a consequence, these sensitive substrates are absent from our comparisons.
Figure 7.
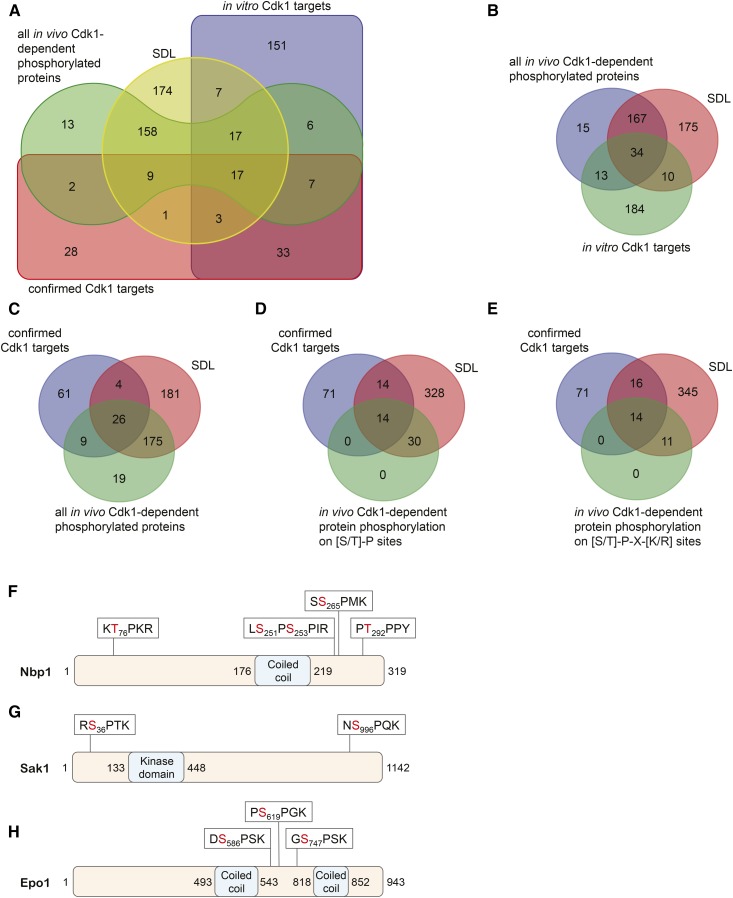
The SDL dataset is enriched for proteins that are phosphorylated in a Cdc28-dependent manner. (A) Venn diagram depicting overlap between the SDL dataset and proteins that are phosphorylated in a Cdc28-dependent manner in vivo and in vitro, and which have previously been confirmed to be Cdc28 substrates. (B) Most proteins that have been found to be phosphorylated in a Cdc28-dependent manner in vivo cause dosage lethality when overexpressed in a cdc28-as1 mutant. The Venn diagram shows overlap between the SDL dataset and proteins that are phosphorylated in a Cdc28-dependent manner in vitro and in vivo. (C) The SDL dataset is enriched for known Cdc28 targets. The Venn diagram shows overlap between the SDL screen, proteins that have been found to be phosphorylated in a Cdc28-dependent manner in vivo, and previously confirmed Cdc28 substrates. (D and E) Overexpression of proteins phosphorylated on potential Cdc28 sites cause an SDL phenotype. The Venn diagrams show overlap between the SDL dataset, confirmed Cdc28 targets, and proteins that are known to be phosphorylated in vivo on either minimal Cdc28 sites (D) or on full Cdc28 consensus sites (E) in a Cdc28-dependent manner. (F–H) Domain structures of Nbp1 (F), Sak1 (G), and Epo1 (H), and location of sites known to be phosphorylated in vivo in a Cdc28-dependent manner. CDK, cyclin-dependent kinase; SDL, synthetic dosage lethality.
We also compared the SDL dataset with the list of all known Cdc28 targets, as well as with the list of proteins shown to be phosphorylated in vivo in a Cdc28-dependent manner. There was substantial overlap between these datasets (Figure 7C and Table S9). To be able to pinpoint potential novel, direct Cdc28 substrates, we then focused only on those proteins that are phosphorylated in a Cdc28-dependent manner in vivo on either minimal Cdc28 sites (S/T-P; Figure 7D and Table S10) or optimal Cdc28 consensus sites (S/T-X-K/R; Figure 7E and Table S11). Interestingly, this revealed that all these proteins were also present in the SDL dataset. We believe that it is likely that these proteins are direct Cdc28 substrates. One example of a likely direct Cdc28 target is the spindle pole body component Nbp1 (Figure 7F), because it induces SDL in a cdc28-as1 background, it is an efficient in vitro Cdc28 target (Ubersax et al. 2003), it is phosphorylated in vivo on optimal Cdc28 consensus sites in a Cdc28-dependent manner (Bodenmiller et al. 2010; Holt et al. 2009), and it physically interacts with Cdc28 (Ear et al. 2013). Nbp1 is essential for bipolar spindle formation, and it is required for the efficient insertion of the spindle pole body into the nuclear membrane. The effect of Nbp1 phosphorylation by Cdc28 will be the subject of future studies.
Another example of a potential Cdc28 target is Sak1 (Figure 7G), a kinase with similarity to mammalian LKB. Sak1 controls the Swi/Snf1 complex and has been found to be phosphorylated on two optimal Cdc28 consensus sites, S36PTK and S966PQK, in a Clb2–Cdc28-dependent manner, although the functional significance of these phosphorylations has not been established (Holt et al. 2009). SWI/Snf has an important function in cell cycle regulation, and conversely its activity is regulated by the cell cycle (Sif et al. 1998; Ruijtenberg and van den Heuvel 2015). Thus, one mechanism by which Cdc28 might regulate SWI/Snf1 activity could be through phosphorylation of Sak1.
Finally, Epo1 may also be a direct Cdc28 target (Figure 7H). Epo1 encodes a protein involved in ER-septin tethering by binding the septin Shs1 and the ER protein Scs2, which is important for creating a diffusion barrier between the mother and daughter cell (Chao et al. 2014). Epo1 has been found to be phosphorylated on multiple residues in large-scale phosphoproteomic studies, three of which are optimal Cdc28 consensus sites: S586PSK, S619PGK, and S747PSK. The phosphorylation of at least one of these sites, i.e., S586, is dependent upon Cdc28 activity in vivo (Holt et al. 2009). Interestingly, the interaction between Epo1 and Scs2 is cell cycle-dependent (Neller et al. 2015), indicating that Cdc28 may play an important role in the regulation of the tethering of the ER to the vacuole by regulating the interaction between Epo1 and Scs2.
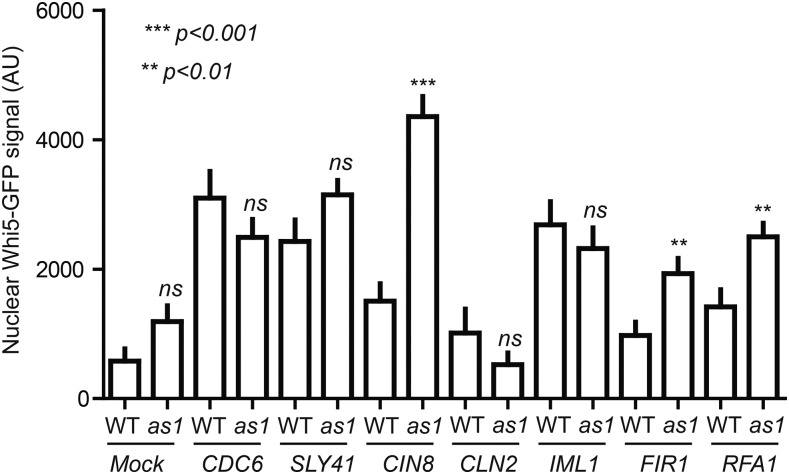
It is not entirely clear why Cdc28 substrates are enriched in the SDL screen, but one explanation could be that overexpression of these substrates results in competition with other Cdc28 targets for phosphorylation by Cdc28. We tested this hypothesis by monitoring phosphorylation of the direct Cdc28 target Whi5, which is a transcriptional repressor of the G1 transcriptional program (de Bruin et al. 2004; Costanzo et al. 2004). In late G1 phase, phosphorylation of Whi5 by Cdc28 results in its release from the transcription factors SBF and MBF to accelerate the G1/S transition (Costanzo et al. 2004). Phosphorylated Whi5 is transported out of the nucleus and only reenters the nucleus at the end of M phase when Cdc28 is inactivated (Costanzo et al. 2004; Taberner et al. 2009), and nuclear localization of Whi5-GFP is a highly sensitive read-out for Cdc28 activity (Talia et al. 2007; Skotheim et al. 2008). We nocodazole-arrested cells in M phase, when Whi5 is hyperphosphorylated by Cdc28 (Taberner et al. 2009), and overexpressed the known Cdc28 targets CDC6, SLY41, CIN8, and FIR1, as well as IML1 and RFA1, which are not currently known to be Cdc28 targets, and monitored cellular localization of Whi5-GFP (Figure 8; see Figure S4 for examples). We overexpressed the mock ORFs YEL074W and CLN2 as controls; as expected, overexpression of neither of these genes resulted in nuclear localization of Whi5 (Figure 8). However, increased nuclear localization of Whi5-GFP could be observed in the nucleus of cells overexpressing CDC6, SLY41, and IML1, although there was no significant difference between WT and cdc28-as1 cells (Figure 8). In contrast, overexpression of CIN8, FIR1, and RFA1 did result in significantly increased nuclear localization of Whi5-GFP in cdc28-as1 mutants compared to WT cells (Figure 8). These data indicate that overexpression of certain proteins, including known and potential Cdc28 substrates, can interfere with normal Cdc28 signaling. Thereby, at least some of these proteins may effectively function as Cdc28 inhibitors when overexpressed, providing an explanation for the SDL phenotype.
Figure 8.
Aberrant nuclear localization of Whi5 in M phase upon overexpression of several SDL genes. Overexpression of CIN8, FIR1, and RFA1 results in a significant increase in nuclear localization of Whi5-GFP in M phase cells. Whi5-GFP-expressing cells containing YEL074W (“Mock”), CDC6, SLY41, CIN8, CLN2, IML1, FIR1, and RFA1 expression plasmids were grown to log phase in the presence of glucose and arrested in M phase with nocodazole. Cells were then washed and incubated in nocodazole-containing medium supplemented with galactose to induce expression for 3 hr, after which nuclear localization of Whi5-GFP was assessed by fluorescence microscopy. Error bars indicate SD. ns, not significant; **P < 0.001, ***P < 0.0001. GFP, green fluorescent protein; SDL, synthetic dosage lethality; WT, wild-type.
In this study, we have interrogated the SDL network of CDC28. Our data indicate that the underlying cause for the SDL phenotype is likely to be heterogeneous and dependent on the overexpressed gene. For instance, overexpression of a number of genes involved in the organization and orientation of the mitotic spindle revealed that overexpression of FIN1 significantly altered the assembly of the mitotic spindle in the cdc28-as1 mutant. Overexpression of several other genes did not result in an obvious defect in cdc28-as1 mutant cells compared with WT cells. This suggests that the spindle defect in the cdc28-as1 mutant cells might have been too subtle to be observed by microscopy. Alternatively, Cdc28 activity may be required for cell survival in the presence of mitotic spindle problems, which is supported by our previous studies that showed that loss of mitotic checkpoint activity results in lethality of cdc28-as1 mutants (Enserink et al. 2009; Zimmermann et al. 2011).
Another reason for dosage lethality can be the activation of certain cell cycle events in the wrong phase of the cell cycle. For example, we found that overexpression of CLN3 in 1-NM-PP1-treated cdc28-as1 mutants, which are arrested in early M phase, resulted in increased transcription of two G1/S-specific cyclin genes, i.e., CLN2 and CLB6, which may further disturb synchrony of the cell cycle. In addition, it is possible that the overexpressed Cln3 titrates away Clb cyclins from the Cdc28 holoenzyme; our genetic data indicate that overexpressed Cln3 is indeed able to activate Cdc28 in vivo, as demonstrated by rescue of the temperature-sensitive phenotype of the cdc28-4 mutant by overexpression of CLN3 (Figure 4B).
Finally, overexpression of a given Cdc28 target may result in competition with other Cdc28 substrates for phosphorylation by Cdc28, thereby disturbing normal cell cycle progression. This is likely to be particularly toxic in the cdc28-as1 mutant background, in which Cdc28 activity is limiting. Supporting this hypothesis, we found that overexpression of several known Cdc28 targets in the cdc28-as1 mutant background resulted in the precocious nuclear entry of Whi5 during M phase; Whi5 is normally localized to the cytoplasm in M phase, because it is phosphorylated by Cdc28, leading to its nuclear export by Msn5 (de Bruin et al. 2004; Costanzo et al. 2004; Wagner et al. 2009; Taberner et al. 2009). Nuclear reentry of Whi5 only occurs when it is dephosphorylated (Wagner et al. 2009). Thus, the SDL phenotype of at least some genes may result from competition with endogenous Cdc28 substrates, thereby effectively functioning as an inhibitor of Cdc28. In addition to deregulation of cell cycle processes and competition for Cdc28 substrates, it is likely that more explanations exist why certain genes induce dosage lethality in cdc28-as1 mutants.
We believe that the results from our screen may lead to the identification of additional targets and processes controlled by Cdc28, which is needed to better understand how the cell cycle regulates cell proliferation and homeostasis.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.042317/-/DC1.
Acknowledgments
This project was supported by grants to J.M.E. from the Norwegian Research Council (221694 and 261936) and the Norwegian Cancer Society (project numbers 3311782 and 4487303). P.C. is supported by a grant from the Norwegian Research Council (221920) and the Norwegian Health Authority South-East (2017064). I.G. is supported by a grant from the Norwegian Health Authority South-East (2017072).
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Aylon Y., Liefshitz B., Kupiec M., 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23(24): 4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Stillman B., 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357(6374): 128–134. [DOI] [PubMed] [Google Scholar]
- Bender A., Pringle J. R., 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11(3): 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B. K., Tinkelenberg A. H., Jean D., Plump S. D., Cross F. R., 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12(13): 5267–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., et al. , 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407(6802): 395–401. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B., Aebersold R., 2011. Phosphoproteome resource for systems biology research. Methods Mol. Biol. 694: 307–322. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., et al. , 2010. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3(153): rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2): 115–132. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Stillman B., 1991. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5(9): 1589–1600. [DOI] [PubMed] [Google Scholar]
- Bueno A., Russell P., 1992. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11(6): 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J. T., Wong A. K., Tavassoli S., Young B. P., Chruscicki A., et al. , 2014. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158(3): 620–632. [DOI] [PubMed] [Google Scholar]
- Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., et al. , 2017. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45(D1): D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P., Enserink J. M., 2013. The cell cycle rallies the transcription cycle: Cdc28/Cdk1 is a cell cycle-regulated transcriptional CDK. Transcription 4(1): 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P., Eldholm V., Lorenz S., Zimmermann C., Lindvall J. M., et al. , 2012. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc. Natl. Acad. Sci. USA 109(26): 10450–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J. L., Tang X., Millman J. S., Schub O., et al. , 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117(7): 899–913. [DOI] [PubMed] [Google Scholar]
- Cvrckova F., Nasmyth K., 1993. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 12(13): 5277–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C., Diffley J. F., Nasmyth K. A., 1995. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 5(11): 1257–1269. [DOI] [PubMed] [Google Scholar]
- de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., III, Wittenberg C., 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117(7): 887–898. [DOI] [PubMed] [Google Scholar]
- Dixon S. J., Costanzo M., Baryshnikova A., Andrews B., Boone C., 2009. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 43: 601–625. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1946. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics 31: 269–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ear P. H., Booth M. J., Abd-Rabbo D., Kowarzyk Moreno J., Hall C., et al. , 2013. Dissection of Cdk1-cyclin complexes in vivo. Proc. Natl. Acad. Sci. USA 110(39): 15716–15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eluere R., Varlet I., Bernadac A., Simon M. N., 2012. Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell Cycle 11(1): 151–158. [DOI] [PubMed] [Google Scholar]
- Enserink J. M., 2012. Chemical genetics: budding yeast as a platform for drug discovery and mapping of genetic pathways. Molecules 17(8): 9258–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Kolodner R. D., 2010. An overview of Cdk1-controlled targets and processes. Cell Div. 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Hombauer H., Huang M. E., Kolodner R. D., 2009. Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae. J. Cell Biol. 185(3): 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P., 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14(2): 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., et al. , 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19(23): 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik K. J., Li X., Pautz A., Burgers P. M., 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273(31): 19747–19755. [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A., 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91(1): 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Mendenhall M. D., Reed S. I., 1989a The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol. 9(5): 2034–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Richardson H. E., de Barros Lopes M., Reed S. I., 1989b A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 86(16): 6255–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. K., Bogomolnaya L. M., Totten J. M., Blank H. M., Dangott L. J., et al. , 2005. Bem1p, a scaffold signaling protein, mediates cyclin-dependent control of vacuolar homeostasis in Saccharomyces cerevisiae. Genes Dev. 19(21): 2606–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., 1976. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol. 104(4): 803–817. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M., 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74(2): 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. L., Charlet A., Haas W., Gygi S. P., Kellogg D. R., 2005. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 122(3): 407–420. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D., 1985. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41(2): 553–563. [DOI] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., et al. , 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325(5948): 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. S., Lew D. J., 2012. Morphogenesis and the cell cycle. Genetics 190(1): 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Friesen H., Andrews B., 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 66(2): 303–314. [DOI] [PubMed] [Google Scholar]
- Hunter T., Pines J., 1994. Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell 79(4): 573–582. [DOI] [PubMed] [Google Scholar]
- Indjeian V. B., Stern B. M., Murray A. W., 2005. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307(5706): 130–133. [DOI] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., et al. , 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431(7011): 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto H., Sugino A., Araki H., 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18(10): 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424(6952): 1078–1083. [DOI] [PubMed] [Google Scholar]
- Korinek W. S., Copeland M. J., Chaudhuri A., Chant J., 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287(5461): 2257–2259. [DOI] [PubMed] [Google Scholar]
- Kota J., Melin-Larsson M., Ljungdahl P. O., Forsberg H., 2007. Ssh4, Rcr2 and Rcr1 affect plasma membrane transporter activity in Saccharomyces cerevisiae. Genetics 175: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll E. S., Hyland K. M., Hieter P., Li J. J., 1996. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar T., Scheglmann D., Gallwitz D., 2002. A novel phospholipid-binding protein from the yeast Saccharomyces cerevisiae with dual binding specificities for the transport GTPase Ypt7p and the Sec1-related Vps33p. Eur. J. Cell Biol. 81(12): 635–646. [DOI] [PubMed] [Google Scholar]
- Lengronne A., Schwob E., 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol. Cell 9(5): 1067–1078. [DOI] [PubMed] [Google Scholar]
- Liu Q., Larsen B., Ricicova M., Orlicky S., Tekotte H., et al. , 2011. SCFCdc4 enables mating type switching in yeast by cyclin-dependent kinase-mediated elimination of the Ash1 transcriptional repressor. Mol. Cell. Biol. 31(3): 584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D. M., Mahanty S. K., Choi K. Y., Manandhar M., Elion E. A., 1996. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol. Cell. Biol. 16(8): 4095–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons N. A., Morgan D. O., 2011. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol. Cell 42(3): 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T. A., Yates J. R., III, et al. , 2007. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 9(5): 506–515. [DOI] [PubMed] [Google Scholar]
- McCusker D., Royou A., Velours C., Kellogg D., 2012. Cdk1-dependent control of membrane-trafficking dynamics. Mol. Biol. Cell 23(17): 3336–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Baetz K., Guzzo J., Yuen K., Kwok T., et al. , 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102(39): 13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall M. D., Hodge A. E., 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62(4): 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91(1): 35–45. [DOI] [PubMed] [Google Scholar]
- Miller R. K., Rose M. D., 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140(2): 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Nurse P., 1990. Substrates for p34cdc2: in vivo veritas? Cell 61(4): 549–551. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Hirata A., Ogawa Y., Yonehara T., Yoda K., et al. , 1991. A cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. J. Cell Biol. 113(2): 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neller J., Dunkler A., Rosler R., Johnsson N., 2015. A protein complex containing Epo1p anchors the cortical endoplasmic reticulum to the yeast bud tip. J. Cell Biol. 208(1): 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Nurse P., 1995. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83(3): 397–405. [DOI] [PubMed] [Google Scholar]
- Nugroho T. T., Mendenhall M. D., 1994. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 14(5): 3320–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Sternglanz R., 1999. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast 15(1): 35–41. [DOI] [PubMed] [Google Scholar]
- Patil A., Nakamura H., 2005. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics 6: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T., 1990. p34cdc2: the S and M kinase? New Biol. 2(5): 389–401. [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., et al. , 2005. Global analysis of protein phosphorylation in yeast. Nature 438(7068): 679–684. [DOI] [PubMed] [Google Scholar]
- Reed S. I., 1980. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95(3): 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Wittenberg C., 1990. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87(15): 5697–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X., Thattikota Y., Wang F., Wee T. L., Pascariu M., et al. , 2015. A high-sensitivity phospho-switch triggered by Cdk1 governs chromosome morphogenesis during cell division. Genes Dev. 29(4): 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D., 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8(11): 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Murray A. W., 2000. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 149(7): 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Hardwick K. G., Murray A. W., 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149(7): 1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S., 2015. G1/S inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell 162(2): 300–313. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Breitkreutz B. J., Stark C., Su T. C., Dahabieh M., et al. , 2013. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database (Oxford) 2013: bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M. A., 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell 8(6): 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A., 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70(3): 451–458. [DOI] [PubMed] [Google Scholar]
- Schwann T., 1837. Vorläufige Mittheilung, betreffend Versuche über die Weingährung und Fäulniss. Ann. Phys. 117(5): 184–193. [Google Scholar]
- Schwob E., Bohm T., Mendenhall M. D., Nasmyth K., 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79(2): 233–244. [DOI] [PubMed] [Google Scholar]
- Sharifpoor S., van Dyk D., Costanzo M., Baryshnikova A., Friesen H., et al. , 2012. Functional wiring of the yeast kinome revealed by global analysis of genetic network motifs. Genome Res. 22(4): 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S., Stukenberg P. T., Kirschner M. W., Kingston R. E., 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12(18): 2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim J. M., Di Talia S., Siggia E. D., Cross F. R., 2008. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454(7202): 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21(3): 319–330. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9(12): 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9(5): 573–586. [DOI] [PubMed] [Google Scholar]
- Surana U., Robitsch H., Price C., Schuster T., Fitch I., et al. , 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65(1): 145–161. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Sako K., Akiyama K., Isoda M., Senoo C., et al. , 2015. Identification of non-Ser/Thr-Pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger proteins and Ect2. Sci. Rep. 5: 7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., et al. , 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43(Database issue): D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberner F. J., Quilis I., Igual J. C., 2009. Spatial regulation of the start repressor Whi5. Cell Cycle 8(18): 3010–3018. [PubMed] [Google Scholar]
- Talia S. D., Skotheim J. M., Bean J. M., Siggia E. D., Cross F. R., 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448(7156): 947–951. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Diffley J. F., 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 16(20): 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R., Chen H., Gao C., Xue P., Yang F., et al. , 2011. Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast. Cell Res. 21(11): 1619–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe B. T., Kitazono A. A., Garcia J. S., Gerber R. A., Bevis B. J., et al. , 2009. Morphogenesis signaling components influence cell cycle regulation by cyclin dependent kinase. Cell Div. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294(5550): 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Pohl M. O., Zhou Y., Rodriguez-Frandsen A., Wang G., et al. , 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18(6): 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Futcher B., 1993. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol. Cell. Biol. 13(9): 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., et al. , 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425(6960): 859–864. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Nasmyth K., 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8(20): 1095–1101. [DOI] [PubMed] [Google Scholar]
- van Hemert M. J., Lamers G. E., Klein D. C., Oosterkamp T. H., Steensma H. Y., et al. , 2002. The Saccharomyces cerevisiae Fin1 protein forms cell cycle-specific filaments between spindle pole bodies. Proc. Natl. Acad. Sci. USA 99(8): 5390–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A., 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278(5337): 460–463. [DOI] [PubMed] [Google Scholar]
- von Mering C., Krause R., Snel B., Cornell M., Oliver S. G., et al. , 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417(6887): 399–403. [DOI] [PubMed] [Google Scholar]
- Wagner M. V., Smolka M. B., de Bruin R. A., Zhou H., Wittenberg C., et al. , 2009. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One 4(1): e4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C., Reed S. I., 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24(17): 2746–2755. [DOI] [PubMed] [Google Scholar]
- Woodbury E. L., Morgan D. O., 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9(1): 106–112. [DOI] [PubMed] [Google Scholar]
- Yaglom J., Linskens M. H., Sadis S., Rubin D. M., Futcher B., et al. , 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15(2): 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Chen Y., Carey L., Futcher B., 2016. Cyclin-dependent kinase co-ordinates carbohydrate metabolism and cell cycle in S. cerevisiae. Mol. Cell 62(4): 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O., Strand N. L., Redmond J. M., Boman A. L., 2001. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast 18(1): 1–18. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Mori S., Oshiumi H., Matsuzaki K., Shinohara M., et al. , 2010. Cyclin-dependent kinase promotes formation of the synaptonemal complex in yeast meiosis. Genes Cells 15(10): 1036–1050. [DOI] [PubMed] [Google Scholar]
- Zimmermann C., Chymkowitch P., Eldholm V., Putnam C. D., Lindvall J. M., et al. , 2011. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc. Natl. Acad. Sci. USA 108(46): 18748–18753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.