Abstract
Swainsonine—a cytotoxic fungal alkaloid and a potential cancer therapy drug—is produced by the insect pathogen and plant symbiont Metarhizium robertsii, the clover pathogen Slafractonia leguminicola, locoweed symbionts belonging to Alternaria sect. Undifilum, and a recently discovered morning glory symbiont belonging to order Chaetothyriales. Genome sequence analyses revealed that these fungi share orthologous gene clusters, designated “SWN,” which included a multifunctional swnK gene comprising predicted adenylylation and acyltransferase domains with their associated thiolation domains, a β-ketoacyl synthase domain, and two reductase domains. The role of swnK was demonstrated by inactivating it in M. robertsii through homologous gene replacement to give a ∆swnK mutant that produced no detectable swainsonine, then complementing the mutant with the wild-type gene to restore swainsonine biosynthesis. Other SWN cluster genes were predicted to encode two putative hydroxylases and two reductases, as expected to complete biosynthesis of swainsonine from the predicted SwnK product. SWN gene clusters were identified in six out of seven sequenced genomes of Metarhzium species, and in all 15 sequenced genomes of Arthrodermataceae, a family of fungi that cause athlete’s foot and ringworm diseases in humans and other mammals. Representative isolates of all of these species were cultured, and all Metarhizium spp. with SWN clusters, as well as all but one of the Arthrodermataceae, produced swainsonine. These results suggest a new biosynthetic hypothesis for this alkaloid, extending the known taxonomic breadth of swainsonine producers to at least four orders of Ascomycota, and suggest that swainsonine has roles in mutualistic symbioses and diseases of plants and animals.
Keywords: comparative genomics, dermatophytes, locoweed endophyte, pathogenic fungi, symbiotic fungi
Swainsonine is known to be produced by fungal symbionts of plants (endophytes), plant pathogens, and insect pathogens, but has not previously been reported from pathogens of humans and other mammals (Figure 1). This indolizidine alkaloid—a mannofuranose analog—specifically inhibits α-mannosidase II in the Golgi apparatus, disrupting the endomembrane system of the cell (Dorling et al. 1980; Tulsiani et al. 1982; Winchester et al. 1993), and is under consideration as a component of chemotherapeutic treatments for some cancers (Santos et al. 2011; Li et al. 2012). Swainsonine-producing endophytes belonging to Alternaria sect. Undifilum (Braun et al. 2003) can occur in certain legumes in the related genera Astragalus, Oxytropis (“locoweeds”), and Swainsona, in semiarid regions of Asia, the Americas, and Australia. Wildlife and livestock that feed on these plants can exhibit toxicosis (“locoism” or “pea struck”) characterized by weight loss, altered behavior, depression, decreased libido, infertility, abortion, birth defects, and death. Swainsonine is also produced by a recently discovered endophyte of the morning glory species, Ipomoea carnea (Cook et al. 2013). This I. carnea endophyte (ICE) has phylogenetic affinity to the order Chaetothyriales, but is of an undescribed species. Swainsonine is also produced by diverse fungi with other ecological functions; namely, the plant pathogen, Slafractonia leguminicola (Alhawatema et al. 2015), and the root-associated insect pathogen, Metarhizium robertsii, which is commonly used for organic plant protection (St. Leger and Wang 2010). Here, we identify and demonstrate function of orthologous swainsonine biosynthesis gene clusters (SWN) in these and other fungi, including all available members of the Arthrodermataceae—a family of dermatophytic human and animal pathogens.
Figure 1.
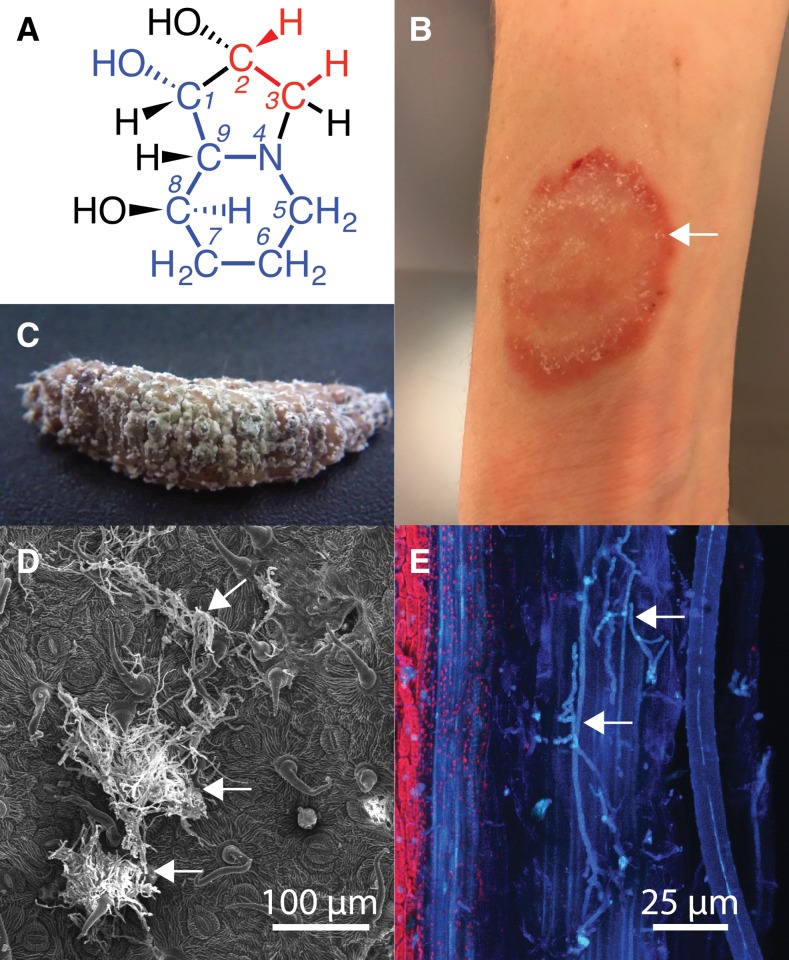
Swainsonine and swainsonine producers in their natural environments. (A) Structure of the indolizidine alkaloid, swainsonine. Atoms indicated in blue are known or predicted to be derived from pipecolic acid, and those in red from mevalonic acid. (B) Clinical symptoms of ringworm caused by Trichophyton benhamiae (Nenoff et al. 2014) (photograph provided by Dr. Pietro Nenoff, Laboratory for Medical Microbiology, Mölbis, Germany, and Dr. Ina Schulze, Markkleeberg near Leipzig, Germany). (C) Insect larva mummified by Metarhizium sp. White fungal mycelium is visible over the surface of the larva. (D) Scanning electron micrograph of ICE on the adaxial leaf surface. Arrows show masses of fungal hyphae (micrograph from Aziza Noor, New Mexico State University). (E) Confocal micrograph of endobiotic A. oxytropis (micrograph from Aziza Noor). Arrows indicate endobiotic hyphae.
Materials and Methods
Biological materials
The source and culture conditions for Alternaria oxytropis (=Undifilum oxytropis) are described in Reyna et al. (2012). The source and culturing conditions for the Ipomoea carnea endophyte (ICE) are described in Cook et al. (2013). Slafractonia leguminicola (=Rhizoctonia leguminicola) ATCC 26280 was obtained from the American Type Culture Collection (ATCC), and cultured as described by Alhawatema et al. (2015). All Metarhizium species were obtained from the ARSEF Collection of Entomopathogenic Fungi. All the dermatophytes were obtained from ATCC with the exception of Trichophyton rubrum, which was obtained from Theodore C. White at the University of Missouri-Kansas City.
Genome sequencing and analysis
Fungal DNA was prepared using the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, Irvine, CA). Genome sequencing and assembly was performed at the Advanced Genetic Technologies Center (AGTC) of the University of Kentucky. The ICE genome was sequenced by pyrosequencing (Roche Diagnostics/454 Life Sciences Corp.) of sheared DNA fragments. A total of 1,334,638 pyrosequencing reads gave 995,708,063 bases, of which Newbler 2.8 (Roche Diagnostics/454 Life Sciences Corp.) aligned 1,284,087 reads totaling 949,835,583 aligned bases, to give an assembly of 32,767,887 bp in 307 contigs, with N50 = 380,768 bp.
Genomes of A. oxytropis and S. leguminicola were sequenced on the MiSeq platform (Illumina, San Diego, CA). For A. oxytropis a total of 40,814,896 paired MiSeq reads gave 8,826,667,915 bases, of which CLC Genomics Workbench 8.0.2 (Qiagen, Valencia, CA) matched 39,668,025 reads totaling 8,584,276,934 aligned bases, and paired 25,326,112 reads with an average paired read length = 426 bp, to give a genome assembly of 112,671,691 bp in 57,645 scaffolds, with N50 = 3841 bp. For S. leguminicola, a total of 27,839,555 paired MiSeq reads gave 6,027,678,056 bases, of which CLC Genomics Workbench 8.0.2 (Qiagen) aligned 27,189,730 reads totaling 5,880,240,986 aligned bases, and paired 25,491,776 reads with an average paired read length = 228 bp, to give a genome assembly of 49,495,572 bp in 24,662 scaffolds, with N50 = 18,922 bp.
Chemical analysis
All isolates were grown on potato dextrose agar and were inoculated from an actively growing culture at a single point, and grown for 14 d in the dark. The Metarhizium species were grown at 25°, and the dermatophytes were grown at 29°. Cultures were air-dried and extracted with 2% acetic acid. Swainsonine was analyzed by LC-MS using methods described by Gardner and Cook (2016).
Genetic manipulations of Metarhizium robertsii
A double crossover gene replacement construct (Figure 2), targeting the swnK gene, was assembled using two gene-specific DNA fragments (flank A and flank B) intercalated by the bar selection marker, which confers resistance to glufosinate ammonium (Donzelli et al. 2016). Gene-specific DNA fragments were produced by standard PCR reactions using primers listed in Supplemental Material, Table S3 in File S1, and M. robertsii ARSEF 2575 genomic DNA as the template. The bar selection marker was amplified from the pBARKS1 derivative pUCAPbarNOSII (Pall and Brunelli 1993; Donzelli et al. 2012) using primers indicated in Figure 2 and Table S3 in File S1. These three fragments were assembled into pBDU vector by the USER method (Nour-Eldin et al. 2006; Geu-Flores et al. 2007; Donzelli et al. 2012). A swnK complementation vector was produced by cloning a 9277 bp PCR product that included 1675 bp of the swnK promoter region, the entire swnK coding region (7467 bp) and 121 bp of the 3′ UTR region, into pBDUN binary vector. The pBDUN vector is a pPK2 (Covert et al. 2001) derivative carrying the nourseothricin resistance gene driven by the Aspergillus nidulans trpC promoter and compatible with the USER cloning method. Both gene replacement and gene complementation vectors were mobilized into Agrobacterium tumefaciens EHA105 by electroporation. Agrobacterium tumefaciens-mediated transformation of M. robertsii ARSEF 2575 was conducted as described (Moon et al. 2008) with the modification that selection of complemented transformants was with nourseothricin at 500 mg/liter.
Figure 2.
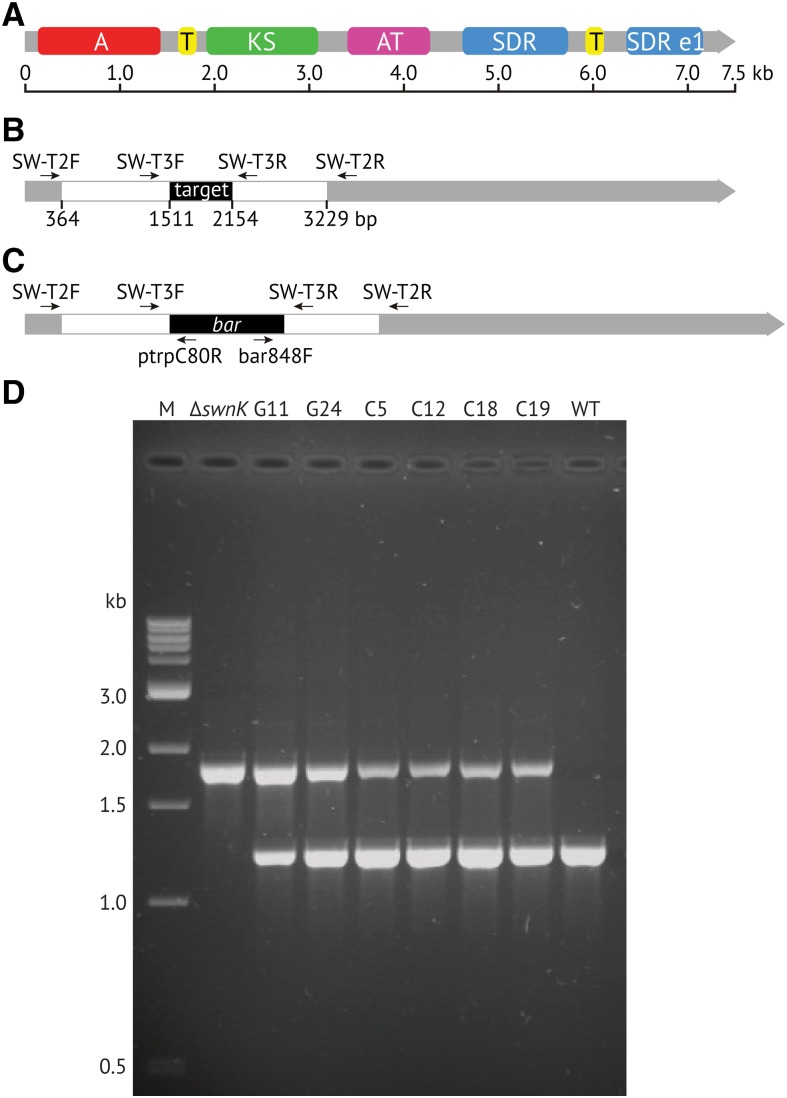
Tests for swnK gene knockouts and complemented strains of M. robertsii ARSEF 2575. (A) Domain structure of SwnK (see text and caption of Figure 3) mapped over the length of the gene. (B) Scale map of sequences cloned into the double crossover gene replacement construct (white), which flank the segment targeted for replacement (black). (C) Map of the ∆swnK mutation in which the target region is replaced with the bar gene. Labeled arrows indicate primers used for PCR screens (see Table S3 in File S1). (D) Electrophoretic analysis of PCR products generated with primers SW-T3F and SW-T3R and template DNAs from ∆swnK, ectopic integrant controls G11 and G24, ∆swnK/swnK complemented strains C5, C12, C18, and C19, and untransformed wild type (WT). PCR on intact and bar-disrupted loci is expected to generate products of 1240 and 1850 bp, respectively. Lane M contains molecular size markers.
Identification of M. robertsii ARSEF 2575 transformants carrying the bar gene at the targeted locus was conducted by PCR using primers SW-T2F and SW-T2R, annealing immediately outside the targeted region, in conjunction with primers ptrpc80R and bar848F, annealing within the bar selection cassette (Figure 2 and Table S3 in File S1). Single conidial progenies derived from putative homologous integrants were tested by PCR with primers SW-T3F and SW-T3R for integration of the complementation fragment in transformants displaying resistance to both glufosinate ammonium and nourseothricin.
Virulence tests
Assays were conducted on Drosophila suzukii flies. Conidia (spores) of each respective M. robertsii strain were suspended in water with 0.01% Silwet L77 to give 2 ☓ 107 conidia/ml. Batches of 48 adult females were dipped in the conidial suspension, and then incubated at 25°, 15 hr:9 hr light:dark cycle for 10 d. Treated insects were provided with preservative-free diet which was replaced every 24–48 hr. Differences in survival rates among treatments were quantified using survival analysis and Cox’s proportional hazards in JMP PRO 11 (SAS Institute Inc., Cary, NC).
Data availability
Biological materials are available upon request. Figure S1 in File S1 shows the survival curves generated from two independent virulence assays where D. suzukii was used as the insect host. Table S1 in File S1 lists SWN gene homologs and swainsonine production in fungal cultures. Table S2 in File S1 shows results of assays for effects of swainsonine on virulence of M. robertsii on D. suzukii. Table S3 in File S1 shows sequences of oligonucleotide primers used in this study. Sequence data are available at GenBank and the accession numbers are listed in Table S1 in File S1.
Results
Swainsonine precursors include pipecolic acid (Guengerich et al. 1973), an analog of the amino acid proline, and mevalonic acid (Clevenstine et al. 1979), a common building block for polyketides. Therefore, we hypothesized that a multifunctional enzyme with an amino-acid adenylylation (A) domain, an acyltransferase domain (AT), a β-ketoacyl synthase (KS) domain, and two phosphopantetheine-binding/thiolation domains (T) would catalyze the condensation of these two precursors. Based on studies of Harris et al. (1988a,b) with isotopically labeled precursors, we expected that additional domains or enzymes would be required for several reduction steps and two hydroxylations. Considering that genes for any particular specialized (secondary) metabolite tend to be closely linked in genomes of filamentous fungi (Spatafora and Bushley 2015), we used comparative genomics to identify orthologous clusters of genes for appropriate biosynthetic enzymes in the known swainsonine-producing fungi.
The genome of ICE (Cook et al. 2013) was sequenced and annotated to model its genes. Inferred protein sequences from this genome and the published genome of M. robertsii ARSEF 23 (Gao et al. 2011) were searched by hidden Markov models (HMM) with InterProScan to identify putative functional domains. The sequences were also submitted as BLASTp queries against fungal protein sequences in the nonredundant GenBank database. Results were displayed in a modified GBrowse version 1.70 for visual inspection to identify any gene models with similar structures that included A, AT, KS, and T domains, which were then compared by reciprocal best-BLASTp between the two genomes. The results indicated that ICE and M. robertsii shared orthologous candidate genes, one of which, with an appropriate multi-domain structure, was designated swnK (Figure 3). Genes closely linked with swnK were also inspected for putative functions, and compared by best-BLASTp analysis to identify shared genes of the putative swainsonine-biosynthesis gene cluster, designated SWN (GenBank accessions KY365740 and JELW01000031.1). In addition to swnK, the SWN clusters included swnN and swnR, putatively encoding reductases of different Rossmann-fold families, and swnH1 and swnH2, putatively encoding related 2-oxoglutarate-dependent nonheme-iron dioxygenases. There were no widely shared regulatory genes in the orthologous SWN clusters, although several of the clusters have adjacent genes that may be regulatory.
Figure 3.
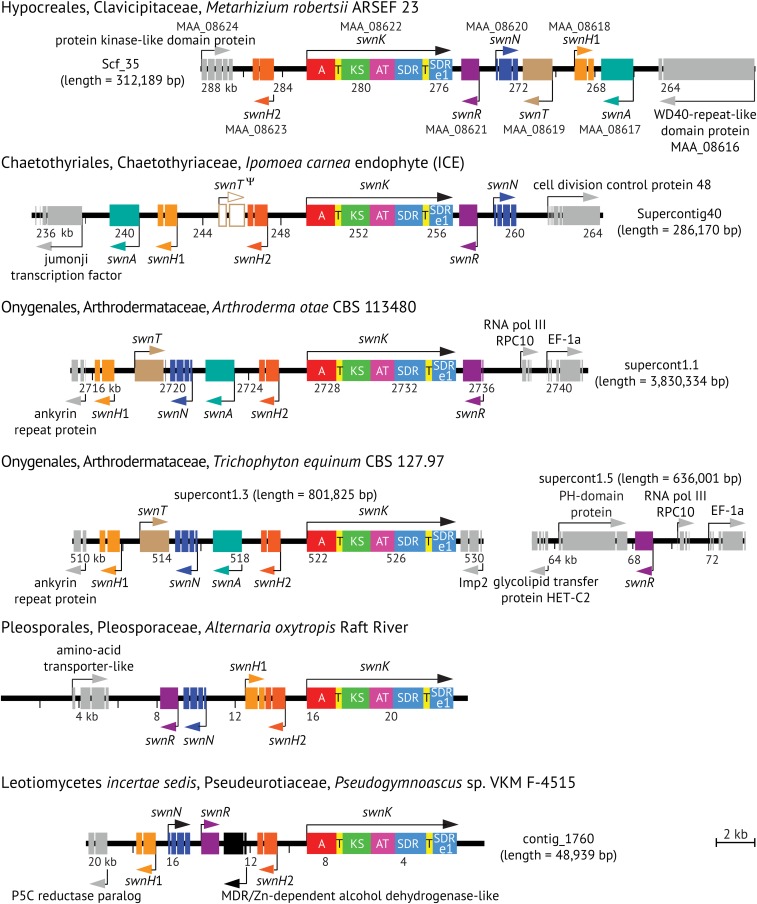
Structures of SWN gene clusters in representatives of five orders of fungi with diverse ecological roles. Taxa are listed by order, family, genus, species, and strain, and the scale on each map indicates the position of the gene cluster on the scaffold (supercontig) or contig indicated. Predicted functions of the gene products are: SwnA, an aromatic amino transferase; SwnH1 and SwnH2, 2-oxoglutarate- and Fe(II)-dependent dioxygenases; SwnN, an NmrA-like, NADB Rossmann-fold reductase; SwnR, an NADB Rossmann-fold reductase; SwnT, a transmembrane transporter; and SwnK, a multifunctional protein with adenylylation (A), phosphopantetheine-binding/thiolation (T), β-ketoacyl synthase (KS), acyltransferase (AT), reductase (SDR), and thioester reductase (SDR e1) domains. SWN genes and swnK domains are color-coded, flanking genes are shown in gray, and swnT of ICE is represented as a likely pseudogene.
As a definitive test of the role of swnK in swainsonine biosynthesis, we disrupted this gene in M. robertsii ARSEF 2575 by introducing the selectable bar gene for bialaphos resistance in place of a segment that included the T domain adjacent to the A domain (Figure 2). The resulting ∆swnK mutant failed to produce swainsonine at detectable levels. This mutant was then complemented by reintroduction of wild-type swnK, restoring swainsonine production in several independent transformants (Table 1). Most of the complemented transformants produced higher levels of swainsonine in culture than did the wild-type strain.
Table 1. Results of molecular genetic tests for the role of swnK in swainsonine biosynthesis.
| Strain | Genotype | Comment | Swainsonine Concentration (µg/g Dry Mass ± SD) |
|---|---|---|---|
| ARSEF 2575 | WT | Wild-type strain | 653 ± 119 |
| ∆swnK | ∆swnK | swnK disruption mutant | Not detected |
| G11 | WT + bar | Ectopic transformant | 620 ± 103 |
| G24 | WT + bar | Ectopic transformant | 633 ± 81 |
| C5 | ∆swnK + swnK | Complemented mutant | 1268 ± 470 |
| C12 | ∆swnK + swnK | Complemented mutant | 1141 ± 497 |
| C18 | ∆swnK + swnK | Complemented mutant | 145 ± 46 |
| C19 | ∆swnK + swnK | Complemented mutant | 2573 ± 1408 |
Exhaustive BLASTp and tBLASTn searches of published genome sequences revealed SWN-cluster orthologs in five different orders of filamentous Ascomycota (Figure 3 and Table S1 in File S1). These included the Arthrodermataceae (order Onygenales), which are skin pathogens that cause ringworm and athlete’s foot in humans and other mammals. All Arthrodermataceae possessed swnH1, swnH2, swnK, swnN, and swnR, but in the Trichophyton species swnR was present in a separate locus (Figure 3 and Table S1 in File S1). In two of the Trichophyton genome sequences, swnR was not annotated, but their apparently complete swnR gene orthologs could be identified by tBLASTn. (These are listed in Table S1 in File S1 by their nucleotide sequence accessions AOKT01000327.1 and LHPM01000018.1.) Representatives of six species of Arthrodermataceae—namely, Arthroderma otae (=Microsporum canis), Nannizzia gypsea (=Microsporum gypseum), Trichophyton benhamiae (=Arthroderma benhamiae), Trichophyton interdigitale, Trichophyton equinum (=Trichophyton tonsurans), and Trichophyton rubrum—were cultured and tested for swainsonine, and all except Ar. otae produced the alkaloid. Considering that symbiotic fungi such as Epichloë species tend to express alkaloids only in their hosts and not in culture (Chujo and Scott 2014), it is possible that Ar. otae produces swainsonine only in its host, or that it requires other culture conditions. SWN clusters were also identified in all sequenced isolates of Metarhizium species except Metarhizium album ARSEF 1941. Cultures of all eight available isolates of six Metarhizium species, but not of M. album, produced detectable levels of swainsonine (Table S1 in File S1), showing a strong correlation of SWN cluster presence and swainsonine production.
Whole genome shotgun sequencing was also conducted on the genomes of the clover black patch pathogen, Slafractonia leguminicola (Alhawatema et al. 2015), and the endophyte, Alternaria oxytropis (Pryor et al. 2009), both of which are known swainsonine producers in the order Pleosporales. Putative orthologs of swnK, swnN, swnR, swnH1, and swnH2 were identified in both, but not necessarily on shared scaffolds. In the A. oxytropis genome assembly, the apparent orthologs were on three scaffolds, but scaffold ends overlapped within coding sequences of swnN and swnH1 to permit manual assembly of the entire cluster (Figure 3; GenBank accession KY365741). For S. leguminicola (GenBank accessions KY365742–KY365746), swnK, swnN, swnR, and swnT assembled uniquely and apparently completely on separate scaffolds, swnH1 and swnH2 assembled together in a scaffold as convergently transcribed genes, similar to the arrangement in A. oxytropis. Therefore, there was no evidence for or against clustering of SWN genes in S. leguminicola. Also like A. oxytropis, no swnA ortholog was found in S. leguminicola.
S. leguminicola produces two distinct indolizidines, swainsonine and slaframine, of which the latter causes slobbers in livestock that graze infected legume foliage (Harris et al. 1988b; Croom et al. 1995). Given the structural similarity of swainsonine and slaframine, a second swnK homolog was expected in S. leguminicola. In fact, two swnK paralogs were identified (GenBank accessions KY365747 and KY365748), both encoding proteins with the SwnK domain structure. The paralogs had similar levels of divergence from each other and from SwnK, with 53.1–55.4% identity to SwnK of S. leguminicola, A. oxytropis, M. robertsii, and ICE. In contrast, SwnK orthologs all shared >70% identity with each other. Presumably one SwnK paralog is involved in biosynthesis of slaframine, and conceivably the other is involved in biosynthesis of a related alkaloid yet to be identified.
The genes designated swnA and swnT were present in SWN clusters of some but not all swainsonine producers. (Figure 3 and Table S1 in File S1). SwnA was a putative aminotransferase, and SwnT was related to a transmembrane choline transporter. Orthologs of swnT were present in all except the two endophytes, ICE, which had a swnT pseudogene, and A. oxytropis, which completely lacked swnT. It is noteworthy that an orthologous swnT gene was present in S. leguminicola, which is in the same order as A. oxytropis, indicating that the gene was lost in an ancestor of the endophyte A. oxytropis. The possibility deserves further investigation that loss of swnT is related to evolution of mutualistic symbionts, perhaps by altering the localization of swainsonine.
BLASTp searches of sequenced genomes also revealed gene clusters orthologous to SWN in Pseudogymnoascus sp. VKM F-4515 from permafrost soil (Figure 3), and in a recently published genome from a fungus designated fungal sp. No.14919 (Table S1 in File S1). As in the Arthrodermataceae, Metarhizium species, A. oxytropis and ICE, these species had apparent swnK orthologs closely linked to swnH1, swnH2, swnN, and swnR. In fungal sp. No.14919 the arrangement of these genes, plus a swnT ortholog, was similar to that in Metarhizium robertsii ARSEF 23 (Figure 3). Pseudogymnoascus sp. VKM F-4515 and fungal sp. No.14919 had distinct additional genes for putative biosynthetic enzymes adjacent to the cluster. It seems likely that these two species also produce swainsonine or a related metabolite.
We employed ∆swnK, swnK-complemented strains, and wild-type M. robertsii to test the role of swainsonine in pathogenicity to the insect, D. suzukii. Survivorship curves indicated no significant reduction in virulence for the ∆swnK strain (Figure S1 and Table S2 in File S1). In fact, in most tests swainsonine-producing strains exhibited slightly less virulence than did the knockout strain.
Discussion
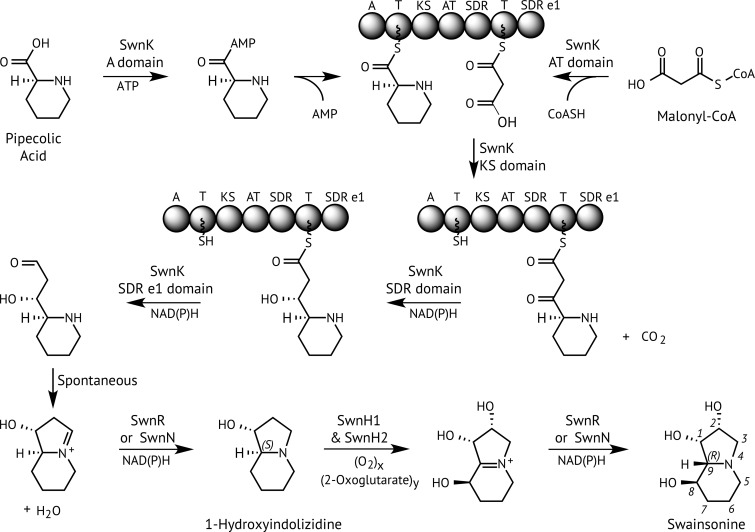
Our comparative genomic analysis proved fruitful in revealing a gene cluster that was common among swainsonine producers, and that included a ketide synthase gene (which we designated swnK) with appropriate domains for the first steps of swainsonine biosynthesis. Subsequent inactivation of swnK in M. robertsii, followed by complementation of the ∆swnK mutant, provided for confirmation that this gene was required for swainsonine production. The arrangement of domains in the inferred SwnK polypeptide sequence, together with published results of isotope labeling experiments (Harris et al. 1988b), suggested biosynthetic roles for SwnK, as well as for the SwnR and SwnN reductases, and the SwnH1 and SwnH2 hydroxylases encoded by nearby genes (Figure 4). Based on the presence and positions in SwnK of the two reductase domains, SDR (β-ketoacyl reductase) and SDR e1 (thioester reductase) (Figure 2 and Figure 4), we predict that the intermediate released from this enzyme has a hydroxyl group instead of the keto group proposed by Harris et al. (1988b). This intermediate should cyclize to generate a C3=N+4 iminium ion that we propose to be reduced by the action of SwnR or SwnN, giving 1-hydroxyindolizidine, which has been reported from the locoweed species, Astragalus oxyphysus (Harris et al. 1988a). Subsequent oxygenations must occur at carbons 2 and 8, consistent with predicted activities of SwnH1 and SwnH2. Furthermore, Harris et al. (1988b) present strong evidence for epimerization at carbon 9, suggesting a second iminium ion intermediate, C9=N+4, perhaps generated by the action of SwnH1 or SwnH2. Thus, the presence of the two putative reductase genes, swnN and swnR, is consistent with a requirement to reduce both a C3=N+4 and a C9=N+4 iminium ion in the proposed pathway.
Figure 4.
Proposed swainsonine biosynthetic pathway based on predicted gene functions. Predicted functions of SwnK domains and other enzymes are listed in the caption of Figure 3.
The diversity of fungi that produce swainsonine suggests multiple ecological roles for the alkaloid among symbionts and parasites of plants and animals. So far, indications have been surprising. In the case of locoweeds, grazing mammals can develop a preference despite the devastating cytotoxic and neurological effects of swainsonine (Ralphs et al. 1990; Pfister et al. 2003). Thus, the alkaloid does not seem to deter these herbivores. Also, our tests of M. robertsii on the insect model, D. suzukii, indicate that swainsonine does not contribute, and may actually moderate, virulence. It is possible that swainsonine may have a deterrent or toxic effect on other herbivores or other microbes, and that it may have a role in maintaining some stable symbioses of fungi with host plants. Furthermore, the ubiquity of SWN genes in the Arthrodermataceae raises the possibility that swainsonine has a role in the etiology of skin infections by these dermatophytic fungi—an important consideration for veterinary and human medicine.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041384/-/DC1.
Acknowledgments
The authors thank Jennifer S. Webb for genome sequencing, Jolanta Jaromczyk for genome assembly, Daniel R. Harris for InterproScan analysis, and Jessie Roper and Richard H. Vaughan for technical support. This work was funded by the United States Department of Agriculture Multistate project W1193, the United States Department of Agriculture–Agricultural Research Service project number 8062-21000-035-00D, and in part by the New Mexico State University Experiment Station. This is publication number 17-12-029 of the Kentucky Agricultural Experiment Station, published with approval of the director.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Alhawatema M. S., Sanogo S., Baucom D. L., Creamer R., 2015. A search for the phylogenetic relationship of the ascomycete Rhizoctonia leguminicola using genetic analysis. Mycopathologia 179: 381–389. [DOI] [PubMed] [Google Scholar]
- Braun K., Romero J., Liddell C., Creamer R., 2003. Production of swainsonine by fungal endophytes of locoweed. Mycol. Res. 107: 980–988. [DOI] [PubMed] [Google Scholar]
- Chujo T., Scott B., 2014. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte–plant symbiosis. Mol. Microbiol. 92: 413–434. [DOI] [PubMed] [Google Scholar]
- Clevenstine E. C., Broquist H. P., Harris T. M., 1979. Biosynthesis of slaframine, (1S,6S,8aS)-1-acetoxy-6-aminooctahydroindolizine, parasympathomimetic alkaloid of fungal origin. 3. Origin of the pyrrolidine ring. Biochemistry 18: 3658–3663. [DOI] [PubMed] [Google Scholar]
- Cook D., Beaulieu W. T., Mott I. W., Riet-Correa F., Gardner D. R., et al. , 2013. Production of the alkaloid swainsonine by a fungal endosymbiont of the ascomycete order Chaetothyriales in the host Ipomoea carnea. J. Agric. Food Chem. 61: 3797–3803. [DOI] [PubMed] [Google Scholar]
- Covert S. F., Kapoor P., Lee M.-h., Briley A., Nairn C. J., 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105: 259–264. [Google Scholar]
- Croom W. J., Hagler W. M., Froetschel M. A., Johnson A. D., 1995. The involvement of slaframine and swainsonine in slobbers syndrome: a review. J. Anim. Sci. 73: 1499–1508. [DOI] [PubMed] [Google Scholar]
- Donzelli B. G. G., Krasnoff S. B., Sun-Moon Y., Churchill A. C. L., Gibson D. M., 2012. Genetic basis of destruxin production in the entomopathogen Metarhizium robertsii. Curr. Genet. 58: 105–116. [DOI] [PubMed] [Google Scholar]
- Donzelli B. G., Turgeon B. G., Gibson D. M., Krasnoff S. B., 2016. Disruptions of the genes involved in lysine biosynthesis, iron acquisition, and secondary metabolisms affect virulence and fitness in Metarhizium robertsii. Fungal Genet. Biol. 98: 23–34. [DOI] [PubMed] [Google Scholar]
- Dorling P. R., Huxtable C. R., Colegate S. M., 1980. Inhibition of lysosomal α-mannosidase by swainsonine, an indolizidine alkaloid isolated from Swainsona canescens. Biochem. J. 191: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Jin K., Ying S.-H., Zhang Y., Xiao G., et al. , 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7: e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D. R., Cook D., 2016. Analysis of swainsonine and swainsonine N-oxide as trimethylsilyl derivatives by liquid chromatography–mass spectrometry and their relative occurrence in plants toxic to livestock. J. Agric. Food Chem. 64: 6156–6162. [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Nour-Eldin H. H., Nielsen M. T., Halkier B. A., 2007. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P., DiMari S. J., Broquist H. P., 1973. Isolation and characterization of a l-pyrindine fungal alkaloid. J. Am. Chem. Soc. 95: 2055–2056. [Google Scholar]
- Harris C. M., Campbell B. C., Molyneux R. J., Harris T. M., 1988a Biosynthesis of swainsonine in the diablo locoweed (Astragalus oxyphysus). Tetrahedron Lett. 29: 4815–4818. [Google Scholar]
- Harris C. M., Schneider M. J., Ungemach F. S., Hill J. E., Harris T. M., 1988b Biosynthesis of the toxic indolizidine alkaloids slaframine and swainsonine in Rhizoctonia leguminicola: metabolism of 1-hydroxyindolizidines. J. Am. Chem. Soc. 110: 940–949. [Google Scholar]
- Li Z., Huang Y., Dong F., Li W., Ding L., et al. , 2012. Swainsonine promotes apoptosis in human oesophageal squamous cell carcinoma cells in vitro and in vivo through activation of mitochondrial pathway. J. Biosci. 37: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Moon Y.-S., Donzelli B. G. G., Krasnoff S. B., McLane H., Griggs M. H., et al. , 2008. Agrobacterium-mediated disruption of a nonribosomal peptide synthetase gene in the invertebrate pathogen Metarhizium anisopliae reveals a peptide spore factor. Appl. Environ. Microbiol. 74: 4366–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenoff P., Uhrlaß S., Krüger C., Erhard M., Hipler U.-C., et al. , 2014. Trichophyton species of Arthroderma benhamiae—a new infectious agent in dermatology. J. Dtsch. Dermatol. Ges. 12: 571–581. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Hansen B. G., Nørholm M. H. H., Jensen J. K., Halkier B. A., 2006. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall M. L., Brunelli J. P., 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 40: 58. [Google Scholar]
- Pfister J. A., Stegelmeier B. L., Gardner D. R., James L. F., 2003. Grazing of spotted locoweed (Astragalus lentiginosus) by cattle and horses in Arizona. J. Anim. Sci. 81: 2285–2293. [DOI] [PubMed] [Google Scholar]
- Pryor B. M., Creamer R., Shoemaker R. A., McLain-Romero J., Hambleton S., 2009. Undifilum, a new genus for endophytic Embellisia oxytropis and parasitic Helminthosporium bornmuelleri on legumes. Botany 87: 178–194. [Google Scholar]
- Ralphs M. H., Panter K. E., James L. F., 1990. Feed preferences and habituation of sheep poisoned by locoweed. J. Anim. Sci. 68: 1354–1362. [DOI] [PubMed] [Google Scholar]
- Reyna R., Cooke P., Grum D., Cook D., Creamer R., 2012. Detection and localization of the endophyte Undifilum oxytropis in locoweed tissues. Botany 90: 1229–1236. [Google Scholar]
- Santos F. M., Latorre A. O., Hueza I. M., Sanches D. S., Lippi L. L., et al. , 2011. Increased antitumor efficacy by the combined administration of swainsonine and cisplatin in vivo. Phytomedicine 18: 1096–1101. [DOI] [PubMed] [Google Scholar]
- Spatafora J. W., Bushley K. E., 2015. Phylogenomics and evolution of secondary metabolism in plant-associated fungi. Curr. Opin. Plant Biol. 26: 37–44. [DOI] [PubMed] [Google Scholar]
- St. Leger R. J., Wang C., 2010. Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl. Microbiol. Biotechnol. 85: 901–907. [DOI] [PubMed] [Google Scholar]
- Tulsiani D. R. P., Harris T. M., Touster O., 1982. Swainsonine inhibits the biosynthesis of complex glycoproteins by inhibition of golgi mannosidase II. J. Biol. Chem. 257: 7936–7939. [PubMed] [Google Scholar]
- Winchester B., al Daher S., Carpenter N. C., Cenci di Bello I., Choi S. S., et al. , 1993. The structural basis of the inhibition of human α-mannosidases by azafuranose analogues of mannose. Biochem. J. 290: 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Biological materials are available upon request. Figure S1 in File S1 shows the survival curves generated from two independent virulence assays where D. suzukii was used as the insect host. Table S1 in File S1 lists SWN gene homologs and swainsonine production in fungal cultures. Table S2 in File S1 shows results of assays for effects of swainsonine on virulence of M. robertsii on D. suzukii. Table S3 in File S1 shows sequences of oligonucleotide primers used in this study. Sequence data are available at GenBank and the accession numbers are listed in Table S1 in File S1.