Abstract
Fungal endophytes were isolated from leaf, bark and stem of Tectona grandis Linn.f. sampled at four geographical locations in winter, summer and monsoon seasons. The recovered 5089 isolates were assigned to 45 distinct morphotypes based on morphology. The sequences of the internal transcribed spacers (ITS) of the nrDNA of some morphotypes were identical, but morphological differences were strong enough to consider these morphotypes as separate species. Forty-three morphotypes were assigned to ascomycotina and two to basidiomycotina. Ascomycotina was the predominating group with 99.7% of total isolates followed by basidiomycotina with only 0.3% of total isolates. Diaporthe (Phomopsis) species dominated the communities independently on tissue type, location or season. More than 60% of the examined tissue pieces were colonized by members of this species complex. While these endophytes are ubiquitous others were tissue or location specific. Tissue type had the strongest effect on the species evenness of the endophytic assemblage followed by geographical location and season. However, Shannon-Wiener index (H’) significantly (p ≤ 0.001) varied with all three factors i.e. season, location and tissue type. Leaves supported the highest diversity across all the seasons and locations. In conclusion, all the three factors together determined the structure of endophytic mycobiota assemblage of T. grandis.
Introduction
The start of endophytes research dates back to year 1866 when a German botanist Anton de Bary, first introduced the term “Endophytes” for any in planta microorganism1. In strict sense, fungal endophytes are fungi that spend either full or a considerable part of their life inside living plant tissues without causing any visible harm2. After decades of research on fungal endophytes, it is now clear that they are unexceptionally present in all taxonomic groups of the plant kingdom, vegetation types (alpine to tropical) and ecological types (hydrophytes to xerophytes) in great diversity3–5. Discovery of high endophyte diversity in trees3, 6 has led to a surge in research efforts in this direction especially on trees growing in tropical regions. Some tropical tree species have been reported to host a hyper diverse endophyte community3, 7. Exploration of tropical woody tree endophyte diversity will definitely improve global fungal diversity estimates. Due to a plethora of factors concomitantly affecting endophyte assemblage in a tree, it is hard to assign specific role to one particular factor in this context3, 8. But the studies covering different regulating factors with appropriate sampling size will definitely improve our understanding about their roles. Isolation of endophytes is also important in view of their potential as prolific source of novel compounds with applications in pharmaceutical, agricultural and food industries9–11. Due to their unique and diverse niche different bioactive secondary metabolites can be obtained from the same endophyte species as their metabolite spectrum changes with change in environment, geography, host and tissue type12. Consequently, the study of endophyte communities in different tissues and plants is essential to harness the maximum benefits of this great resource.
Due to diverse ecological communities in wide-ranging environmental regimes, India is a suitable place for the assessment of endophytes and factors responsible for shaping their assemblage. Tectona grandis Linn.f. (Teak), family Verbenaceae, is a major tree of tropical regions and native to India. Teak can grow under a wide range of climatic and edaphic conditions. All parts of teak, including, bark, leaf, stem, root, flower and seed, are traditionally used in folklore medicine to cure various diseases. Its good pharmacological potentials, such as, cytotoxic (ellagic acid), antibacterial (juglone), antifungal (deoxylapachol and tectoquinine), antioxidant (gallic acid, ferulic acid and quercetin), anti-diabetes, anti-inflammatory and tocolytic effects have been well documented13, 14. The above mentioned outstanding properties of T. grandis and the possibility of horizontal gene transfer from host to endophytes motivated us to select this plant as a study material. Teak is one of the most studied tropical trees with regard to its ecology and application in forestry, however, it has rarely been subjected to endophytic study15, 16. Hitherto, there is not a single report available on Tectona which includes such disparate tissue types and sampling sites with different climatic conditions and seasonal variability.
The aim of the present work was to study geographical and seasonal variations in fungal endophyte assemblage of T. grandis in three different tissues. Fungal endophytes were identified using both morphological and molecular tools.
Results
Diversity of endophytic mycobiota in Tectona grandis
The 5089 fungal isolates obtained from the 8100 tissue segments could be assigned to 45 OTUs (operational taxonomic units referring to different morphological types) based on culture morphology. All, except for two OTUs, sporulated in culture and could be identified to the genus or species level. Molecular identification based on ITS sequences corroborated morphological identification in almost all cases (Table 1). ITS sequencing revealed names for non-sporulating OTUs too. Interestingly, many OTUs possessed almost identical ITS sequences (i.e. Tectona grandis endophytes TGE1 and TGE16, TGE2 and TGE3, TGE8 and TGE27, TGE9 and TGE10, TGE11 and TGE20, TGE18 and TGE19, TGE28 and TGE30), although the OTUs were distinctly different in micromorphology, culture morphology and matched with different reference strains of NCBI database. Due to these differences TGEs with almost identical ITS sequences were considered separate species. To minimize confusion between morphological and molecular identity bellow species level, ITS based name is given in parenthesis wherever required.
Table 1.
Morphological and molecular (based on nrITS1 sequencing) identification of different OTUs, their closest match from NCBI database with their accession number, similarity (%ID) and query coverage (%QC).
| OTU acronym | GenBank accession No.2 | Morphological identification1,2 | Closest NCBI match1 | ID/QC (%) | Reference accession No. |
|---|---|---|---|---|---|
| TGE1 | KF688116 | Paecilomyces sp.1 | Paecilomyces variotii | 100/99 | KY019238 |
| TGE2 | KF688118 | Alternaria sp.1 | Alternaria sp. | 100/100 | HQ914881 |
| TGE3 | KF910776 | Alternaria sp.2 | Alternaria sp. | 99/99 | KP027305 |
| TGE4 | NS | Aspergillus flavus | |||
| TGE5 | NS | Aspergillus fumigatus | |||
| TGE6 | NS | Aspergillus niger | |||
| TGE7 | KU519494 | Aspergillus sp.1 | Aspergillus viridinutans | 98/100 | NR135409 |
| TGE8 | KF688119 | Aspergillus sp.2 | Aspergillus versicolor | 100/100 | KX776009 |
| TGE9 | JX951183 | Chaetomium globosum | Chaetomium globosum | 100/99 | JF826006 |
| TGE10 | KF688114 | Chaetomium sp. | Chaetomium sp. | 100/100 | HM535379 |
| TGE11 | KF688120 | Colletotrichum gloeosporioides | Colletotrichum gloeosporioides | 99/100 | KX347456 |
| TGE12 | KF688125 | Corynespora cassiicola | Corynespora cassiicola | 100/100 | KY290563 |
| TGE13 | KU519495 | Emericella sp. | Aspergillus quadrilineatus | 100/100 | KX683008 |
| TGE14 | KF688115 | Gibberella baccata | Fusarium sp. | 100/99 | KM066585 |
| TGE15 | KU519496 | Fusarium sp.1 | Albonectria rigidiuscula | 100/100 | KX788159 |
| TGE16 | KF688112 | Paecilomyces sp.2 | Paecilomyces variotii | 99/100 | JX231004 |
| TGE17 | KF688126 | Ceratobasidium sp. | Ceratobasidium sp. | 99/100 | HM623627 |
| TGE18 | KF910777 | MS1 | Pseudofusicoccum adansoniae | 99/100 | KF766220 |
| TGE19 | KF910778 | MS2 | Lasiodiplodia theobromae | 99/100 | KM508495 |
| TGE20 | KF688122 | Colletotrichum sp. | Colletotrichum siamense | 100/100 | KY242353 |
| TGE21 | NS | Penicillium sp. | |||
| TGE22 | KF688130 | Phomopsis sp.1 | Diaporthe sp. | 100/100 | KU663500 |
| TGE23 | KF910772 | Phomopsis sp.2 | Diaporthe amygdali | 99/100 | KF308686 |
| TGE24 | KF910766 | Phomopsis sp.3 | Phomopsis longicolla | 99/100 | KP174768 |
| TGE25 | KF910765 | Lasiodiplodia theobromae | Lasiodiplodia theobromae | 100/100 | KX506786 |
| TGE26 | KU512642 | Fusarium sp.2 | Fusarium incarnatum | 100/100 | KX184815 |
| TGE27 | KF910774 | Aspergillus sp.3 | Aspergillus versicolor | 99/100 | LT594460 |
| TGE28 | KF910773 | Cercospora sp.1 | Cercospora gerberae | 99/100 | KX347479 |
| TGE29 | KF688117 | Phomopsis sp.4 | Diaporthe sp. | 99/100 | KC145847 |
| TGE30 | KF688113 | Cercospora sp.2 | Cercospora nicotianae | 99/100 | KM485926 |
| TGE31 | KF688123 | Periconia sp.1 | Periconia sp. | 100/100 | JF817312 |
| TGE32 | KF910767 | Botryosphaeria sp. | Lasiodiplodia pseudotheobromae | 100/100 | KY583266 |
| TGE33 | KF910769 | Diaporthe sp. | Diaporthe sp. | 99/100 | KU375727 |
| TGE34 | KF688129 | Guignardia sp. | Phyllosticta elongata | 100/100 | KX424992 |
| TGE35 | KF910770 | Glomerella sp. | Colletotrichum acutatum | 99/100 | KX347475 |
| TGE36 | KF688121 | Phomopsis sp.5 | Phomopsis sp. | 99/100 | DQ235667 |
| TGE37 | KU512643 | Lasiodiplodia sp. | Diplodia seriata | 100/100 | KY620311 |
| TGE38 | KF910775 | Fusarium sp.3 | Albonectria rigidiuscula | 100/99 | KY024396 |
| TGE39 | KF688127 | Hypoxylon sp. | Hypoxylon fragiforme | 99/100 | KU317724 |
| TGE40 | KF688128 | Coprinellus sp. | Coprinellus sp. | 99/99 | GQ249274 |
| TGE41 | JX951186 | Periconia sp.2 | Periconia sp. | 98/100 | JF817328 |
| TGE42 | KF910768 | Pyrenochaeta sp. | Pyrenochaeta sp. | 99/99 | GQ895137 |
| TGE43 | KF688124 | Phomopsis sp.6 | Phomopsis sp. | 100/100 | KX401432 |
| TGE44 | KF910771 | Curvularia sp. | Curvularia sp. | 100/100 | HE861827 |
| TGE45 | KF910779 | Alternaria sp.3 | Alternaria sp. | 100/100 | KM051397 |
1Species names according to the Index fungorum (http://www.indexfungorum.org/Names/Names.asp; accessed March 25, 2017). 2MS1 and 2 = Mycelia-Sterilia; NS = not sequenced, TGE = Tectona grandis endophyte.
The dendrogram created using ITS sequences of endophytic OTUs and reference taxa retrieved from NCBI database shows that the endophyte assemblages of T. grandis include representative taxa of the ascomycotina and basidiomycotina (Fig. 1). All The members of ascomycotina and basidiomycotina formed eighteen different clades in the dendrogram.
Figure 1.
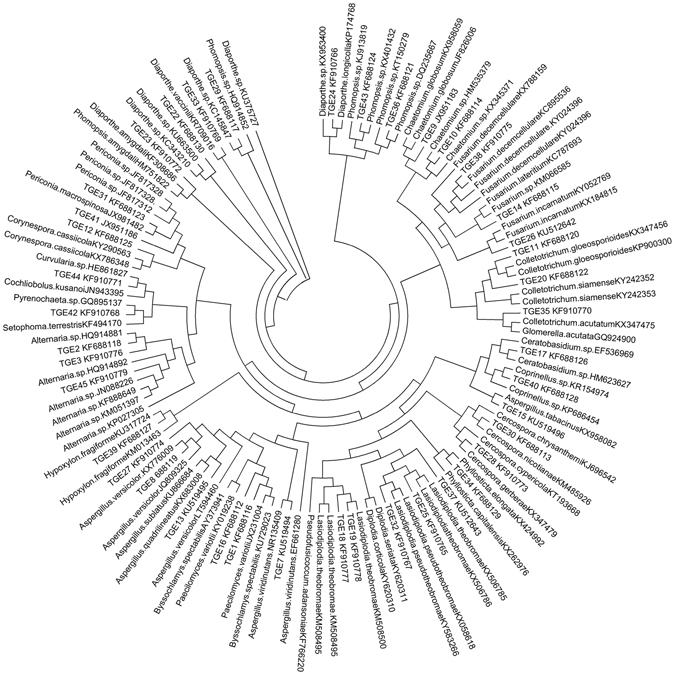
Phylogenetic tree constructed by neighbour-joining method using nrDNA ITS sequences generated during this study (as TGE) and sequences retrieved from GenBank using p-distance for nucleotides and the pairwise gap deletion choice.
Overall frequency of colonization of fungal endophytes in T. grandis tissues was 62.83%. Diaporthe (Phomopsis) spp. dominated the endophytic assemblage in teak tissues. Phomopsis sp.3 (Phomopsis longicolla) was the most frequent species (21.12%) followed by Phomopsis sp.1 (Diaporthe sp.) (12.38%). Colletotrichum gloeosporioides was the third most abundant species with 11.08% of dominance. Eighteen other OTUs showed abundance ranging from 1% to 7.8% and collectively amounted to almost half of the isolates (48.12%). The above mentioned 21 morphotypes with overall share of 92.7% were the predominating taxa in the T. grandis endophyte assemblage. Seven morphotypes with relative abundance from ≥0.5% to <1% were considered frequent taxa while the remaining 17 morphotypes whose relative abundance was <0.5% were considered as rare. Interestingly, Chaetomium globosum and Guignardia sp. (Phyllosticta elongata) were among frequent OTUs but were never isolated from bark and stem. Similarly, Lasiodiplodia theobromae was limited to location 2 (Loc2, Hathinala, Uttar Pradesh) only. Detail of colonization pattern of fungal endophytes in different seasons and tissue types of T. grandis at four geographical locations is given in Table 2 . Among 45 fungal morphotypes, 43 (95.6%) belonged to ascomycotina and 2 (4.4%) to basidiomycotina. Of the total isolates, numbering 5089, 99.7% belonged to ascomycotina and the rest 0.3% to basidiomycotina.
Table 2.
Number, percent colonization frequency (%CF) and percent dominance (%D) of different fungal endophytes of T. grandis.
| Endophyte | Winter | Summer | Monsoon | Total | % CF | % D | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loc1 | Loc2 | Loc3 | Loc4 | Loc1 | Loc2 | Loc3 | Loc4 | Loc1 | Loc2 | Loc3 | Loc4 | ||||||||||||||||||||||||||||
| B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | B | L | S | ||||
| Phomopsis sp.3 | 21 | 36 | 30 | 24 | 30 | 27 | 26 | 36 | 34 | 36 | 40 | 37 | 17 | 26 | 23 | 21 | 27 | 27 | 23 | 29 | 27 | 30 | 34 | 30 | 25 | 34 | 29 | 29 | 32 | 32 | 29 | 35 | 34 | 33 | 37 | 35 | 1075 | 13.2716 | 21.1240 |
| Phomopsis sp.1 | 0 | 26 | 23 | 0 | 29 | 27 | 0 | 29 | 26 | 0 | 33 | 29 | 0 | 23 | 17 | 0 | 24 | 16 | 0 | 26 | 20 | 0 | 24 | 30 | 0 | 28 | 24 | 0 | 30 | 27 | 0 | 32 | 28 | 0 | 30 | 29 | 630 | 7.7778 | 12.3796 |
| Colletotrichum gloeosporioides | 0 | 26 | 20 | 0 | 28 | 23 | 0 | 29 | 24 | 0 | 30 | 23 | 0 | 20 | 17 | 0 | 22 | 14 | 0 | 28 | 19 | 0 | 33 | 21 | 0 | 24 | 22 | 0 | 26 | 24 | 0 | 25 | 24 | 0 | 22 | 20 | 564 | 6.9630 | 11.0827 |
| MS1 | 10 | 13 | 11 | 7 | 12 | 8 | 11 | 12 | 14 | 13 | 14 | 19 | 7 | 10 | 8 | 5 | 12 | 6 | 7 | 10 | 13 | 10 | 13 | 14 | 13 | 15 | 15 | 7 | 14 | 10 | 14 | 12 | 14 | 5 | 6 | 14 | 398 | 4.9136 | 7.8208 |
| Gibberell baccata | 22 | 5 | 0 | 29 | 0 | 0 | 35 | 0 | 0 | 30 | 4 | 0 | 20 | 4 | 0 | 22 | 2 | 0 | 26 | 3 | 0 | 26 | 4 | 0 | 24 | 8 | 0 | 26 | 3 | 0 | 30 | 10 | 0 | 36 | 7 | 0 | 376 | 4.6420 | 7.3885 |
| MS2 | 5 | 13 | 9 | 6 | 9 | 8 | 8 | 9 | 9 | 9 | 11 | 10 | 5 | 7 | 6 | 6 | 8 | 8 | 8 | 9 | 8 | 8 | 10 | 10 | 11 | 14 | 9 | 6 | 12 | 8 | 10 | 14 | 9 | 9 | 13 | 11 | 325 | 4.0123 | 6.3863 |
| Alternaria sp.1 | 4 | 6 | 7 | 0 | 7 | 9 | 0 | 6 | 9 | 5 | 7 | 8 | 3 | 4 | 5 | 0 | 4 | 6 | 0 | 5 | 7 | 4 | 5 | 6 | 6 | 8 | 8 | 0 | 8 | 9 | 5 | 8 | 10 | 7 | 8 | 10 | 204 | 2.5185 | 4.0086 |
| Colletotrichum sp. | 4 | 9 | 3 | 0 | 11 | 7 | 8 | 9 | 9 | 8 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 8 | 5 | 5 | 10 | 8 | 8 | 12 | 12 | 9 | 12 | 9 | 190 | 2.3457 | 3.7335 |
| Corynespora cassiicola | 9 | 0 | 6 | 21 | 24 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 7 | 26 | 31 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 162 | 2.0000 | 3.1833 |
| Fusarium sp.1 | 6 | 0 | 4 | 6 | 0 | 6 | 8 | 0 | 7 | 9 | 0 | 7 | 3 | 0 | 2 | 2 | 0 | 1 | 5 | 0 | 4 | 8 | 0 | 6 | 8 | 0 | 5 | 6 | 0 | 6 | 10 | 0 | 9 | 12 | 0 | 7 | 147 | 1.8148 | 2.8886 |
| Cercospora sp.2 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | 6 | 9 | 7 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 4 | 7 | 6 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 8 | 7 | 8 | 9 | 7 | 121 | 1.4938 | 2.3777 |
| Phomopsissp.2 | 0 | 4 | 0 | 0 | 7 | 0 | 3 | 9 | 0 | 6 | 11 | 0 | 0 | 4 | 0 | 0 | 3 | 0 | 0 | 5 | 0 | 0 | 7 | 0 | 0 | 7 | 0 | 0 | 5 | 0 | 0 | 9 | 0 | 10 | 10 | 0 | 100 | 1.2346 | 1.9650 |
| Fusarium sp.2 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 7 | 0 | 7 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 9 | 0 | 10 | 10 | 0 | 83 | 1.0247 | 1.6310 |
| Chaetomium globosum | 0 | 4 | 0 | 0 | 3 | 0 | 0 | 7 | 0 | 0 | 5 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 4 | 0 | 0 | 5 | 0 | 0 | 7 | 0 | 0 | 6 | 0 | 0 | 7 | 0 | 0 | 8 | 0 | 60 | 0.7407 | 1.1790 |
| Aspergillus flavus | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 3 | 2 | 4 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 3 | 1 | 1 | 3 | 0 | 1 | 2 | 52 | 0.6420 | 1.0218 |
| Penicillium sp. | 1 | 0 | 1 | 0 | 4 | 0 | 2 | 1 | 3 | 2 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 0 | 2 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 42 | 0.5185 | 0.8253 |
| Aspergillus fumigatus | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 0 | 1 | 3 | 0 | 2 | 3 | 1 | 1 | 2 | 1 | 0 | 2 | 1 | 2 | 3 | 2 | 1 | 40 | 0.4938 | 0.7860 |
| Guignardia sp. | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 5 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 5 | 0 | 0 | 4 | 0 | 38 | 0.4691 | 0.7467 |
| Alternaria sp.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 6 | 0 | 5 | 5 | 0 | 37 | 0.4568 | 0.7271 |
| Aspergillus niger | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 5 | 1 | 0 | 2 | 0 | 3 | 0 | 0 | 2 | 2 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | 3 | 0 | 1 | 37 | 0.4568 | 0.7271 |
| Botryosphaeria sp. | 0 | 0 | 0 | 7 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 0.4568 | 0.7271 |
| Phomopsis sp.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 26 | 0.3210 | 0.5109 |
| Hypoxylon sp. | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 9 | 0 | 0 | 24 | 0.2963 | 0.4716 |
| Periconia sp.1 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 0.2716 | 0.4323 |
| Pyrenochaeta sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 22 | 0.2716 | 0.4323 |
| Curvularia sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 7 | 5 | 22 | 0.2716 | 0.4323 |
| Glomerella sp. | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 0.2593 | 0.4127 |
| Phomopsis sp.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 21 | 0.2593 | 0.4127 |
| Alternaria sp.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 6 | 5 | 21 | 0.2593 | 0.4127 |
| Periconia sp.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 19 | 0.2346 | 0.3734 |
| Aspergillus sp.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 17 | 0.2099 | 0.3341 |
| Paecilomyces sp.1 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 16 | 0.1975 | 0.3144 |
| Cercospora sp.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 16 | 0.1975 | 0.3144 |
| Fusarium sp.3 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 16 | 0.1975 | 0.3144 |
| Aspergillus sp.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 3 | 15 | 0.1852 | 0.2948 |
| Chaetomium sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 15 | 0.1852 | 0.2948 |
| Emericella sp. | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0.1852 | 0.2948 |
| Phomopsis sp.5 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0.1605 | 0.2555 |
| Lasiodiplodia sp. | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0.1605 | 0.2555 |
| Diaporthe sp. | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0.1358 | 0.2162 |
| Ceratobasidium sp. | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 10 | 0.1235 | 0.1965 |
| Lasiodiplodia theobromae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0.0741 | 0.1179 |
| Coprinellus sp. | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 6 | 0.0741 | 0.1179 |
| Paecilomyces sp.2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0.0370 | 0.0590 |
| Aspergillus sp.1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0123 | 0.0197 |
| Total | 94 | 153 | 119 | 116 | 171 | 131 | 135 | 175 | 155 | 149 | 201 | 181 | 70 | 110 | 85 | 73 | 114 | 90 | 93 | 137 | 108 | 109 | 166 | 143 | 120 | 170 | 132 | 133 | 199 | 149 | 150 | 212 | 170 | 171 | 215 | 190 | 5089 | 62.8272 | 100.0 |
Loc 1, 2, 3, 4 denotes location 1 to 4; B, L and S denotes bark, leaf and stem respectively.
Season, site and tissue type significantly influenced diversity of fungal endophytes (p ≤ 0.001) as summarised in Table 3. Shannon-Wiener index varied significantly (p ≤ 0.001) with season, location and tissue type (Fig. 2 and Table 3). Tissue type had the strongest effect (p ≤ 0.001) on species evenness of the endophytic fungal assemblage followed by location (p ≤ 0.05) (Fig. 3 and Table 3). CF (colonization frequency) increased from the driest location 1 (Loc1, Banaras Hindu Uunivesrsity campus, Uttar Pradesh) to the wettest location 4 (Loc4, Rajiv Gandhi University campus, Arunachal Pradesh) in all season. CF was significantly (p ≤ 0.001) lower in bark than in leaves (Fig. 2 and Table 4).
Table 3.
Summary of MANOVA of different diversity indices.
| Indices | Season (F2, 72) | Location (F3, 72) | Tissue (F2, 72) | Season × Location (F6, 72) | Season × Tissue (F4, 72) | Location × Tissue (F6, 72) | Season × Location × Tissue (F12, 72) |
|---|---|---|---|---|---|---|---|
| Shannon (H′) | 180.857*** | 80.500*** | 59.126*** | 1.808 ns | 2.481 ns | 1.764 ns | 0.651 ns |
| Evenness | 1.463 ns | 3.715* | 19.131*** | 1.282 ns | 0.817 ns | 2.047 ns | 1.524 ns |
| Species richness | 187.583*** | 104.233*** | 91.291*** | 4.087*** | 3.621** | 1.738 ns | 1.350 ns |
***Significant at p ≤ 0. 001, **Significant at p ≤ 0.01, *Significant at p ≤ 0.05, nsnon-significant; where F is degree of freedom for different variables.
Figure 2.
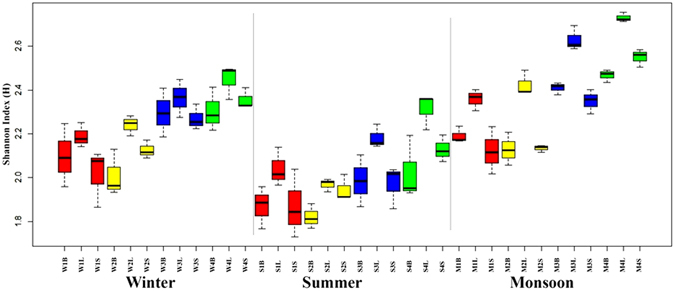
Boxplot representation of variability of species diversity (H) with respect to location, season and tissue type. The first letter of the three-letter acronyms denotes the season (W, winter; S, summer; M, monsoon), the second the location and the third the tissue type (B, barks; L, leaf; S, stem).
Figure 3.
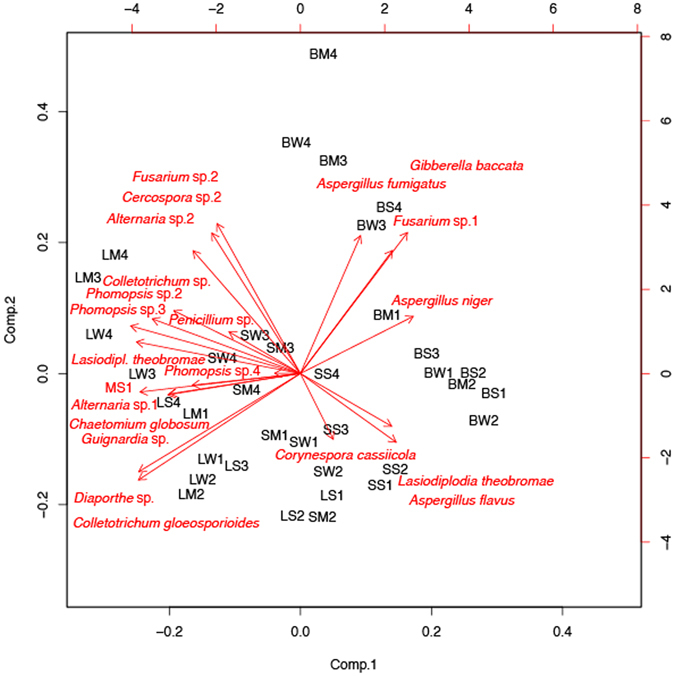
Biplot (Principal components analysis) depicting the relationship between fungal endophyte taxa (arrows) and sampling units (tissue type, season and location). The longer the arrow the more influence has the fungus connected with it, and the further removed a sampling unit gets positioned from the centre the stronger its influence. The closer a sampling unit and a fungal taxon the stronger is their relationship. The first letter of the three-letter acronyms denotes the tissue type (B, barks; L, leaf; S, stem), the second the season (W, winter; S, summer; M, monsoon), and the third the location.
Table 4.
Diversity parameters for aggregate data based on location, season and tissue.
| Diversity Parameters | Loc1 | Loc2 | Loc3 | Loc4 | Winter | Summer | Monsoon | Bark | Leaf | Stem |
|---|---|---|---|---|---|---|---|---|---|---|
| Morphotypes (S)/Species richness | 23 | 23 | 28 | 29 | 44 | 31 | 43 | 27 | 30 | 23 |
| Individuals | 1053 | 1176 | 1335 | 1525 | 1780 | 1298 | 2011 | 1413 | 2023 | 1653 |
| Dominance (D) | 0.1132 | 0.1055 | 0.09357 | 0.08807 | 0.09672 | 0.1198 | 0.08182 | 0.124 | 0.1073 | 0.1242 |
| Shannon (H′) | 2.51 | 2.56 | 2.74 | 2.81 | 2.79 | 2.54 | 2.95 | 2.59 | 2.63 | 2.44 |
| Evenness (e^H/S) | 0.5345 | 0.5619 | 0.5526 | 0.5723 | 0.369 | 0.4074 | 0.4441 | 0.4918 | 0.4613 | 0.5003 |
| Colonization frequency (% CF) | 75.3 | 65.9 | 58.07 | 52. 0 | 65.9 | 48.1 | 74.5 | 52.3 | 74.9 | 61.2 |
A more detailed consideration of each factor is given below
Effects of geographical location
Highest number of endophytic isolates were recovered from samples of Loc4 (1525 isolates, 29 morphotypes) followed by location 3 (Loc3, Ranibagh, Uttarakhand) (1335 isolates, 28 morphotypes), Loc2 (1176 isolates, 23 morphotypes) and Loc1 (1053 isolates, 23 morphotypes) (Table 4). In term of colonization frequency Loc4 showed 75.3% followed by Loc3 with 65.9%, Loc2 with 58.07% and minimum at Loc1 with 52.0% (Table 4). Loc4 achieved the highest Shannon-Wiener (H′) index (2.81), followed by Loc3 (2.74), Loc2 (2.56) and Loc1 (2.51) (Fig. 2, Table 4). Different diversity parameters recorded in different locations, seasons and tissues is given in Table 4.
Effects of season
Seasonal effect was prominent with the maximum colonization occurring in monsoon (2011 isolates, 43 morphotypes) followed by winter (1780 isolates, 44 morphotypes) and the minimum in summer (1298 isolates, 31 morphotypes) (Table 4). CF (%) value varied with season with the highest in monsoon (74.5%) followed by winter (65.9%) and summer (48.1%) (Table 4). The relative abundance of most species was the highest in monsoon followed by winter and summer. Shannon-Wiener index was found as high as 2.95 in monsoon. Even the lowest index value recorded in summer (2.54) indicates good endophyte diversity in teak tree (Table 4).
Effects of tissue type
Of the different tissues sampled, leaf harboured maximum endophytes in terms of both total isolates (2023) and morphotypes (30). From stem tissue, 1653 isolates belonging to 23 different OTUs were recovered, whereas 1413 isolates and 27 morphotypes were obtained from bark. Leaf showed highest CF (74.9%) followed by stem (61.2%) and bark (52.3%). Leaf scored highest Shannon-Wiener index (2.63) followed by bark (2.59) and stem (2.44) (Table 4). Leaf also scored highest Shannon-Wiener index across all seasons and sites (Fig. 2).
Discussion
Diversity of endophytic mycobiota of Tectona grandis
The ITS1-5.8 S-ITS2 region of the rDNA is the most widely used marker for bar coding of fungi because it is considered to appropriately discriminate species17. However, in this study some of the morphotypes had identical sequences, although morphological differences were strong enough to keep them as separate species. Insufficiency of ITS sequences to separate fungal species has also been reported for other fungal species complexes18, 19. In contrast, intra-generic diversity of the ITS region of Diaporthe (Phomopsis) was high leading to the recognition of seven different species. High species diversity in this genus is in accordance with previous findings20, 21. Diversity of endophytic fungi in Tectona was found to be lower than in other tropical woody trees22–24. Inhibition of slow growing endophytes by the fast growing Phomopsis species might be one of the reasons for the low number of species detected in this study. The use of next generation sequencing (NGS) would probably lead to the discovery of many other species25–27. Predominance of ascomycotina in teak is in accordance with most studies done about endophytic assemblages in other plant species, so far28, 29, including tropical trees, e.g. Terminalia arjuna 24 and Madhuca indica 30. Interestingly, overall CF (62.83%) in teak tree was found low while many previous studies reported very high level of colonization frequency ranging from 95 to 100% in other trees20, 25, 31. The low CF and morphotypes might be due to the fact that the present study was carried out in different seasons, site and tissue types, unlike earlier studies limited to foliar tissue or favourable season which support highest CF. The reason for fewer morphotypes recovery may be also due to inability of fungi to overcome the physical and chemical barriers of the host plant32. Fungi easily colonise low-density wood compared to solid and high density wood, as found in teak. Teak wood also has high lignin content as another barrier for fungal colonization. Furthermore, flavonoids (rutin and quercitin) present in teak leaves play important roles in resisting fungal invasion33. The extract of hardwood sawdust of T. grandis has shown great inhibitory activity against several brown rot and white rot fungi. The naphthoquinone derivatives found in teak heartwood and leaves act as strong anti fungal agent34. Again, in a study teak leaf extract suppressed significantly the growth and spore formation of Arthrinium phaeospermum exhibiting its anti fungal property35. Furthermore, culture-dependent methods make it extremely difficult to isolate and enumerate biotrophic and unculturable species36 leading to lower CF and morphotypes estimate. This might be one of the reasons for the isolation of fewer basidiomycotina in the endophytes of T. grandis. Currently researchers prefer culture independent next generation sequencing (NGS) approaches to reveal the real endophytes diversity. However such methods are not error proof and have been blamed for overestimation21, 36–38. Serious doubts arise when predominating species or communities found in culture based isolation are completely absent39 or differ26 in NGS methods. Some studies which compared the traditional culture based endophyte diversity data with NGS data of the same host tree revealed that culture-based technique alone can reveal real qualitative picture of fungal endophytes27, 36. Furthermore, culture technique is the only way to get isolates for future wet lab use and to improve reference taxonomic database. In view of the above, it can be concluded that a combination of culture-dependent and culture-independent approaches need to be used for reliable estimate of endophyte diversity37. Colletotrichum gleosporioides, Colletotrichum sp. (Colletotrichum siamense), Phomopsis spp. and Fusarium sp.1 (Albonectria rigidiuscula) were ubiquitous and dominant at all four sites. Their dominance increased from Loc1 to Loc4. Perhaps to more favourable abiotic and biotic conditions for fungal growth, such as the prolonged duration of rain fall, greater annual precipitation, favourable temperature (Table 5), higher relative humidity, lesser anthropogenic perturbation and higher inoculum dose, tend towards optimum from Loc1 to Loc4. Above species along with Fusarium spp., Alternaria spp., C. globosum, Aspergillus spp., Corynespora cassiicola, Lasiodiplodia spp., Hypoxylon fragiforme and Curvularia sp. are well established endophytes of teak and other host plants15, 16, 30, 39, 40. Species diversity and dominance of Diaporthe (Phomopsis) species in T. grandis is similar to the findings of Mekkamol15 and Chareprasert et al.16. This finding added in establishing the Diaporthe (Phomopsis) spp. as consistent teak endophytes which might have co-evolved with teak41. However, Diaporthe (Phomopsis) spp. have also been reported as the dominant endophyte in other trees like Azadirachta indica 42, Luehea divaricata 43 and Madhuca indica 30. Dominance of Diaporthe (Phomopsis) spp. in Trichilia elegans in a recent report also supported our finding20. Of 45 morphotypes, 17 showed dominance <0.5%. Presence of such a large number of species in low colonization frequency clearly suggests the adequacy of sample size as well as isolation protocol. The presence of rarely reported fungal endophytes, such as, Coprinellus, Cercospora, Ceratobasidium, Periconia and Pyrenochaeta also suggests that sampling and isolation methods were satisfactorily employed. These endophytes seem to be reported for the first time from the Indian subcontinent. Previous studies have shown that colonization of endophytes is strongly affected by geographic location, climatic conditions, seasonal changes, host and host tissues44, 45. Keeping in view the suggestions of Arnold et al.3 that host preference and spatial orientation of tropical tree mycobiota can be better explained by their relative colonization frequency rather than presence or absence of morphospecies, we analysed our data for the same. Significant statistical differences in diversity measures were observed along locations, seasons and tissue types in MANOVA (p ≤ 0.001), based on Shannon-Wiener index (H′) and species richness data (Table 3). Biplot analysis done to analyse interactive effect of all the three variables on dominant endophytes (%D ≥ 0.5) showed tissue type based grouping of isolates indicating its strongest effect (Fig. 3).
Table 5.
Geographic coordinates, climatic conditions and unique characteristics of study sites.
| Site | Geographic coordinates | *Seasonal rainfall during the study period in mm (Jan 2011–Dec 2011) | *Monsoon period in study year | *Seasonal mean of minimum and maximum T (in °C) | Uniqueness and reasons for selection as study site | ||||
|---|---|---|---|---|---|---|---|---|---|
| Winter | Summer | Monsoon | Onset | Withdrawal | Mean Min. | Mean Max. | |||
| Loc1: (BHU) Banaras Hindu University, Uttar Pradesh | 25°16′34.93″N 82°59′20.22″E | 8.5 | 247.3 | 844.5 | 17 June | 30 Sept. | 8.8 (W), 16.3 (S), 20.7 (M) | 29.3 (W), 40.9 (S), 33.8 (M) | Located in the highly fertile Gangetic plain of India |
| Loc2: (HN) Hathinala, Uttar Pradesh | 24°18′0.97″N 83°6′29.96″E | 9.5 | 267.8 | 1094 | 17 June | 30 Sept. | 11.1 (W), 22.69 (S), 22.65 (M) | 25.82 (W), 37.44 (S), 31.24 (M) | Located in the dry deciduous tropical forest of Vindhayan region |
| Loc3: (RB) Ranibagh, Uttrakhand | 29°17′31.97″N 79°32′34.23″E | 97.8 | 462.90 | 1460.5 | 20 June | 26 Sept. | 1.7 (W), 7.5 (S), 9.7 (M) | 15.4 (W), 23.5 (S), 21.6 (M) | Located in the biodiversity rich Himalayan foot hills with prevailing low temperature |
| Loc4: (RGU) Rajiv Gandhi University, Arunachal Pradesh | 27°8′52.34″N 93°45′53.44″E | 51.5 | 1158.1 | 1948.3 | 8 June | 13 Oct. | 15 (W), 20.4 (S), 22 (M) | 21(W), 34.2 (S), 30.2 (M) | Located in one of the world’s top mega diversity centre, world’s northernmost tropical rain forest, receives almost heaviest annual precipitation worldwide |
(*Source: National Data Centre, IMD, Pune, India). W = Winter, S = Summer, M = Monsoon.
Effects of spatial variation
Earlier studies have supported the view that climate and rainfall pattern of the investigation area strongly affect the fungal assemblage45. Loc4 in Arunachal Pradesh registered highest annual precipitation followed in decreasing order by the Loc3 in Uttrakhand, Loc2 at Mirzapur in Uttar Pradesh and Loc1 at Varanasi in Uttar Pradesh during the year of study. The decreasing patterns of annual rainfall from Loc4 to Loc1 exactly matched the decreasing trend for diversity and species richness at these sites. Diversity index and species richness were maximum at Loc4, is in concordance with the favourable conditions found there for fungal growth and dispersion as region is attributed with tropical climate accompanied with heavy and comparatively long spanned annual rain fall46. Greater species richness of pine trees in autumn compared to spring due to higher rainfall in the former also highlights the role of rainfall in shaping fungal assemblage47. Vaz et al.48 found significant statistical difference in colonization of fungal endophytes with increase in precipitation and decrease in temperature. Higher diversity at Loc3 in comparison to Loc1 and Loc2 might be due to higher relative humidity, as reported in the case of Trichilia elegans 20. No doubt, factors other than rainfall and humidity also affect the diversity of endophytic assemblage. We also observed location specific distribution of certain endophytes at least at species level. Aspergillus sp.1 (Aspergillus viridinutans) was specific to Loc1, likewise Coprinellus sp., Ceratobasidium sp., and Fusarium sp.2 (Fusarium incarnatum) were exclusively isolated from Loc3 whereas Cercospora sp.1 (Cercospora gerberae), and Curvularia sp. were restricted to Loc4. Space limited distribution of these taxa indicates spatial structuring of endophytic communities.
Effects of temporal variation
Rain splashes help in dissemination of inoculum materials, and high humidity and low temperature support fungal spore germination and multiplication. These conditions lead to high infection rate and fungal establishment in monsoon and winter seasons. Contrary to this, ambient environment exhausted of potential inocula in summer resulted in little horizontal transfer of endophytes38. Further, under stress conditions host plants develop different structural modifications and defense chemicals leaving lesser access to intruders. We found colonization frequency of most endophytic species maximum in monsoon followed by winter and summer. Further, increase in colonization of A. flavus, Aspergillus niger and Guignardia sp., which are considered as xerophilic fungi, in summer indicates seasonal effect on mycobiota assemblage. These fungi face lesser competition from other humidity requiring fungi resulting in greater colonization during dry period. On the other hand, Phomopsis producing a large number of slimy spores in rainy season may reduce the chances of xerophilic fungal growth during monsoon period. The present seasonal findings are in accord with the earlier observations16, 40. The highest CF in monsoon and the greatest species richness in the winter is agree with the findings of Chareprasert et al.16 in T. grandis. Guo et al.49 also reported highest endophyte CF in spring in needle of Pinus tabulaeformis. On the other hand, Collado et al.50 reported highest species richness in spring and Helander et al.51 reported no seasonal effect on colonization frequency of endophytes in old needles of Scots pine but in young needle CF increased in summer.
Effects of tissue type
Maximum colonization frequency and species richness in leaves might be due to their large size (20–50 cm × 15–40 cm) which offers considerably great inoculum capture area, abundance of natural openings in the form of stomata, hydathodes and glandular openings as easy entry points, and hairy surface helpful in inoculum landing and attachment. Tenderness of leaf, in contrast to bark and stem tissues, helps endophytes in easy access to internal tissue. The present findings of maximum colonization frequency and species richness in leaves corroborates the earlier findings15, 16. The highest diversity in Tectona leaf is supported by the presence of as many as 22 species of endophytic fungi in a single leaf of tropical tree Manilkara bidentata 52. Indian medicinal herb Tinospora cordifolia 40 tissues also harbours endophytes like Tectona (leaf > stem > bark) while A. Indica 42 showed slightly different results where endophytes recovered maximally in leaf followed by bark and stem. Gond et al.53 reported maximum endophyte colonization in bark of Aegel marmelos while Verma et al.30 reported greatest recovery of endophytes from stem tissue of M. indica. The above findings lead to the conclusion that different tissues of various plants support endophytes colonization at varying rate. Reports on strict host specificity and preference are very rare44, 54 however, isolation of C. globosum strictly from leaf tissue across all four sites in all three seasons, indicated its remarkable specificity towards leaf in the present study. This finding is also supported by host specificity of Chaetomium for T. arjuna 24. Besides C. globosum, Guignardia sp. (Phyllosticta elongata) and Paecilomyces sp.1 (Paecilomyces variotii) also showed strong affinity towards leaf tissue as they were isolated uniformly but exclusively from leaf sample of at least three distantly located sites in different seasons. Sun et al.55 reported significant host and tissue preference in Betula platyphylla, Quercus liaotungensis and Ulmus macrocarpa. Ek-Ramos et al.56 also reported tissue specificity of some endophytes in Gossypium hirsutum. Fungal endophytes associated with three South American Myrtaceae members also exhibited preferences in the colonization at leaf level48. In this study stem comes second in terms of CF but with less species richness and diversity than bark. Minimum endophytes CF in bark might be due to its dead rhytidome but direct exposure to the external environment supports it to harbour greater species richness than stem. Since different plant tissues have distinct anatomy and functions, these may have influenced the dominance of dissimilar fungal groups in different tissue types in the present study. Effect of tissue type on endophyte colonisation has also been highlighted in many earlier studies40, 55–57. Host-dependent structuring of endophytic components can also be explained by the fact that being inside the host they never experience external environmental fluctuations directly. But we can not wholly discount the role of external environment, e.g. during infection, as it would influence the fungal inocula type and density.
Since bark and stem are permanent organs of the plant and do not show significant changes in their shape and size within a year, the age of these organs during the study does not seem to have exerted significant influence. However, the leaves are deciduous and their age, toughness, shape and chemistry tremendously vary within a year. New leaves of T. grandis appear in March (spring), grow and mature up to September (monsoon) and senesce thereafter until partial fall in January (winter). For the leaf, minimum diversity was recorded in summer and maximum in monsoon. But it cannot be ascertained that this trend was solely due to leaf age and its chemistry as Arnold and Herre58 found no observable effect of leaf age on endophyte colonization in Theobroma cacao with all young and mature leaves equally colonisable. However, leaf age may be considered as an additional factor along with seasonal and geographic factors for variations in diversity.
In conclusion, all the three variables, namely, tissue type, season and location, effected the fungal endophyte composition of T. grandis with tissue type having the predominant effect. T. grandis has supported moderate endophytic fungal diversity. This diversity estimate can further be augmented by using both culture-dependent and culture-independent approaches.
Methods
Sites and sample acquisition
Four different sampling sites in northern and north-eastern India differing in climatic and geographic conditions were selected for the study (Table 5). Loc1 is situated in the sub-tropical eastern Gangetic plains, Loc2 is situated in the dry deciduous tropical forest of the Vindhayan region, Loc3 is situated in the foot hills of Himalaya with mild summer and low winter (mean minimum temperature 1.7 °C) temperature and Loc4 is atop Rono hills situated in one of the global biodiversity hot spot region with heavy annual precipitation.
Mature, green, asymptomatic leaves in triplicate were randomly collected from the lowermost branches of five T. grandis trees. Bark and stem samples were taken at chest height (1.37 m, to maintain sampling consistency). Sampling was done at every study sites in three season namely winter (January), summer (May) and monsoon (September) in 2011. At every occasion sampling was done from the same trees. Collected plant parts were packed separately in polybags and kept in icebox (4 °C) for transportation and further processing.
Surface sterilization and endophyte recovery
The surface sterilization method proposed by Petrini et al.59 was adopted to T. grandis tissues and verified by ‘leaf imprint method’ of Schulz et al.60 Briefly, 5 cm × 5 cm piece from each leaf, and 5 cm × 2 cm section from each bark and stem sample were cut out and cleansed in running tap water followed by sequential dipping in 70% ethanol (2 min), 4% sodium hypochlorite (4 min for bark and stem, 2 min for leaf), 70% ethanol (2 min) and finally rinsed thrice with sterile deionised water to remove the surface sterilents. After air drying in laminar flow, the samples were cut into small pieces (5 mm × 5 mm) under strict aseptic conditions. About 4–5 segments were placed on each Petri dish containing potato dextrose agar medium (PDA, HiMedia) supplemented with 150 μgml−1 of streptomycin sulphate (HiMedia). Selection of PDA, which supported maximum endophytes recovery in terms of both number and types, was done over two other media namely Czapek Dox agar (CDA) and malt extract agar (MEA) after screening 50 segments of each tissue types as details given in Table 6. A total of 225 segments of each tissue type in each season from each location were screened for endophyte presence. Thus, seasonally 2700 segments were examined in this study, amounting to a total of 8100 segments. The Petri dishes were sealed with Parafilm (Bemis Flexible Packaging, USA) and incubated at 27 ± 2 °C in BOD cum humidity incubator (Calton, NSW, New Delhi, India) under 12 h light and dark cycle and monitored every day for fungal emergence up to one month. Different endophytes emerging from the explants were transferred and maintained on fresh PDA plates as axenic cultures.
Table 6.
Preliminary fungal endophytes isolation for suitable culture medium selection.
| Endophytes | Czapek Dox agar | Malt extract agar | Potato dextrose agar | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Bark | Leaf | Stem | Bark | Leaf | Stem | Bark | Leaf | Stem | |
| Alternaria sp. | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 2 | 2 |
| Chaetomium globosum | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 |
| Colletotrichum gloeosporioides | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 3 | 0 |
| Fusarium baccata | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 |
| Fusarium sp. | 0 | 0 | 1 | 1 | 0 | 3 | 4 | 0 | 6 |
| Pseudofusicoccum adansoniae | 0 | 1 | 0 | 0 | 3 | 0 | 2 | 4 | 2 |
| Diaporthe sp. | 2 | 2 | 1 | 3 | 4 | 3 | 4 | 8 | 6 |
| Diaporthe longicola | 3 | 5 | 4 | 4 | 7 | 6 | 6 | 9 | 9 |
| Phomopsis sp. | 0 | 0 | 0 | 2 | 0 | 0 | 6 | 0 | 0 |
| Total isolates | 5 | 9 | 6 | 11 | 16 | 13 | 27 | 31 | 25 |
| Grand total isolates | 20 | 40 | 83 | ||||||
| Total morphotypes | 5 | 8 | 9 | ||||||
Macroscopic and microscopic identification of recovered fungal endophytes
Based on the culture characteristic, such as, shape, size, colour, texture, growth pattern and back side colour of colony and microscopic details, all the fungal isolates were grouped into 45 distinct morphotypes/OTUs. Sporulating cultures were examined using a camera-coupled Nikon Trinocular Light Microscope (Model E-600). Standard taxonomic manuals were used for the morphological identification and grouping of fungal isolates61, 62. After identification, representatives of each morphotype were preserved and stored in triplicates at the Department of Botany, Banaras Hindu University, Varanasi, India.
Molecular identification of endophytes
Total genomic DNA extraction
The total genomic DNA of all morphotypes, except four well known, was extracted following the lab developed amended SDS-CTAB protocol to get high throughput fungal DNA extraction. In short, 0.3 g pure mycelium was pulverized with plastic micro-pestle in 500 µl lysis buffer in a microcentrifuge tube. If needed, more lysis buffer was added to maintain viscosity of the paste. This step was followed by the addition of 75 µl of 10% SDS solution. After gentle mixing, the tube was placed in shaking water bath at 37 °C for 1.5 h with intermittent gentle inversion followed by addition of 80 µl of each 5 M NaCl and CTAB/NaCl solutions and incubation at 65 °C in water bath for 45 min at 60 rpm. DNA was extracted by addition of equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) and centrifugation at 12000 rpm for 15 min. Aqueous supernatant was transferred to fresh centrifuge tube and DNA was precipitated by addition of equal volume of ice chilled isopropanol at −20 °C. After 12 hr of incubation, the tube was centrifuged at 12000 rpm for 15 min. The pellet obtained was washed with 70% ethanol and air dried in laminar flow before re-suspending in 20–30 µl of TE buffer. Isolated DNA was electrophoresed on 0.8% (w/v) agarose gel stained with ethidium bromide (0.5 μgml−1) and visualized under UV transilluminator.
PCR amplification and DNA sequencing
The universal primer pair ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) were used to amplify the fungal nrITS regions of representative isolate of each morphotype63. PCR amplifications64 (modified) were performed on programmable S1000 Thermal Cycler (BIO-RAD) in 50 µl reaction volumes, each having 0.4 µl of 5 Uµl−1 Taq DNA polymerase (Genei, India), 1 µl of 10 mM dNTPs (Genei, India), 2 µl of each primer (10 pM, Eurofins genomics, India), 5 µl of 10X PCR buffer with MgCl2, 37.6 µl ultrapure water and 2 µl extracted DNA template. Optimized condition used for amplification was pre-denaturation at 95 °C for 5 min followed by 35 cycles of each denaturation at 95 °C for 1 min, primer annealing at 54 °C for 1 min, extension at 72 °C for 1 min and at last 1 cycle of final extension at 72 °C for 5 min. A non-template negative control was also run each time. Integrity and quality of resulting PCR products were examined on 1.5% (w/v) agarose gel impregnated with ethidium bromide (0.5 μgml−1) and visualized under a UV transilluminator. Amplified ITS fragments were cleaned by HiYield Gel/PCR DNA mini kit (Real Biotech Corporation) strictly following manufacturer manual and sequenced (Applied Biosystems 3130 Genetic Analyser) at Interdisciplinary School of Life Sciences (ISLS), BHU, India using ITS1 (forward) primer and BigDye Terminator v3.1 Cycle Sequencing Kit.
Blast search and phylogeny analysis
For phylogenetic analysis sequences were nBLAST searched against the NCBI database (blast.ncbi.nlm.nih.gov) and trimmed to cover the entire region of closest reference sequence. These trimmed sequences were further used to get the closest taxonomic match and phylogenetic tree construction. Multiple sequence alignments of 41 OTUs and 2 closest named reference sequences for each morphotype were done online by Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/). The neighbor-joining phylogenetic and molecular evolutionary analyses were conducted using MEGA5. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site65. The obtained ITS sequences (unedited) were submitted to GenBank for accession numbers (www.ncbi.nlm.nih.gov/genbank). A detailed account of the sequenced OTUs with their respective GenBank accession number and the closest match (based on maximum % identity) are listed in Table 1.
Fungal diversity analysis
The percent colonization frequency (%CF) was calculated66 as %CF = (Ncol/Nt) × 100, where, Ncol = number of plant tissue segments colonized and Nt = total number of plant tissue segments examined. Percentage dominance (%D) of each OTU was calculated as %D = n/N × 100 where, n = total isolates of an OTU, N = total isolates of all OTUs. Shannon-Wiener index (H′) was calculated for each season, location and tissue type using PAST software (http://folk.uio.no/ohammer/past/). Variation in Shannon-Wiener index (H′) with season, site and tissue is depicted in the form of boxplots drawn with the help of “R” statistical software (R Development Core Team 2009)67. Effects of season, site and tissue type on distribution and diversity of fungal endophytes were analysed by multivariate linear model analysis (MANOVA) and bi-plot analysis using SPSS v16 and R v2.15.1 (R Development Core Team 2009)67 software, respectively. For MANOVA, season, site and tissue type were considered as independent factors while Shannon–Wiener index, species richness and evenness were treated as dependent factors or response variables. Principal components analysis was done to reveal the interaction between fungal endophytes with sampling variables (tissue type, season and location) using R statistical software.
Acknowledgements
Authors are thankful to Prof. R. K. Singh, Dept. of Botany, Rajiv Gandhi University, Doimukh, Arunachal Pradesh, India, for his help in sampling at Loc4; to Head and Coordinator, CAS and DST-FIST in Botany, Institute of Science, BHU, Varanasi, India, for providing essential research facilities and technical supports. Authors appreciably acknowledge the helps of ISLS and DST-PURSE, UGC-UPE, BHU, Varanasi, India for rDNA sequencing and financial help, respectively. This work was supported by Indian Council of Medical Research, New Delhi, [JRF and SRF to DKS] and Department of Science and Technology, New Delhi [Project SB/EMEQ-121/2014 to RNK].
Author Contributions
Hypothesised and designed the experiments: D.K.S., S.K.V. and R.N.K., Performed the experiments: D.K.S., V.K.S., A.M. and J.K., Analyzed the data: D.K.S., R.N.K., V.K.S., J.K. and T.N.S., Contributed reagents/materials/analysis tools: R.N.K., D.K.S., V.K.S. and T.N.S. Wrote the paper: D.K.S., R.N.K., and T.N.S.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Bary. Morphologie und Physiologie der Pilze, Flechten und Myxomyceten. Hofmeister’s Hand Book of Physiological Botany. Vol. 2 (Leipzig, 1866).
- 2.Bacon, C. W. & White, J. F. Microbial endophytes (eds Bacon, C. W. & White, J. F.) 487 (Marcel-Dekker, New York, USA, 2000).
- 3.Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 2000;3:267–274. doi: 10.1046/j.1461-0248.2000.00159.x. [DOI] [Google Scholar]
- 4.Rodriguez R, Redman R. More than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008;59:1109–1114. doi: 10.1093/jxb/erm342. [DOI] [PubMed] [Google Scholar]
- 5.Persoh D. Factors shaping community structure of endophytic fungi-evidence from the Pinus-Viscum-system. Fungal Divers. 2013;60:55–69. doi: 10.1007/s13225-013-0225-x. [DOI] [Google Scholar]
- 6.Porras-Alfaro A, Bayman P. Hidden Fungi, Emergent Properties: Endophytes and Microbiomes. Annu. Rev. Phytopathol. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- 7.Frohlich J, Hyde KD. Biodiversity of palm fungi in the tropics: Are global fungal diversity estimates realistic? Biodivers. Conserv. 1999;8:977–1004. doi: 10.1023/A:1008895913857. [DOI] [Google Scholar]
- 8.Unterseher M, Schnittler M. Dilution-to-extinction cultivation of leaf-inhabiting endophytic fungi in beech (Fagus sylvatica L.) – different cultivation techniques influence fungal biodiversity assessment. Mycol. Res. 2009;113:645–654. doi: 10.1016/j.mycres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Strobel GA, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma VC, Kharwar RN, Strobel GA. Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat. Prod. Commun. 2009;4(11):1511–1532. [PubMed] [Google Scholar]
- 11.Kharwar RN, et al. Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat. Prod. Rep. 2011;28:1208–1228. doi: 10.1039/c1np00008j. [DOI] [PubMed] [Google Scholar]
- 12.Schulz B, Boyle C. The endophytic continuum. Mycol. Res. 2005;109(6):661–686. doi: 10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- 13.Neamatallah A, Yan L, Dewar SJ, Austin B. An extract from teak (Tectona grandis) bark inhibited Listeria monocytogenes and methicillin resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2005;41(1):94–6. doi: 10.1111/j.1472-765X.2005.01680.x. [DOI] [PubMed] [Google Scholar]
- 14.Krishna MS, Jayakumaran NA. Antibacterial, cytotoxic and antioxidant potential of different extracts from leaf, bark and wood of Tectona grandis. Int. J. Pharm. Sci. Res. 2010;2:155–158. [Google Scholar]
- 15.Mekkamol, S. Endophytic fungi in Tectona grandis L. (Teak). PhD thesis, John Moores University, Liverpool, England (1998).
- 16.Chareprasert S, Piapukiew J, Thienhirun S, Whalley AJS, Sihanonth P. Endophytic fungi of teak leaves Tectona grandis L. and rain tree leaves Samanea saman Merr. World. J. Microbiol. Biotechnol. 2006;22:481–486. doi: 10.1007/s11274-005-9060-x. [DOI] [Google Scholar]
- 17.Schoch CL, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grünig CR, McDonald BA, Sieber TN, Rogers SO, Holdenrieder O. Evidence for subdivision of the root-endophyte Phialocephala fortinii into cryptic species and recombination within species. Fungal Genet. Biol. 2004;41:676–87. doi: 10.1016/j.fgb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Harder CB, et al. Three-gene phylogeny of the Mycena pura complex reveals 11 phylogenetic species and shows ITS to be unreliable for species identification. Fungal Biol. 2013;117:764–75. doi: 10.1016/j.funbio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Rhoden SA, Garcia A, Filho CJR, Azevedo JL, Pamphile JA. Phylogenetic diversity of endophytic leaf fungus isolates from the medicinal tree Trichilia elegans (Meliaceae) Genet. Mol. Res. 2012;11(3):2513–2522. doi: 10.4238/2012.June.15.8. [DOI] [PubMed] [Google Scholar]
- 21.Ovaskainen O, et al. Identifying wood-inhabiting fungi with 454 sequencing-what is the probability that BLAST gives the correct species? Fungal Ecol. 2010;3:274–283. doi: 10.1016/j.funeco.2010.01.001. [DOI] [Google Scholar]
- 22.Gazis R, Chaverri P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 2010;3:240–254. doi: 10.1016/j.funeco.2009.12.001. [DOI] [Google Scholar]
- 23.Kumaresan V, Suryanarayanan TS. Occurrence and distribution of endophytic fungi in a mangrove community. Mycol. Res. 2001;105:1388–1391. doi: 10.1017/S0953756201004841. [DOI] [Google Scholar]
- 24.Tejesvi MV, et al. Endophytic fungal assemblages from inner bark and twig of Terminalia arjuna W. and A. (Combretaceae) World. J. Microbiol. Biotechnol. 2005;21:1535–1540. doi: 10.1007/s11274-005-7579-5. [DOI] [Google Scholar]
- 25.Arnold AE, Henk DA, Eells RL, Lutzoni F. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 2007;99:185–206. doi: 10.1080/15572536.2007.11832578. [DOI] [PubMed] [Google Scholar]
- 26.Guo LD, Hyde KD, Liew ECY. Detection and identification of endophytic fungi within frond tissues of Livistona chinensis based on rDNA sequence. Mol. Phylogenet. Evol. 2001;20:1–13. doi: 10.1006/mpev.2001.0942. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Yao YF. Endophytic fungal communities associated with vascular plants in the high arctic zone are highly diverse and hostplant specific. PLoS ONE. 2015;10(6):e0130051. doi: 10.1371/journal.pone.0130051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieber TN. Endophytic fungi in forest trees: are they mutualists? Fungal. Biol. Rev. 2007;21:75–89. [Google Scholar]
- 29.Bullington LS, Larkin BG. Using direct amplification and next-generation sequencing technology to explore foliar endophyte communities in experimentally inoculated western white pines. Fungal Ecol. 2015;17:170–178. doi: 10.1016/j.funeco.2015.07.005. [DOI] [Google Scholar]
- 30.Verma SK, et al. Impact of environmental variables on the isolation, diversity and antibacterial activity of endophytic fungal communities from Madhuca indica Gmel. at different locations in India. Ann. Microbiol. 2014;64:721–734. doi: 10.1007/s13213-013-0707-9. [DOI] [Google Scholar]
- 31.Gamboa MA, Bayman P. Communities of endophytic fungi in leaves of a tropical timber tree (Guarea guidonia: Meliaceae) Biotropica. 2001;33:352–360. doi: 10.1111/j.1744-7429.2001.tb00187.x. [DOI] [Google Scholar]
- 32.Rodríguez, J., Elissetche, J. P. & Valenzuela, S. Tree endophytes and wood biodegradation in Pirttilä, A.M. & Frank, A.C. (eds) Endophytes in forest trees: biology and applications., 81–93, (Dortrecht: Springer, 2011).
- 33.Sun S, Zeng X, Zhang D, Guo S. Diverse fungi associated with partial irregular heartwood of Dalbergia odorifera. Sci. Rep. 2015;5:8464. doi: 10.1038/srep08464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumthong P, Romero-Gonz´alez RR, Verpoorte R. Identification of anti-wood rot compounds in teak (Tectona grandis l.f.) sawdust extract. J. Wood Chem. Technol. 2008;28:247–260. doi: 10.1080/02773810802452592. [DOI] [Google Scholar]
- 35.Astiti NPA, Suprapta DN. Antifungal activity of teak (Tectona grandis L.f.) leaf extract against Arthrinium phaeospermum (corda) M.B. Ellis, the cause of wood decay on Albizia falcataria (L.) FOSBERG. J. ISSAAS. 2012;18(1):62–69. [Google Scholar]
- 36.Unterseher M, Gazis R, Chaverri P, Guarniz CFG, Tenorio DFG. Endophytic fungi from Peruvian highland and lowland habitats form distinctive and host plant-specific assemblages. Biodivers. Conserv. 2013;22:999–1016. doi: 10.1007/s10531-013-0464-x. [DOI] [Google Scholar]
- 37.Sun X, Guo LD. Endophytic fungal diversity: review of traditional and molecular techniques. Mycology. 2012;3:65–76. [Google Scholar]
- 38.Tedersoo L, et al. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010;188:291–301. doi: 10.1111/j.1469-8137.2010.03373.x. [DOI] [PubMed] [Google Scholar]
- 39.Tejesvi MV, Kajula M, Mattila S, Pirttilä AM. Bioactivity and genetic diversity of endophytic fungi in Rhododendron tomentosum Harmaja. Fungal Divers. 2011;47:97–107. doi: 10.1007/s13225-010-0087-4. [DOI] [Google Scholar]
- 40.Mishra A, et al. Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb. Ecol. 2012;64:3288–398. doi: 10.1007/s00248-012-0029-7. [DOI] [PubMed] [Google Scholar]
- 41.Saikkonen K, Wali P, Helander M, Faeth SH. Evolution of endophyte-plant symbioses. Trends in Plant Sci. 2004;9(6):275–280. doi: 10.1016/j.tplants.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Verma VC, Gond SK, Kumar A, Kharwar RN, Strobel GA. Endophytic mycoflora from leaf, bark, and stem of Azadirachta indica A Juss. from Varanasi, India. Microb. Ecol. 2007;54:119–125. doi: 10.1007/s00248-006-9179-9. [DOI] [PubMed] [Google Scholar]
- 43.Wenzel JB, Garcia A, Filho CJR, Prioli AJ, Pamphile JA. Evaluation of foliar fungal endophyte diversity and colonization of medicinal plant Luehea divaricata (Martius et Zuccarini) Biol. Res. 2010;43:375–384. doi: 10.4067/S0716-97602010000400001. [DOI] [PubMed] [Google Scholar]
- 44.Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical trees biodiversity hotspots? Ecology. 2007;88(3):541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 45.Wilson D, Carroll GC. Infection studies of Discula quercina, an endophyte of Quercus Zhang garryana. Mycologia. 1994;86(5):635–647. doi: 10.2307/3760534. [DOI] [Google Scholar]
- 46.Banerjee D. Endophytic fungal diversity in tropical and subtropical plants. Res. J. Microbiol. 2011;6:54–62. doi: 10.3923/jm.2011.54.62. [DOI] [Google Scholar]
- 47.Zamora P, Martínez-Ruiz C, Diez JJ. Fungi in needles and twigs of pine plantations from northern Spain. Fungal Divers. 2008;30:171–184. [Google Scholar]
- 48.Vaz ABM, da Costa AGFC, Raad L VV, Góes-Neto A. Fungal endophytes associated with three South American Myrtae (Myrtaceae) exhibit preferences in the colonization at leaf level. Fungal Biol. 2014;118:277–28. doi: 10.1016/j.funbio.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Guo LD, Huang GR, Wang Y. Seasonal and tissue age influences on endophytic fungi of Pinus tabulaeformis (pinaceae) in the dongling mountains, Beijing. J. Int. Plant. Biol. 2008;50(8):997–1003. doi: 10.1111/j.1744-7909.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- 50.Collado J, Platas G, Gonzalez I, Pelaez F. Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol. 1999;144:525–532. doi: 10.1046/j.1469-8137.1999.00533.x. [DOI] [PubMed] [Google Scholar]
- 51.Helander ML, Sieber TN, Petrini O, Neuvonen S. Endophytic fungi in Scots pine needles: spatial variation and consequences of simulated acid rain. Can. J. Bot. 1994;72(8):1108–1113. doi: 10.1139/b94-135. [DOI] [Google Scholar]
- 52.Lodge DJ, Fisher PJ, Sutton BC. Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia. 1996;88:733–738. doi: 10.2307/3760967. [DOI] [Google Scholar]
- 53.Gond SK, Verma VC, Kumar A, Kumar V, Kharwar RN. Study of endophytic fungal community from different parts of Aegle marmelos Correae (Rutaceae) from Varanasi (India) World. J. Microbiol. Biotechnol. 2007;23:1371–1375. doi: 10.1007/s11274-007-9375-x. [DOI] [Google Scholar]
- 54.Cannon PF, Simmons CM. Diversity and host preference of leaf endophytic fungi in the Iwokrama Forest Reserve, Guyana. Mycologia. 2002;94(2):210–220. doi: 10.1080/15572536.2003.11833226. [DOI] [PubMed] [Google Scholar]
- 55.Sun X, Ding Q, Hyde KD, Guo LD. Community structure and preference of endophytic fungi of three woody plants in a mixed forest. Fungal Ecol. 2012;5:624–632. doi: 10.1016/j.funeco.2012.04.001. [DOI] [Google Scholar]
- 56.Ek-Ramos MJ, Zhou W, Valencia CU, Antwi JB, Kalns LL. Spatial and temporal variation in fungal endophyte communities isolated from cultivated cotton (Gossypium hirsutum) PLoS ONE. 2013;8(6):e66049. doi: 10.1371/journal.pone.0066049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar DSS, Hyde KD. Biodiversity and tissue recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004;17:69–90. [Google Scholar]
- 58.Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae) Mycologia. 2003;95:388–398. doi: 10.1080/15572536.2004.11833083. [DOI] [PubMed] [Google Scholar]
- 59.Petrini O, Sieber TN, Toti L, Viret O. Ecology, metabolite production and substrate utilization in endophytic fungi. Nat. Toxin. 1992;1:185–196. doi: 10.1002/nt.2620010306. [DOI] [PubMed] [Google Scholar]
- 60.Schulz B, Guske S, Dammann U, Boyle C. Endophyte–host interaction II. Defining symbiosis of the endophyte–host interaction. Symbiosis. 1998;25:213–227. [Google Scholar]
- 61.Ainsworth, G. C., Sparrow, F. K. & Sussman, A. S. The fungi: an advanced treatise, taxonomic review with keys. Vol. IV A, Academic Press, New York, USA (1973).
- 62.Barnett, H. L. & Hunter, B. B. Illustrated genera of imperfect fungi, 4th edn. The American Phytopathological Society, St. Paul (1998).
- 63.White, T. F., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR Protocol:A GuideTo Methods And Applications (eds Innis, M. A., Gelfand, D. H., Sninsky, F. S. & White, T. T.) Ch. 38, 315–322 (Academic Press, San Diego, USA, 1990).
- 64.Sim JH, Khoo CH, Lee LH, Cheah YK. Molecular diversity of fungal endophytes isolated from Garcinia mangostana and Garcinia parvifolia. J. Microbiol. Biotechnol. 2010;20(4):651–658. doi: 10.4014/jmb.0909.09030. [DOI] [PubMed] [Google Scholar]
- 65.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hata K, Futai K. Endophytic fungi associated healthy Pine needle infested by Pine needle gall midge Thecodiplosis japonensis. Can. J. Bot. 1995;73:384–390. doi: 10.1139/b95-040. [DOI] [Google Scholar]
- 67.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2009).


