Abstract
Bacterial populations exhibit a range of metabolic states influenced by their environment, intra- and interspecies interactions. The identification of bacterial metabolic states and transitions between them in their native environment promises to elucidate community behavior and stochastic processes, such as antibiotic resistance acquisition. In this work, we employ two-photon fluorescence lifetime imaging microscopy (FLIM) to create a metabolic fingerprint of individual bacteria and populations. FLIM of autofluorescent reduced nicotinamide adenine dinucleotide (phosphate), NAD(P)H, has been previously exploited for label-free metabolic imaging of mammalian cells. However, NAD(P)H FLIM has not been established as a metabolic proxy in bacteria. Applying the phasor approach, we create FLIM-phasor maps of Escherichia coli, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus epidermidis at the single cell and population levels. The bacterial phasor is sensitive to environmental conditions such as antibiotic exposure and growth phase, suggesting that observed shifts in the phasor are representative of metabolic changes within the cells. The FLIM-phasor approach represents a powerful, non-invasive imaging technique to study bacterial metabolism in situ and could provide unique insights into bacterial community behavior, pathology and antibiotic resistance with sub-cellular resolution.
Introduction
Bacteria are found in diverse environments, and their ability to modulate metabolic processes in response to adverse conditions gives them unique survival advantages. In the natural environment, multiple species interact in complex niches and exhibit a range of community behavior1–4. In the medical environment, these metabolic changes make bacterial infections difficult to eliminate. Treatment of bacterial infections are often hindered by emergence of multi-drug resistant phenotypes5, 6. A major source of antibiotic resistance and tolerance are slow growing ‘persister’ phenotypes, which are often associated with biofilm formation and chronic infections7–9. Mapping the metabolic activity of bacteria within natural communities and medical infections, therefore, can provide insights into the role of metabolism in determining bacterial community behavior.
Fluorescence spectroscopy is a non-invasive technique which has been extensively used for metabolic imaging in mammalian cells. Fluorescence lifetime imaging microscopy (FLIM), in particular, is a powerful label-free method to probe the local environment and molecular conformation of endogenous fluorophores10. The FLIM signature of an autofluorescent metabolic coenzyme, reduced nicotinamide adenine dinucleotide (phosphate) (NAD(P)H), has been employed as an endogenous biomarker for metabolic activity of mammalian cells in cancer biology and the detection of stem cell differentiation11–17. NAD(P)H in its free state has a significantly shorter fluorescence lifetime (0.4 ns), due to self-quenching, than its protein bound state and is thus easily discernable by FLIM18. FLIM measurements allow mapping of NAD(P)H lifetimes with sub-cellular resolution. The relationship between the NAD(P)H fluorescence lifetime and metabolic activity, however, has not been established in bacteria.
As in the case of mammalian cells, bacteria produce many endogenous fluorescent molecules with distinct spectral characteristics19, 20, some unique to specific bacteria21, 22, rendering them as promising probes for identification and characterization. Fluorescence spectroscopy of some intrinsic fluorophores has been previously explored in bacteria for the detection, differentiation, and characterization of various species23–27. The relationship between cytosolic concentrations of NAD(P)H in bacteria and their metabolic activity has been previously investigated28–32, and differences in the ratio of NAD+ and NADH are correlated with different metabolic states under aerobic and anaerobic conditions, dissolved oxygen tension, and growth phase33–35. Fluorescence lifetime measurements of bacteria have been previously reported in attempts to detect and differentiate bacterial species34, 36, 37. More recently, NAD(P)H fluorescence lifetime measurements were obtained from bacteria infecting mammalian cells, though no relationship to metabolic activity was determined38.
A previously established phasor approach to FLIM is a fit-free technique which does not require a priori knowledge of the fluorescent species in the sample39, 40. In brief, lifetime decay information from each pixel of the image is analyzed via Fourier transform to obtain the corresponding phasor position and the resulting 2-D histogram is plotted as a distribution on the phasor plot with coordinates (g,s). The g coordinate of the phasor plot extends between 0 and 1 while the s coordinate has values between 0 and 0.5. All single exponential lifetime decays fall on the ‘universal circle’ defined as a semi-circle of radius 0.5 between points (0,0) and (1,0) on the phasor plot. Phasors corresponding to a fluorescence lifetime value of 0 will fall on point (1,0) while longer lifetime signals will shift towards point (0,0) with increasing lifetime. The phasor of a mixture of two molecular species (each with single exponential lifetimes) will lie inside the universal circle on the line joining the phasor position of the two pure species, depending on the fractional contribution of each component. Hence, the position of the FLIM-phasor is directly related to the ratio of free to enzyme-bound NAD(P)H41, 42. A ‘right shift’ of the phasor indicates a larger free to bound ratio while a decrease in the ratio is indicated by a ‘left shift’ to coordinates corresponding to longer lifetimes. Lifetimes within the phasor distribution from a sample can be mapped back onto the acquired image to form the corresponding FLIM map. NAD(P)H FLIM-phasor has been successfully applied to study metabolism and oxidative stress in tumors16, 17, 43, cardiomyocytes16, metabolic fingerprinting of macrophages44, stem cell differentiation15, and in Lactobacillus acidophilus cultures45.
In this work, we demonstrate the potential of the NAD(P)H FLIM-phasor technique to differentiate metabolic states in live bacterial populations at the single-cell level. Phasor fingerprints were generated for immobilized planktonic cell populations of Escherichia coli, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus epidermidis. The phasor positions of the cells, corresponding to the free to bound NAD(P)H ratio, was found to depend on species, growth phase, and history of exposure to antibiotics and nutrient media. The phasor distribution shifted to shorter (free) NAD(P)H fluorescence lifetimes with exposure to antibiotics, and recovered to longer (bound) lifetimes when bacteriostatic antibiotics were administered and washed away. Moreover, the diffraction-limited FLIM maps achieved here display differences in the spread of phasor positions of individual cells within the same population under different environmental treatments. As a function of growth phase, the NAD(P)H lifetime phasor distribution changed non-monotonically. This suggests that the FLIM-phasor position captures information about metabolic activity not simply inferred by cell density measurements. These results demonstrate that FLIM of NAD(P)H in live bacterial cultures can be directly related to changes in metabolic activity, and that this method can provide distinct and nuanced information about metabolic states of bacteria in situ as compared to existing fluorescence spectroscopy and metabolomic techniques.
Results
Fluorescence lifetime phasor fingerprint of different bacterial species
Two-photon FLIM of five clinically relevant species of bacteria, E. coli, S. Typhimurium, P. aeruginosa, B. subtilis and S. epidermidis, was performed on planktonic cells immobilized in agarose. Each species was imaged at a mid-exponential phase of growth to generate a phasor fingerprint (Fig. 1a and Supplementary S1). A comparative analysis of all the phasor coordinates of each bacterial cell, represented by a single data point, within each species population (n = 10) shows variations along both g and s axes of the phasor plot (Fig. 1b). The bacterial phasor distribution displays greater variation along the g axis (between 0.26 to 0.41) than the s axis on the phasor plot (Fig. 1c). Supplementary Fig. S2 shows the fluorescence emission spectra of the five bacterial species (n = 10) excited at 740 nm.
Figure 1.
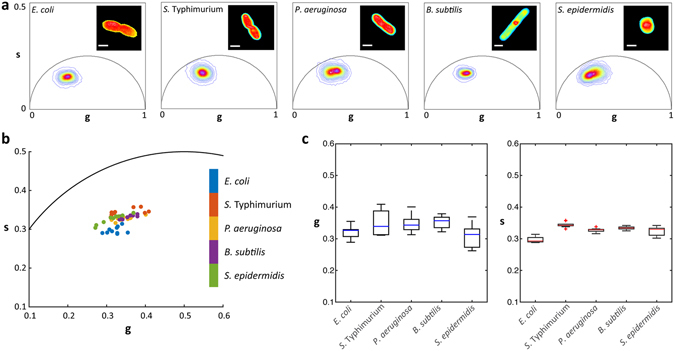
Bacterial phasor fingerprints. (a) Phasor distribution of E. coli, S. Typhimurium, P. aeruginosa, B. subtilis and S. epidermidis. Inset panels are representative fluorescence intensity images of each bacterium. (b) Bacterial phasor fingerprints of individual cells within each bacterial population. (c) Distribution of g and s within each bacterial population. Scale bars are 1 µm.
Response of E. coli phasor to antibiotic exposure
Bacteria encounter various stresses in their natural environments which elicit specific and highly regulated responses via changes in their metabolism7, 9, 46, 47. To determine the relationship between the FLIM phasor and metabolic activity, we probed the phasor response of E. coli to antibiotic treatment. Agarose embedded cells were treated with nalidixic acid or ampicillin, a bacteriostatic antibiotic and bactericidal antibiotic, respectively, then NAD(P)H FLIM was performed. The phasor position of E. coli cells treated with nalidixic acid shifted towards longer fluorescence lifetimes with increasing concentration (light to darker shades of brown dots), as compared to the untreated control population (cyan dots) (Fig. 2a). The g distribution exhibited a sharply increasing mean value at the lowest nalidixic acid concentration, while the mean s values were weakly sensitive to nalidixic acid concentration (Fig. 2b). E. coli cells similarly exposed to increasing concentrations of a bactericidal antibiotic, ampicillin, also exhibited corresponding phasor position shifts to larger g values, while s values remained relatively constant with ampicillin concentration (Fig. 2c and d) when compared with cells in control conditions. These shifts of the phasor position of cells treated with antibiotics corresponds to a concentration-dependent increase in the fraction of dead cells when stained in a live/dead assay (Supplementary Figs S3 and S4). Exposure of E. coli cells to the same volume of water for the same amount of time did not cause a shift and the position of the phasor was maintained at the same position as the control population (Supplementary Fig. S9).
Figure 2.
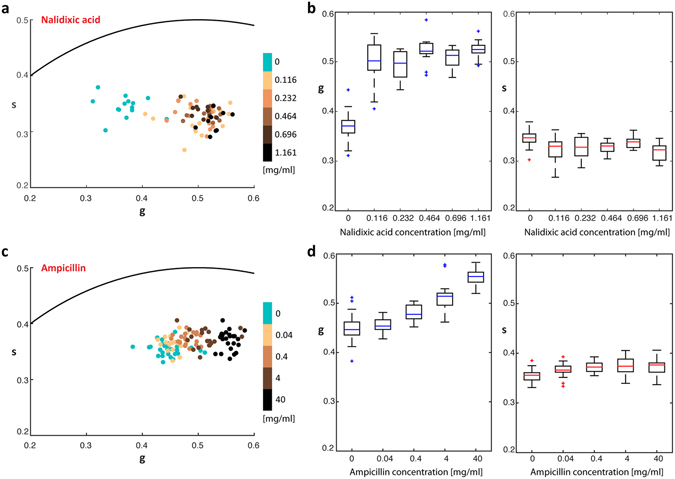
Bacterial phasor response to bacteriostatic and bactericidal antibiotic treatment. (a) Bacterial phasor of E. coli control culture (cyan) and culture treated with increasing concentrations of a bacteriostatic antibiotic, nalidixic acid (light to dark brown). (b) Distribution of g and s within populations of cells at each concentration of nalidixic acid. (c) Bacterial phasor of E. coli control culture (cyan) and culture treated with increasing concentrations of bactericidal antibiotic, ampicillin (light to dark brown). (d) Distribution of g and s within populations of cells at each concentration of ampicillin.
To benchmark the FLIM phasor data with an independent metabolic activity measure, we used a resazurin assay to quantify aerobic respiration in planktonic cultures. Resazurin is a redox indicator, sensitive to the presence of oxygen and reducing equivalents, such as NADH. Under reducing conditions, resazurin is reduced to the fluorescent compound, resorufin, and the fluorescence intensity has been linked to bacterial respiration rates48, 49. Exposure of E. coli to the bacteriostatic nalidixic acid resulted in a significant drop in culture respiration, even at the lowest concentration tested in our study (Supplementary Fig. S5). Exposure of E. coli to bactericidal ampicillin, however, resulted in a slight increase in metabolic activity for cells exposed to 0.04 mg/mL ampicillin, which decreased with further increases in ampicillin concentration. The resazurin data are consistent with results and trends in the FLIM phasor data, which show a large shift to higher free to bound NAD(P)H ratios with exposure to 0.116 mg/mL nalidixic acid, and little to no concentration dependence thereafter (Fig. 2b). The FLIM data do not show a shift to lower free to bound NAD(P)H at 0.04 mg/mL ampicillin, but the data is within error of the starting culture and the change indicated by the resazurin assay is small relative to that observed for nalidixic acid exposure (Fig. 2d). The resazurin assay and FLIM data, however, show a trend to a lower respiration rate and higher free to bound NAD(P)H ratio, respectively, with increasing ampicillin concentration, which is distinct from the cell response to nalidixic acid as assessed by both methods.
Recovery of E. coli cells after exposure to antibiotics
To verify whether the bacterial phasor position correlates with global metabolic activity, E. coli cells were allowed to recover after exposure to both antibiotics. Cells were subjected to 0.464 mg/ml nalidixic acid for 30 min and FLIM data was acquired. The cells were then washed and incubated in growth media for 30 min and imaged again. As observed in the previous section, exposure to nalidixic acid shifted the phasor to the right, indicating a higher free to bound NAD(P)H ratio (Fig. 3a and b). When the cells recovered in media, the phasor distribution of bacteria shifted back towards the left, indicating a lower free to bound NAD(P)H ratio. The phasor of the recovered cells shifted to significantly longer lifetimes than the initial population before exposure to nalidixic acid. Further, the phasor distribution of individual cells within the population along the g axis was much narrower than both the initial control and antibiotic-exposed populations (Fig. 3a and c).
Figure 3.
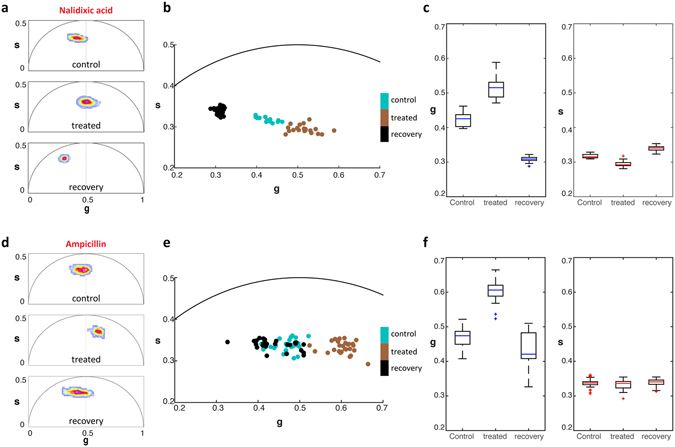
Bacterial phasor response to antibiotic treatment and recovery. (a) Phasor distributions of E. coli (control), E. coli treated with 0.464 mg/ml nalidixic acid for 30 min (treated), and treated E. coli recovered in fresh growth medium for 30 min (recovery). (b) Individual bacterial cell phasor of control, treated, and recovery groups exposed to nalidixic acid and their corresponding g and s distributions (c). (d) Phasor distributions of E. coli (control), E. coli treated with 0.4 mg/ml ampicillin for 30 min (treated) and treated E. coli recovered in fresh growth medium for 30 min (recovery). (e) Individual bacterial cell phasor of control, treated and recovery groups exposed to ampicillin and their corresponding g and s distributions (f).
Ampicillin treated E. coli displayed similar shifts in cell phasors towards shorter lifetimes (Fig. 3d and e). However, recovery of these cells in fresh media showed a limited shift back to smaller g values compared to cells recovered from nalidixic acid exposure (Fig. 3c and f). The number of viable cells in the ampicillin treated samples was also lower by 1.8 times compared to the initial and nalidixic acid treated cells (Supplementary Fig. S6). The phasor distribution of cells in the recovered population was similar to the phasor of the cells in the initial population (Fig. 3f). In both cases, cells exposed to either antibiotic had phasors shifted more significantly along the g axis than along the s axis of the phasor plot in response to treatment (Fig. 3c and f). By comparing these phasor positions, changes in the ratio of free to bound NAD(P)H within E. coli cells were differentiated between initial and antibiotic-exposed cells. Further, the phasor positions and distribution of the recovered populations upon recovery from bacteriostatic and bactericidal antibiotics represent distinguishing metrics captured by FLIM.
Bacterial phasors as a function of culturing time
To track the change in bacterial phasor position at different growth phases, we imaged E. coli, S. Typhimurium, P. aeruginosa and B. subtilis cells from shaking cultures at varied growth times (Fig. 4a and b). The cell density of these bacterial cultures was tracked in parallel using optical density (OD) measurements (Fig. 4c). FLIM data was collected from shaking culture aliquots at regular intervals, starting at 2 h growth time (Supplementary Fig. S7). No cells were found in the agarose-embedded samples at shorter times to acquire FLIM data. Comparing exponential phase cells, at 4 and 6 h, to the 2 h time point, all cultures except P. aeruginosa exhibited a shift in their mean phasor position to the right, i.e. a larger free to bound NAD(P)H ratio (Fig. 4a and b). At 8 h and beyond, the mean phasor position of E. coli, S. Typhimurium, and B. subtilis cells shifted back to smaller values of g and began to oscillate, to differing degrees, along the g axis as cultures reached the stationary phase (Fig. 4b). In E. coli and B. subtilis, these oscillations brought the mean phasor positions to even smaller g values than the cultures sampled at 2 h growth. s positions of the mean phasor were not as sensitive as the g position, but tended towards larger values, also non-monotonically, with culture time. As opposed to the other three species, the mean phasor of P. aeruginosa tended to smaller values in both g and s, and no oscillations were observed in g as a function of growth time.
Figure 4.
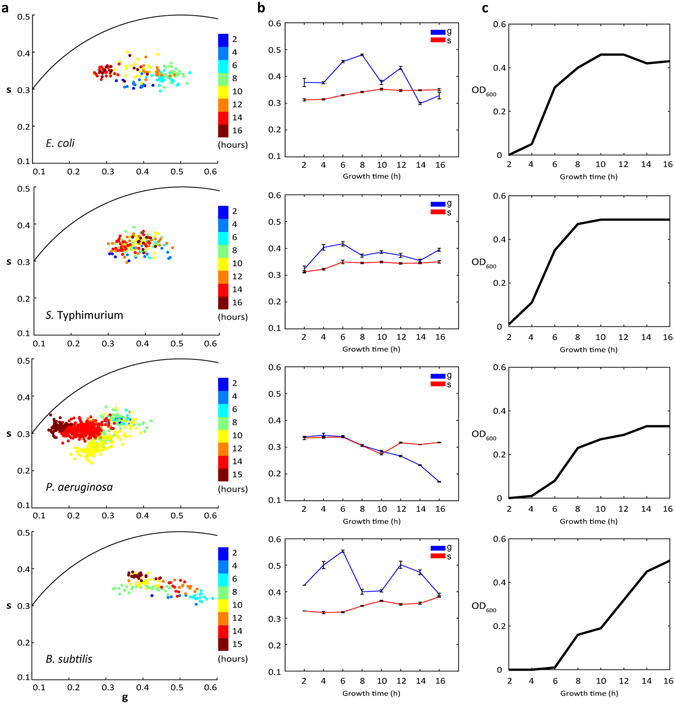
The effect of growth time on the bacterial phasor. (a) Bacterial phasor fingerprint of individual E. coli, S. Typhimurium, P. aeruginosa, and B. subtilis cells from aliquots taken at different shaking culture growth time points. (b) Mean g and s values along the growth curve, and (c) the corresponding optical densities of the shaking cultures.
Growth recovery from extended stationary phase cultures
The FLIM phasors of E. coli cells grown for 15 h, 24 h, 48 h and 307 h (13 days) were compared to those grown for the same amount of time and resuspended in fresh media. Figure 5 shows the cell phasor distribution of each of the growth time groups. We observed an increase in the ratio of free to bound NAD(P)H in the cells resuspended in fresh media when compared to their corresponding spent culture media counterparts at all time points except 13 day old cultures. The phasors of recovered cells remained at about the same position for each growth time point, but the phasors of cells taken directly from the spent culture medium shifted closer to the recovered cell phasor position with growth time. The difference in phasor position between the spent media and recovered cultures decreased with longer growth times. E. coli cells grown for 13 days did not show any change in their phasor position after 2 h recovery in fresh media.
Figure 5.
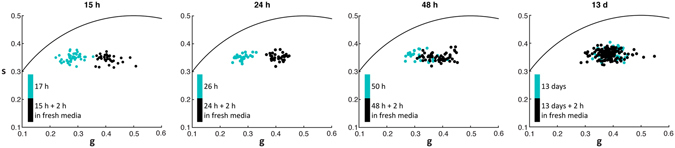
Bacterial phasor recovery after late stationary phase growth. Bacterial phasor of E. coli cells from 15 h, 24 h, 48 h and 13 d cultures recovered for an additional 2 h in spent medium from the same culture (cyan) or fresh growth medium (black).
Discussion
In this work, we employ label-free NAD(P)H FLIM to characterize fluorescence lifetime fingerprint of bacteria with cellular resolution. The phasor approach to FLIM proves to be a powerful tool to differentiate metabolic states of bacteria via the relative quantification of free to bound NAD(P)H ratios. This ratio is expected to be related to metabolic activity, as NAD(P)+ reducing enzymes are central to catabolic pathways such as glycolysis, the citric acid cycle and the pentose phosphate pathway, which are each active in bacteria under different growth conditions. Similarly, enzymes such as NADH dehydrogenases and NAD(P)H oxidases play a central role in various anabolic pathways and show a high level of specificity in binding to NADPH or NADH50–52.
NAD(P)H FLIM of E. coli, P. aeruginosa, B. subtilis, S. Typhimurium and S. epidermidis was used to generate the bacterial species fingerprint phasors as well as those for the antibiotic experiments. We observed small variations between phasors of control populations of E. coli even when grown under the same conditions and harvested at the same time and OD, perhaps due to normal metabolic variability (Supplementary Fig. S8). The phasor fingerprint exhibits a distribution of values of individual cells within a population of same species (Fig. 1b and c). These differences suggest a distribution of metabolic activity within and between species populations at the same time of growth. The observed diversity of phasor positions may be a result of varying rates of NAD(P)+/NAD(P)H reduction/oxidation and shifts in NAD(P)H utilization due to catabolic and anabolic adaptations32, 33, 53, 54.
The shift of the NAD(P)H phasor position towards shorter lifetimes in response to antibiotic exposure suggests a decrease in enzyme binding of NAD(P)H, indicated by a larger free to bound ratio. In case of bacteriostatic antibiotics this shift tracks with lower respiration activity indicated by the resazurin assay (Supplementary Fig. S5). These results are consistent with previously reported data46, 55 which suggest that bacteriostatic antibiotics induce lower oxygen consumption rates (OCR) in cultures by arresting respiration in cells. The increase in respiration activity at low ampicillin concentrations, on the other hand, is consistent with findings that OCR increases upon exposure to bactericidal antibiotics46, 55. While the effects of higher concentrations of ampicillin on respiration have not been studied, to our knowledge, the resazurin and NAD(P)H FLIM data show a shift to a more reducing environment and an increase in the free to bound NAD(P)H ratio in the phasor plots, respectively.
In the event of cell death, the metabolic state is irreversible; however bacterial cells are known to recover from a state of static growth and dormancy induced by bacteriostatic compounds9, 46, 47, 56. The observed return of the mean phasor position to the left of the plot when nalidixic acid was washed away from the E. coli cells (Fig. 3a and b) is consistent with these previous results. In addition to the shift in the mean phasor, the distribution of single-cell phasors about the mean in the recovered population exposed to nalidixic acid was much narrower than the control and treated populations (Fig. 3c). In the context of the above results, the decrease in phasor variance indicates a recovery to a relatively homogeneous metabolic state throughout the population, as compared to the initial and nalidixic acid-treated populations, which exhibit a greater spread in NAD(P)H lifetimes.
In contrast to the recovery results from nalidixic acid, the shift of the phasor position of cells treated with ampicillin and recovered in fresh medium was less pronounced and their distribution was much broader (Fig. 3d). This phasor data was acquired from a population of cells that did not lyse as a result of the ampicillin treatment, so the extent of phasor recovery is of a biased population and characteristic only of cells that survived the treatment. Indeed, the density of cells that survived this treatment was 19.1 times lower than the nalidixic acid treatment (Supplementary Fig. S6). In addition, the phasor distribution of the recovered cell population was much broader than that of cells recovered from nalidixic acid treatment, indicating more metabolic heterogeneity. The response of bacteria to high-stress environments depends on a range of factors, such as the rate of cell division and initial metabolic states9, 57–60. Such factors can direct the fate of part of the population towards persister or dormant states and the rest towards death. Exposure to bactericidal antibiotics also results in generation of reactive oxygen species61, 62 and broad, and ultimately toxic, changes to the activity of central metabolic pathways in E. coli 55, 62. Consequently, the bactericidal effects of ampicillin, in contrast to nalidixic acid, may result in a population with the diversity of metabolic states observed in the recovered populations.
The metabolism of bacterial populations vary drastically over time, resulting in a collection of different metabolic states that cannot be captured by optical density measurements alone63–65. In the case of aerobically grown bacteria, these variations depend on the type of nutrient source, temperature of growth, and rate of aeration, among others. Moreover, the bacterial growth curve extends beyond the conventional lag, exponential and stationary phases of growth66–68. Many interesting physiological phenomena occur during late stationary phase growth, such as the emergence of persister cells and growth advantage in stationary phase (GASP) phenotypes, changes in gene expression and cell morphology, and programmed cell death60, 68–71. Most importantly each bacterium within the population may experience different metabolic and growth rates compared to neighboring cells, yet this information cannot be obtained by cell density measurement or CFU counts.
We employed NAD(P)H FLIM phasor to fingerprint the metabolic states of 4 bacterial species at different stages of growth (Fig. 4a). The earliest time point, 2 h, is already near the end of the lag phase for these cultures. The mean phasor positions of E. coli, S. Typhimurium, and B. subtilis all begin at relatively small g values. In comparison with observed phasors from the antibiotic exposure and recover experiments, these smaller g values suggest a greater metabolic activity. As cells enter their exponential phase, their phasors shift right to larger g values, consistent with a ramp down of metabolic rates as previously determined via indirect methods64, 65. At 8–10 h growth, this phasor trend reverses for these three cultures, suggesting an additional metabolic transition commensurate with the change in growth rate as cultures enter the stationary phase. In the stationary phase, their phasor positions shift back and forth in g in a manner seemingly unrelated to the cell density (Figs 4b and Supplementary S10). Bacterial cultures display an oscillatory behavior of cell density in the culture with rise and fall during the stationary phase due to GASP phenotypes69, 71, 72, but these typically occur at late stationary phase growth, and the observed phasor shifts are not directly correlated with cell density. Instead, these results indicate that the NAD(P)H phasor is sensitive to metabolic changes within the cells not captured by cell density or CFU analyses.
To look at metabolic adaptations in the stationary phase cultures, FLIM data was obtained from E. coli cultures grown for 15 h, 24 h, 48 h, and 13 d (Fig. 5). Interestingly, cells grown for an additional 2 h in the spent culture medium from 15, 24, and 48 h cultures had phasors at longer lifetimes than the early and mid-exponential phase cells in Figures 1, 2, 3 and 4 (g ~ 0.4). The NAD(P)H lifetimes in these populations were also longer than those of cells from those same cultures that recovered for an additional 2 h in fresh growth medium, which were themselves consistent with early exponential phase E. coli cells (Fig. 4), suggesting a growth recovery. The difference in mean phasor position between cells recovered in fresh and spent medium, however, shrinks with increasing culture time at 48 h and 13 d, indicating a diminished capacity of these bacteria to recover (Supplementary Fig. S11). Indeed, cells from the 13 d culture did not show any recovery, suggesting that most cells are dead or in a phenotypic state with an extended lag time to exponential growth. Future experiments will focus on FLIM and independent metabolic rate measurement in late stationary phase cultures to draw quantitative correlations between the two.
In conclusion, we have demonstrated a label-free, two-photon FLIM method for tracking changes in bacterial metabolism. Our results represent the first NAD(P)H FLIM phasor fingerprinting of various bacterial species, and they indicate that the NAD(P)H fluorescence lifetime, as captured in the phasor position, is sensitive to changes in metabolic states within cell populations. The NAD(P)H phasor of cells exposed to antibiotics exhibited the expected shifts due to diminished metabolic capacity, as well as detecting the recovery of cells resistant or tolerant to ampicillin exposure. The distribution of cell phasors within a population also changes as a function of treatment conditions, providing unique insight into single-cell and community behavior. Lastly, NAD(P)H FLIM of bacteria from planktonic cultures exhibited changes in the phasors that did not track with cell density, indicating that this technique can provide additional information about cell metabolism beyond what is inferred from conventional growth rate measurements. This work demonstrates the power of NAD(P)H FLIM to track metabolic states of individual bacteria in situ, and that the bacterial phasor represents a unique and complimentary set of data to conventional metabolism and growth characterization results.
Material and Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table S1. All strains were revived from frozen stocks by streaking on lysogeny broth (LB) agar (1.5%) plates. Shaking cultures of bacterial strains were grown in 2 ml LB at 37 °C for 5 h unless stated otherwise. For imaging, the samples were prepared by mixing LB shaking cultures with 1% agarose in a 3:7 ratio. 100 μl of the resulting solution was cast on a glass coverslip by spin coating at 500 rpm for 10 seconds.
Exposure of agarose-embedded bacteria to antibiotics
Solutions of nalidixic acid or ampicillin (Sigma-Aldrich) were prepared at the indicated concentrations in sterile ultrapure water, well above the minimum inhibitory concentration in E. coli. The agarose-embedded bacterial samples were incubated with a 200 μl of each concentration for 30 min, then washed with sterile ultrapure water and imaged.
Recovery of agarose-embedded bacteria from antibiotic exposure
Agarose-embedded bacterial samples on glass coverslip were incubated in 0.464 mg/ml nalidixic acid or 0.4 mg/ml ampicillin for 30 min. The samples were washed twice with ultrapure water, then incubated in LB media for 30 min and imaged.
Measurement of cellular respiration
E. coli cells were grown shaking in LB media at 37 °C for 5 h. 300 µl of E. coli cells in LB media was diluted in 700 µl of PBS with different final concentrations of nalidixic acid and ampicillin. Each of these solutions were done in triplicates. The E. coli cells were incubated at 22 °C for 30 min. 20 µl of 0.15 mg/ml resazurin was injected into 100 µl of each of these solutions, and sterile control solutions, in a 96 well plate and the plates were incubated at 37 °C for 1 h. Fluorescence intensity from the wells was recorded using a plate reader (BioTek Synergy H1) with a 560 nm excitation/590 nm emission filter set.
Growth curve sample preparation
For each growth curve, 20 ml LB media was inoculated with bacteria to an optical density at 600 nm (OD) of 0.02 and aliquoted into 1 ml cultures in 20 ml glass culture tubes with loose-fitting metal caps, which were kept shaking at 200 rpm and 37 °C. One culture aliquot was taken out for each bacterium every 2 h, and samples for FLIM measurements were prepared as described above.
The OD of 1 ml bacterial shaking cultures was measured every 2 h to generate a growth curve. The OD was measure at 600 nm using a Biowave CO8000 cell density meter (Biochrom, Holliston, MA).
Growth curve recovery sample preparation
For growth curve recovery 1 ml cultures were grown for 15 h, 24 h, 48 h and 13 d in duplicates. 100 μl from one of the duplicate samples was diluted into 900 μl of fresh LB media and shaken for 2 h at 200 rpm and 37 °C. The other set of cultures from each time point was allowed to grow in the same spent culture media for 2 more hours. After 2 h both cultures were taken out of the incubator and samples were prepared for FLIM imaging as described above.
FLIM data acquisition
FLIM was performed on a Zeiss LSM 710 microscope (Carl Zeiss, Jena, Germany) coupled to an 80 MHz multiphoton excitation laser source, Titanium:Sapphire MaiTai laser (Spectra-Physics, Mountain View, CA) with excitation at 740 nm using a 60X, 1.2 N.A. oil immersion objective, (Carl Zeiss, Oberkochen, Germany). The image scan speed was 25.21 µs/pixel with an image size of 256 × 256 pixels. Excitation from emission signal were separated at 690 nm followed by bandpass emission filter 460/80 nm (Semrock, Rochester, NY). Photomultiplier tube (H7422P-40, Hamamatsu, Japan) was used as the microscope external detector port photo-sensor unit. A320 FastFLIM FLIMbox (ISS, Champaign, IL) was employed to acquire frequency domain FLIM data. SimFCS software (LFD, Irvine) was used for FLIM data acquisition. For calibrating the FLIM system, Rhodamine 110 with known lifetime of 4 ns was measured for every experiment. The (g, s) coordinate system used to indicate phasor cursor coordinates in this article used the first harmonic phasor plots at 80 MHz (repetition rate of the laser).
Spectral data acquisition
NAD(P)H in the bacterial samples was excited with two-photon excitation at 740 nm. Fluorescence spectra were collected on a Zeiss LSM 710 microscope spectral detector consisting of 32 channels, between 416–728 nm, each with 9.7 nm bandwidth. For image acquisition, the pixel frame size was set to 256 × 256 and the pixel dwell time was 177 μs/pixel. Spectral data were acquired from the same sample on which FLIM was performed.
Data analysis
FLIM data was analyzed using the SimFCS software developed at the Laboratory for Fluorescence Dynamics (LFD, UC Irvine). To create the bacterial phasors, the average g and s value of phasor distribution from individual bacterial cell were calculated and plotted as a scatter plot. Thus, each point of the bacterial phasor scatter plot represents a single bacterium. Image segmentation, scatter plot and box plots calculations were performed on MATLAB. For Supplementary Fig. S2, the acquired fluorescence spectra were smoothed and normalized to maximum intensity.
Live/dead assay
Agarose-embedded samples were exposed to different antibiotic concentrations for 30 min and stained with Syto 9 and propidium iodide for 20 min. The samples were then washed with ultrapure water and imaged using a Zeiss LSM780 inverted confocal microscope (Carl Zeiss, Jena, Germany) with 63X, NA 1.2 water immersion objective. Dichroic beam splitters were used to reflect laser lines at 488 and 561 nm. Both color channels were imaged using alternating line averaging to avoid blurring of the image due to drifting. These adjustments reduced fluorescence cross-talk to undetectable level in two-color imaging experiments. Confocal images were obtained with a pinhole size of 1 airy unit.
The fraction of dead cells was quantified using the “select objects” from the intensity images from each channel using the Volocity software package after deconvoluting the images by applying the point spread function to each channel.
Colony forming unit (CFU) analysis
1 ml shaking cultures of E. coli were grown for 5 h and exposed to 0.464 mg/ml nalidixic acid or 0.4 mg/ml ampicillin for 30 min. The cells were centrifuged at 2000 × g, resuspended in LB media and incubated at 37 °C and shaking at 250 rpm for 30 min. The control was not exposed to antibiotics and each condition was done in triplicate. Serial dilutions of the shaking cultures were plated to count CFUs to measure recovery from antibiotic exposure.
Electronic supplementary material
Acknowledgements
A.B. acknowledges fellowship support from the Schlumberger Foundation and research support from a 3 M Non-Tenured Faculty award to A.I.H. R.D. and E.G. would like to acknowledge NIH grants P50GM076516 and P41GM103540. Research reported in this publication was supported by the Optical Biology Core Facility at UC, Irvine and by the National Cancer Institute of the National Institutes of Health under Award Number P30CA062203. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
A.B. and A.I.H. designed the experiments. A.B. and R.D. conducted the experiments, and R.D. analyzed the FLIM data. E.G. developed the SimFCS software for the acquisition and analysis of the FLIM data. A.B., R.D., A.I.H. and E.G. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Arunima Bhattacharjee and Rupsa Datta contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04032-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stasulli NM, Shank EA, Gibbs K. Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. FEMS Microbiol. Rev. 2016;40:807–813. doi: 10.1093/femsre/fuw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaltenpoth M, Strupat K, Svatoš A. Linking metabolite production to taxonomic identity in environmental samples by (MA)LDI-FISH. ISME J. 2016;10:527–531. doi: 10.1038/ismej.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louie KB, et al. ‘Replica-Extraction-Transfer’ Nanostructure-Initiator Mass Spectrometry Imaging of Acoustically Printed Bacteria. Anal. Chem. 2013;85:10856–10862. doi: 10.1021/ac402240q. [DOI] [PubMed] [Google Scholar]
- 4.Lanni EJ, et al. MALDI-guided SIMS: Multiscale Imaging of Metabolites in Bacterial Biofilms. Anal. Chem. 2014;86:9139–9145. doi: 10.1021/ac5020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 2007;3:549–556. doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 8.Shah D, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Berezin MY, Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird DK, et al. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 12.Skala MC, et al. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J. Biomed. Opt. 2007;12:24014. doi: 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh AJ, et al. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 2013;73:6164–6174. doi: 10.1158/0008-5472.CAN-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn KP, et al. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci. Rep. 2013;3:3432. doi: 10.1038/srep03432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stringari C, Sierra R, Donovan PJ, Gratton E. Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy. J. Biomed. Opt. 2012;17:46012. doi: 10.1117/1.JBO.17.4.046012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta R, Heylman C, George SC, Gratton E. Label-free imaging of metabolism and oxidative stress in human induced pluripotent stem cell-derived cardiomyocytes. Biomed. Opt. Express. 2016;7:1690–1701. doi: 10.1364/BOE.7.001690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobrino A, et al. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakowicz JR, Szmacinski H, Nowaczyk K, Johnson ML. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. 1992;89:1271–1275. doi: 10.1073/pnas.89.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ammor MS. Recent Advances in the Use of Intrinsic Fluorescence for Bacterial Identification and Characterization. J. Fluoresc. 2007;17:455–459. doi: 10.1007/s10895-007-0180-6. [DOI] [PubMed] [Google Scholar]
- 20.Estes C, et al. Reagentless detection of microorganisms by intrinsic fluorescence. Biosens. Bioelectron. 2003;18:511–519. doi: 10.1016/S0956-5663(03)00008-3. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan NL, Tzeranis DS, Wang Y, So PTC, Newman D. Quantifying the Dynamics of Bacterial Secondary Metabolites by Spectral Multiphoton Microscopy. ACS Chem. Biol. 2011;6:893–899. doi: 10.1021/cb200094w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alimova A, et al. Native fluorescence and excitation spectroscopic changes in Bacillus subtilis and Staphylococcus aureus bacteria subjected to conditions of starvation. Appl. Opt. 2003;42:4080–4087. doi: 10.1364/AO.42.004080. [DOI] [PubMed] [Google Scholar]
- 23.Dartnell LR, Roberts TA, Moore G, Ward JM, Muller J-P. Fluorescence Characterization of Clinically-Important Bacteria. PLOS ONE. 2013;8:e75270. doi: 10.1371/journal.pone.0075270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ammor S, Yaakoubi K, Chevallier I, Dufour E. Identification by fluorescence spectroscopy of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausages. J. Microbiol. Methods. 2004;59:271–281. doi: 10.1016/j.mimet.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Leblanc L, Dufour É. Monitoring the identity of bacteria using their intrinsic fluorescence. FEMS Microbiol. Lett. 2002;211:147–153. doi: 10.1111/j.1574-6968.2002.tb11217.x. [DOI] [PubMed] [Google Scholar]
- 26.Giana HE, Silveira L, Zângaro RA, Pacheco MTT. Rapid Identification of Bacterial Species by Fluorescence Spectroscopy and Classification Through Principal Components Analysis. J. Fluoresc. 2003;13:489–493. doi: 10.1023/B:JOFL.0000008059.74052.3c. [DOI] [Google Scholar]
- 27.Bailey, K. F. R. J. Metabolic Pathway Rates and Culture Fluorescence in Batch Fermentations of Clostridium Acetobutylicum. 3, 153–167 (1987).
- 28.Harrison DE, Loveless JE. The effect of growth conditions on respiratory activity and growth efficiency in facultative anaerobes grown in chemostat culture. J. Gen. Microbiol. 1971;68:35–43. doi: 10.1099/00221287-68-1-35. [DOI] [PubMed] [Google Scholar]
- 29.Armiger WB, Forro JF, Montalvo LM, Lee JF, Zabriskie DW. The Interpretation of on-Line Process Measurements of Intracellular Nadh in Fermentation Processes. Chem. Eng. Commun. 1986;45:197–206. doi: 10.1080/00986448608911383. [DOI] [Google Scholar]
- 30.Rao G, Mutharasan R. NADH levels and solventogenesis in Clostridium acetobutylicum: New insights through culture fluorescence. Appl. Microbiol. Biotechnol. 1989;30:59–66. [Google Scholar]
- 31.Wos M, Pollard P. Sensitive and meaningful measures of bacterial metabolic activity using NADH fluorescence. Water Res. 2006;40:2084–2092. doi: 10.1016/j.watres.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Harrison DE. Undamped oscillations of pyridine nucleotide and oxygen tension in chemostat cultures of Klebsiella aerogenes. J. Cell Biol. 1970;45:514–521. doi: 10.1083/jcb.45.3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimpenny JWT, Firth A. Levels of Nicotinamide Adenine Dinucleotide and Reduced Nicotinamide Adenine Dinucleotide in Facultative Bacteria and the Effect of Oxygen. J. Bacteriol. 1972;111:24–32. doi: 10.1128/jb.111.1.24-32.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalterio RA, et al. The Steady-State and Decay Characteristics of Primary Fluorescence from Live Bacteria. Appl. Spectrosc. 1987;41:234–241. doi: 10.1366/000370287774986804. [DOI] [Google Scholar]
- 35.Wos ML, Pollard PC. Cellular nicotinamide adenine dinucleotide (NADH) as an indicator of bacterial metabolic activity dynamics in activated sludge. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2009;60:783–791. doi: 10.2166/wst.2009.393. [DOI] [PubMed] [Google Scholar]
- 36.Brahma SK, et al. The Rapid Identification of Bacteria Using Time-Resolved Fluorescence and Fluorescence Excitation Spectral Methods. Appl. Spectrosc. 1985;39:869–872. doi: 10.1366/0003702854249916. [DOI] [Google Scholar]
- 37.Awad, F. et al. Optical Fiber-Based Steady State and Fluorescence Lifetime Spectroscopy for Rapid Identification and Classification of Bacterial Pathogens Directly from Colonies on Agar Plates, Int. Sch. Res. Not. Int. Sch. Res. Not. 2014, e430412 (2014). [DOI] [PMC free article] [PubMed]
- 38.Szaszák M, et al. Fluorescence Lifetime Imaging Unravels C. trachomatis Metabolism and Its Crosstalk with the Host Cell. PLOS Pathog. 2011;7:e1002108. doi: 10.1371/journal.ppat.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Digman, M. & Gratton, E. In Fluorescence Lifetime Spectroscopy and Imaging 235–248 (CRC Press, 2014).
- 40.Stringari C, et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta R, Alfonso-García A, Cinco R, Gratton E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Sci. Rep. 2015;5:9848. doi: 10.1038/srep09848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar-Arnal L, et al. Spatial dynamics of SIRT1 and the subnuclear distribution of NADH species. Proc. Natl. Acad. Sci. 2016;113:12715–12720. doi: 10.1073/pnas.1609227113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stringari C, Pate KT, Edwards RA, Waterman ML, Gratton E. Metabolic Imaging of Colon Cancer Tumors In Vivo by Phasor Fluorescence Lifetime Microscopy of NADH. Biophys. J. 2013;104:342a–343a. doi: 10.1016/j.bpj.2012.11.1902. [DOI] [Google Scholar]
- 44.Alfonso-García A, et al. Label-free identification of macrophage phenotype by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2016;21:46005. doi: 10.1117/1.JBO.21.4.046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torno K, et al. Real-time Analysis of Metabolic Activity Within Lactobacillus acidophilus by Phasor Fluorescence Lifetime Imaging Microscopy of NADH. Curr. Microbiol. 2012;66:365–367. doi: 10.1007/s00284-012-0285-2. [DOI] [PubMed] [Google Scholar]
- 46.Lobritz MA, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. dks196 doi:10.1093/jac/dks196 (2012). [DOI] [PubMed]
- 48.Zotta, T. et al. Assessment of Aerobic and Respiratory Growth in the Lactobacillus casei Group. PLoS ONE9 (2014). [DOI] [PMC free article] [PubMed]
- 49.González-Pinzón R, Haggerty R, Myrold DD. Measuring aerobic respiration in stream ecosystems using the resazurin-resorufin system. J. Geophys. Res. Biogeosciences. 2012;117:G00N06. doi: 10.1029/2012JG001965. [DOI] [Google Scholar]
- 50.Spaans, S. K., Weusthuis, R. A., van der Oost, J. & Kengen, S. W. M. NADPH-generating systems in bacteria and archaea. Front. Microbiol. 6 (2015). [DOI] [PMC free article] [PubMed]
- 51.Abrahams GL, Abratt VR. The NADH-dependent glutamate dehydrogenase enzyme of Bacteroides fragilis Bf1 is induced by peptides in the growth medium. Microbiol. Read. Engl. 1998;144(Pt 6):1659–1667. doi: 10.1099/00221287-144-6-1659. [DOI] [PubMed] [Google Scholar]
- 52.Heikal A, et al. Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014;91:950–964. doi: 10.1111/mmi.12507. [DOI] [PubMed] [Google Scholar]
- 53.London J, Knight M. Concentrations of Nicotinamide Nucleotide Coenzymes in Micro-Organisms. Microbiology. 1966;44:241–254. doi: 10.1099/00221287-44-2-241. [DOI] [PubMed] [Google Scholar]
- 54.Graef MR, de Alexeeva S, Snoep JL, de Mattos MJT. The Steady-State Internal Redox State (NADH/NAD) Reflects the External Redox State and Is Correlated with Catabolic Adaptation in Escherichia coli. J. Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 57.Torres-Barceló C, Kojadinovic M, Moxon R, MacLean RC. The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic. Proc R Soc B. 2015;282:20150885. doi: 10.1098/rspb.2015.0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meouche IE, Siu Y, Dunlop MJ. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci. Rep. 2016;6:19538. doi: 10.1038/srep19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 60.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial Persistence as a Phenotypic Switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 61.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- 62.Belenky P, et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rolfe MD, et al. Lag Phase Is a Distinct Growth Phase That Prepares Bacteria for Exponential Growth and Involves Transient Metal Accumulation. J. Bacteriol. 2012;194:686–701. doi: 10.1128/JB.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin DS. The oxygen consumption of escherichia coli during the lag and logarithmic phases of growth. J. Gen. Physiol. 1932;15:691–708. doi: 10.1085/jgp.15.6.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greig ME, Hoogerheide JC. The Correlation of Bacterial Growth with Oxygen Consumption. J. Bacteriol. 1941;41:549–556. doi: 10.1128/jb.41.5.549-556.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Llorens N, María J, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 67.Kolter R. The Stationary Phase of The Bacterial Life Cycle. Annu. Rev. Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 68.Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 69.Zambrano MM, Kolter R. GASPing for Life in Stationary Phase. Cell. 1996;86:181–184. doi: 10.1016/S0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 70.Bacun-Druzina V, Cagalj Z, Gjuracic K. The growth advantage in stationary-phase (GASP) phenomenon in mixed cultures of enterobacteria. FEMS Microbiol. Lett. 2007;266:119–127. doi: 10.1111/j.1574-6968.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 71.Pletnev P, Osterman I, Sergiev P, Bogdanov A, Dontsova O. Survival guide: Escherichia coli in the stationary phase. Acta Naturae. 2015;7:22–33. [PMC free article] [PubMed] [Google Scholar]
- 72.Farrell MJ, Finkel SE. The Growth Advantage in Stationary-Phase Phenotype Conferred by rpoS Mutations Is Dependent on the pH and Nutrient Environment. J. Bacteriol. 2003;185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


