Summary
Introduction
Matrix metalloproteinases (MMPs) and ‘aggrecanase’ a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs) are well established to play key roles in osteoarthritis (OA) through degradation of extracellular matrix (ECM) type II collagen and aggrecan, and are thus potential targets for development of OA therapies.
Objective
This paper aims to provide a comprehensive review of the expression and potential roles of other, lesser-known ADAMTSs and related adamalysins (or a disintegrin and metalloproteinases (ADAMs)) in cartilage, with a view to identifying potentially protective or homeostatic metalloproteinases in the joint and informing consequent selective inhibitor design.
Design
A comprehensive literature search was performed using PubMed terms ‘osteoarthritis’ and ‘ADAMTS’ or ‘ADAM’.
Results
Several ADAMTSs and ADAMs were identified as having reportedly increased expression in OA. These include enzymes likely to play roles in cartilage matrix anabolism (e.g., the procollagen N-proteinases ADAMTS-2, ADAMTS-3 and ADAMTS-14), chondrocyte differentiation and proliferation (e.g., ADAM9, ADAM10, ADAM12), as well as enzymes contributing to cartilage catabolism (e.g., Cartilage oligomeric protein (COMP)-degrading ADAMTS-7 and ADAMTS-12).
Conclusions
In addition to the well-characterised MMPs, ADAMTS-4 and ADAMTS-5, many other ADAMTSs and ADAMs are expressed in cartilage and several show significantly altered expression in OA. Studies aimed at elucidating the pathophysiological roles of these enzymes in cartilage will contribute to our understanding of OA pathogenesis and enable design of targeted inhibitors that effectively target metalloproteinase-mediated cartilage degradation while sparing cartilage repair pathways.
Keywords: Osteoarthritis, Metalloproteinase, ADAMTS, ADAM
Introduction
Osteoarthritis (OA) is a common degenerative joint disease characterised by cartilage loss, subchondral bone remodelling and osteophyte development. These structural changes are accompanied by impaired movement, stiffness and chronic joint pain. Primary risk factors for OA include age, obesity and joint injury, which alter the mechanical loading of the joint and initiate dysregulated cellular signalling and activation of catabolic pathways.
The role of matrix metalloproteinases (MMPs) in osteoarthritic degradation of the extracellular matrix (ECM) has been well documented. In particular, the collagenase matrix metalloproteinase 13 (MMP-13) plays a central role in degrading type II collagen1, 2, and the two ‘aggrecanases’, namely a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-4 and -5, degrade aggrecan3, 4. Collagen and aggrecan are the primary structural components of the cartilage ECM, and their degradation correlates with progression of OA. Collagenases and aggrecanases are thus potential targets for the development of disease-modifying OA drugs (DMOADs). For such an approach to be successful, it is vital that we learn lessons from previous attempts to develop metalloproteinase inhibitors as anti-cancer therapies5. These drugs failed due to limited specificity and consequent off-target inhibition of other metalloproteinases with homologous catalytic domains. Only by understanding the full spectrum of metalloproteinases expressed in the joint and their biological function(s) in this location will it be possible to design strategies to selectively target pathological tissue destruction. Several ADAMTSs other than ADAMTS-4 and -5 are expressed in cartilage, but little is known about whether they are required for joint health or whether they contribute to OA pathogenesis. The roles of the related adamalysin (a disintegrin and metalloproteinase, ADAM) family of metalloproteinases in cartilage are similarly poorly understood. Here, we review studies examining the role of ADAMTSs and ADAMs in cartilage, and compare microarray studies examining their expression in murine models of OA6, 7, 8 and human normal and osteoarthritic cartilage9, 10, 11, 12, 13, 14, 15, 16, 17. This review thus highlights ADAMTSs and ADAMs that are expressed in cartilage and whose expression is altered in OA, with a view to developing a broader understanding of the contribution of the metalloproteinase family to joint health and disease.
ADAMTSs
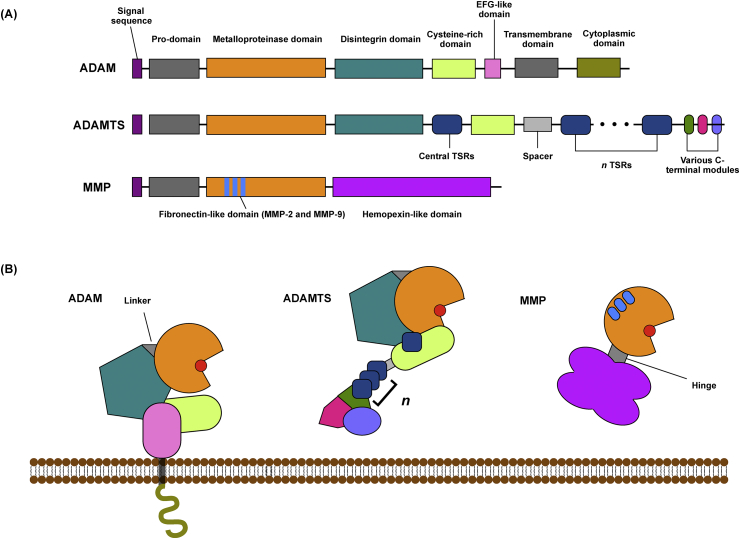
The ADAMTSs are a family of 19 secreted metalloproteinases (Fig. 1) involved in various developmental and homeostatic processes18. The ‘aggrecanases’ ADAMTS-4 and -5 have been extensively reviewed elsewhere19, 20, and will not be covered here. Several other ADAMTSs are expressed in cartilage, and have emerging roles in joint pathophysiology.
Fig. 1.
Schematic representation of ADAM and ADAMTS topography. ADAMs and ADAMTSs are metzincin metalloproteinases whose catalytic domains share homology with those of the MMPs, and contain a zinc ion (red circle) that is essential for their proteolytic activity. All three groups of enzymes have a prodomain that keeps them in an inactive zymogen form until they are activated. The families differ in their C-terminal ancillary domains, which mediate interaction with substrates and other proteins. ADAM ancillary domains: ADAMs contain C-terminal disintegrin-like domains, thought to regulate cell–cell and cell–matrix adhesion, as well as conserved cysteine-rich domains and EGF-like domains102. The cytoplasmic domains are the most diverse, and vary in sequence and length. Some ADAM cytoplasmic domains contain proline-rich Src homology (SH)-2 and/or SH-3 binding sites, indicating that they may participate in intracellular signalling. Some also contain potential serine–threonine and/or tyrosine phosphorylation sites, making them plausible adaptors for conveying signals between the cell and its surroundings. ADAMTS ancillary domains: In contrast to the ADAMs, ADAMTSs are secreted metalloproteinases that lack transmembrane and cytoplasmic domains. In addition to their catalytic and pro-domains, the enzymes contain a variable number of thrombospondin type 1 sequence repeat (TSR) motifs, which are homologous to thrombospondins18, as well as a cysteine-rich domain and spacer domain. Some members of the family contain additional C-terminal domains18. For example, ADAMTS-9 and -20 contain GON-1 domains, ADAMTS-2, -3 and -14 contain a procollagen N-propeptidase (PNP) domain, and ADAMTS-7 and -12 contain a PLAC domain.
ADAMTS-1
ADAMTS-1 is expressed in cartilage and synovium18 and has been shown to cleave aggrecan and versican21. Several studies show that ADAMTS-1 expression is significantly upregulated in OA cartilage7, 10, 11, 12, 14, 22, though some studies indicate reduced expression in late-stage human OA9, 13, 17 (Fig. 2). Immunohistochemical analysis indicates that in normal cartilage, ADAMTS-1 is primarily expressed in the superficial zone, with OA cartilage showing increased staining in the middle zone and in osteophytes22.
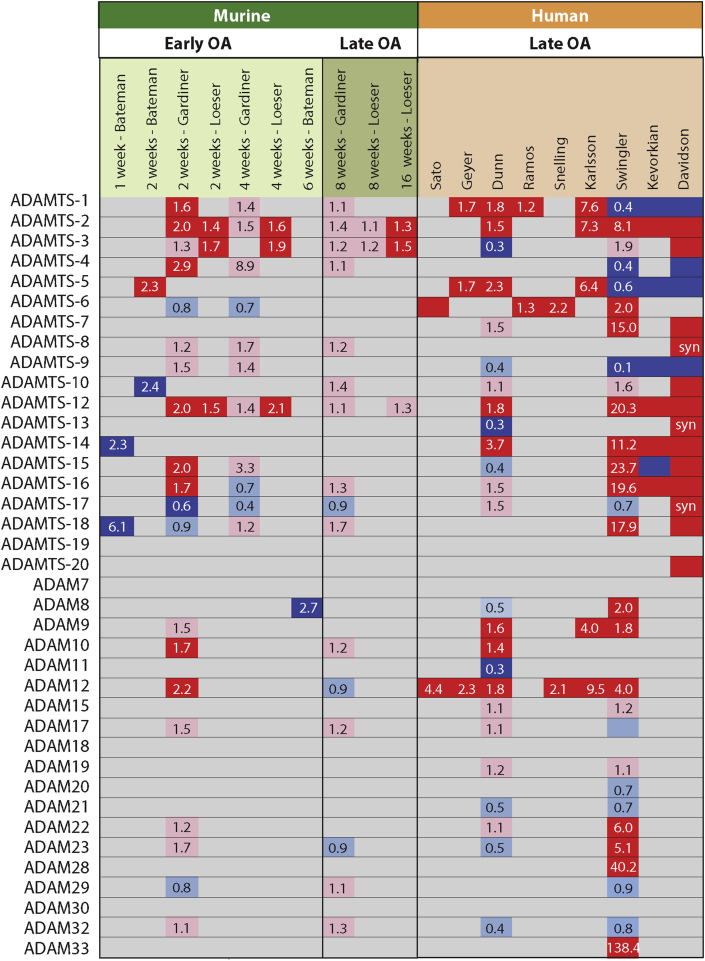
Fig. 2.
Fold-change in expression of ADAMTSs and ADAMs in OA compared to normal cartilage. Upregulated genes are marked in red (statistically significant, P < 0.05) or pink (not statistically significant, P > 0.05), while down-regulated genes are shown in dark blue (statistically significant, P < 0.05) or light blue (not statistically significant, P > 0.05). Bateman6, Gardiner7 and Loeser8 analysed murine knee cartilage at various time points after DMM. Sato15, Geyer11, Dunn10, Ramos14, and Snelling16 analysed paired samples from intact and OA lesion areas of the same patients. Karlsson12 compared knee OA samples with healthy controls. Swinger17, Kevorkian13 and Davidson9 compared femoral head cartilage from OA patients with that of fracture patients. Syn, synovium.
Adamts1-null mice display unaltered susceptibility in the antigen-induced model of inflammatory arthritis and there is also no change in the level of aggrecan degradation in response to interleukin 1 (IL-1) stimulation of cartilage explants in vitro23. The susceptibility of these mice to surgical destabilisation of the medial meniscus (DMM), a model that more closely resembles human OA, has not been reported. Given that Adamts1-null mice show developmental abnormalities24, conditional deletion may be required to establish the role of the enzyme in cartilage homeostasis.
ADAMTS-2, ADAMTS-3 and ADAMTS-14
ADAMTS-2, ADAMTS-3 and ADAMTS-14 are procollagen N-proteinases, responsible for removing the N-terminal propeptide of type I, II, III and V pro-collagen25, 26, 27, and thereby enabling collagen fibril formation. Lack of procollagen N-proteinase activity is associated with defective collagen fibrilogenesis and connective tissue dysfunctions such as skin fragility26. Since type II collagen is a critical structural component of the cartilage ECM, these enzymes are likely to be important for cartilage homeostasis and repair. Bekhouche et al.28 recently identified additional substrates for this group of enzymes, including several proteins involved in transforming growth factor β (TGFβ) signalling.
Several studies indicate statistically significant increased expression of ADAMTS-2 in OA cartilage7, 8, 9, 10, 12, 13, 17. No cartilage or joint abnormalities have been identified in Adamts2-deficient mice26, although they exhibit skin fragility reminiscent of Ehlers-Danlos syndrome in humans, which is also caused by mutation of ADAMTS-2. Partial processing of collagen in these mice appears to indicate some redundancy between the procollagen N-proteinases.
ADAMTS-3 is more potent than ADAMTS-2 at processing type II pro-collagen29, suggesting that it may be more relevant than ADAMTS-2 in cartilage. As with ADAMTS-2, ADAMTS-3 expression is reportedly increased in OA cartilage8, 9, although this reaches statistical significance in fewer studies than ADAMTS-2, and Dunn et al.10 report a reduction in expression in late stage human OA cartilage. Adamts3 knockout mice are not viable after E15.030, so generation of conditional knockout mice will be necessary to investigate the role of this enzyme in adult cartilage homeostasis and OA pathogenesis.
ADAMTS-14, the third of the procollagen N-proteinases to be discovered31, is also significantly up-regulated in human OA cartilage9, 10, 13, 17. Single nucleotide polymorphisms (SNPs) of ADAMTS14 have been associated with an increased risk of knee OA in two female cohorts32, 33. The enzyme may be differently regulated in murine OA models, since little regulation is observed in most murine microarray studies, other than reduced expression observed by Bateman et al.6 1 week after surgical induction of OA in mice.
The increased expression of all three of these enzymes in human OA cartilage may reflect an attempted anabolic repair response of osteoarthritic chondrocytes, and it would be desirable to spare these enzymes when designing metalloproteinase inhibitors for potential OA therapy.
ADAMTS-7 and ADAMTS-12
ADAMTS-734 and ADAMTS-1235 are thought to contribute to OA pathogenesis by degrading cartilage oligomeric protein (COMP), an important regulator of cartilage ECM assembly and a potential biomarker for cartilage degradation.
Several studies have observed increased expression of ADAMTS-7 in OA cartilage9, 17, 23, 36, 37, 38. OA progression and COMP degradation were increased in mice over-expressing ADAMTS-7 and decreased in Adamts7−/− mice37. COMP degradation could also be inhibited in vitro by addition of ADAMTS-7 neutralising antibodies or siRNA39. Expression of the enzyme can be stimulated by pro-inflammatory cytokines such as tumour necrosis factor (TNF)39. A positive feedback loop has been proposed between ADAMTS-7 and TNF, since TNF expression is elevated in transgenic mice over-expressing ADAMTS-7 in chondrocytes37. The molecular mechanism underpinning this feedback loop is not known.
Increased expression of ADAMTS-12 is also consistently observed in OA cartilage7, 8, 9, 10, 13, 17, 35, 38. COMP degradation in OA cartilage explants could be additively inhibited by neutralising antibodies against ADAMTS-7 and ADAMTS-1239, suggesting that ADAMTS-12 may also contribute to pathological COMP degradation. These enzymes may also work together to process substrates in vivo. Systematic analysis of single and combined Adamts7-and Adamts12-null mice would help to clarify the relative contributions these enzymes make to COMP degradation in OA.
ADAMTS-8
ADAMTS-8 is expressed in normal cartilage and has been reported to have aggrecanase activity40. Expression of the enzyme in cartilage is not significantly altered in murine models of OA,7, 15 although Davidson et al.9 reported increased expression in human OA synovium.
ADAMTS-9 and ADAMTS-20
ADAMTS-9 and ADAMTS-20 are the largest members of the ADAMTS family, with 15 thrombospondin domains and a C-terminal GON-1 domain.
ADAMTS-9 is expressed in normal cartilage, and highly induced in response to pro-inflammatory cytokines (e.g., IL-1β and TNF41) and adipokines (e.g., leptin42). Expression is reduced in late-stage OA cartilage9, 13, 17. ADAMTS-9 is able to cleave aggrecan, although a truncated form of the enzyme (comprising the catalytic, disintegrin and first TS domain) exhibits low aggrecanase activity compared to a similarly truncated form of ADAMTS-543. It is not known whether the full-length enzyme has higher aggrecanase activity, since this large enzyme is difficult to express and purify.
ADAMTS-20 is expressed in cartilage and reportedly shows increased expression in OA9, but the enzyme is primarily considered to be important for versican cleavage during development.
ADAMTS-15
ADAMTS-15 is also thought to participate in developmental versican cleavage44, but is expressed in cartilage and can cleave aggrecan45. Three microarray studies found increased expression of ADAMTS-15 in OA cartilage7, 9, 17, while two10, 13 found reduced expression. The contribution of the enzyme to cartilage homeostasis and disease is unknown.
ADAMTS-16
ADAMTS-16 expression is increased in OA cartilage7, 9, 13, 17 and a truncated form of the enzyme shows some aggrecanase activity43. To our knowledge, the aggrecanase activity of the full-length enzyme has not been characterised. Surridge et al.46 observed that over-expression of ADAMTS-16 in SW1353 chondrosarcoma cells decreased MMP13 expression, cell migration and proliferation, raising the possibility that ADAMTS-16 may have a protective role. Mechanistic investigation of these observations may shed light on the role of this enzyme in cartilage.
Other ADAMTSs
Microarray studies have reported increased expression of ADAMTS-614, 15, 16, 17, ADAMTS-109, and ADAMTS-189, 17 in OA cartilage. These are ‘orphan’ enzymes without known substrates. Studies on their wider biological functions have implicated ADAMTS-6 and -10 in regulations of cell–cell junctions47 and ADAMTS-18 in development48.
ADAM metalloproteinases
The A Disintegrin and Metalloproteinase (ADAM, or adamalysin) family are conserved type-I transmembrane metzincin metalloproteinases related to the MMPs and ADAMTSs (Fig. 1). ADAMs are widely expressed and have been shown to participate in a wide variety of biological processes49, 50.
Among the 34 known ADAMs, 20 are present in the human genome and 12 of these (ADAM8, ADAM9, ADAM10, ADAM12, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28, ADAM30 and ADAM33) are predicted to be proteolytically active based on the presence of a conserved HEXGHXXGXXHD motif and downstream ‘Met turn’ in the catalytic domain49. The proteolytically active ADAMs mainly function as ‘sheddases’, cleaving the juxta-membrane region of their trans-membrane substrates (reviewed by Edwards et al.49) to release the soluble ectodomain of the substrate to the extracellular space. This activity enables them to regulate the extracellular availability of autocrine and paracrine signalling molecules, such as transmembrane cytokines and growth factors and their receptors. The remaining eight human ADAMs (ADAM2, ADAM7, ADAM11, ADAM18, ADAM22, ADAM23, ADAM29, ADAM32) are predicted to lack proteolytic activity, but still play important biological roles49.
The roles of ADAMs in development of the musculoskeletal system have been well documented, and the roles of some of the ADAMs in rheumatoid arthritis have been examined. The roles of these enzymes in adult joint tissues and in OA have been less well characterised. We review the emerging evidence for roles of these enzymes in joint homeostasis and OA pathogenesis.
ADAM8
Expression of ADAM8 is reportedly elevated in human OA cartilage17, 51, 52, and the enzyme has been postulated to contribute to OA pathogenesis by cleaving fibronectin and generating cryptic pro-catabolic fibronectin fragments52. Zack et al.52 showed that addition of recombinant ADAM8 to human OA cartilage explants increases aggrecan cleavage and cartilage degradation in vitro. Expression of ADAM8 was significantly reduced 6 weeks after surgical induction of OA in a murine model6, indicating either species-specific differences in regulation or that the enzyme is dynamically regulated during OA progression.
ADAM8 is also expressed in cells of the mononuclear phagocyte lineage53. ADAM8 promotes osteoclast formation54, 55 and is thought to contribute to the bone erosion associated with aseptic loosening of hip replacement prostheses56. Increased expression of ADAM8 has been observed in rheumatoid arthritis pannus tissue55, suggesting that ADAM8 may also contribute to osteoclast formation and bone erosion in rheumatoid arthritis.
ADAM9 (meltrin-Ɣ)
Several studies consistently report significant upregulation of ADAM9 in human OA cartilage10, 12, 17 and the enzyme is non-significantly upregulated in mice 2 weeks after DMM surgery7. ADAM9 substrates include growth factor precursors49, and the enzyme has been suggested to play a role in chondrogenesis57. Its expression is decreased in response to IL-1 or retinoic acid58. Reminiscent of ADAM8, ADAM9 is expressed in mononuclear phagocytes and osteoclasts, and has been suggested to contribute to bone resorption associated with aseptic loosening of hip prostheses59.
ADAM10
ADAM10 is one of the best characterised of the ADAMs, and is crucial for embryonic development through its shedding of Notch receptor and consequent regulation of downstream Notch signalling49. ADAM10 also has pivotal roles in cell migration and adhesion, mediated through its cleavage of transmembrane precursors of growth factors (e.g., epidermal growth factor, EGF), chemokines (e.g., CX3CL1 and CXCL16) and adhesion molecules (e.g., E-cadherin and VE-cadherin)49.
Expression of ADAM10 is low in normal adult cartilage, but increased during development60, in OA7, 10, 60 and in response to pro-inflammatory cytokines60, 61. In OA or IL-1α-stimulated cartilage, highest expression of ADAM10 was observed in regions with greatest damage and proteoglycan loss60, leading Chubinskaya et al.60 to suggest the enzyme may contribute to cartilage damage. ADAM10 substrates in normal or OA cartilage have not been characterised.
ADAM10 expression is also increased in synovium lining and endothelial cells of rheumatoid arthritis patients61. Silencing of ADAM10 in endothelial cells reduced migration and tubule formation, suggesting the enzyme may contribute to angiogenesis in the rheumatoid synovium61.
ADAM12 (meltrin-α)
ADAM12 is the ADAM most consistently reported to display increased expression in human OA cartilage10, 11, 12, 15, 16, 17, 62. Its expression is reported to correlate with Mankin score62, and increased expression has also been reported in mice 2 weeks after DMM surgery7. Several studies have linked ADAM12 SNPs (e.g., rs1278279, rs3740199, rs1044122, and rs1871054) with an increased risk of OA63, 64, 65, 66, 67, 68, 69, 70, although post-hoc stratification of data was often required to establish a significant association. A large case-controlled study of over 1000 UK OA patients and an equal number of matched controls failed to find any association between ADAM12 SNPs and OA71.
Substrates of ADAM12 in cartilage have not been directly investigated. The enzyme is widely expressed and has been shown to promote cell proliferation, differentiation and migration through its ability to shed transmembrane ligands of the EGF receptor and thus stimulate EGF receptor signalling72. ADAM12 also modulates insulin-like growth factor (IGF) signalling by cleavage of IGF binding proteins72. ADAM12 has similarly been shown to promote chondrocyte proliferation and maturation during development62, 73, although it is proposed to act by promoting IGF-1 activity by degrading IGF binding protein 562, rather than by an EGF receptor-dependent pathway.
ADAM15 (Metargidin)
ADAM15 is the only ADAM so far demonstrated to have a protective role in cartilage, with Adam15-deficient mice developing accelerated spontaneous OA with age74. Bohm et al. reported that expression of the enzyme is increased in OA cartilage using in situ hybridisation75, but subsequent microarrays have reported either no change7 or slight but statistically non-significant increased expression in OA10, 17.
Understanding of how ADAM15 protects cartilage is still in its infancy. Böhm et al. proposed that ADAM15 increases chondrocyte survival by reinforcing adhesion to collagen types II and VI74, promoting outside-in pro-survival signalling76, 77 and up-regulating anti-apoptotic molecules such as X-linked inhibitor of apoptosis (XIAP)78.
ADAM15 has been shown to affect cell–cell and cell–matrix adhesion in other cell types79, 80, 81, potentially also through its ability to interact with integrins. ADAM15 is the only ADAM that contains an integrin-binding Arginine-glycine-aspartic acid (RGD) motif in its disintegrin domain82, enabling RGD-dependent interaction with αvβ3 and α5β183, and ADAM15 is also able to bind to α9β3 in an RGD-independent manner84. ADAM15 over-expression been shown to promote migration of mesangial cells85, possibly by disrupting integrin-ECM interactions or by proteolytic cleavage of adhesion molecules. N- and E-cadherin86, 87 are among the few ADAM15 substrates described, and their degradation may underpin the effect of ADAM15 on cell migration. The relevance of these studies to ADAM15's protective role in cartilage is unclear, especially since many were done using overexpression systems that may not accurately reflect the physiological activity of the enzyme.
It is not yet known how these different activities of ADAM15 are regulated or coordinated. Additionally, ADAM15 substrates in chondrocyte have not been identified. Further studies to investigate the protective role of this enzyme in the joint are required.
ADAM17 (TNF-α-converting enzyme, TACE)
ADAM17, or TACE, is the most extensively studied ADAM, with important roles in development and inflammation through its shedding of EGF receptor ligands and the membrane-bound precursor of the pleiotropic cytokine TNF88.
ADAM17 deletion leads to perinatal lethality, so conditional and tissue-specific deletion have been studied to evaluate the function of the enzyme in specific tissues and in the adult. Deletion of ADAM17 in chondrocytes retarded expansion of hypertrophic chondrocytes in the growth plate and impaired bone growth89, 90. This indicates that ADAM17 is important for musculoskeletal development, most likely through its role in EGF receptor signalling.
ADAM17 is also expressed in adult chondrocytes17, but chondrocyte-specific inducible knockout of ADAM17 has not been reported, so the role of the enzyme in adult cartilage is not known. Microarray studies indicate that ADAM17 expression is not significantly altered in OA7, 10, 17, but such studies do not take account of the fact that ADAM17 activity is largely regulated post-translationally.
Oldefest et al.91 showed that ADAM17 shedding of the IL-6 receptor (IL-6R) can be inhibited by secreted Frizzled-related protein 3 (sFRP3), but not by variants of sFRP3 that have previously been associated with an increased risk of OA. This raises the possibility that the sFRP3 variants promote cartilage damage by failing to control ADAM17 pro-inflammatory activity in cartilage.
ADAM19
ADAM19 is expressed during chondrogenesis, with increased expression during the later stages of the process57. Expression of the enzyme is reportedly not significantly altered in OA cartilage10, 17.
ADAM23
Like ADAM19, ADAM23 is also up-regulated at the late stage of chondrogenesis57, 92. ADAM23 is up-regulated in OA cartilage17, but its function in cartilage has not been investigated.
ADAM28
ADAM28 is expressed at low levels in normal cartilage, with increased expression in OA17. The enzyme has been suggested to promote retinoic acid-stimulated proteoglycan degradation93, although the molecular mechanism for this observation has not been established. Known substrates of ADAM28 include TNF94 and growth factors such as IGF binding protein-395 and connective tissue growth factor (CTGF)96. In osteoblasts, IL-1β-induced MMP-13 expression is dependent on ADAM2897, suggesting the enzyme may also affect bone remodelling.
Other ADAMs
Swinger et al.17 report that expression of the ADAM22 and ADAM33 is increased in OA cartilage, while expression of ADAM2, ADAM7, ADAM11, ADAM18, ADAM20, ADAM21, ADAM29, ADAM30 and ADAM32 is not significantly altered in OA17.
Concluding remarks and future study directions
Collagenases and aggrecanases have received attention as potential targets for development of DMOADs. In order for such an approach to be successful, we should learn the lessons of previous attempts to design MMP inhibitors to treat cancer, namely that it is crucial to understand the full spectrum of metalloproteinases expressed in a target tissue. Many ADAMTSs and ADAMS are expressed in cartilage and several are differentially regulated in OA, but their roles in cartilage health and disease are largely unexplored and the full spectrum of their substrates is not yet known. Further research into the functions and substrates of these enzymes in the joint and in OA pathogenesis is required to evaluate therapeutic potential. Unbiased proteomic analysis will assist in defining the range of substrates cleaved by each enzyme. Additionally, enzymes may be important at different temporal stages in the disease process, with implications for timing of effective inhibitor therapy. Inducible knockout strategies may be useful in defining relevant windows of activity.
It is especially important to understand which enzymes may serve protective functions in the joint, since inhibiting their activity is likely to further impair cartilage homeostasis. For example, the procollagen N-proteinases ADAMTS-2, ADAMTS-3 and ADAMTS-14 are likely to promote matrix anabolism, and several studies have demonstrated that their expression is increased in OA. Inducible and combined knockout of these enzymes are required to investigate their contribution to cartilage repair in OA and to investigate potential redundancy among the enzymes. Similarly, ADAMs play important roles in signalling in other tissues, and are likely to modulate a diverse range of signalling pathways in chondrocytes. For example, ADAMs can activate signalling pathways by mobilising transmembrane cytokine and growth factor precursors, or through interaction of their cytoplasmic domains with intracellular signalling machinery. Conversely, ADAM-mediated shedding of cytokine and growth factor receptors can suppress their downstream signalling. ADAMs thus fine-tune cellular responses and contribute to maintenance of tissue homeostasis, and changes in ADAM expression may contribute to the shift in balance from cartilage repair to cartilage degradation. Studies on ADAM15, one of the few ADAMs to be studied in the context of OA, have shown that Adam15-null animals develop accelerated OA74, although the molecular mechanism of this protection is poorly understood. Increased expression of ADAM9, ADAM10 and ADAM12 in OA cartilage has been consistently reported in multiple studies. The activities of these enzymes in other tissues indicate that they may promote chondrocyte differentiation and proliferation, and so may contribute to repair pathways in OA, but direct evidence for this is lacking. Since several of the ADAMs are critically involved in embryogenesis and development, inducible knockout systems will be necessary to expand our understanding of their pathophysiological roles in the joint.
Mechanisms regulating ADAM and ADAMTS expression and activity in the joint also require further study. Several ADAM and ADAMTS genes displayed altered methylation in OA. These included ADAMTS2, ADAMTS8 (hypermethylated in OA) and ADAMTS4, ADAMTS5, ADAMTS10 and ADAMTS17 (hypomethylated in OA)98. How these differences in methylation influence expression is unclear. Expression is also affected by microRNAs, as has been reviewed elsewhere99. Additionally, enzymatic activity is often regulated post-translationally. ADAMTS-4 and -5 are post-translationally regulated by endocytosis100, and other ADAMTSs may be similarly regulated. Activity of several ADAMs is also regulated post-translationally, through mechanisms such as conformational change, or regulation of substrate or enzyme localisation101.
It is important to keep in mind that altered expression of ADAMTSs and ADAMs in OA does not necessarily mean that a particular enzyme is mechanistically involved in disease pathogenesis. This is particularly true in late-stage OA, where expression may be modified by changes in cell homeostasis or altered substrate availability occurring due to matrix catabolism. Knock-out mice studies are required to validate roles in OA pathogenesis. Inducible knock-out models will enable analysis of enzyme functions at different stages of OA progression. Murine studies are potentially complicated by species differences and the question of whether acute surgical models accurately reflect the human disease, but they have the advantage of enabling analysis of early OA, which is difficult to achieve with human clinical samples.
ADAMTSs and ADAMs have the potential to modulate multiple aspects of cell behaviour and tissue homeostasis. Understanding their roles in cartilage is therefore essential for the development of successful therapies to target osteoarthritic cartilage degradation.
Contributions
CYY and LT performed the literature review and wrote the manuscript. AC analysed microarray data sets.
Conflict of interest statement
The authors have no financial or personal conflicts of interest.
Funding
This work was supported by Arthritis Research UK (grants 19466, 20205 and 20887) and the Kennedy Trust for Rheumatology Research.
Acknowledgements
We thank Hideaki Nagase for helpful discussions and critical editing of the manuscript.
Abbreviations
- ADAM
a disintegrin and metalloproteinase
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- COMP
cartilage oligomeric protein
- CTGF
connective tissue growth factor
- DMM
destabilisation of the medial meniscus
- DMOAD
disease-modifying OA drug
- ECM
extracellular matrix
- EGF
epidermal growth factor
- IGF
insulin-like growth factor
- IL-1
interleukin 1
- IL-16R
interleukin 6 receptor
- MMP
matrix metalloproteinase
- OA
osteoarthritis
- PNP
procollagen N-propeptidase
- SH
Src homology
- SNP
single nucleotide polymorphism
- sFRP3
secreted Frizzled-related protein 3
- Syn
synovium
- TACE
TNF-α-converting enzyme
- TGFβ
transforming growth factor β
- TNF
tumour necrosis factor
- TSR
thrombospondin type 1 sequence repeat
- XIAP
X-linked inhibitor of apoptosis protein
References
- 1.Neuhold L.A., Killar L., Zhao W., Sung M.L., Warner L., Kulik J. Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J Clin Invest. 2001;107:35–44. doi: 10.1172/JCI10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M., Sampson E.R., Jin H., Li J., Ke Q.H., Im H.J. MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res Ther. 2013;15:R5. doi: 10.1186/ar4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanton H., Rogerson F.M., East C.J., Golub S.B., Lawlor K.E., Meeker C.T. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 4.Glasson S.S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H.L. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 5.Coussens L.M., Fingleton B., Matrisian L.M. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 6.Bateman J.F., Rowley L., Belluoccio D., Chan B., Bell K., Fosang A.J. Transcriptomics of wild-type mice and mice lacking ADAMTS-5 activity identifies genes involved in osteoarthritis initiation and cartilage destruction. Arthritis Rheum. 2013;65:1547–1560. doi: 10.1002/art.37900. [DOI] [PubMed] [Google Scholar]
- 7.Gardiner M.D., Vincent T.L., Driscoll C., Burleigh A., Bou-Gharios G., Saklatvala J. Transcriptional analysis of micro-dissected articular cartilage in post-traumatic murine osteoarthritis. Osteoarthritis and Cartilage. 2015;23:616–628. doi: 10.1016/j.joca.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeser R.F., Olex A.L., McNulty M.A., Carlson C.S., Callahan M., Ferguson C. Disease progression and phasic changes in gene expression in a mouse model of osteoarthritis. PLoS One. 2013;8:e54633. doi: 10.1371/journal.pone.0054633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson R.K., Waters J.G., Kevorkian L., Darrah C., Cooper A., Donell S.T. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn S.L., Soul J., Anand S., Schwartz J.M., Boot-Handford R.P., Hardingham T.E. Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses. Osteoarthritis and Cartilage. 2016;24:1431–1440. doi: 10.1016/j.joca.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer M., Grassel S., Straub R.H., Schett G., Dinser R., Grifka J. Differential transcriptome analysis of intraarticular lesional vs intact cartilage reveals new candidate genes in osteoarthritis pathophysiology. Osteoarthritis and Cartilage. 2009;17:328–335. doi: 10.1016/j.joca.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson C., Dehne T., Lindahl A., Brittberg M., Pruss A., Sittinger M. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis and Cartilage. 2010;18:581–592. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Kevorkian L., Young D.A., Darrah C., Donell S.T., Shepstone L., Porter S. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50:131–141. doi: 10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 14.Ramos Y.F., den Hollander W., Bovee J.V., Bomer N., van der Breggen R., Lakenberg N. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLoS One. 2014;9:e103056. doi: 10.1371/journal.pone.0103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T., Konomi K., Yamasaki S., Aratani S., Tsuchimochi K., Yokouchi M. Comparative analysis of gene expression profiles in intact and damaged regions of human osteoarthritic cartilage. Arthritis Rheum. 2006;54:808–817. doi: 10.1002/art.21638. [DOI] [PubMed] [Google Scholar]
- 16.Snelling S., Rout R., Davidson R., Clark I., Carr A., Hulley P.A. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthritis and Cartilage. 2014;22:334–343. doi: 10.1016/j.joca.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swingler T.E., Waters J.G., Davidson R.K., Pennington C.J., Puente X.S., Darrah C. Degradome expression profiling in human articular cartilage. Arthritis Res Ther. 2009;11:R96. doi: 10.1186/ar2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelwick R., Desanlis I., Wheeler G.N., Edwards D.R. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondeson J., Wainwright S., Hughes C., Caterson B. The regulation of the ADAMTS4 and ADAMTS5 aggrecanases in osteoarthritis: a review. Clin Exp Rheumatol. 2008;26:139–145. [PubMed] [Google Scholar]
- 20.Verma P., Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J Cell Biochem. 2011;112:3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Manzaneque J.C., Westling J., Thai S.N., Luque A., Knauper V., Murphy G. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun. 2002;293:501–508. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 22.Wachsmuth L., Bau B., Fan Z., Pecht A., Gerwin N., Aigner T. ADAMTS-1, a gene product of articular chondrocytes in vivo and in vitro, is downregulated by interleukin 1beta. J Rheumatol. 2004;31:315–320. [PubMed] [Google Scholar]
- 23.Little C.B., Mittaz L., Belluoccio D., Rogerson F.M., Campbell I.K., Meeker C.T. ADAMTS-1-knockout mice do not exhibit abnormalities in aggrecan turnover in vitro or in vivo. Arthritis Rheum. 2005;52:1461–1472. doi: 10.1002/art.21022. [DOI] [PubMed] [Google Scholar]
- 24.Shindo T., Kurihara H., Kuno K., Yokoyama H., Wada T., Kurihara Y. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest. 2000;105:1345–1352. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colige A., Li S.W., Sieron A.L., Nusgens B.V., Prockop D.J., Lapiere C.M. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc Natl Acad Sci USA. 1997;94:2374–2379. doi: 10.1073/pnas.94.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S.W., Arita M., Fertala A., Bao Y., Kopen G.C., Langsjo T.K. Transgenic mice with inactive alleles for procollagen N-proteinase (ADAMTS-2) develop fragile skin and male sterility. Biochem J. 2001;355:271–278. doi: 10.1042/0264-6021:3550271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bekhouche M., Colige A. The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. 2015;44–46:46–53. doi: 10.1016/j.matbio.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Bekhouche M., Leduc C., Dupont L., Janssen L., Delolme F., Vadon-Le Goff S. Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-β signaling as primary targets. FASEB J. 2016;30:1741–1756. doi: 10.1096/fj.15-279869. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes R.J., Hirohata S., Engle J.M., Colige A., Cohn D.H., Eyre D.R. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J Biol Chem. 2001;276:31502–31509. doi: 10.1074/jbc.M103466200. [DOI] [PubMed] [Google Scholar]
- 30.Janssen L., Dupont L., Bekhouche M., Noel A., Leduc C., Voz M. ADAMTS3 activity is mandatory for embryonic lymphangiogenesis and regulates placental angiogenesis. Angiogenesis. 2016;19:53–65. doi: 10.1007/s10456-015-9488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colige A., Vandenberghe I., Thiry M., Lambert C.A., Van Beeumen J., Li S.W. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J Biol Chem. 2002;277:5756–5766. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 32.Poonpet T., Honsawek S., Tammachote N., Kanitnate S., Tammachote R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol Res. 2013;12:5301–5309. doi: 10.4238/2013.November.7.5. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Lopez J., Pombo-Suarez M., Loughlin J., Tsezou A., Blanco F.J., Meulenbelt I. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis and Cartilage. 2009;17:321–327. doi: 10.1016/j.joca.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Liu C.J., Kong W., Ilalov K., Yu S., Xu K., Prazak L. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006;20:988–990. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C.J., Kong W., Xu K., Luan Y., Ilalov K., Sehgal B. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J Biol Chem. 2006;281:15800–15808. doi: 10.1074/jbc.M513433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo F., Lai Y., Tian Q., Lin E.A., Kong L., Liu C. Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis Rheum. 2010;62:2023–2036. doi: 10.1002/art.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai Y., Bai X., Zhao Y., Tian Q., Liu B., Lin E.A. ADAMTS-7 forms a positive feedback loop with TNF-alpha in the pathogenesis of osteoarthritis. Ann Rheum Dis. 2014;73:1575–1584. doi: 10.1136/annrheumdis-2013-203561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., Ji Q., Wang X., Kang L., Fu Y., Yin Y. SOX9 is a regulator of ADAMTSs-induced cartilage degeneration at the early stage of human osteoarthritis. Osteoarthritis and Cartilage. 2015;23:2259–2268. doi: 10.1016/j.joca.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Luan Y., Kong L., Howell D.R., Ilalov K., Fajardo M., Bai X.H. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthritis and Cartilage. 2008;16:1413–1420. doi: 10.1016/j.joca.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins-Racie L.A., Flannery C.R., Zeng W., Corcoran C., Annis-Freeman B., Agostino M.J. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004;23:219–230. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Demircan K., Hirohata S., Nishida K., Hatipoglu O.F., Oohashi T., Yonezawa T. ADAMTS-9 is synergistically induced by interleukin-1beta and tumor necrosis factor alpha in OUMS-27 chondrosarcoma cells and in human chondrocytes. Arthritis Rheum. 2005;52:1451–1460. doi: 10.1002/art.21010. [DOI] [PubMed] [Google Scholar]
- 42.Yaykasli K.O., Hatipoglu O.F., Yaykasli E., Yildirim K., Kaya E., Ozsahin M. Leptin induces ADAMTS-4, ADAMTS-5, and ADAMTS-9 genes expression by mitogen-activated protein kinases and NF-kB signaling pathways in human chondrocytes. Cell Biol Int. 2015;39:104–112. doi: 10.1002/cbin.10336. [DOI] [PubMed] [Google Scholar]
- 43.Zeng W., Corcoran C., Collins-Racie L.A., Lavallie E.R., Morris E.A., Flannery C.R. Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs: comparative analyses with ADAMTS-5, -9, -16 and -18. Biochim Biophys Acta. 2006;1760:517–524. doi: 10.1016/j.bbagen.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Dancevic C.M., Fraser F.W., Smith A.D., Stupka N., Ward A.C., McCulloch D.R. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. J Biol Chem. 2013;288:37267–37276. doi: 10.1074/jbc.M112.418624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelwick R., Wagstaff L., Decock J., Roghi C., Cooley L.S., Robinson S.D. Metalloproteinase-dependent and -independent processes contribute to inhibition of breast cancer cell migration, angiogenesis and liver metastasis by a disintegrin and metalloproteinase with thrombospondin motifs-15. Int J Cancer. 2015;136:E14–E26. doi: 10.1002/ijc.29129. [DOI] [PubMed] [Google Scholar]
- 46.Surridge A.K., Rodgers U.R., Swingler T.E., Davidson R.K., Kevorkian L., Norton R. Characterization and regulation of ADAMTS-16. Matrix Biol. 2009;28:416–424. doi: 10.1016/j.matbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cain S.A., Mularczyk E.J., Singh M., Massam-Wu T., Kielty C.M. ADAMTS-10 and -6 differentially regulate cell-cell junctions and focal adhesions. Sci Rep. 2016;6:35956. doi: 10.1038/srep35956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ataca D., Caikovski M., Piersigilli A., Moulin A., Benarafa C., Earp S.E. Adamts18 deletion results in distinct developmental defects and provides a model for congenital disorders of lens, lung, and female reproductive tract development. Biol Open. 2016;5:1585–1594. doi: 10.1242/bio.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards D.R., Handsley M.M., Pennington C.J. The ADAM metalloproteinases. Mol Asp Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber S., Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139:3693–3709. doi: 10.1242/dev.076398. [DOI] [PubMed] [Google Scholar]
- 51.Aigner T., Fundel K., Saas J., Gebhard P.M., Haag J., Weiss T. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 52.Zack M.D., Malfait A.M., Skepner A.P., Yates M.P., Griggs D.W., Hall T. ADAM-8 isolated from human osteoarthritic chondrocytes cleaves fibronectin at Ala(271) Arthritis Rheum. 2009;60:2704–2713. doi: 10.1002/art.24753. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida S., Setoguchi M., Higuchi Y., Akizuki S., Yamamoto S. Molecular cloning of cDNA encoding MS2 antigen, a novel cell surface antigen strongly expressed in murine monocytic lineage. Int Immunol. 1990;2:585–591. doi: 10.1093/intimm/2.6.585. [DOI] [PubMed] [Google Scholar]
- 54.Choi S.J., Han J.H., Roodman G.D. ADAM8: a novel osteoclast stimulating factor. J Bone Min Res. 2001;16:814–822. doi: 10.1359/jbmr.2001.16.5.814. [DOI] [PubMed] [Google Scholar]
- 55.Ainola M., Li T.F., Mandelin J., Hukkanen M., Choi S.J., Salo J. Involvement of a disintegrin and a metalloproteinase 8 (ADAM8) in osteoclastogenesis and pathological bone destruction. Ann Rheum Dis. 2009;68:427–434. doi: 10.1136/ard.2008.088260. [DOI] [PubMed] [Google Scholar]
- 56.Mandelin J., Li T.F., Hukkanen M.V., Liljestrom M., Chen Z.K., Santavirta S. Increased expression of a novel osteoclast-stimulating factor, ADAM8, in interface tissue around loosened hip prostheses. J Rheumatol. 2003;30:2033–2038. [PubMed] [Google Scholar]
- 57.Djouad F., Delorme B., Maurice M., Bony C., Apparailly F., Louis-Plence P. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res Ther. 2007;9:R33. doi: 10.1186/ar2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flannery C.R., Little C.B., Caterson B., Hughes C.E. Effects of culture conditions and exposure to catabolic stimulators (IL-1 and retinoic acid) on the expression of matrix metalloproteinases (MMPs) and disintegrin metalloproteinases (ADAMs) by articular cartilage chondrocytes. Matrix Biol. 1999;18:225–237. doi: 10.1016/s0945-053x(99)00024-4. [DOI] [PubMed] [Google Scholar]
- 59.Ma G.F., Liljestrom M., Ainola M., Chen T., Tiainen V.M., Lappalainen R. Expression of ADAM9 (meltrin-gamma) around aseptically loosened total hip replacement implants. Rheumatology (Oxford) 2006;45:808–814. doi: 10.1093/rheumatology/kel003. [DOI] [PubMed] [Google Scholar]
- 60.Chubinskaya S., Mikhail R., Deutsch A., Tindal M.H. ADAM-10 protein is present in human articular cartilage primarily in the membrane-bound form and is upregulated in osteoarthritis and in response to IL-1alpha in bovine nasal cartilage. J Histochem Cytochem. 2001;49:1165–1176. doi: 10.1177/002215540104900910. [DOI] [PubMed] [Google Scholar]
- 61.Isozaki T., Rabquer B.J., Ruth J.H., Haines G.K., 3rd, Koch A.E. ADAM-10 is overexpressed in rheumatoid arthritis synovial tissue and mediates angiogenesis. Arthritis Rheum. 2013;65:98–108. doi: 10.1002/art.37755. [DOI] [PubMed] [Google Scholar]
- 62.Okada A., Mochizuki S., Yatabe T., Kimura T., Shiomi T., Fujita Y. ADAM-12 (meltrin alpha) is involved in chondrocyte proliferation via cleavage of insulin-like growth factor binding protein 5 in osteoarthritic cartilage. Arthritis Rheum. 2008;58:778–789. doi: 10.1002/art.23262. [DOI] [PubMed] [Google Scholar]
- 63.Valdes A.M., Hart D.J., Jones K.A., Surdulescu G., Swarbrick P., Doyle D.V. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004;50:2497–2507. doi: 10.1002/art.20443. [DOI] [PubMed] [Google Scholar]
- 64.Kerna I., Kisand K., Tamm A.E., Lintrop M., Veske K., Tamm A.O. Missense single nucleotide polymorphism of the ADAM12 gene is associated with radiographic knee osteoarthritis in middle-aged Estonian cohort. Osteoarthritis and Cartilage. 2009;17:1093–1098. doi: 10.1016/j.joca.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 65.Kerna I., Kisand K., Laitinen P., Tamm A.E., Kumm J., Lintrop M. Association of ADAM12-S protein with radiographic features of knee osteoarthritis and bone and cartilage markers. Rheumatol Int. 2012;32:519–523. doi: 10.1007/s00296-010-1717-6. [DOI] [PubMed] [Google Scholar]
- 66.Shin M.H., Lee S.J., Kee S.J., Song S.K., Kweon S.S., Park D.J. Genetic association analysis of GDF5 and ADAM12 for knee osteoarthritis. Jt Bone Spine. 2012;79:488–491. doi: 10.1016/j.jbspin.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 67.Kerna I., Kisand K., Tamm A.E., Kumm J., Tamm A.O. Two single-nucleotide polymorphisms in ADAM12 gene are associated with early and late radiographic knee osteoarthritis in Estonian population. Arthritis. 2013;2013 doi: 10.1155/2013/878126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lou S., Zhao Z., Qian J., Zhao K., Wang R. Association of single nucleotide polymorphisms in ADAM12 gene with susceptibility to knee osteoarthritis: a case-control study in a Chinese Han population. Int J Clin Exp Pathol. 2014;7:5154–5159. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L., Guo L., Tian F., Hao R., Yang T. Analysis of single nucleotide polymorphisms within ADAM12 and risk of knee osteoarthritis in a Chinese Han population. Biomed Res Int. 2015;2015 doi: 10.1155/2015/518643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poonpet T., Tammachote R., Tammachote N., Kanitnate S., Honsawek S. Association between ADAM12 polymorphism and knee osteoarthritis in Thai population. Knee. 2016;23:357–361. doi: 10.1016/j.knee.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 71.Limer K.L., Tosh K., Bujac S.R., McConnell R., Doherty S., Nyberg F. Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case-control population (the GOAL study) Osteoarthritis and Cartilage. 2009;17:782–789. doi: 10.1016/j.joca.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 72.Kveiborg M., Albrechtsen R., Couchman J.R., Wewer U.M. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40:1685–1702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 73.Kveiborg M., Albrechtsen R., Rudkjaer L., Wen G., Damgaard-Pedersen K., Wewer U.M. ADAM12-S stimulates bone growth in transgenic mice by modulating chondrocyte proliferation and maturation. J Bone Min Res. 2006;21:1288–1296. doi: 10.1359/jbmr.060502. [DOI] [PubMed] [Google Scholar]
- 74.Böhm B.B., Aigner T., Roy B., Brodie T.A., Blobel C.P., Burkhardt H. Homeostatic effects of the metalloproteinase disintegrin ADAM15 in degenerative cartilage remodeling. Arthritis Rheum. 2005;52:1100–1109. doi: 10.1002/art.20974. [DOI] [PubMed] [Google Scholar]
- 75.Böhm B.B., Aigner T., Gehrsitz A., Blobel C.P., Kalden J.R., Burkhardt H. Up-regulation of MDC15 (metargidin) messenger RNA in human osteoarthritic cartilage. Arthritis Rheum. 1999;42:1946–1950. doi: 10.1002/1529-0131(199909)42:9<1946::AID-ANR21>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 76.Böhm B.B., Schirner A., Burkhardt H. ADAM15 modulates outside-in signalling in chondrocyte-matrix interactions. J Cell Mol Med. 2009;13:2634–2644. doi: 10.1111/j.1582-4934.2008.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Böhm B.B., Freund I., Krause K., Kinne R.W., Burkhardt H. ADAM15 adds to apoptosis resistance of synovial fibroblasts by modulating focal adhesion kinase signaling. Arthritis Rheum. 2013;65:2826–2834. doi: 10.1002/art.38109. [DOI] [PubMed] [Google Scholar]
- 78.Böhm B., Hess S., Krause K., Schirner A., Ewald W., Aigner T. ADAM15 exerts an antiapoptotic effect on osteoarthritic chondrocytes via up-regulation of the X-linked inhibitor of apoptosis. Arthritis Rheum. 2010;62:1372–1382. doi: 10.1002/art.27387. [DOI] [PubMed] [Google Scholar]
- 79.Herren B., Garton K.J., Coats S., Bowen-Pope D.F., Ross R., Raines E.W. ADAM15 overexpression in NIH3T3 cells enhances cell-cell interactions. Exp Cell Res. 2001;271:152–160. doi: 10.1006/excr.2001.5353. [DOI] [PubMed] [Google Scholar]
- 80.Charrier L., Yan Y., Driss A., Laboisse C.L., Sitaraman S.V., Merlin D. ADAM-15 inhibits wound healing in human intestinal epithelial cell monolayers. Am J Physiol Gastrointest Liver Physiol. 2005;288:G346–G353. doi: 10.1152/ajpgi.00262.2004. [DOI] [PubMed] [Google Scholar]
- 81.Chen Q., Meng L.H., Zhu C.H., Lin L.P., Lu H., Ding J. ADAM15 suppresses cell motility by driving integrin alpha5beta1 cell surface expression via Erk inactivation. Int J Biochem Cell Biol. 2008;40:2164–2173. doi: 10.1016/j.biocel.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 82.Kratzschmar J., Lum L., Blobel C.P. Metargidin, a membrane-anchored metalloprotease-disintegrin protein with an RGD integrin binding sequence. J Biol Chem. 1996;271:4593–4596. doi: 10.1074/jbc.271.9.4593. [DOI] [PubMed] [Google Scholar]
- 83.Nath D., Slocombe P.M., Stephens P.E., Warn A., Hutchinson G.R., Yamada K.M. Interaction of metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on different haemopoietic cells. J Cell Sci. 1999;112(Pt 4):579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- 84.Eto K., Puzon-McLaughlin W., Sheppard D., Sehara-Fujisawa A., Zhang X.P., Takada Y. RGD-independent binding of integrin alpha9beta1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J Biol Chem. 2000;275:34922–34930. doi: 10.1074/jbc.M001953200. [DOI] [PubMed] [Google Scholar]
- 85.Martin J., Eynstone L.V., Davies M., Williams J.D., Steadman R. The role of ADAM 15 in glomerular mesangial cell migration. J Biol Chem. 2002;277:33683–33689. doi: 10.1074/jbc.M200988200. [DOI] [PubMed] [Google Scholar]
- 86.Najy A.J., Day K.C., Day M.L. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Najy A.J., Day K.C., Day M.L. ADAM15 supports prostate cancer metastasis by modulating tumor cell-endothelial cell interaction. Cancer Res. 2008;68:1092–1099. doi: 10.1158/0008-5472.CAN-07-2432. [DOI] [PubMed] [Google Scholar]
- 88.Scheller J., Chalaris A., Garbers C., Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 89.Hall K.C., Hill D., Otero M., Plumb D.A., Froemel D., Dragomir C.L. ADAM17 controls endochondral ossification by regulating terminal differentiation of chondrocytes. Mol Cell Biol. 2013;33:3077–3090. doi: 10.1128/MCB.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saito K., Horiuchi K., Kimura T., Mizuno S., Yoda M., Morioka H. Conditional inactivation of TNFalpha-converting enzyme in chondrocytes results in an elongated growth plate and shorter long bones. PLoS One. 2013;8:e54853. doi: 10.1371/journal.pone.0054853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oldefest M., Dusterhoft S., Desel C., Thysen S., Fink C., Rabe B. Secreted Frizzled-related protein 3 (sFRP3)-mediated suppression of interleukin-6 receptor release by A disintegrin and metalloprotease 17 (ADAM17) is abrogated in the osteoarthritis-associated rare double variant of sFRP3. Biochem J. 2015;468:507–518. doi: 10.1042/BJ20141231. [DOI] [PubMed] [Google Scholar]
- 92.James C.G., Appleton C.T., Ulici V., Underhill T.M., Beier F. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell. 2005;16:5316–5333. doi: 10.1091/mbc.E05-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hikichi Y., Yoshimura K., Takigawa M. All-trans retinoic acid-induced ADAM28 degrades proteoglycans in human chondrocytes. Biochem Biophys Res Commun. 2009;386:294–299. doi: 10.1016/j.bbrc.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 94.Jowett J.B., Okada Y., Leedman P.J., Curran J.E., Johnson M.P., Moses E.K. ADAM28 is elevated in humans with the metabolic syndrome and is a novel sheddase of human tumour necrosis factor-alpha. Immunol Cell Biol. 2012;90:966–973. doi: 10.1038/icb.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mitsui Y., Mochizuki S., Kodama T., Shimoda M., Ohtsuka T., Shiomi T. ADAM28 is overexpressed in human breast carcinomas: implications for carcinoma cell proliferation through cleavage of insulin-like growth factor binding protein-3. Cancer Res. 2006;66:9913–9920. doi: 10.1158/0008-5472.CAN-06-0377. [DOI] [PubMed] [Google Scholar]
- 96.Mochizuki S., Tanaka R., Shimoda M., Onuma J., Fujii Y., Jinno H. Connective tissue growth factor is a substrate of ADAM28. Biochem Biophys Res Commun. 2010;402:651–657. doi: 10.1016/j.bbrc.2010.10.077. [DOI] [PubMed] [Google Scholar]
- 97.Ozeki N., Kawai R., Yamaguchi H., Hiyama T., Kinoshita K., Hase N. IL-1beta-induced matrix metalloproteinase-13 is activated by a disintegrin and metalloprotease-28-regulated proliferation of human osteoblast-like cells. Exp Cell Res. 2014;323:165–177. doi: 10.1016/j.yexcr.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 98.Rushton M.D., Reynard L.N., Barter M.J., Refaie R., Rankin K.S., Young D.A. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66:2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nugent M. MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthritis and Cartilage. 2016;24:573–580. doi: 10.1016/j.joca.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 100.Yamamoto K., Owen K., Parker A.E., Scilabra S.D., Dudhia J., Strickland D.K. Low density lipoprotein receptor-related protein 1 (LRP1)-mediated endocytic clearance of a disintegrin and metalloproteinase with thrombospondin motifs-4 (ADAMTS-4): functional differences of non-catalytic domains of ADAMTS-4 and ADAMTS-5 in LRP1 binding. J Biol Chem. 2014;289:6462–6474. doi: 10.1074/jbc.M113.545376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hartmann M., Herrlich A., Herrlich P. Who decides when to cleave an ectodomain? Trends Biochem Sci. 2013;38:111–120. doi: 10.1016/j.tibs.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 102.Przemyslaw L., Boguslaw H.A., Elzbieta S., Malgorzata S.M. ADAM and ADAMTS family proteins and their role in the colorectal cancer etiopathogenesis. BMB Rep. 2013;46:139–150. doi: 10.5483/BMBRep.2013.46.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]