Abstract
Modern intensive care units (ICUs) collect large volumes of data in monitoring critically ill patients. Clinicians in the ICUs face the challenge of interpreting large volumes of high-dimensional data to diagnose and treat patients. In this work, we explore the use of Hierarchical Dirichlet Processes (HDP) as a Bayesian nonparametric framework to infer patients’ states of health by combining multiple sources of data. In particular, we employ HDP to combine clinical time series and text from the nursing progress notes in a probabilistic topic modeling framework for patient risk stratification. Given a patient cohort, we use HDP to infer latent “topics” shared across multimodal patient data from the entire cohort. Each topic is modeled as a multinomial distribution over a vocabulary of codewords, defined over heterogeneous data sources. We evaluate the clinical utility of the learned topic structure using the first 24-hour ICU data from over 17,000 adult patients in the MIMIC-II database to estimate patients’ risks of in-hospital mortality. Our results demonstrate that our approach provides a viable framework for combining different data modalities to model patient’s states of health, and can potentially be used to generate alerts to identify patients at high risk of hospital mortality.
I. Introduction
Modern intensive care units (ICUs) provide continuous monitoring of critically ill patients, collecting large volumes of clinical and physiological data. Furthermore, detailed information about the patients’ disease progression, symptoms and medications is carefully documented by the clinical staff in the form of progress notes. Despite the rich sources of clinical data, existing severity scores, such as the Simplified Acute Physiology (SAPS) [1] and APACHE [2], use snapshot observations of commonly measured clinical variables to assess the severity of patients’ illness. Recently, a body of research surrounding the analysis of patient data using machine learning techniques has arisen, broadening the array of methods that can be used to analyze a variety of clinical and physiological data [3], [4], [5], [6], [7], [8].
In this work, we explore the use of Hierarchical Dirichlet Processes (HDP) [9] as a probabilistic topic modeling framework to estimate patient’s health state by combining multiple sources of information, including nursing notes, clinical variable ranges, and clustering analysis on physiological time series data. Our goal is to identify patterns from heterogeneous sources of patient data to alert clinicians of early warning signs of patient deterioration. We use HDP to automatically discover groups of co-occurring patterns, or “topics”, shared across multimodal patient data from the entire cohort. We characterize each patient by a k-dimensional vector, defined as the distribution of the patient data over the learned k topics. In this study, we apply HDP to the first 24 hour ICU data from the MIMIC II database [10], and evaluate the clinical utility of our approach in predicting patient in-hospital mortality.
Topic modeling is a Bayesian learning method often used for analyzing documents. The method represents documents using an un-ordered collection of words, and then finds patterns of co-occurring words across multiple documents. This enables grouping of words by themes, which are called topics [9], [11], [12]. Several recent works have analyzed patient data in a topic modeling framework [6], [13], [14], [15]. Saria et al. [13] developed a method of analyzing time series data by applying topic modeling to find patterns in time series data. In [6], [14], physiological variables were combined with topic models of clinical text for patient mortality prediction. However, these previous works did not combine features of physiological time series with clinical text in the same generative topic modeling framework to characterize patients’ states of health.
In [16], Lehman et al. used a Bayesian nonparametric switching state space approach to model dynamics of time series for outcome prediction. In [17], a method based on generalized linear dynamic models was proposed to estimate patients’ mortality risks in the ICU by combining heterogeneous data. In contrast, our approach focuses on pattern discovery and uses HDP to combine clinical time series with text in a Bayesian nonparametric mixture modeling framework for patient phenotyping and risk stratification.
The rest of the paper is organized as follows. First, we describe the data used in the study. We then describe HDP as a topic modeling technique, and detail the methods used to encode data from clinical text and time series data as input to the HDP framework. Finally, we evaluate the clinical utility of our approach and present its predictive performance in hospital mortality prediction.
II. Methodology
A. Patient and Variable Selection
Data for this study was obtained from the Multi-Parameter Intelligent Monitoring in Intensive Care (MIMIC- II) [10] database available from PhysioNet [18]. The creation, maintenance, and use of the MIMIC-II database was approved by the institutional review boards of the Massachusetts Institute of Technology (MIT) and Beth Israel Deaconess Medical Center (BIDMC). The database contains records from 24,581 ICU patients admitted to Bostons Beth Israel Deaconess Medical Center between 2001–2008. MIMIC-II contains information from bedside monitors, validated by ICU nurses.
We analyzed patient data from the MIMIC-II database, using only adult patients whose stay in the ICU lasted for at least 24 hours. The measured clinical variables included heart rate (HR), mean arterial blood pressure, respiration rate, temperature, Glasgow Coma Score (GCS), glucose, sodium, potassium, hematocrit, creatinine, urine output, white blood cell count, HCO3, lactate, pH, and PCO2. The data was processed to consist of hourly averages of the patient data for each variable. Only the first 24 hours of data for each patient were used in this study.
B. Topic Modeling Using HDP
We used HDP to automatically discover topics from multimodal patient data. A topic is a multinomial distribution over words from a finite, known vocabulary. In this study, the vocabulary of words were defined based on combining “codewords” extracted from both time series and clinical text; a topic represents shared groups of co-occurring time series features and clinical concepts extracted from the nursing progress notes. HDP models documents with multiple Dirichlet Processes (DP), one for each document, to enable document-specific mixing proportions. It uses a non-parametric prior to enable mixture models to share components [9]. The number of topics is assumed to be unknown a priori, and is inferred from the data. Running HDP results in a topic model containing an inferred number of discovered topics.
For HDP parameter settings, we used the same notations as in [9]. A two-level hierarchical Dirichlet process implementation was used to build our topic models. We used a symmetric Dirichlet distribution with parameters of 0.2 for the prior H over topic distributions. We used fixed concentration parameters 0.1 and 1 for γ and α respectively. Results presented were output of the model after 1000 iterations of Gibbs sampling.
C. Data Preparation
We generated our input for HDP by combining features from both nursing notes and the time series. In order to incorporate time series data into the HDP framework, we used two different methods to encode features of time series data as codewords. We describe the two encoding procedures in the following sections.
1) Static Features of Clinical Time Series
In the first method, we quantized each time series using the clinician-defined static ranges in SAPS-I[1]. For variables without listed SAPS-I variable ranges, we used the percentiles at 2.5%, 25%, 50%, 75%, and 97.5% as the edges for our clinical variable ranges. In total, 103 unique codewords corresponding to different clinical variable ranges were defined. Our data for generating the clinical variable range input iterated through the data for each patient, counting the number of times that the average hourly data for each patient fell into the various ranges for the clinical variables. Each variable range was turned into a codeword for HDP.
2) Dynamic Features of Clinical Time Series
In the second method, we used K-means clustering to group time series into clusters with similar trajectories and dynamic features for generating codewords for the topic modeling algorithm. For this we used hourly averaged clinical time series from the last 18 hours of the first 24 hours of any given ICU stay for clustering procedure, and only patients with at least two data points in this range for the variable in question were included in the clustering.
We extracted feature vectors from time series of each patient. These feature vectors included the minimum and maximum values, the slope and y-intercept of the corresponding linear regression, and the sum of the squared residuals of the linear regression. We chose these features because they succinctly captured the dynamic features of the time series, including threshold values, overall trend, and variability. Our K-means clustering algorithm used a standard Euclidean distance function, and was run once for each variable, with one feature vector representing each patients data for the given variable. We used the silhouette values [19] to determine the number of clusters for each variable. K-means was then run on the feature matrix for each variable, and the results were turned into word counts by having a codeword to represent each cluster, resulting in 100 unique codewords covering all the clusters for the 16 variables. Word counts amounted to one word for each cluster that a given patient had a feature vector assigned to.
3) Clinical Concepts from Nursing Progress Notes
Using natural language processing techniques [20], the nursing progress notes were parsed into clinical concepts, defined by the Unified Medical Language System (UMLS) codes. We generated word counts using the UMLS clinical concepts extracted from nursing notes. These word counts were determined using 5792 unique codewords which ranged from documented procedures to patient symptoms to noted diseases. The number of occurrences for each of these codewords in the nurses notes for each patient were totaled and included in the input for HDP.
D. Evaluation and Statistical Methods
Using the word to topic assignments, we constructed a topic proportion vector for each patient, which is a k-dimensional vector that contains the proportion of words belonging to each of the k topic. Patient data was divided into development (70%) and test sets (30%) in order to evaluate prediction performance. Our primary metric for measuring the success of the algorithm is area under the receiver operator characteristic curve (AUC). To evaluate the in-hospital mortality prediction performance, the topic proportion of each document (defined as the proportion of words assigned to each topic) was used as input to logistic regression for hospital mortality prediction. The nursing progress notes and time series of the 70% development set patients were used as the development set to build the topic model and train the logistic regression model; data from the remaining 30% testset patients were used as the held-out data set to test the mortality prediction performance.
In analyzing the development set, we performed association analysis on each time series cluster identified. Univariate logistic regression analysis produced p-values for association with patient mortality of clusters from the K-means clustering results. We measured the p-value, cluster size, and odds ratio for each cluster. Odds ratios greater than one indicate that patients in the given cluster have a higher odds to die in the hospital than the average patient, and odds ratios less than one indicate that patients in the given cluster have a lower odds for in-hospital mortality than the average patient.
III. Results
We analyzed over 17,000 adult patients who had stayed in the ICU for at least 24 hours. The development set contained 12,091 patients and the test set contained 5,183 patients. We encoded the first 24-hour patient data (nursing progress notes and 16 clinical variables) using a total of 5995 unique words; the number of unique UMLS terms in this corpus was 5792; the number of unique time series “codewords” was 203 (103 from clinician-defined static ranges, and 100 from K-means clustering). Figure 1 shows an example progress notes where bolded texts were extracted as UMLS codewords for formulating the word count input to HDP.
Fig. 1.

An example clinical text for a patient from the MIMIC-II database, where bolded texts were extracted as UMLS concepts, representing either a disease, symptom, medication, procedure, or finding from the patients nursing notes. A total of 5792 unique UMLS codes were used to capture clinical concepts from the first 24-hour nursing notes of the patient cohort.
A. Clustering Results
The optimal number of clusters learned based on the silhouette function was between 5 and 11 clusters, depending on the variable. In Figure 2, we generated plots of example patient time series nearest the cluster centroids for example variable. Figure 2a shows an example HR cluster (N=360) with a p-value of 0.0451, and an odds ratio of 0.6833. Figure 2b shows a GCS cluster (N=687) with a p-value of 0.0094, and an odds ratio of 1.33 with a 95% confidence interval of (1.07 1.65). Figure 2c shows a sodium cluster (N=277) with a p-value of 0.0006 and an odds ratio of 1.6976. Figure 2d shows a HCO3 cluster (N=162) with a p-value of 0.0154 and an odds ratio of 1.6147. These results demonstrate that there is value in phenotyping the patients based on the trajectories and dynamic features of their vital signs and lab results, and in particular there are significant patterns with predictive value for mortality.
Fig. 2.
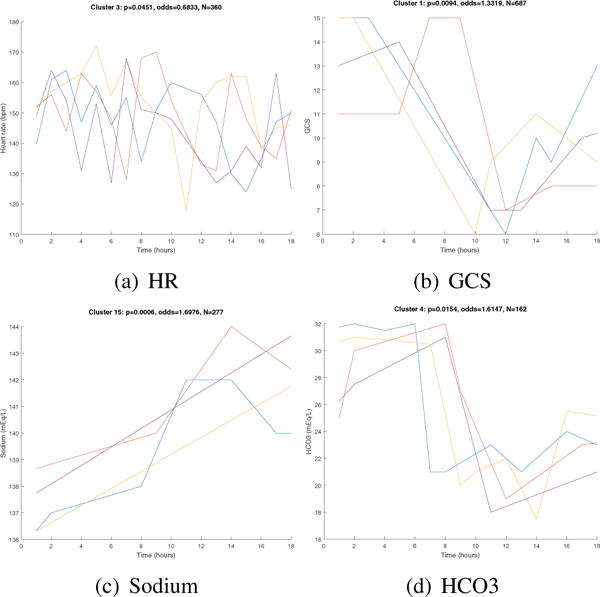
Example time series clusters. Each plot shows time series of four different patients from the same cluster. For each cluster plot, we show the p-value and odds ratios from association analysis and the number of patients N in each cluster. A total of 100 unique “words” were defined, each corresponding to a unique cluster of similar time series segment. Note that GCS works on a scale of 3–15 to indicate how responsive the patient is; three denotes a completely unresponsive patient, whereas fifteen indicates a fully responsive patient.
B. Hospital Mortality Prediction Performance from HDP Combining Text and Time Series
To evaluate the in-hospital mortality prediction performance, the topic proportion of each patient was used as input to logistic regression for mortality prediction. HDP generated 49 topics; after pruning rare topics, the remaining 40 topics were used to construct topic proportions for the prediction task. We performed 10-fold cross validation using the development set, producing a median AUC of 0.80 with an interquartile range of (0.79 0.81). We then apply the model with the best AUC from the development set on the test set, an AUC of 0.80 was achieved. This exceeds the performance when using an algorithm based on the SAPS-I acuity score, which produces an average AUC of 0.72.
IV. Discussion
We have conducted a proof-of-concept study and demonstrated that HDP provided a viable framework for combining different data modalities for patient risk stratification. Using HDP, related clinical concepts and time series patterns that tended to co-occur across patient data were grouped together to form topics. Topic learning was done in a completely unsupervised manner; no prior medical knowledge of disease associations were used. Our clustering results have identified time series patterns (e.g., from Glasgow Coma Scores) that were clinically meaningful and statistically significantly associated with patient hospital mortality, demonstrating the potential use of a dynamics based approach in patient phenotyping.
We have chosen to use clinician-defined thresholds to quantize physiological variables so as to learn clinically interpretable “codewords” from time series data. While we have used hand-crafted features to characterize trends and variability patterns in clinical time series, future work will include automated feature learning and a dynamics-based approach in modeling physiological time series [3], [4], [16]. We employed K-means clustering as a simple and computationally efficient approach to identify patterns in clinical time series. Future work will investigate clustering techniques that can better capture patterns and trajectories of sparse and irregularly sampled clinical time series [21], [22]. We chose HDP, as opposed to the most prevalent topic modeling framework, Latent Dirichlet Allocation (LDA) [23], because HDP infers the number of topics from the data, and therefore is more suited for applications in pattern discovery, whereas LDA assumes a fixed number of topics.
V. Conclusions
We have presented a Bayesian non-parametric approach based on HDP that combined data from different modalities for estimating patient’s health state. We demonstrated that the learned topic structure of time series and clinical text contained prognostic values in stratifying patients’ in-hospital mortality risks. Future work involves incorporating other sources of data (such as waveforms or images) into the HDP framework to develop a more comprehensive view of each patient’s condition. As part of our ongoing and future work, we plan to investigate the clinical utility of our approach in identifying disease phenotypes [24] for patient prognosis and treatment decision support.
Acknowledgments
The authors thank Eric Lehman for his contribution in deriving and evaluating time series features, Yang Dai for her assistance with data analysis, and Dr. Mohammed Saeed for his insightful comments. This work was supported by the National Institutes of Health (NIH) grant R01-EB017205, R01-EB001659 and R01GM104987 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIBIB or the NIH.
References
- 1.Le Gall JR, Loirat P, Alperovitch A, Glaser P, Granthil C, Mathieu, Mercier P, Thomas R, Villers D. A simplified acute physiology score for ICU patients. Crit Care Med. 1984 Nov;12(11):975977. doi: 10.1097/00003246-198411000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman JE, Kramer AA, McNair DS, Malila FM. Acute Physiology and Chronic Health Evaluation (APACHE) IV: hos pital mortality assessment for todays critically ill patients. Crit Care Med. 2006 May;34(5):12971310. doi: 10.1097/01.CCM.0000215112.84523.F0. [DOI] [PubMed] [Google Scholar]
- 3.Lasko T, Denny J, Levy M. Computational Phenotype Discovery Using Unsupervised Feature Learning over Noisy, Sparse, and Irregular Clinical Data. PLOS ONE. 2013 doi: 10.1371/journal.pone.0066341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehman LH, Adams RP, Mayaud L, Moody GB, Malhotra A, Mark RG, Nemati S. A Physiological Time Series Dynamics-Based Approach to Patient Monitoring and Outcome Prediction. IEEE Journal of Biomedical and Health Informatics. 2015 May;19(3):1068–1076. doi: 10.1109/JBHI.2014.2330827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehman LH, Mark RG, Nemati S. A Model-Based Machine Learning Approach to Probing Autonomic Regulation from Nonstationary Vital-Sign Time Series. IEEE Journal of Biomedical and Health Informatics. 2016 doi: 10.1109/JBHI.2016.2636808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehman LH, Saeed M, Long W, Lee J, Mark RG. Risk stratification of ICU patients using topic models inferred from unstructured progress notes. Proceedings of the AMIA Annual Symposium. 2012 Nov;:505–511. [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AEW, Clifford GD. Journal of Critical Care. Vol. 28. Elsevier; 2013. Risk-adjustment of patient subpopulations in the intensive care unit using OASIS, a novel severity score; pp. 1 e20–e21. [Google Scholar]
- 8.Miotto R, Kidd BA, Dudley JT. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Scientific Reports. 2016;6 doi: 10.1038/srep26094. Article number: 26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teh Y, Jordan M, Beal MJ, Blei D. Hierarchical Dirichlet Processes. Journal of the American Statistical Association. 2006;101:1566–1581. [Google Scholar]
- 10.Saeed M, Villarroel M, Reisner A, Clifford G, Lehman LH, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. Multiparameter Intelligent Monitoring in Intensive Care II: A public-access intensive care unit database. Crit Care Med. 2011;39:952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blei D, Lafferty J. Topic Models. In: Srivastava A, Sahami M, editors. Text Mining: Classification, Clustering, and Applications. Chapman & Hall/CRC; 2009. (Data Mining and Knowledge Discovery Series). [Google Scholar]
- 12.Blei D, Carin L, Dunson D. Probabilistic Topic Models. IEEE Signal Processing Magazine. 2010 Nov;:5565. doi: 10.1109/MSP.2010.938079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saria S, Koller D, Penn A. Learning individual and population level traits from clinical temporal data. NIPS Predictive Models in Personalized Medicine. 2010 [Google Scholar]
- 14.Ghassemi M, Naumann T, Doshi-Velez F, Brimmer N, Joshi R, Rumshisky A, Szolovits P. Unfolding Physiological State: Mortality Modelling in Intensive Care Units. NIH; Aug, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman LH, Long W, Saeed M, Mark RG. Latent Topic Discovery of Clinical Concepts from Hospital Discharge Summaries of a Heterogeneous Patient Cohort. Proceedings of the 36th International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Chicago. August 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman LH, Johnson M, Nemati S, Adams RP, Mark RG. In: Bayesian nonparametric learning of switching dynamics in cohort physiological time series: application in critical care patient monitoring, Chapter 11 in Advanced State Space Methods for Neural and Clinical Data. Chen Z, editor. Cambridge University Press; 2015. pp. 257–282. [Google Scholar]
- 17.Caballero K, Akella R. Dynamically Modeling Patient’s Health State from Electronic Medical Records: A Time Series Approach. Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. :69–78. [Google Scholar]
- 18.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):e215e220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 19.Ng AY, Jordan MI, Weiss Y, et al. On spectral clustering: Analysis and an algorithm. Advances in neural information processing systems. 2002;2:849856. [Google Scholar]
- 20.Long W. Extracting Diagnoses from Discharge Summaries. Proc AMIA 2005 Symposium. 2005:470–474. [PMC free article] [PubMed] [Google Scholar]
- 21.Pimentel M, Clifton D, Tarassenko L. Gaussian process clustering for the functional characterisation of vital-sign trajectories, 2013. IEEE International Workshop on Machine Learning for Signal Processing (MLSP) 2013 [Google Scholar]
- 22.Marlin B, Kale D, Khemani R, et al. Unsupervised Pattern Discovery in Electronic Health Data Using Probabilistic Clustering Models. Proceedings of IHI. 2012 [Google Scholar]
- 23.Blei D, Ng A, Jordan M. Latent Dirichlet Allocation. The Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 24.Dai Y, Lokhandwala S, Long W, Mark RG, Lehman LH. Proceedings of IEEE International Conference on Biomedical and Health Informatics (BHI) Orlando FL: 2017. Phenotyping Hypotensive Patients in Critical Care Using Hospital Discharge Summaries. [DOI] [PMC free article] [PubMed] [Google Scholar]


