Abstract
P0-Cre and Wnt1-Cre mouse lines have been widely used in combination with loxP-flanked mice to label and genetically modify neural crest (NC) cells and their derivatives. Wnt1-Cre has been regarded as the gold standard and there have been concerns about the specificity of P0-Cre because it is not clear about the timing and spatial distribution of the P0-Cre transgene in labeling NC cells at early embryonic stages. We re-visited P0-Cre and Wnt1-Cre models in the labeling of NC cells in early mouse embryos with a focus on cranial NC. We found that R26-lacZ Cre reporter responded to Cre activity more reliably than CAAG-lacZ Cre reporter during early embryogenesis. Cre immunosignals in P0-Cre and reporter (lacZ and RFP) activity in P0-Cre/R26-lacZ and P0-Cre/R26-RFP embryos were detected in the cranial NC and notochord regions in E8.0–9.5 (4–19 somite) embryos. P0-Cre transgene expression was observed in migrating NC cells and was more extensive in the forebrain and hindbrain but not apparent in the midbrain. Differences in the Cre distribution patterns of P0-Cre and Wnt1-Cre were profound in the midbrain and hindbrain regions, i.e., extensive in the midbrain of Wnt1-Cre and in the hindbrain of P0-Cre embryos. The difference between P0-Cre and Wnt1-Cre in labeling cranial NC may provide a better explanation of the differential distributions of their NC derivatives and of the phenotypes caused by Cre-driven genetic modifications.
Keywords: P0-Cre, neural crest, notochord, lineage tracing, derivation, mouse
Introduction
Neural crest (NC) is a multipotent cell population derived from the lateral ridges of the neural plate in early vertebrate embryos (Leikola, 1976; Trainor, 2015). After delamination from the neural folds, NC cells migrate ventrally and extensively, giving rise to a wide variety of differentiated cell types, e.g., neurons, glia, bone, cartilage, and connective tissue of the head (Bronner-Fraser, 2004; Crane and Trainor, 2006; Meulemans and Bronner-Fraser, 2004; Munoz and Trainor, 2015; Trainor, 2005b). Labeling and tracking NC have been essential for understanding the NC contribution to different tissue and cell types, which is fundamental for organogenesis.
Recently, cell lineage tracing using NC-specific promoter driven Cre transgenic mouse lines in combination with Cre reporter mouse lines has facilitated genetic marking of NC cells and their derivatives. Multiple Cre transgenic mouse lines have been generated using a NC marker gene promoter, e.g., Wnt1-Cre (Echelard et al., 1994; Jiang et al., 2002; McMahon et al., 1992), P0-Cre (Wang et al., 2011; Yamauchi et al., 1999; Zhang et al., 1995), Dhh-Cre (Gershon et al., 2009; Wang et al., 2007), Pax3-Cre (Jarad and Miner, 2009), HtPA-Cre (Lee et al., 2013), Sox10-Cre (Simon et al., 2012), Mef2c-F10N-Cre (Aoto et al., 2015). Use of these models has yielded new data pertaining to NC cell specifications in mice, e.g., distinct genesis of skin-derived precursors in craniofacial and dorsal skin from NC and mesoderm, respectively (Jinno et al., 2010), NC and placodal derivation of the otic vesicle (Freyer et al., 2011), and a dual origin of sensory organs that include olfactory epithelium (Katoh et al., 2011), tooth bud epithelium (Wang et al., 2011), and taste bud cells (Boggs et al., 2016; Liu et al., 2012).
However, variations/discrepancies in the different models that label NC cell lineage have been reported, such as in tooth (Wang et al., 2011); olfactory epithelium (Suzuki et al., 2013); heart (Cavanaugh et al., 2015; Milgrom-Hoffman et al., 2014; Nakamura et al., 2006; Tomita et al., 2005), and taste organs (Boggs et al., 2016; Liu et al., 2012). In comparative studies on NC contributions to specific lineages, the Wnt1-Cre and P0-Cre lines have been widely used (Katoh et al., 2011; Liu et al., 2012; Morikawa et al., 2009; Nagoshi et al., 2011; Nagoshi et al., 2008; Nakamura et al., 2006; Yoshida et al., 2006; Yoshida et al., 2008). Wnt1-Cre has been regarded as the gold standard for NC lineage, however, Wnt1-Cre also labels cells in the neural tube that makes researchers question the specificity of this line and other models when a difference is observed compared to Wnt1-Cre (Trainor, 2005a). In fact, a difference between Wnt1-Cre and P0-Cre in labeling NC has been found in multiple organs (Freem et al., 2010; Kawakami et al., 2011; Lewis et al., 2013; Liu et al., 2012; Wang et al., 2011). In contrast to Wnt1-Cre, which has been reported to trace NC cells as early as E8.5 using a ROSA26 (hereafter, R26-lacZ) Cre reporter in mice (Chai et al., 2000; Ikeya et al., 1997; Jiang et al., 2002; McMahon et al., 1992), P0-Cre has been reported to label NC derivatives at stages later than E9.0 using CAAG-CAT-lacZ (hereafter, CAAG-lacZ) as a reporter system (Yamauchi et al., 1999). Although in situ hybridization for P0-Cre transgene expression was performed (Yamauchi et al., 1999), there is no detailed information available regarding the labeling of P0-Cre in NC cells at early stages.
In the present study, the expression of two commonly used reporter transgenes, CAAG-lacZ and R26-lacZ, were compared in early embryos, and we found that DNA recombination was not consistent in the CAAG-lacZ reporter when driven by Meox2-Cre in early embryos. To further evaluate the timing, specificity and spatial distribution of P0-Cre and Wnt1-Cre activity, we used the R26-lacZ and R26-tdTomato (R26-RFP) reporters and carried out immunoreactions for Cre. Subsequently, we thoroughly compared the presence of Cre recombinase in pre- and post-migratory NC cells in P0-Cre and Wnt1-Cre embryos. Our data indicate that the P0-Cre transgene expression is specifically distributed in migrating cranial NC in addition to the notochord, and Cre immunosignals were especially extensive in the forebrain and hindbrain regions. In contrast, Wnt1-Cre transgene expression was obvious in forebrain and midbrain NC cells but was sparse in migrating NC in the hindbrain regions.
Materials and Methods
Animals
Animals were maintained and used in compliance with institutional animal care protocols of the University of Georgia, the University of Michigan and the National Institute of Environmental Health, and in accordance with the National Institutes of Health Guidelines for Care and Use of Animals in research.
Mice carrying Cre recombinase driven by the protein zero (P0) promoter (P0-Cre mouse, C57BL/6J-Tg(P0-Cre)94Imeg (ID 148)), were provided by CARD, Kumamoto University, Japan (Yamauchi et al., 1999) and were mated with C57BL/6 wild type (WT) or Cre-reporter mice R26-lacZ (Soriano, 1999) or ZEG (LacZ/EGFP) (Novak et al., 2000) or R26-tdTomato (RFP) (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, Jackson Lab, Stock #007914) to generate P0-Cre/WT, P0-Cre/R26-lacZ, P0-Cre/ZEG, and P0-Cre/RFP mice. Meox2-Cre mice, which express Cre in the epiblast from E5.5 (Tallquist and Soriano, 2000), were also bred with R26-lacZ or Cre-reporter CAAG-lacZ mouse line (Sakai and Miyazaki, 1997). Wnt1-Cre mice (B6.Cg-Tg(Wnt1-cre)11Rth Tg(Wnt1-GAL4)11Rth/J, Jackson Lab, Stock# 009107) were bred with C57BL/6 WT or Cre-reporter mice R26-lacZ (Soriano, 1999) or R26-RFP to generate Wnt1-Cre/WT or Wnt1-Cre/R26-lacZ or Wnt1-Cre/RFP. R26-lacZ reporter mice were maintained on a mixed background of C57BL/6 (majority) and 129S6.
Tissue collection
Timed pregnant mice were euthanized using CO2 followed by cervical dislocation to ensure death. The uterus with embryos was removed and embryos were retrieved under a microscope. The tissues were further dissected and processed for analysis, as described below.
Embryos from P0-Cre mice were harvested at embryonic day (E) 8.0, E8.5, E9.5, and E10.5. Newborn P0-Cre/ZEG mice were harvested immediately after birth. Meox2-Cre mouse embryos carrying a Cre-reporter were obtained from E6.5 to E11.5. Wnt1-Cre mouse embryos were collected at E8.0–E8.5. Noon of the day on which the dam was positive for vaginal plug was designated as E0.5. The stages of these embryos were confirmed by comparing the sizes and morphologies of multiple organs. The number of pairs of somites was counted under a microscope at the time of dissection.
Genotyping
Specific primers were used to genotype the mice. Primers A (5′-CTC GTG ATC TGC AAC TCC AGTC-3′) and B (5′-GAG ACT AGT GAG ACG TGC TACT-3′) were used to amplify fragments from the un-recombined R26-lacZ reporter allele (approx. 550 bp). Primers A and C (5′-TGT GAG CGA GTA GTA ACA ACC-3′) were used to detect the Cre-recombined allele (approx. 680 bp). Primers E (5′-CAG TCA GTT GCT CAA TGT ACC-3′) and F (5′-ACT GGT GAA ACT CAC CCA-3′) were used to amplify fragments from the un-recombined CAAG-lacZ allele (approx. 450 bp), whereas primers D (5′-GTG CTG GTT ATT GTG CTG TCTC-3′) and C were used to detect Cre-dependent deletion of the floxed region (approx. 680 bp). To detect a Cre transgene, Cre1 (5′-GGA CAT GTT CGG GAT CGC CAG GCG-3′) and Cre2 (5′-GCA TAA CCA GTG AAA CAG CAT TGC TG-3′) primers were used.
X-Gal staining
After dissection, the embryos were fixed with 4% paraformaldehyde for 5 min at 4°C, and washed three times with 0.1 M sodium phosphate buffer (PBS, pH 7.5), then stained with 1 mg/ml of 5-bromo-4-chloro-3-indolyl-beta-D-galactoside (X-gal, Sigma, Cat# B-4252) in 0.1 M PBS, 0.1% sodium deoxycholate, 0.2% Nonie P-40, 2 mM magnesium chloride, 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide at 37°C.
Immunohistochemistry
Embryos were fixed with 4% paraformaldehyde in 0.1 M PBS (pH 7.3) for 2 hr at 4°C, rinsed in PBS and then infiltrated with 20–30% sucrose prior to embedding in OCT/Tissue Tek (Sakura Finetek). Serial sections were cut at 10-μm thickness. Sections were air dried, rehydrated and blocked with 10% normal donkey serum for 1 hr at room temperature. Primary antibodies against the following proteins were applied: Cre recombinase (1:500, MAB3120, EMD Millipore, Billerica, MA), FOXA2 (1:500, 07-633, EMD Millipore, Billerica, MA), GFP (1:100, ab13970, abcam, Cambridge, MA), p75 (1:200, sc-6188, Santa Cruz Biotechnology, Dallas, TX), SOX9 (1:1000, sc-20095, Santa Cruz Biotechnology, Dallas, TX; 1:1000, ab185966, abcam, Cambridge, MA). Primary antibodies were diluted with 0.1 M PBS that contained 0.3% Triton X-100 and 1% normal donkey serum. After incubation with primary antibody overnight at 4°C, sections were washed three times with 0.1 M PBS, and then incubated with secondary antibodies that were conjugated with Alexa Fluor 488, Alexa Fluor 546 or 647 (1:500, Invitrogen) for 1 hr at room temperature in 0.1 M PBS containing 0.3% Triton X-100 and 1% normal donkey serum. Sections were counterstained with DAPI solution for 5–10 min to visualize nuclei and were then rinsed in PBS. After air drying, the slides were mounted with ProLong® Gold antifade medium (Invitrogen, Eugene, OR), and the sections were examined thoroughly using a light microscope (EVOS FL, Life Technologies). Co-localization of different immunosignals was confirmed and photographed using a laser-scanning confocal microscope (Zeiss LSM 710).
To confirm the specificity of immunosignals, slides without the addition of primary antibodies were used as negative staining controls for all immunostainings in the present study. For determining specificity of Cre immuno-labeling, Cre negative littermate embryos were sectioned and immunoreacted with the same concentration of primary and secondary antibodies. Additionally, Western blotting was conducted using E9.5 P0-Cre embryos (Cre+ embryos and Cre− littermate control) to further investigate the specificity of antibodies against Cre recombinase (1:5000, MAB3120, EMD Millipore, Billerica, MA). A ubiquitously expressed protein, IκB kinase complex associated protein (IKAP) (1:5000, sc-8336, Santa Cruz Biotechnology) was used as a loading control. Equal amounts of protein were used for electrophoresis in sodium dodecyl sulfate-polyacrylamide gel and transferred to a nitrocellulose membrane. Procedures for blocking and antibody probing were performed as described (Li et al., 2011). A single band at the expected molecular weight was detected in a genotype-dependent manner with the Cre antibody (Supplemental Fig. 1).
Results
Meox2-Cre driven R26-lacZ reporter expression emerges earlier and is more consistent than CAAG-lacZ
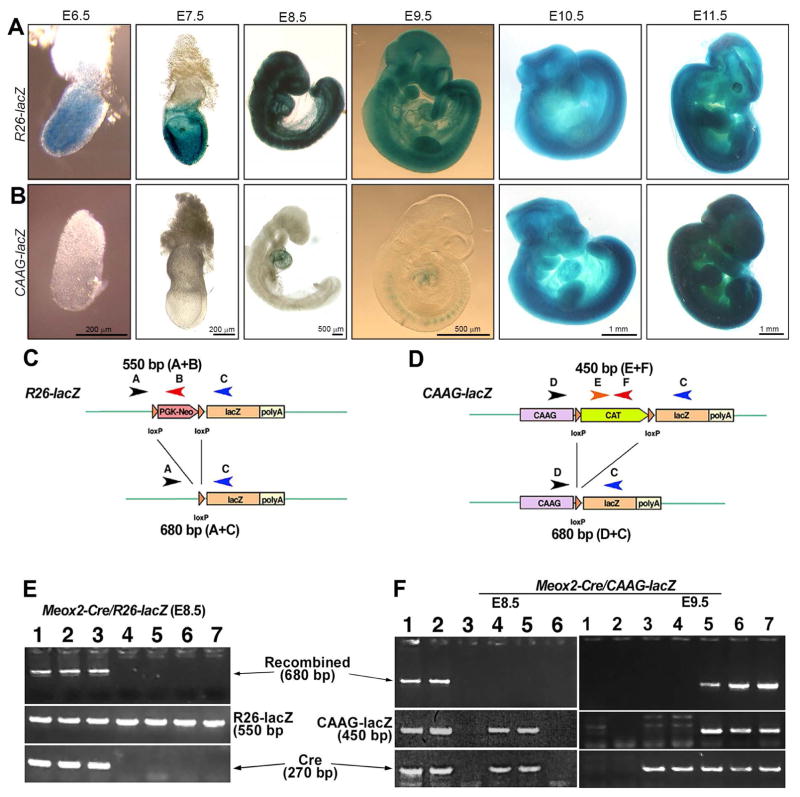
ROSA26 Cre reporter (hereafter, R26-lacZ) and CAAG-CAT-lacZ Cre reporter (hereafter, CAAG-lacZ) mice have been widely used for cell fate mapping. First, we compared the reporter sensitivity of these two lines at early embryo stages (E6.5-E11.5, Fig. 1) using the Meox2-Cre transgenic line in which Cre activity emerges as early as E5.5 in the epiblast (Tallquist and Soriano, 2000). In Meox2-Cre/R26-lacZ mice, X-gal staining of whole embryos showed that signals appeared in the earliest stage examined (E6.5) and that labeled cells were extensively distributed in the whole embryos throughout all the stages examined (Fig. 1A) similarly to the previous report (Tallquist and Soriano, 2000). In contrast, β-galactosidase (β-gal) activity in Meox2-Cre/CAAG-lacZ embryos was not detected until E7.5 (n=5 or more for each stage). Weak signals were detected in the cardiac region at E8.5 (2/5) and in the somites and heart region at E9.5 (2/5) (Fig. 1B), but were not apparent in head and other tissues in the trunk regions. At E10.5 and E11.5 (6/6 for each stage), the distribution of CAAG-lacZ expression in Meox2-Cre/CAAG-lacZ embryos was similar to that of Meox2-Cre/R26-lacZ throughout the entire embryo (Fig. 1B).
Fig. 1.
A–B: X-Gal visualization of the lacZ gene product β-galactosidase in R26-lacZ (A) and CAAG-lacZ (B) reporters driven by Meox2-Cre at different mouse embryonic stages. β-gal positive signals (blue) were ubiquitous in R26-lacZ embryos at all stages examined from E6.5 to E11.5. In the CAAG-lacZ embryos, positive signals were not detected until E9.5 and the embryos were not ubiquitously labeled until E10.5. At least two litters were dissected to collect 5 or more embryos at each stage. Scale bars in B also apply to A. C–F: Schematic diagrams (C–D) and representative genotyping data (E–F) of the R26-lacZ (C, E) and CAAG-lacZ (D, F) reporter systems. Meox2-Cre was used to drive reporter gene activation. In the E8.5 Meox2-Cre/R26R-lacZ embryos, Cre recombined bands were consistently detected when both R26-lacZ and Cre were present (E, lanes 1–3). However, recombined bands were detected in only 50% of the E8.5 Meox2-Cre/CAAG-lacZ embryos (F, lane 1–2). Although both CAAG-lacZ and Cre were present, Cre recombination products were not detected in half of the E8.5 embryos (F, E8.5, lane 4–5). At E9.5, the detection of Cre recombination products was coincident with the presence of both CAAG-lacZ and Cre (F, E9.5, lane 5–7). n=5 or more for each stage and genotype.
To examine whether the discrepancy between Meox2-Cre driven R26-lacZ and CAAG-lacZ reporter activity was caused by DNA recombination efficiency, promoter silencing, or both, specific primers were designed to detect un-recombined and recombined reporter alleles (Fig. 1C, 1D). A fragment of approximately 550 bp was produced using primers A and B to detect the un-recombined R26-lacZ alleles. A Cre-recombined R26-lacZ allele was detected using primers A and C, and the product was approximately 680 bp (Fig. 1C). For the CAAG-lacZ reporter system, primers E and F were used to generate an approximately 450 bp fragment from un-recombined allele whereas primers D and C produced an approximately 680 bp fragment from the Cre-mediated allele (Fig. 1D). Both Cre transgenes were detected using primers Cre1 and Cre2 to generate an approximately 270 bp fragment (Fig. 1C, 1D).
In E8.5 Meox2-Cre/R26-lacZ embryos, the recombined allele was reliably detected when both Cre and R26-lacZ transgenes were present (Fig. 1E, lane 1–3) compared to the absence of bands without Cre transgene (Fig. 1E, lane 4–6). In contrast, in Meox2-Cre/CAAG-lacZ mice only half of the double transgenic embryos showed detectable recombined allele at E8.5 (Fig. 1F, E8.5, lane 1–2 versus 4–5). The recombined allele was reliably detected at E9.5 (Fig. 1F, E9.5, lane 5–7). These data suggest that Cre-dependent DNA recombination is inconsistent in CAAG-lacZ early stage (≤E8.5) embryos.
P0-Cre/R26-lacZ labels NC cells and derivatives in early embryos
P0-Cre/CAAG-lacZ has been reported to label NC cells and derivatives after E9.0 (Yamauchi et al., 1999). Because the R26-lacZ reporter is more accessible to Cre-mediated DNA recombination than the CAAG-lacZ described above, we used the R26-lacZ reporter to re-examine P0-Cre transgene activity in early embryos at different somite stages using X-gal staining in whole embryo tissue.
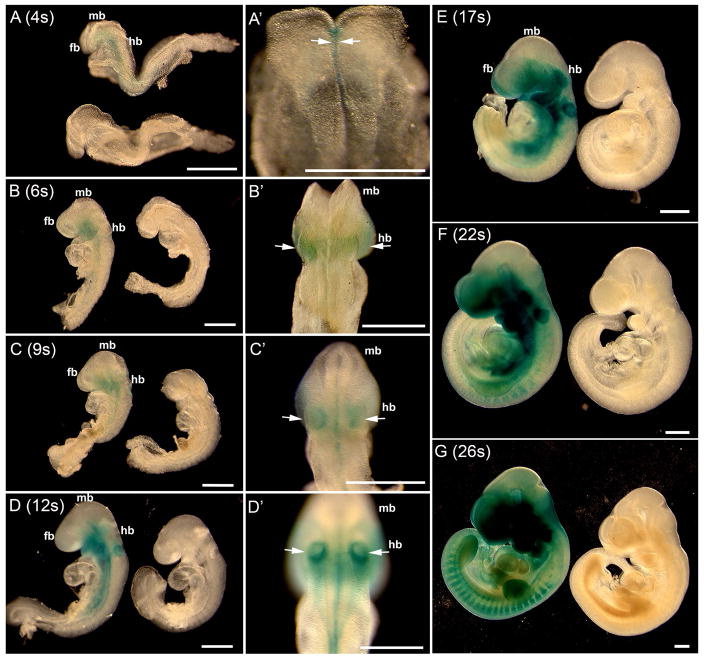
At the 4-somite stage (E8.5), signals emerged in the midline of the neural groove region (Fig. 2A, A′, arrows). In the 6-somite embryo (E8.5) (Fig. 2B, B′), β-gal positive signals were found in the forebrain, hindbrain (arrows, Fig. 2B′), first branchial arch and midline of the trunk neural groove region (Fig. 2B, B′). The staining pattern in the 9-somite embryo was similar to that of the 6-somite stage but extended more caudally and β-gal signals were obvious in the midline and otic placodes (arrows, Fig. 2C, C′). At the 12-somite (Fig. 2D, D′), intense staining was found in the hindbrain region, branchial arches, otic placodes (arrows, Fig. 2D′), and optic eminences. In the trunk region, signals were apparent in the neural groove area and in lateral tissues (Fig. 2D, D′). At the 17-somite stage, positive signals were extensively distributed in the embryo, adding more labeled tissue regions that included the heart, frontonasal, and eye regions (Fig. 2E). At the 22–26 somite stages, β-gal positive signals were extensively distributed in NC derivatives throughout the body, e.g., frontonasal and eye regions, branchial arches, heart, and dorsal root ganglia (Fig. 2F, G). Notably, signals in the midbrain region were not apparent throughout the examined stages (Fig. 2A–G). In littermate control embryos, no positive signals were observed within the embryos at the stages examined (Fig. 2A–G).
Fig. 2.
Whole mount X-Gal visualization of the lacZ gene product β-galactosidase in P0-Cre/R26-lacZ mouse embryos at different somite (s) stages (E8–E9, 4s–26s). The somite number is indicated in each panel. At the 4-somite stage, β-gal positive signals (blue) emerged (A top, A′, arrows) in the midline ventral to the neural tube. At later stages, signals (arrows) extended to the forebrain (fb), hindbrain (hb), and trunk regions of NC and NC derivatives. Signals were not obvious in the midbrain (mb) region. Cre negative control littermates (A, bottom; B–G, right) were devoid of β-gal signal. Scale bars: 500 μm.
P0-Cre transgene expression is restricted to migrating NC cells and notochord in early embryos
To analyze P0-Cre expression in cranial NC, we used a monoclonal antibody against Cre to examine Cre immunoreactivity at different early somite stages of embryos. Specificity of Cre immunosignals were validated by three different methods; (1) Western blot analysis to demonstrate the presence of a single band of expected size in a genotype-dependent manner (Supplemental Figure 1), (2) immunohistochemistry using Cre− embryos labeled with primary antibody to show no signals (Supplemental Figure 2), and (3) immunohistochemistry using Cre+ embryos without the primary antibody to determine background levels (Figs. 3, 4).
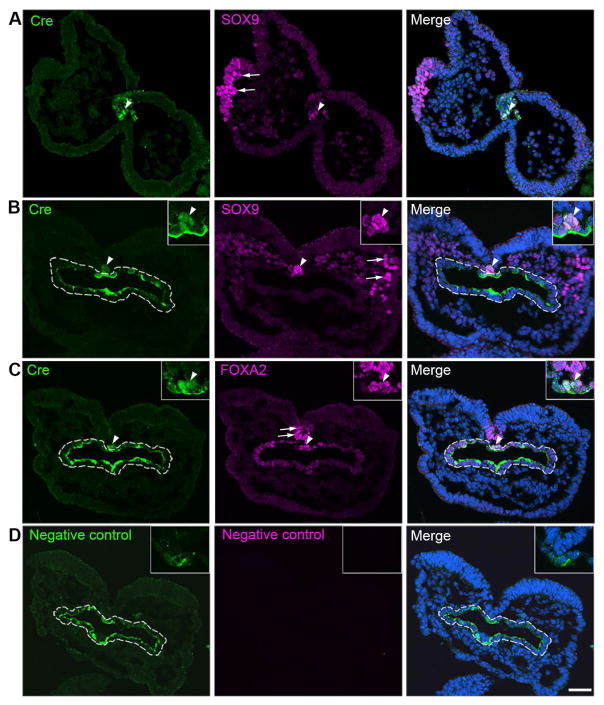
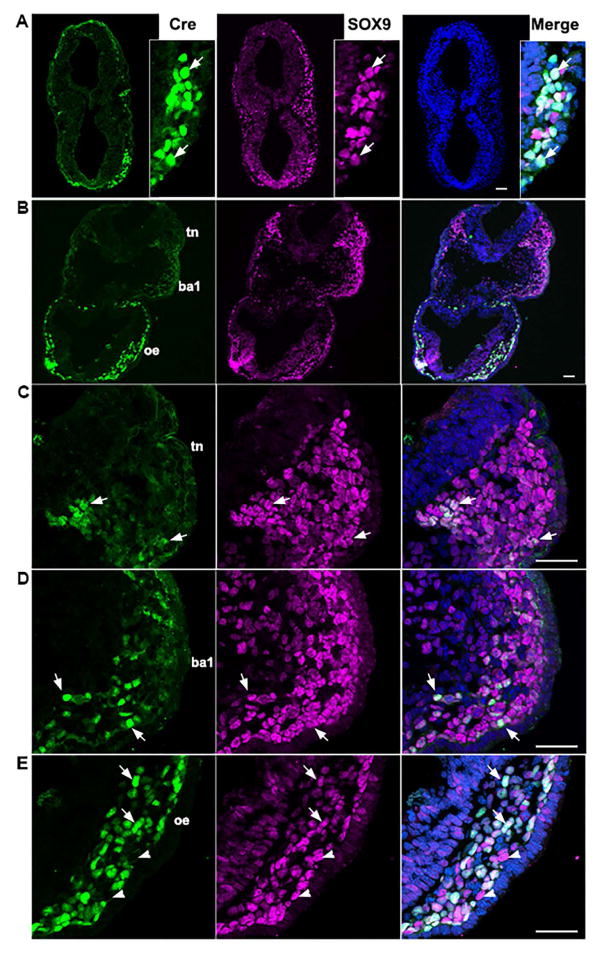
Fig. 3.
Single-plane laser scanning confocal photomicrographs of transverse sections of a 4-somite P0-Cre embryo. Sections were immunostained using an antibody against Cre (green) and were double labeled with SOX9 (purple, A–B) or FOXA2 (purple, C). Arrowheads point to Cre immunoreactive cells co-labeled with SOX9 (A–B) or FOXA2 (C) in the notochord region. Arrows point to single labeled SOX9+ cells in the NC cell region (A–B) and FOXA2+ cells in the floor plate of neural tube (C). White dashed lines outline the foregut diverticulum (B–D). In the negative control slide (D), primary antibodies were omitted. Scale bar: 50 μm for all images.
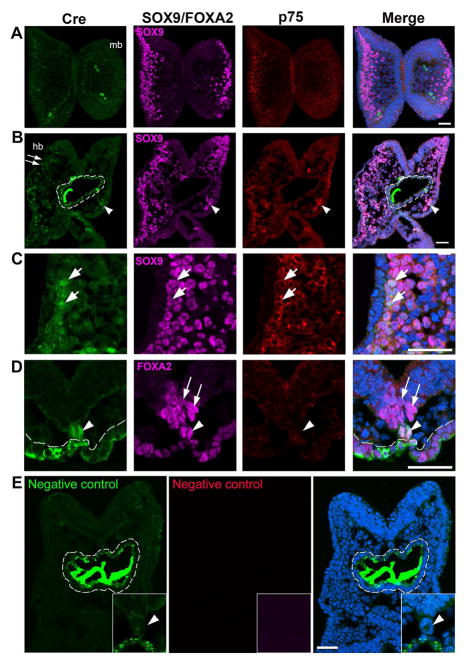
Fig. 4.
Immunoreactivity of Cre (green, A–D), SOX9 (purple, A–C) or FOXA2 (purple, D), and p75 (red, A–D) in transverse sections of a 7-somite P0-Cre embryo at the midbrain (A) and hindbrain-forebrain (B–E) levels. Arrows in C (high magnification of squared region in B) point to some of the triple labeled Cre+SOX9+p75+ cells in the NC cell region. Arrowheads in D point to Cre+ cells co-labeled with FOXA2 in the notochord region and arrows to FOXA2+ cells in the floor plate of neural tube (D). White dashed lines outline the foregut diverticulum (B, D, and E). In the negative control slide (E), primary antibodies were omitted and only secondary antibodies were applied. Scale bar: 50 μm for all images (single-plane laser scanning confocal).
β-gal signals in the P0-Cre/R26-lacZ embryo emerged in the neural groove region during the 4-somite stage (Fig. 2A). We further identified the location and types of cells that express P0-Cre transgene through investigating immunoreactivity for Cre recombinase and co-localization with a NC cell marker SOX9 (Fig. 3A, B) and a notochord marker FOXA2 (Fig. 3C) in serial sections of 4-somite embryos (n=7). Cre+ signals were seen in a cluster of cells ventrally adjacent to the neural groove at the midbrain (Fig. 3A, white arrowheads) and hindbrain levels (Fig. 3B, C, white arrowheads), which is consistent with the P0-Cre/R26-lacZ whole mount staining for the reporter activity (Fig. 2A). Cre immunosignals in clustered cells in the midline and ventrally adjacent to floor plate co-localized with SOX9 (arrowheads, Fig. 3A, B) and FOXA2 (arrowheads, Fig. 3C). However, Cre immunosignals were not detected in SOX9+ cells in the neural plate border or mesoderm layer (arrows, Fig. 3A, B) nor in the FOXA2+ cells seen in the floor plate (arrows, Fig. 3C). Non-specific immunosignals from the secondary antibody were seen in the foregut diverticulum (dashed outlines, Fig. 3B–C), which were confirmed by staining while omitting primary antibody (dashed outlines, Fig. 3D).
At the 7–8 somite stage (n=3), another NC marker p75+ or/and SOX9+ cells were seen at all brain levels examined, i.e., forebrain, midbrain (Fig. 4A), and hindbrain (Fig. 4B). No Cre immunosignals were seen in the midbrain (Fig. 4A). In the hindbrain region Cre immunosignals were detected in a small population of p75+SOX9+ cells in the mesodermal layer (arrows, Fig. 4B), and in the mesenchyme of prospective branchial arch 1 (white arrowheads, Fig. 4B). Cre immunoreactivity was also apparent in FOXA2+ cells in the notochord region, i.e., clustered cells ventrally adjacent to the neural epithelium (white arrowheads, Fig. 4D) but not in the FOXA2 + cells seen in the floor plate (arrows, Fig. 4D). Again, non-specific signals were seen in the foregut diverticulum when using secondary antibody for Cre immunoreactions, which was confirmed by staining while omitting primary antibody (white dashed outlines, Fig. 4B, D, and E).
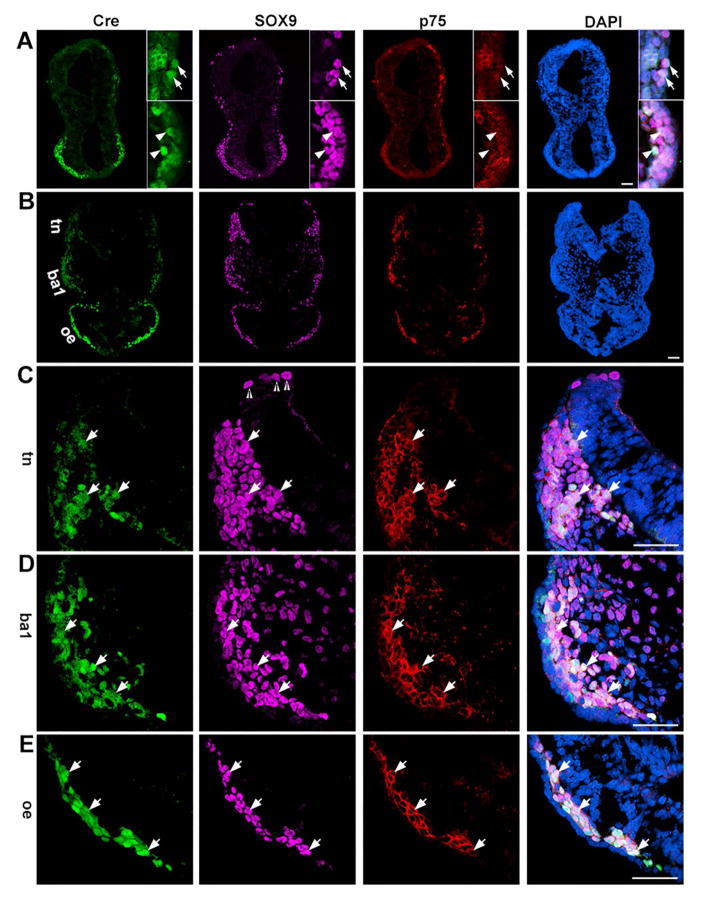
In 14-somite embryos (n=3), Cre+ cells were abundantly distributed in the forebrain region and the immunosignals co-localized with SOX9 and p75 (Fig. 5A, arrowheads). Additionally, Cre signals were seen in some of the SOX9+p75+ cells in the anterior hindbrain region (Fig. 5A, arrows). At the posterior hindbrain level (Fig. 5B), an increasing intensity gradient of Cre immunoreactivity was observed from the dorsal to ventral region, i.e., no immunosignals in the dorsal most part, faint immunosignals in the trigeminal NC region (Fig. 5B, tn), obvious immunosignals in the branchial arch 1 region (Fig. 5B, ba1), and intense immunosignals in the optic eminence (Fig. 5B, oe). Similarly, the proportions of Cre+SOX9+p75+ cells relative to total SOX9+ or/and p75+ cells in different regions were different. In the dorsal most part, some SOX9+ cells in the neural fold region, presumably pre-migratory NC cells, were observed and negative for Cre immunoreactivity (open arrowheads, Fig. 5C). In the trigeminal NC region, a subpopulation of SOX9+p75+ cells were Cre+ (white arrows, Fig. 5C). In the primordium of branchial arch 1, Cre immunosignals were apparent in a significant population of SOX9+p75+ cells (arrows, Fig. 5D). In the optic eminence most, if not all, SOX9+p75+ cells were brightly labeled with Cre (white arrows, Fig. 5E). Importantly, Cre+ cells were not seen in the Sox9− and p75− cells in any tissue regions. In the negative control slides (Cre− embryo sections with primary antibody staining and Cre+ embryo sections without primary antibody), no immunosignals were seen in the 14-somite embryo tissue (Supplemental Fig. 2).
Fig. 5.
Photomicrographs of transverse sections of a P0-Cre embryo at the 14-somite stage. Sections were immunostained for Cre (green), SOX9 (purple), and p75 (red) at the anterior hindbrain-forebrain level (A) and posterior hindbrain-forebrain level (B). C–D are higher magnification images of trigeminal NC (C, tn), branchial arch 1 (D, ba1), and optic eminence (D, oe). Arrows (A) point to Cre+ cells that were co-labeled with SOX9 and p75 immunosignals in the anterior hindbrain regions, and arrowheads (A) point to triple labeled Cre+SOX9+p75+ cells in the forebrain regions. Open arrowheads (C) point to SOX9+ cells in the neural fold region, presumably pre-migratory NC cells. Arrows (C–E) point to Cre+ cells co-labeled with SOX9+ and p75+, presumably migratory NC, in trigeminal NC regions (tn, C), first branchial arch 1 (ba1, D) and optic eminence (oe, E). Scale bars: 50 μm for all images (single-plane laser scanning confocal).
Compared to the 14-somite stage, 19-somite embryos (n=4) had a similar distribution pattern of Cre immunosignals, i.e., an increasing intensity gradient from the dorsal to ventral regions of the tissue (Fig. 6A, B). Additionally, increasing proportions of Cre+SOX9+ cells relative to the total number of SOX9+ cells were observed from dorsal to ventral regions (Fig. 6C–E). However, the proportions of Cre+SOX9+ cells relative to total SOX9+ cells in all regions were lower than those at the 14-somite stage. At the anterior hindbrain level, Cre+ immunosignals were seen in some of the SOX9+ cells in the hindbrain and intense signals were distributed in the majority of SOX9+ cells in the forebrain (Fig. 6A, arrows). At the posterior hindbrain level, Cre+ cells were seen in the trigeminal NC (Fig. 6B–C, tn), branchial arch 1 (Fig. 6B and D, ba1), and optic eminence (Fig. 6B and E, oe). The intensity of Cre immunosignals in the optic eminence was diverse in the SOX9+ cells, some of which were negative for Cre (arrowheads, Fig. 6E), which is in contrast to almost all of SOX9+ cells that showed presence of Cre immunosignals in the 14-somite embryos (Fig. 5E).
Fig. 6.
Single-plane laser scanning confocal photomicrographs in transverse sections of a 19-somite P0-Cre embryo. Sections were immunostained using an antibody against Cre (green) and were double labeled with SOX9 (purple, A–E). C–D are higher magnification images of trigeminal NC (C, tn), branchial arch 1 (D, ba1), and optic eminence (D, oe). Arrows (A) point to Cre+ cells that were co-labeled with SOX9 in the forebrain. Arrows (C–E) point to Cre+ cells co-labeled with SOX9 in trigeminal NC regions (tn, C), branchial arch 1 (ba1, D), and optic eminence (oe, E). Arrowheads in E point to SOX9+ cells without Cre immunosignals. Scale bars: 50 μm, also applies to other images in the same panel.
P0-Cre labeled cells are distributed in cranial NC and derivatives in early embryos and newborn mice
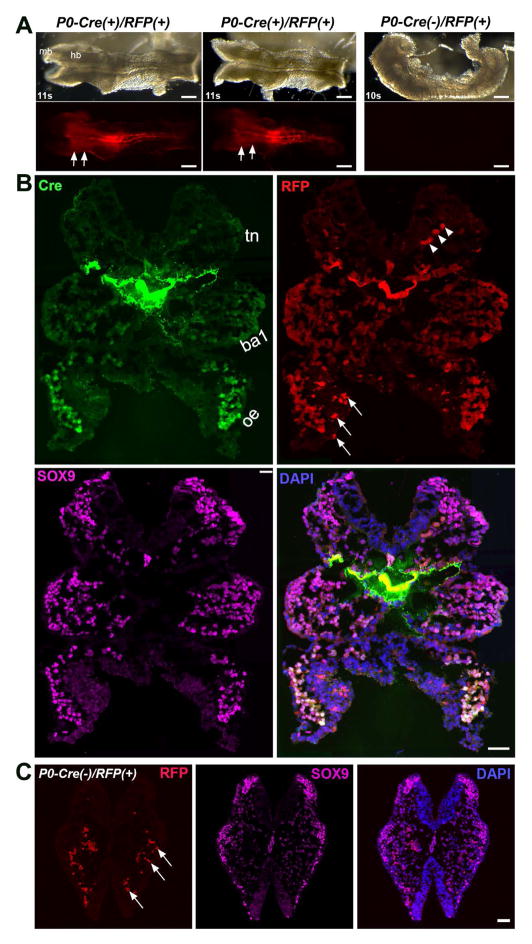
To confirm the specific labeling of P0-Cre/R26-lacZ and Cre immunosignals, we used another Cre reporter line (R26-RFP) and found that the distribution pattern of P0-Cre/R26-RFP signals (11s, Fig. 7) was the same as P0-Cre/R26-lacZ (Fig. 2D, 2D′), i.e., signals were found in the hindbrain region, branchial arches, in the neural groove area and lateral tissues in the trunk region (Fig. 7A). Similar to lacZ expression in P0-Cre/R26-lacZ embryos, RFP signals were not apparent in the midbrain region (mb). On tissue sections at the posterior hindbrain level, Cre immunosignals and RFP signals largely co-localized (Fig. 7B). Again, an increasing gradient of Cre and RFP signal intensity and proportions of labeled cells was observed from the dorsal to ventral region, i.e., no signals in the dorsal-most part, faintly labeled cells were sparsely found in the trigeminal NC region (Fig. 7B, tn), obviously and frequent labeled cells (Cre+RFP+) in the branchial arch 1 region (Fig. 7B, ba1), and intensely and mostly labeled in the optic eminence (Fig. 7B, oe). Auto-fluorescent blood cells and non-cellular RFP+ fragments were also seen (Fig. 7B, arrowheads) and confirmed in the Cre- tissue sections (Fig. 7C, arrowheads).
Fig. 7.
Consistent distribution of Cre and RFP signals in P0-Cre/R26-RFP embryos with that in P0-Cre and P0-Cre/R26-lacZ embryos. A: Bright-field (top panel) and fluorescent (bottom panel) images of whole P0-Cre/R26-RFP embryos at 10–11 somite stages. Arrowheads point to the midbrain (mb) region that was devoid of RFP signals. Arrows point to the hindbrain region labeled with RFP. B: Single-plane laser scanning confocal photomicrographs of a transverse section of an 11-somite P0-Cre(+)/R26-RFP(+) embryo at the hindbrain level. An increasing intensity gradient of Cre and RFP signals from the dorsal to ventral region, i.e., faint and sparse in the trigeminal NC (tn), clear and frequent in the branchial arch 1 (ba1), bright and almost all in the optic eminence (oe). Arrowheads point to the autofluorescent blood cells. Arrows point to some non-cellular RFP+ fragments. C: Confocal images of a transverse section of a 10-somite Cre− embryo to illustrate the autofluorescent blood cells and non-cellular RFP+ fragments (arrows). Scale bars: 200 μm in A; 50 μm in B and C.
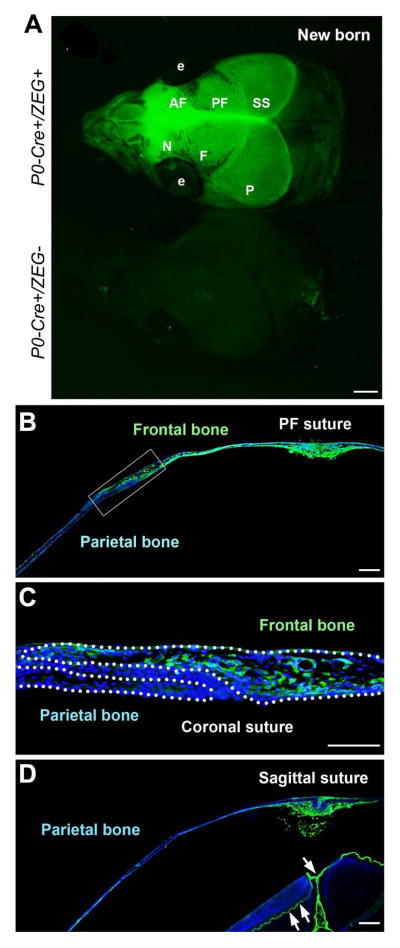
To verify the specificity of P0-Cre in labeling NC cells and derivatives, we examined whether P0-Cre faithfully labels the nasal and frontal bones in the skull that are well-characterized cranial NC derivatives. Consistent with a previous report using Wnt1-Cre mice (Jiang et al., 2002), P0-Cre successfully labeled both nasal and frontal bones along with dura mater (Fig. 8A–C). Importantly, P0-Cre did not label the parietal bones that are known as paraxial mesoderm derivatives (Fig. 8C, D) (Chai and Maxson, 2006; Noden and Trainor, 2005). The fluorescent signals in the parietal bone region of whole mount tissue (Fig. 8A) were confirmed on sections to be from the underlying meninges (arrows, Fig. 8D), which are NC derivatives (Jiang et al., 2002).
Fig. 8.
P0-Cre marks cranial NC and derived cells in the skull. A: GFP signals in the newborn P0-Cre+/ZEG+ (upper) and control littermate (bottom) mice. B: GFP signals on the coronal tissue sections illustrate that the frontal bones (B–C), but not the parietal bone (C–D), were labeled. C is the high magnification image of the squared tissue region in B. Abbreviations: AF, anterior frontal suture; e, eye; F, frontal bone; N, nasal bone; P, parietal bone; PF, posterior frontal suture; SS, sagittal suture. Scale bars: 500 μm in A, 200 μm in B and D; 100 μm in C.
Different distribution pattern of Wnt1-Cre labeling from P0-Cre in cranial regions
In P0-Cre/R26-lacZ embryos (Fig. 2), reporter labeling was not apparent in the midbrain region. To further examine our observation, R26-lacZ was crossed with Wnt1-Cre, which is well known for labeling midbrain.
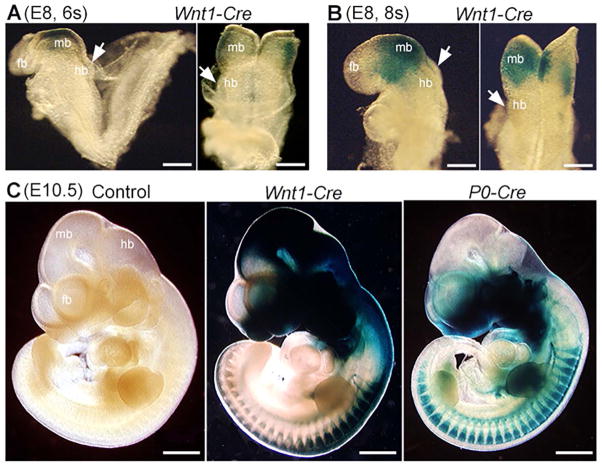
At the 6-somite stage (Fig. 9A), the signals in Wnt1-Cre/R26-lacZ mouse embryos were intense in the midbrain and along the midline of the neural groove (Fig. 9A). In the 8-somite embryo (Fig. 9B), β-gal positive signals were extensively distributed in the midbrain (Fig. 8B). However, signals in the hindbrain were absent at the 6-somite stage (Fig. 9B, arrows) and were sparse at the 8-somite stage (Fig. 9B, arrows). At E10.5, the signals extended to the forebrain (fb), midbrain (mb), hindbrain (hb) and trunk regions of NC and NC derivatives in Wnt1-Cre/R26-lacZ embryos (Fig. 9C), e.g., frontonasal and eye regions, branchial arches, heart, and dorsal root ganglia. In contrast to the extensive labeling of Wnt1-Cre in the midbrain, P0-Cre labeling in midbrain was not obvious (Fig. 9C), which is consistent with our earlier observations (Fig. 2).
Fig. 9.
Whole mount X-Gal visualization of lacZ gene product β-galactosidase in Wnt1-Cre/R26-lacZ mouse embryos at different stages (E8–E10.5). β-gal positive signals (blue) were extensive in the midbrain (mb) at the 6s (A) and 8s (B) stages, but were sparse in the hindbrain. At later stages, signals extended to the forebrain (fb), midbrain (mb), hindbrain (hb) and trunk regions of NC and NC derivatives in Wnt1-Cre/R26-lacZ tissue at E10.5 (C). In E9 P0-Cre/R26-lacZ mouse embryos (C), β-gal positive signals (blue) were found in the forebrain (fb), hindbrain (hb) and trunk regions of NC and NC derivatives, but were not apparent in the midbrain (mb). Scar bars: 200 μm in A and B; 1 mm in C.
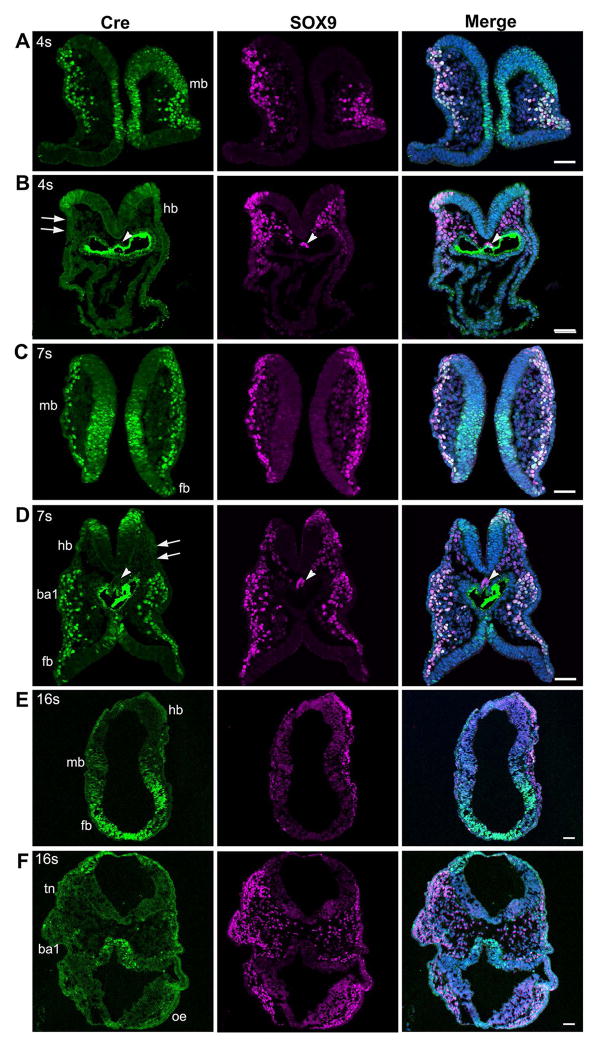
Next, we further analyzed the expression patterns of Wnt1-Cre transgene in cranial NC using the antibody for Cre recombinase. In the 4-somite embryos (Fig. 10A), Cre immunosignals were found in SOX9+ pre- and post-migratory NC cells and in the neural epithelium at the midbrain level (Fig. 10A). In contrast, SOX9+ migrating NC cells were negative for Cre in the hindbrain region (arrows, Fig. 10B). At the 7-somite stage (Fig. 10C), Cre immunosignals were extensively distributed in migrating NC cells in the midbrain and forebrain regions, in addition to labeling of part of the neuroepithelium. Again, in hindbrain regions, Cre immunosignals were observed only in the neural fold and were not apparent in SOX9+ migrating NC cells (arrows, Fig. 10D). This is an interesting contrast to the abundant Cre immunosignals found in the hindbrain region of P0-Cre embryos at the 7-somite stage (arrows, Fig. 4B, C). At the presumptive notochord regions, no Cre immunosignals were seen in Wnt1-Cre embryos (arrowheads, Fig. 10B, D). In the 16-somite embryos, Cre immunosignals were found in the midbrain and forebrain but were not apparent in the hindbrain (Fig. 10E), which is in contrast to the distribution of Cre immunosignals in the 16–19 somite P0-Cre embryos (Fig. 5, 6). In the trigeminal NC regions (tn), first branchial arch 1 (ba1) and optic eminence region (oe), Cre immunosignals were sparsely seen in migratory NC cells (Fig. 10F).
Fig. 10.
Photomicrographs from sections of E8.5 Wnt1-Cre embryos at different somite stages immunostained for the Cre and NC cell marker SOX9. At the 4–7 somite stages (A–D), Cre immunosignals (green) were bright in the midbrain (A, C) and derived NC cells and co-localized with SOX9 (purple). In contrast, in the hindbrain-forebrain levels (B, D) Cre immunosignals were seen in SOX9+ pre-migratory NC cells but were absent in migrating NC cells. In 16-somite embryos (E, F), Cre immunosignals were seen in migrating NC cells in the midbrain and forebrain regions (E). In the trigeminal NC regions (tn), first branchial arch 1 (ba1), and optic eminence region (oe), Cre immunosignals were sparse in migratory NC cells. Scale bars: 50 μm (single-plane laser scanning confocal).
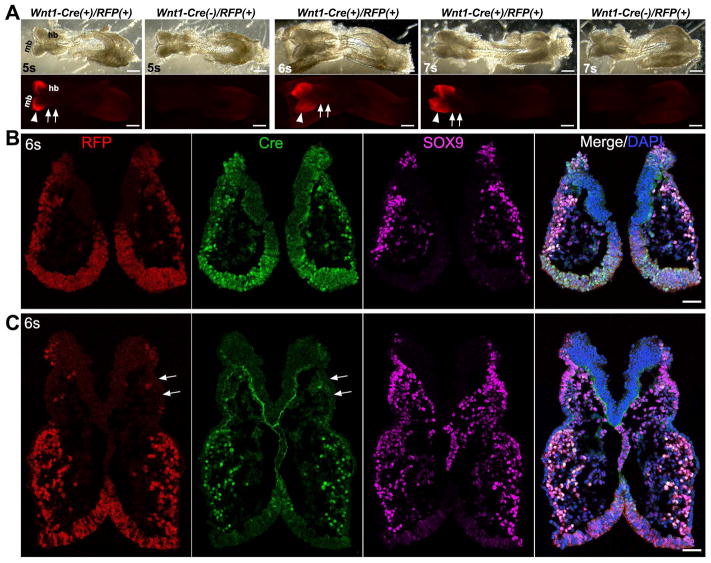
Also, R26-RFP Cre reporter was used to confirm the specific labeling of Wnt1-Cre driven R26-lacZ and Cre immunoreactivity. The distribution pattern of signals in Wnt1-Cre/R26-RFP embryos (Fig. 11A, 5–7s) was identical to that of Wnt1-Cre/R26-lacZ embryos (Fig. 9A–B, 6–8s). The signals in Wnt1-Cre/R26-RFP mouse embryos were intense in the midbrain (Fig. 11A, arrowheads), and signals in the hindbrain were not obvious at the 6- and 7-somite stages (Fig. 11A, arrows). On tissue sections of a 6-somite Wnt1-Cre/RFP embryo, the distribution of RFP+ cells was coincident with that of Cre+ cells (Fig. 11B–C), such as abundance within migrating NC cells in the midbrain and in the forebrain neuroepithelium (Fig. 11B). The labeling was sparse among migrating NC cells in the hindbrain region (arrows, Fig. 11C).
Fig. 11.
Distribution of Cre and RFP signals in Wnt1-Cre/R26-RFP embryos. A: Bright-field (top panel) and fluorescent (bottom panel) images of whole Wnt1-Cre/R26-RFP embryos at 5–7 somite stages. In contrast to the P0-Cre/R26-RFP embryos, RFP signals were detected in midbrain (mb) region (arrowheads), but not obvious in the hindbrain (hb, arrows). B and C: Single-plane laser scanning confocal photomicrographs of a transverse section of a 6-somite Wnt1-Cre(+)/R26-RFP(+) embryo at the midbrain (B) and hindbrain (C) level. Arrows point to the trigeminal NC region that was devoid of RFP and Cre signals. Scale bars: 250 μm in A; 50 μm in B and C.
Taken together (Table 1 and Supplemental Fig. 3), both P0-Cre and Wnt1-Cre label cranial NC cells, peaked at 7-somite stage in Wnt1-Cre and 14-somite in P0-Cre. An overlap (in forebrain) but distinct (Wnt1-Cre in midbrain and P0-Cre in hindbrain) distribution patterns of the Cre activities were observed. In addition to NC, both P0-Cre and Wnt1-Cre labeled other cells, e.g., notochord was labeled by P0-Cre and neuroepithelium by Wnt1-Cre.
Table 1.
Comparison between P0-Cre and Wnt1-Cre in labeling cranial neural crest (NC), neural epithelium (NE), and notochord to various extent (++ extensively; + clearly; ± rarely; − negatively).
| Cranial regions | P0-Cre | Wnt1-Cre | |
|---|---|---|---|
| Forebrain | NC | ++ | ++ |
| NE | − | ++ | |
| Midbrain | NC | − | ++ |
| NE | − | + | |
| Hindbrain | NC | ++ | ± |
| NE | − | ± | |
| Notochord | + | − | |
DISCUSSION
The P0-Cre transgenic mouse line has been widely used for NC cell fate mapping (Feltri et al., 1999b; Ono et al., 2015; Sommer and Suter, 1998) and genetic modifications of NC-derived cells (Hu et al., 2014; Liu and Xiao, 2011). However, questions remain about how early, how specifically, and to what extent P0-Cre transgene labels NC cells in early embryos. Moreover, concerns about the specificity of P0-Cre and other Cre models in labeling NC lineage have been raised (Trainor, 2005a, 2005b). Therefore, a detailed analysis of P0-Cre transgene activity is essential for the use and data interpretation of this model. In the present study, we found that the CAAG-CAT-Z (CAAG-lacZ) Cre reporter mice used in the original report (Yamauchi, 1999) did not appropriately reflect Cre activity in early stage embryos. We then investigated Cre activity using a R26-lacZ reporter and Cre immunoreactivity to find P0-Cre expression as early as the 4-somite stage in notochord labeled with FOXA2 and in the migrating NC cells at the 7-somite stage using the commonly used NC cell markers SOX9 (Mori-Akiyama et al., 2003; Sahar et al., 2005; Spokony et al., 2002) and p75 (Young, 2000; Young et al., 1999). Cre immunosignals were especially extensive in the forebrain and hindbrain but not apparent in the midbrain. Importantly, Cre immunosignals were not seen outside of the NC cell population and notochord. Given the fact that notochord derivatives are well-known to be restricted to the intervertebral disc (McCann et al., 2012; Yamauchi et al., 1999), our data demonstrated that P0-Cre can serve as a valuable tool for studies on NC cell lineage, especially for forebrain and hindbrain NC derivatives. In contrast, Wnt1-Cre extensively labeled pre- and post-migratory NC cells at the midbrain level, but only sparsely in the trigeminal NC region at the hindbrain level. Additionally, Wnt1-Cre labeled a large population of non-NC cells in the neural plate, which is consistent with previous reports (Echelard et al., 1994; Rowitch et al., 1998).
P0-Cre specifically labels a significant population of, if not all, migrating NC cells and notochord
The specificity of Cre expression in the NC cell population was examined with two commonly used NC cell markers, p75 (Menendez et al., 2011) to label migrating NC and SOX9 (Nakanishi et al., 2007; Pomp et al., 2005) to label both pre- and post-migratory NC cells. Our data support the idea that P0-Cre specifically labels migrating NC cells and notochord based on the following observations: (1) Cre immunosignals were only detected in p75+SOX9+ double labeled cells, and were more intense in the NC cells that were close to their destinations; (2) importantly, Cre immunosignals were not seen outside of the p75+SOX9+ double labeled cells; (3) Cre immunosignals were not seen in SOX9+ cells in the neural fold region, which presumably pre-migratory NC cells, and Cre immunosignals were rarely detectable in the immediately delaminated NC cell region; and (4) Cre immunosignals only co-localized with FOXA2 in the notochord region, and were absent in FOXA2+ cells in the floor plate. These data demonstrate that P0-Cre is specific in labeling migrating NC cells and notochord. An increasing gradient of intensity of Cre immunosignals and RFP signals was observed from the dorsal to ventral regions suggesting that the expression of P0-Cre transgene in the NC cells is obtained during migration. Our data are valuable in demonstrating the specificity of P0-Cre in labeling NC cells in early embryos. The specificity of P0-Cre in labeling cranial NC cells was further confirmed by the distribution of P0-Cre labeled cells in the well-known NC derivatives in the skull, e.g., frontal bones and sutures, in contrast to the absence of labeled cells in the parietal bone that is known to be non-NC derived.
The notochord is a rod-shaped structure in the midline that is ventrally adjacent to the neural tube (Jurand, 1974). Studies using a Cre mouse line driven by the notochord-specific homeobox gene Noto that traced the cell fate of notochord demonstrated that notochord serves as a source of embryonic precursors of cells within the nucleus pulposus of the mature intervertebral disc (McCann et al., 2012). In our study, P0-Cre transgene expression was detected in the notochord region in the 4–7 somite stage embryos, which is consistent with a previous report demonstrating that P0-Cre transgene labels notochord in addition to NC cells and derivatives (Yamauchi et al., 1999). With exception to the labeling of notochord, P0-Cre labels migrating NC cells specifically. Indeed, our data using P0-Cre to mark cranial bones, i.e., nasal and frontal but not parietal bone, further demonstrated the specificity of P0-Cre in labeling cranial NC derivatives and that P0-Cre mice can be considered as a useful tool to trace NC lineages, at least cranial NC-derived bones (Komatsu et al., 2013), during mouse embryonic development.
However, P0-Cre did not label all of the NC cells in all of the tissue regions. The proportions of Cre+ cells relative to the total number of p75+SOX9+ cells were different in different tissue regions. NC cells migrate ventrally to their target tissue/organs and follow certain paths after delamination. Thus, the increasing intensity gradient of Cre immunosignals and RFP signals and increasing cell proportions of Cre+ and RFP+ cells from dorsal to ventral regions suggest that P0-Cre transgene expression emerged during NC migration. Although Cre immunosignals were not detected in all the p75+SOX9+ cells, we cannot exclude the possibility that P0-Cre does indeed label all NC cells. It would be difficult to evaluate the exact proportion of P0-Cre transgene in labeling migrating NC cells because the expression is transient and because P0-Cre can be expressed in a NC cell for a short time frame at any time point. Our observation that detection of Cre immunosignals in all the p75+SOX9+ cells in the optic eminence and the majority of mesenchymal cells immediately under the epithelium of prospective branchial arch 1 at the 14 somite stage supports the idea that P0-Cre transgene is expressed in a significant population of, if not all, migrating NC cells in the forebrain and hindbrain regions.
Several Cre reporter lines have been generated and are widely used for cell fate mapping (Ikeya et al., 1997; Kawamoto et al., 2000; McMahon et al., 1992; Novak et al., 2000; Weissman and Pan, 2015; Weissman et al., 2011). Among these reporter lines, ROSA26 and CAAG are two commonly used promoters for driving reporter gene expression (Araki et al., 1995; Kawamoto et al., 2000). In the present study, the efficiency and reliability of two Cre reporters, R26-lacZ and CAAG-lacZ, were compared using Mexo2-Cre, in which Cre is expressed as early as E5.5 in epiblast (Tallquist and Soriano, 2000). We found that R26-lacZ reporter expression emerges earlier than CAAG-lacZ reporter expression and that DNA recombination in R26-lacZ was reliable and consistent with Cre expression. One interesting observation is that although all E9.5 Meox2-Cre/CAAG-lacZ embryos showed evidence of DNA recombination, the lacZ signals were weak, and were restricted to the heart and somites. It is possible that the promoter activity to drive lacZ expression in the CAAG-lacZ cassette was silenced or weaker than that of R26-lacZ in early stage embryos. Together, the data suggest that less efficient and inconsistent DNA recombination, and variable CAAG promotor activity might be the major reasons for this discrepancy.
P0-Cre labels cranial NC cells in a brain region-specific manner
It has been reported that NC cells are generated by the 7-somite stage at all levels of the forebrain, midbrain, and hindbrain (Chan and Tam, 1988; Nichols, 1981; Serbedzija et al., 1992). By detecting both R26-lacZ reporter expression and Cre immunoreactivity we found that P0-Cre transgene expression emerges at the 7-somite stage in hindbrain and forebrain migratory NC cells. In the 14-somite embryos, we found strong Cre immunosignals in a considerable subpopulation of migrating NC cells labeled with SOX9 and p75 in the forebrain and hindbrain NC regions.
However, P0-Cre expression was rarely detected in the midbrain region, which is in contrast to extensive distribution of Cre immunosignals and RFP signals in the midbrain crest of Wnt1-Cre (Huang et al., 2010; Lewis et al., 2013). At E8.5, when cranial crest cells emerge, Wnt1-Cre labels NC cells extensively in midbrain but many fewer cells were seen in hindbrain (Danielian et al., 1998; Echelard et al., 1994). Wnt1 and Wnt1-Cre expression is not restricted to the NC domain but also labels dorsal neural stem cells that contribute to both the central nervous system and to neural progenitors (Barriga et al., 2015; McMahon et al., 1992). In the present study, we crossed the same R26-lacZ reporter mouse with P0-Cre and Wnt1-Cre and confirmed the labeling pattern (Fig. 9). The difference between P0-Cre and Wnt1-Cre in labeling NC cells was profound in the midbrain and hindbrain, which may be why P0-Cre and Wnt1-Cre label different populations of cells in the orofacial organs, e.g., tooth buds (Wang et al., 2011) and taste buds (Boggs et al., 2016; Liu et al., 2012).
It has been reported that forebrain crest cells appear by the 8–14 somite stages and migrate dorsally over the presumptive eye where they meet the ventrally migrating midbrain crest cells. Although we did not see much Cre reporter activity or Cre immunoreactivity in the midbrain, Cre immunoreactivity was detected in all the SOX9+p75+ cells in the optic eminence, which suggests that forebrain is the source of NC cells that contribute to optic eminence development.
Conclusions
The P0-Cre transgenic mouse model was first generated to label NC derivatives (Yamauchi et al., 1999) and has been widely used for NC lineage tracing (Boggs et al., 2016; Kawakami et al., 2011; Liu et al., 2012; Ogawa et al., 2015; Wang et al., 2011) and genetic modification (Feltri et al., 1999a; Komatsu et al., 2013; Nomura-Kitabayashi et al., 2009; Ogawa et al., 2015). Differences have been found between P0-Cre and Wnt1-Cre in labeling NC lineage. Here, we revisited the P0-Cre model at early embryonic stages and demonstrated that P0-Cre transgene specifically labels migrating cranial NC cells in the forebrain and hindbrain as early as the 7-somite stage and reaches its peak expression at the 14-somite stage. Additionally, P0-Cre labels notochord at the 4–7 somite stages in early embryos. Moreover, the distribution pattern of P0-Cre was different from Wnt1-Cre in labeling NC cells, especially in the midbrain and hindbrain regions. Our data indicate that the P0-Cre mouse line is a valuable model for studies of NC and notochord lineage, and that careful attention needs to be paid in choosing promoters to drive Cre and its reporters for studies on NC lineage in early embryos.
Supplementary Material
Supplemental Fig. 1. Western blotting data to show the single, specific band of Cre with the primary antibody (1:5000, MAB3120, EMD Millipore) that was used to generate immunosignals in embryonic tissue sections. Embryos carrying P0-Cre transgene were collected at E9.5. A littermate that did not carry the transgene was used as negative control (Cre−). Tissue lysates were prepared and immunoblotted with the Cre antibody indicating their specificity against Cre protein (38-kD). IKAP (150-kD) antibody was also used as a loading control. Bands of expected proteins were visualized with DAB reaction.
Supplemental Fig. 2. Representative photomicrographs taken with a fluorescent light microscope illustrate the absence of Cre immunoreactivity in transverse sections of a 14-somite P0-Cre negative embryo. Sections were immunostained using an antibody against Cre (green) and triple labeled with Sox9 (purple, A–C) and p75 (red, A–C). Scale bar: 50 μm for all images.
Supplemental Fig. 3. Representative photomicrographs shown previously are assembled to compare P0-Cre and Wnt1-Cre in labeling cranial neural crest, neuroepithelium, and notochord. In brief, in bright-field (top) and fluorescent (bottom) images of whole P0-Cre/RFP and Wnt1-Cre/RFP embryos (A), the midbrain (mb) region was labeled by Wnt1-Cre signals but not by P0-Cre (arrowheads), while the hindbrain was labeled by P0-Cre but sparse in Wnt1-Cre (Arrows). On sections (B), P0-Cre activities, peaked at 14-somite stage, were distributed in post-migratory NC cells in trigeminal neural crest (tn, B1) at posterior hindbrain level, in the optic eminence (oe, B2), in anterior hindbrain (hb, B3), and the forebrain (fb, B3). Wnt1-Cre activities were extensive at 7-somite stage (Wnt1-Cre, B1-2), in the pre- and post-migratory neural crest cells in midbrain (mb, B1) and forebrain (fb, B2-3) regions, optic eminence (oe, B2) and in the neuroepithelium (mb, hb, fb, B1-3). Scale bars: 250 μm in A and 50 μm in B.
Acknowledgments
We thank Dr. Kenichi Yamamura for providing P0-Cre mice, Dr. Shigeto Miura for his contribution to the early stage of the work, especially Fig. 1. We also thank Drs. Andrew McMahon, Michelle Tallquist, Junichi Miyazaki and Phillipe Soriano for genetically modified mouse lines. The study was supported by NIH Grant R01DE020843 and ES071003-11 to YM and R01DC012308 to HXL.
Footnotes
Author Contribution
Experimental design: GC, MI, JY, SC, YK, YM and HXL.
Experiment conduction and data analysis: GC, MI, JY, SK, TF, GS, MKR, CS, YK, YM and HXL.
Data interpretation: GC, YK, YM and HXL.
Writing manuscript: GC, YM and HXL.
Approving final version of manuscript: GC, MI, JY, SK, TF, GS, MKR, CS, SC, YK, YM and HXL.
HXL and YM take responsibility for the integrity of the data analysis.
References
- Aoto K, Sandell LL, Butler Tjaden NE, Yuen KC, Watt KE, Black BL, Durnin M, Trainor PA. Mef2c-F10N enhancer driven beta-galactosidase (LacZ) and Cre recombinase mice facilitate analyses of gene function and lineage fate in neural crest cells. Dev Biol. 2015;402:3–16. doi: 10.1016/j.ydbio.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga EH, Trainor PA, Bronner M, Mayor R. Animal models for studying neural crest development: is the mouse different? Development. 2015;142:1555–1560. doi: 10.1242/dev.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs K, Venkatesan N, Mederacke I, Komatsu Y, Stice S, Schwabe RF, Mistretta CM, Mishina Y, Liu HX. Contribution of Underlying Connective Tissue Cells to Taste Buds in Mouse Tongue and Soft Palate. PLoS One. 2016;11:e0146475. doi: 10.1371/journal.pone.0146475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. Development. Making sense of the sensory lineage. Science. 2004;303:966–968. doi: 10.1126/science.1094732. [DOI] [PubMed] [Google Scholar]
- Cavanaugh AM, Huang J, Chen JN. Two developmentally distinct populations of neural crest cells contribute to the zebrafish heart. Dev Biol. 2015;404:103–112. doi: 10.1016/j.ydbio.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chan WY, Tam PP. A morphological and experimental study of the mesencephalic neural crest cells in the mouse embryo using wheat germ agglutinin-gold conjugate as the cell marker. Development. 1988;102:427–442. doi: 10.1242/dev.102.2.427. [DOI] [PubMed] [Google Scholar]
- Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Feltri ML, D’Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci. 1999a;883:116–123. [PubMed] [Google Scholar]
- Feltri ML, D’Antonio M, Quattrini A, Numerato R, Arona M, Previtali S, Chiu SY, Messing A, Wrabetz L. A novel P0 glycoprotein transgene activates expression of lacZ in myelin-forming Schwann cells. Eur J Neurosci. 1999b;11:1577–1586. doi: 10.1046/j.1460-9568.1999.00568.x. [DOI] [PubMed] [Google Scholar]
- Freem LJ, Escot S, Tannahill D, Druckenbrod NR, Thapar N, Burns AJ. The intrinsic innervation of the lung is derived from neural crest cells as shown by optical projection tomography in Wnt1-Cre;YFP reporter mice. J Anat. 2010;217:651–664. doi: 10.1111/j.1469-7580.2010.01295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L, Aggarwal V, Morrow BE. Dual embryonic origin of the mammalian otic vesicle forming the inner ear. Development. 2011;138:5403–5414. doi: 10.1242/dev.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon TR, Shiraz A, Qin LX, Gerald WL, Kenney AM, Cheung NK. Enteric neural crest differentiation in ganglioneuromas implicates Hedgehog signaling in peripheral neuroblastic tumor pathogenesis. PLoS One. 2009;4:e7491. doi: 10.1371/journal.pone.0007491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla PH, Bronner ME. Epigenetic regulation in neural crest development. Dev Biol. 2014;396:159–168. doi: 10.1016/j.ydbio.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jarad G, Miner JH. The Pax3-Cre transgene exhibits a rostrocaudal gradient of expression in the skeletal muscle lineage. Genesis. 2009;47:1–6. doi: 10.1002/dvg.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, Rudnicki MA, Chai Y, Rossi F, Marra MA, Miller FD. Convergent genesis of an adult neural crest-like dermal stem cell from distinct developmental origins. Stem Cells. 2010;28:2027–2040. doi: 10.1002/stem.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurand A. Some aspects of the development of the notochord in mouse embryos. J Embryol Exp Morphol. 1974;32:1–33. [PubMed] [Google Scholar]
- Katoh H, Shibata S, Fukuda K, Sato M, Satoh E, Nagoshi N, Minematsu T, Matsuzaki Y, Akazawa C, Toyama Y, Nakamura M, Okano H. The dual origin of the peripheral olfactory system: placode and neural crest. Mol Brain. 2011;4:34. doi: 10.1186/1756-6606-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami M, Umeda M, Nakagata N, Takeo T, Yamamura K. Novel migrating mouse neural crest cell assay system utilizing P0-Cre/EGFP fluorescent time-lapse imaging. BMC Dev Biol. 2011;11:68. doi: 10.1186/1471-213X-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Yu PB, Kamiya N, Pan H, Fukuda T, Scott GJ, Ray MK, Yamamura K, Mishina Y. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res. 2013;28:1422–1433. doi: 10.1002/jbmr.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RT, Nagai H, Nakaya Y, Sheng G, Trainor PA, Weston JA, Thiery JP. Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development. 2013;140:4890–4902. doi: 10.1242/dev.094680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikola A. The neural crest: migrating cells in embryonic development. Folia Morphol (Praha) 1976;24:155–172. [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xie WB, Escano CS, Asico LD, Xie Q, Jose PA, Chen SY. Response gene to complement 32 is essential for fibroblast activation in renal fibrosis. J Biol Chem. 2011;286:41323–41330. doi: 10.1074/jbc.M111.259184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Komatsu Y, Mishina Y, Mistretta CM. Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev Biol. 2012;368:294–303. doi: 10.1016/j.ydbio.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiao A. Epigenetic regulation in neural crest development. Birth Defects Res A Clin Mol Teratol. 2011;91:788–796. doi: 10.1002/bdra.20797. [DOI] [PubMed] [Google Scholar]
- McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73–82. doi: 10.1242/dmm.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Menendez L, Yatskievych TA, Antin PB, Dalton S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc Natl Acad Sci U S A. 2011;108:19240–19245. doi: 10.1073/pnas.1113746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Milgrom-Hoffman M, Michailovici I, Ferrara N, Zelzer E, Tzahor E. Endothelial cells regulate neural crest and second heart field morphogenesis. Biol Open. 2014;3:679–688. doi: 10.1242/bio.20148078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci U S A. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S, Mabuchi Y, Niibe K, Suzuki S, Nagoshi N, Sunabori T, Shimmura S, Nagai Y, Nakagawa T, Okano H, Matsuzaki Y. Development of mesenchymal stem cells partially originate from the neural crest. Biochem Biophys Res Commun. 2009;379:1114–1119. doi: 10.1016/j.bbrc.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Munoz WA, Trainor PA. Neural crest cell evolution: how and when did a neural crest cell become a neural crest cell. Curr Top Dev Biol. 2015;111:3–26. doi: 10.1016/bs.ctdb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Nagoshi N, Shibata S, Hamanoue M, Mabuchi Y, Matsuzaki Y, Toyama Y, Nakamura M, Okano H. Schwann cell plasticity after spinal cord injury shown by neural crest lineage tracing. Glia. 2011;59:771–784. doi: 10.1002/glia.21150. [DOI] [PubMed] [Google Scholar]
- Nagoshi N, Shibata S, Kubota Y, Nakamura M, Nagai Y, Satoh E, Morikawa S, Okada Y, Mabuchi Y, Katoh H, Okada S, Fukuda K, Suda T, Matsuzaki Y, Toyama Y, Okano H. Ontogeny and multipotency of neural crest-derived stem cells in mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem Cell. 2008;2:392–403. doi: 10.1016/j.stem.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Chan YS, Ito K. Notch signaling is required for the chondrogenic specification of mouse mesencephalic neural crest cells. Mech Dev. 2007;124:190–203. doi: 10.1016/j.mod.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Nichols DH. Neural crest formation in the head of the mouse embryo as observed using a new histological technique. J Embryol Exp Morphol. 1981;64:105–120. [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura-Kitabayashi A, Phoon CK, Kishigami S, Rosenthal J, Yamauchi Y, Abe K, Yamamura K, Samtani R, Lo CW, Mishina Y. Outflow tract cushions perform a critical valve-like function in the early embryonic heart requiring BMPRIA-mediated signaling in cardiac neural crest. Am J Physiol Heart Circ Physiol. 2009;297:H1617–1628. doi: 10.1152/ajpheart.00304.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Ogawa Y, Eto A, Miyake C, Tsuchida N, Miyake H, Takaku Y, Hagiwara H, Oishi K. Induced Pluripotent Stem Cells Generated from P0-Cre;Z/EG Transgenic Mice. PLoS One. 2015;10:e0138620. doi: 10.1371/journal.pone.0138620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Suzawa T, Takami M, Yamamoto G, Hosono T, Yamada A, Suzuki D, Yoshimura K, Watahiki J, Hayashi R, Arata S, Mishima K, Nishida K, Osumi N, Maki K, Kamijo R. Localization and osteoblastic differentiation potential of neural crest-derived cells in oral tissues of adult mice. Biochem Biophys Res Commun. 2015 doi: 10.1016/j.bbrc.2015.07.106. [DOI] [PubMed] [Google Scholar]
- Pomp O, Brokhman I, Ben-Dor I, Reubinoff B, Goldstein RS. Generation of peripheral sensory and sympathetic neurons and neural crest cells from human embryonic stem cells. Stem Cells. 2005;23:923–930. doi: 10.1634/stemcells.2005-0038. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Echelard Y, Danielian PS, Gellner K, Brenner S, McMahon AP. Identification of an evolutionarily conserved 110 base-pair cis-acting regulatory sequence that governs Wnt-1 expression in the murine neural plate. Development. 1998;125:2735–2746. doi: 10.1242/dev.125.14.2735. [DOI] [PubMed] [Google Scholar]
- Sahar DE, Longaker MT, Quarto N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev Biol. 2005;280:344–361. doi: 10.1016/j.ydbio.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Simon C, Lickert H, Gotz M, Dimou L. Sox10-iCreERT2 : a mouse line to inducibly trace the neural crest and oligodendrocyte lineage. Genesis. 2012;50:506–515. doi: 10.1002/dvg.22003. [DOI] [PubMed] [Google Scholar]
- Sommer L, Suter U. The glycoprotein P0 in peripheral gliogenesis. Cell Tissue Res. 1998;292:11–16. doi: 10.1007/s004410051029. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Yoshizaki K, Kobayashi T, Osumi N. Neural crest-derived horizontal basal cells as tissue stem cells in the adult olfactory epithelium. Neurosci Res. 2013;75:112–120. doi: 10.1016/j.neures.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, Ieda M, Kanakubo S, Shimazaki T, Ogawa S, Osumi N, Okano H, Fukuda K. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005;170:1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA. Specification and patterning of neural crest cells during craniofacial development. Brain Behav Evol. 2005a;66:266–280. doi: 10.1159/000088130. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin Cell Dev Biol. 2005b;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Neural crest and placodes. Preface. Curr Top Dev Biol. 2015;111:xv–xvi. doi: 10.1016/S0070-2153(15)00010-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kumar S, Mitsios N, Slevin M, Kumar P. Investigation of downstream target genes of PAX3c, PAX3e and PAX3g isoforms in melanocytes by microarray analysis. Int J Cancer. 2007;120:1223–1231. doi: 10.1002/ijc.22316. [DOI] [PubMed] [Google Scholar]
- Wang SK, Komatsu Y, Mishina Y. Potential contribution of neural crest cells to dental enamel formation. Biochem Biophys Res Commun. 2011;415:114–119. doi: 10.1016/j.bbrc.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Pan YA. Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics. 2015;199:293–306. doi: 10.1534/genetics.114.172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Sanes JR, Lichtman JW, Livet J. Generation and imaging of Brainbow mice. Cold Spring Harb Protoc. 2011;2011:851–856. doi: 10.1101/pdb.prot5632. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shimmura S, Nagoshi N, Fukuda K, Matsuzaki Y, Okano H, Tsubota K. Isolation of multipotent neural crest-derived stem cells from the adult mouse cornea. Stem Cells. 2006;24:2714–2722. doi: 10.1634/stemcells.2006-0156. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Young HM. Increased expression of p75NTR by neural crest-derived cells in vivo during mitosis. Neuroreport. 2000;11:725–728. doi: 10.1097/00001756-200003200-00014. [DOI] [PubMed] [Google Scholar]
- Young HM, Ciampoli D, Hsuan J, Canty AJ. Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut. Dev Dyn. 1999;216:137–152. doi: 10.1002/(SICI)1097-0177(199910)216:2<137::AID-DVDY5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Zhang SM, Marsh R, Ratner N, Brackenbury R. Myelin glycoprotein P0 is expressed at early stages of chicken and rat embryogenesis. J Neurosci Res. 1995;40:241–250. doi: 10.1002/jnr.490400213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Western blotting data to show the single, specific band of Cre with the primary antibody (1:5000, MAB3120, EMD Millipore) that was used to generate immunosignals in embryonic tissue sections. Embryos carrying P0-Cre transgene were collected at E9.5. A littermate that did not carry the transgene was used as negative control (Cre−). Tissue lysates were prepared and immunoblotted with the Cre antibody indicating their specificity against Cre protein (38-kD). IKAP (150-kD) antibody was also used as a loading control. Bands of expected proteins were visualized with DAB reaction.
Supplemental Fig. 2. Representative photomicrographs taken with a fluorescent light microscope illustrate the absence of Cre immunoreactivity in transverse sections of a 14-somite P0-Cre negative embryo. Sections were immunostained using an antibody against Cre (green) and triple labeled with Sox9 (purple, A–C) and p75 (red, A–C). Scale bar: 50 μm for all images.
Supplemental Fig. 3. Representative photomicrographs shown previously are assembled to compare P0-Cre and Wnt1-Cre in labeling cranial neural crest, neuroepithelium, and notochord. In brief, in bright-field (top) and fluorescent (bottom) images of whole P0-Cre/RFP and Wnt1-Cre/RFP embryos (A), the midbrain (mb) region was labeled by Wnt1-Cre signals but not by P0-Cre (arrowheads), while the hindbrain was labeled by P0-Cre but sparse in Wnt1-Cre (Arrows). On sections (B), P0-Cre activities, peaked at 14-somite stage, were distributed in post-migratory NC cells in trigeminal neural crest (tn, B1) at posterior hindbrain level, in the optic eminence (oe, B2), in anterior hindbrain (hb, B3), and the forebrain (fb, B3). Wnt1-Cre activities were extensive at 7-somite stage (Wnt1-Cre, B1-2), in the pre- and post-migratory neural crest cells in midbrain (mb, B1) and forebrain (fb, B2-3) regions, optic eminence (oe, B2) and in the neuroepithelium (mb, hb, fb, B1-3). Scale bars: 250 μm in A and 50 μm in B.