Abstract
Evolutionary theory of ageing maintains that increased allocation to early-life reproduction results in reduced somatic maintenance, which is predicted to compromise longevity and late-life reproduction. This prediction has been challenged by the discovery of long-lived mutants with no loss of fecundity. The first such long-lived mutant was found in the nematode worm Caenorhabditis elegans. Specifically, partial loss-of-function mutation in the age-1 gene, involved in the nutrient-sensing insulin/insulin-like growth factor signalling pathway, confers longevity, as well as increased resistance to pathogens and to temperature stress without appreciable fitness detriment. Here, we show that the long-lived age-1(hx546) mutant has reduced fecundity and offspring production in early-life, but increased fecundity, hatching success, and offspring production in late-life compared with wild-type worms under standard conditions. However, reduced early-life performance of long-lived mutant animals was not fully compensated by improved performance in late-life and resulted in reduced individual fitness. These results suggest that the age-1(hx546) allele has opposing effects on early-life versus late-life fitness in accordance with antagonistic pleiotropy (AP) and disposable soma theories of ageing. These findings support the theoretical conjecture that experimental studies based on standing genetic variation underestimate the importance of AP in the evolution of ageing.
Keywords: ageing, senescence, life history
1. Introduction
Ageing is a progressive deterioration of organismal function leading to reduced reproduction and/or increased probability of death with increasing age [1]. Despite the presence of cellular repair systems, ageing affects nearly all organisms and reduces Darwinian fitness in natural populations [2–4]. The evolutionary theory of ageing rests on the basic assumption that the lifespan of individual organisms in nature is inescapably curtailed by environmental hazards (e.g. predation, parasitism, starvation, drought, or extreme temperatures) and accidents. Because of such environment-driven mortality, deleterious mutations whose effects are concentrated in late-life are partially shielded from selection and can accumulate in the population to cause an age-specific decline in organismal performance (mutation accumulation (MA) theory; [5–7]). Moreover, alleles with beneficial effects on early-life can be positively selected despite their negative effects on late-life (antagonistic pleiotropy (AP) theory; [8]). The ‘disposable soma’ (DS) theory of ageing [9–11], which can be seen as a physiological account of quantitative genetic AP theory, specifically posits that organisms evolve to optimally allocate their limited metabolic resources between reproduction and somatic maintenance.
Empirical evidence suggests that standing genetic variation for ageing and longevity can be dominated by mutations with positively pleiotropic effects across the life stages supporting the MA theory (reviewed in [12]). Nevertheless, it has often been argued that quantitative genetic approaches may underestimate the importance of AP because AP alleles would rapidly go to fixation [1]. Increased somatic maintenance manifested in high-fidelity DNA repair, better clearance of damaged proteins from the cells, and stronger immune response can safeguard the organism against genome and proteome damage, as well as against parasites, and extend its projected lifespan (reviewed in [13]). However, when such measures hinder growth and impair reproduction, increased longevity can be detrimental to fitness, resulting in rapid fixation of alleles that boost early-life performance at the cost of reduced longevity and accelerated ageing. Because of the possible rapid fixation of adaptive AP alleles, there could be relatively little standing AP genetic variation for fitness in populations [1]. However, the occurrence of de novo mutations that increase somatic maintenance leading to improved longevity and stress resistance but result in reduced early-life growth and/or reproductive performance and compromised net individual fitness would provide support for AP and DS theories of ageing. There are few examples of such mutations in model organisms, including mutations in daf-2 [14], nuo-6, and isp-1 [15] genes in Caenorhabditis elegans and three mutations in the insulin/insulin-like growth factor signalling (IIS) pathway in mice [16], where increased lifespan is associated with reduced reproductive performance (reviewed in [17,18]).
The discovery of long-lived mutants that combine longevity and stress resistance with normal reproduction challenged the inevitability of a resource allocation trade-off between reproduction and somatic maintenance [18–21]. The first long-lived mutant was discovered in the nematode worm C. elegans, where partial loss-of-function mutation in the age-1 gene encoding a catalytic subunit of the Pl3 kinase results in downregulation of the nutrient-sensing IIS pathway [19,22]. Initially, the longevity of age-1 mutants was associated with severe reduction in fecundity [22]. However, it was later discovered that fecundity deficit was caused by a different co-segregating allele, while fecundity of age-1(hx546) worms was considered normal [23]. Because the finding of a long-lived mutant with normal fecundity contradicted the prediction from AP and DS theories, later studies focused on testing the performance of age-1(hx546) mutant animals under different environmental conditions. Thus, mutant worms were found to perform worse than wild-type under nutritional stress signifying that the trade-off between longevity and starvation resistance could explain the prevalence of the age-1(+) wild-type allele [24]. However, age-1(hx546) worms outperform age-1(+) wild-type counterparts in resistance to a range of bacterial pathogens [25], as well as in resistance to cold stress [26] and heat stress [27,28] suggesting that the age-1(+) allele may not necessarily confer advantage under ecologically relevant environmental conditions [28,29]. Moreover, a recent study combined the effects of starvation and heat stress to show that the age-1(hx546) mutation confers a selective advantage when periodic starvation is encountered together with temperature fluctuations, suggesting that environmental heterogeneity can favour long-lived mutants over their short-lived wild-type counterparts [28]. Thus, while several long-lived mutants in different model organisms were found to perform poorly under more natural conditions [17], age-1(hx546) animals seem to thrive under a range of ecologically relevant environments.
Here, we studied age-specific reproduction and survival of the long-lived and stress-resistant age-1(hx546) mutant to test two non-mutually exclusive hypotheses that could explain the reported lack of fitness loss in response to increased investment into somatic maintenance under standard conditions. First, we hypothesized that the long-lived genotype has reduced early-life reproductive performance, which can be compensated by improved performance in late-life. If so, individual rate-sensitive fitness of long-lived worms will be reduced. The rate-sensitive estimates of fitness should be particularly relevant for longevity evolution under natural conditions where high environmental mortality due to predation and the ephemeral nature of their food sources can place higher priority on early-life reproduction than in the laboratory selection experiments.
Second, it is possible that long-lived worms invest less into protection and repair of their germline reducing the quality of their gametes rather than their quantity. Germline maintenance is costly, and a recent study has found that increased investment into germline stem cells can accelerate somatic ageing in nematodes [30]. To this end, we used an established technique to test the ability of the worms to repair the germline damage by subjecting them to low-dose ionizing radiation [31]. Because somatic cells of C. elegans nematodes are post-mitotic, they are highly resistant to radiation, while their germline stem cells are easily affected [31–33]. The germline that is damaged by exposure to low-dose ionizing radiation has the capacity to recover, either partially or fully [31], as long as some of the germline stem cells survive. We used both hatching success and total offspring production following low-dose ionizing radiation relative to baseline levels of these variables in untreated control animals as an indicator of investment into germline protection, repair, and recovery.
2. Material and methods
(a). Nematode strains and experimental procedures
We used C. elegans strains Bristol N2 wild-type [34] and TJ1052 age-1(hx546) [35]. We confirmed the genotype of the age-1(hx546) mutant by PCR using primers 261 age1F, 5′-GATGTTATCCATAACTTCGA-3′, and 262 age1R, 5′-TTACCTCCGTGGAAATGAAG-3′, followed by sequencing. The populations were recovered and maintained under standard laboratory conditions at 20°C and 60% RH on nematode growth medium (NGM) agar plates [34] that also contained kanamycin, streptomycin, and nystatin following Lionaki & Tavernarakis [36], to prevent bacterial and fungal growth and fed with OP50-1 E. coli obtained from J. Ewbank from Centre d'Immunologie de Marseille-Luminy, which are resistant to these antibiotics. The population was synchronized by bleaching [37].
For life-history assays (lifespan and fecundity), we only used nystatin to prevent fungal contamination and worms were fed standard OP50 E. coli in these assays. In lifespan assays, 50 individual L4 worms per plate were placed on 35 mm agar plates and checked daily until death. Some worms were lost resulting in slightly reduced final sample sizes (see the electronic supplementary material, table S4). The worms were transferred every second day to a new plate seeded with 0.1 ml E. coli OP50 grown in Luria-Bertani (LB) broth supplemented with 10 µg ul−1 nystatin. In reproduction assays, we followed the same procedure but individually kept worms were transferred to new plates every day until day 7, when reproduction has virtually ceased. Plates were saved for counting developed larvae and eggs 2.5 days later. Offspring were killed by placing them in a 40°C chamber for 2 h prior to counting.
Gamma-irradiation treatment was conducted using a caesium source at Rudbeck laboratory, Uppsala University. All worms (including non-irradiated controls) were transported simultaneously to Rudbeck and back. Half of the plates were irradiated at 90 Gy (97.7 min at estimated 0.9214 Gy min−1) by being randomly placed in one layer inside the caesium source.
(b). Statistical analyses
Survival was analysed in the Cox proportional hazard models with Gaussian random effects implemented in the coxph package for R v. 3.2.2. The two worm strains (wild-type N2 and TJ1052 age-1(hx546)) and the treatments (control and irradiation) were modelled as crossed fixed factors.
We fitted the mortality data to the Gompertz family of four nested models (Gompertz, Gompertz–Makeham, Logistic, and Logistic–Makeham) using a maximum-likelihood approach in WinModest software and the best fit was decided based on likelihood ratio tests [38]. The fullest model is Logistic–Makeham, where μx = c + αeβx/[1 + (αs/β) (eβx − 1)], where μx is the mortality hazard at age x, c is the constant mortality, α is the baseline mortality rate, β is the rate of increase in mortality with age (Gompertz rate-of-senescence), and s is the rate of deceleration in mortality late in life (late-life deceleration). When c and s parameters equal zero, the model is reduced to the simplest two-parameter Gompertz equation (μx = αeβx).
Fecundity (eggs produced) and offspring production (larvae alive after 2 days) were analysed in separate mixed models using the lme4 package in R. Response variables were log transformed after adding a small constant (0.01) to meet the assumptions of normality and avoid infinite values. Strain, treatment, age, and age2 were modelled as crossed fixed factors, and female ID as a random factor, to control for repeated measures. To investigate the effects of early and late reproduction, we also analysed the total reproduction between days 1–3 and 4–6 of adulthood for every individual that survived this time period. These data were analysed in separate ANOVAs with strain and treatment as crossed fixed factors.
Hatching success was analysed in a generalized mixed model with binomial error structure. Strain, treatment, age, and age2 were modelled as crossed fixed factors, and female ID as a random factor, to control for repeated measures. To test and control for overdispersion, we also added an individual-level observation effect (ILOE) as a random factor. We compared models with and without the ILOE using the DHARMa package in R. Inspection of the scaled residuals and testing for overdispersion showed that none of the models was significantly overdispersed (with ILOE: p = 1.0, without ILOE: p = 0.46) The dispersion parameters of the models were calculated using the blmeco package in R (with ILOE: 0.595, without ILOE: 0.995).
Individual fitness (λind) is a rate-sensitive measure of fitness and was calculated from the life table of age-specific daily reproduction from the Euler–Lotka equation following Brommer et al. [39]. Individual fitness and lifetime reproductive success (LRS) was only calculated for individuals surviving at least 6 days (13 individuals excluded), which encompasses the absolute majority of reproduction. Individual fitness and total reproduction were analysed in separate ANOVAs, using strain and treatment as crossed fixed factors. LRS was log-transformed prior to the analysis.
3. Results and discussion
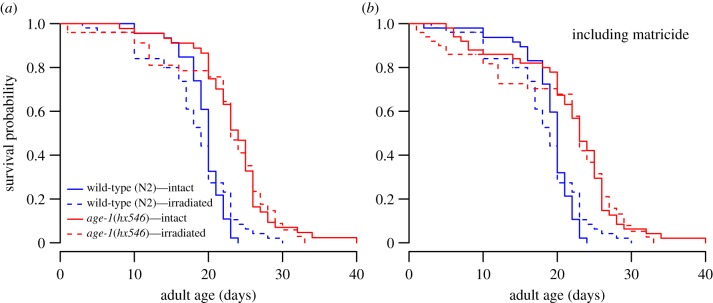
Mutant age-1(hx546) worms indeed lived longer than wild-type, while there was no significant effect of ionizing radiation on longevity and no interaction between strain and radiation effects. Similar results were also obtained if worms dying of matricide (internal hatching of eggs) were treated as dead instead of excluded (figure 1 and table 1). However, radiation increased early-life mortality resulting in a difference in age-specific mortality rates (figure 1 and electronic supplementary material, table S1). The age-1(hx546) mutant had significant late-life deceleration in mortality with the Logistic–Makeham model being the best fit (electronic supplementary material, table S1), while the best fit for wild-type worms was the Gompertz–Makeham model suggesting zero late-life deceleration in mortality rate in this cohort. Irradiated mutant animals lived longer than control wild-type individuals (figure 1; χ2 = 25.2, d.f. = 1, p < 0.001) further supporting increased stress resistance and longevity of age-1(hx546) worms. Interestingly, the cohort of irradiated mutant worms had lower rate-of-senescence than non-irradiated controls (−2*ΔLLR = 8.201, d.f. = 1, p = 0.004; electronic supplementary material, table S1).
Figure 1.
Survival curves of Bristol N2 wild-type and TJ1052 age-1(hx546) strains when kept under standard conditions (intact) or gamma-irradiated at 90 Gy (irradiated). Panel (a) shows survival when worms that died by matricide (bagging) are excluded, while (b) shows survival when all worms are included.
Table 1.
Survival when matricidal worms (died of internal hatching of eggs early in life) were either excluded or counted as dead.
| matricide excluded |
matricide as dead |
|||||
|---|---|---|---|---|---|---|
| z | d.f. | p-value | z | d.f. | p-value | |
| strain | −5.36 | 3 | <0.001 | −4.70 | 3 | <0.001 |
| treatment | −0.98 | 3 | 0.33 | −0.91 | 3 | 0.36 |
| strain × treatment | 0.86 | 3 | 0.39 | 0.87 | 3 | 0.38 |
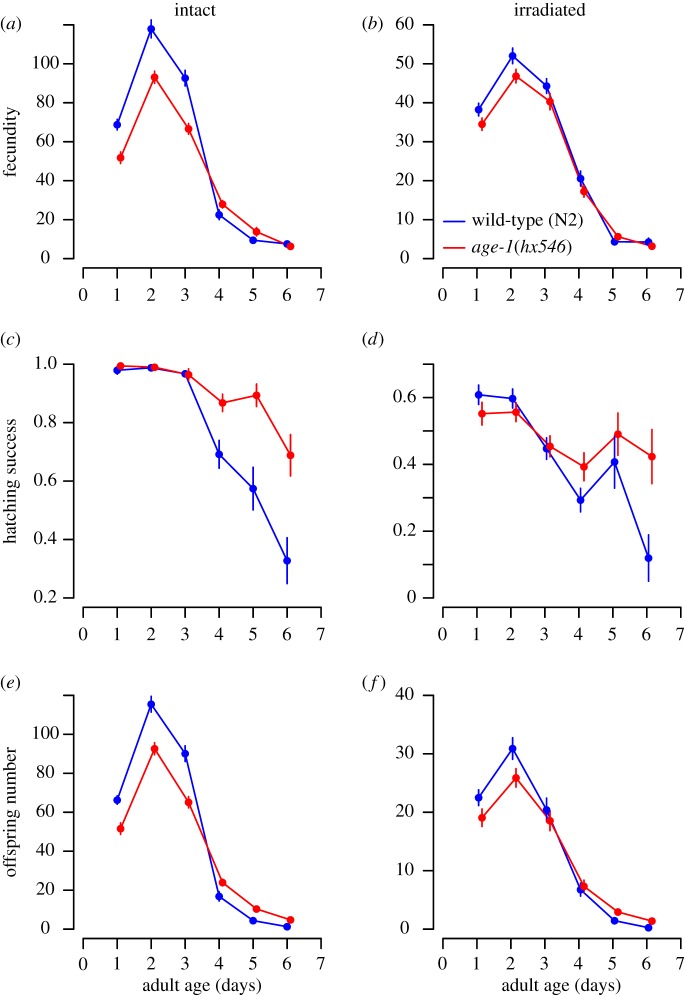
Long-lived age-1(hx546) mutant animals laid fewer eggs than wild-type worms both under irradiated and control conditions (table 2), and this difference in fecundity was driven by lower egg-laying rates during the first 3 days of life (figure 2a,b; electronic supplementary material, table S2). However, age-1(hx546) worms had higher hatching success than wild-type worms during the second part of their reproductive life (days 4–6; figure 2c,d and table 3). Because the age-1(hx546) long-lived mutant had similar fecundity but higher hatching success in late-life, they also had higher offspring production during the last 3 days of reproduction (figure 2e,f and electronic supplementary material, table S3).
Table 2.
Total fecundity (eggs laid), viable offspring produced, and hatching success. Residual variance is not calculated for binomial mixed effect models (hatching success).
| parameter | fecundity |
offspring |
hatching success |
||||||
|---|---|---|---|---|---|---|---|---|---|
| χ2 | d.f. | p | χ2 | d.f. | p | χ2 | d.f. | p | |
| strain | 7.23 | 1 | 0.007 | 17.38 | 1 | <0.001 | 3.50 | 1 | 0.061 |
| treatment | 31.09 | 1 | <0.001 | 34.98 | 1 | <0.001 | 538.9 | 1 | <0.001 |
| age | 4 684 | 1 | <0.001 | 5 522 | 1 | <0.001 | 63.61 | 1 | <0.001 |
| age2 | 1 981 | 1 | <0.001 | 2 724 | 1 | <0.001 | 0.01 | 1 | 0.944 |
| strain × treatment | 0.06 | 1 | 0.799 | 3.73 | 1 | 0.054 | 1.49 | 1 | 0.222 |
| strain × age | 12.28 | 1 | <0.001 | 34.40 | 1 | <0.001 | 8.14 | 1 | 0.004 |
| treatment × age | 4.99 | 1 | 0.026 | 4.75 | 1 | 0.029 | 102.3 | 1 | <0.001 |
| strain × age2 | 16.46 | 1 | <0.001 | 47.07 | 1 | <0.001 | 48.41 | 1 | <0.001 |
| treatment × age2 | 14.69 | 1 | <0.001 | 0.46 | 1 | 0.496 | 29.17 | 1 | <0.001 |
| strain × treatment × age | 2.00 | 1 | 0.158 | 8,57 | 1 | 0.003 | 28.99 | 1 | <0.001 |
| strain × treatment × age2 | 1.41 | 1 | 0.235 | 9.51 | 1 | 0.002 | 58.37 | 1 | <0.001 |
| random effects | Var. | s.d. | Var. | s.d. | Var. | s.d. | |||
| female ID (intercept) | 0.629 | 0.793 | 0.893 | 0.945 | 5.661 | 2.379 | |||
| female ID (age slope) | 0.002 | 0.048 | 0.004 | 0.063 | 0.518 | 0.713 | |||
| residual | 2.446 | 1.564 | 2.165 | 1.471 | |||||
Figure 2.
Age-specific reproductive performance (fecundity, hatching success, total number of offspring produced) of Bristol N2 wild-type and TJ1052 age-1(hx546) strains when kept under standard conditions (a,c,e) or gamma-irradiated at 90 Gy (b,d,f). N2 worms reproduced better during the first three days, while TJ1052 worms reproduced better during the last three days (table 2 and electronic supplementary material, tables S2 and S3).
Table 3.
Total reproduction and individual fitness (λind).
| parameter | LRS |
individual fitness (λind) |
||||||
|---|---|---|---|---|---|---|---|---|
| MS | d.f. | F | p-value | MS | d.f. | F | p-value | |
| strain | 1.14 | 1 | 8.68 | 0.004 | 3.60 | 1 | 29.05 | <0.001 |
| treatment | 88.81 | 1 | 677 | <0.001 | 83.98 | 1 | 678 | <0.001 |
| strain × treatment | 0.006 | 1 | 0.04 | 0.835 | 0.11 | 1 | 0.89 | 0.346 |
| residuals | 24.02 | 183 | 22.67 | 183 | ||||
Thus, long-lived age-1(hx546) mutants have lower fecundity and lower offspring production in early-life, but higher hatching success and higher offspring production in late-life, while irradiation has a negative effect on reproduction in both strains. The interactions between irradiation, strain, and age for offspring production (table 2) suggest that the effect of irradiation on this trait was somewhat stronger in wild-type worms in early-life (figure 2e,f). Because the worms produce most of their eggs and offspring during the first 3 days of life (figure 2a,b,e,f and electronic supplementary material, tables S2 and S3), it is not surprising that both LRS and rate-sensitive individual fitness (λind) of wild-type worms are higher than those of the long-lived strain (figure 3a,b and table 3).
Figure 3.
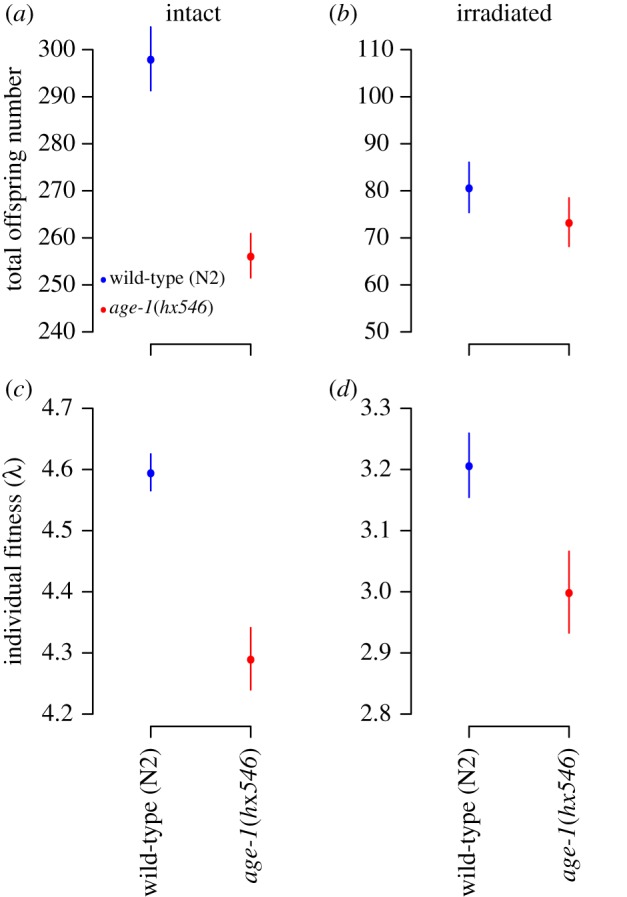
LRS and individual fitness (rate-sensitive fitness) of Bristol N2 wild-type and TJ1052 age-1(hx546) strains when kept under standard conditions (intact) or gamma-irradiated at 90 Gy (irradiated). N2 worms have higher LRS and individual fitness both when intact and irradiated (table 3).
These results are fully in line with AP and DS theories of ageing, and suggest that age-1(hx546) is an antagonistically pleiotropic allele that increases longevity, stress resistance, and late-life reproductive performance at the cost of early-life reproduction and net fitness. These findings thus provide support for the hypothesis that ageing evolved as an optimal life-history strategy [1], where alleles that increase fitness are fixed by selection even when they accelerate ageing and reduce lifespan.
Several previous studies exploited standing genetic variation for ageing to test for the trade-offs between early-life and late-life fitness but produced mixed results (reviewed in [12]). The general problem with such studies is that standing genetic variation for age-specific fitness is not very informative as a test of the evolutionary theories of ageing, because beneficial alleles will become fixed by selection, while detrimental alleles can become fixed by drift [1]. Thus, even if AP alleles do play a key role in shaping the evolution of ageing in any given species or population, it may be difficult to reveal this using quantitative genetic approaches based on standing genetic variation. MA studies can provide a useful alternative but they are also not conclusive because AP alleles can be rare. Indeed, several mutations that increase longevity in model organisms also cause major reductions in fitness [17,40,41], suggesting that they would be rapidly selected against if occurred under natural conditions. Therefore, the discovery of an age-1(hx546) allele that substantially increased longevity, as well as stress resistance, of C. elegans nematodes that maintain normal development, locomotion, and fecundity [24,28] was surprising and remains one of the hallmarks of the idea that lifespan extension can be relatively cost-free [19,42]. Moreover, recent studies suggest that heat-shock resistance can confer substantial evolutionary advantage to age-1(hx546) worms under starvation [28], the only condition to date where these long-lived mutants were shown to suffer in comparison to the wild-type [24]. Here, we analysed survival and age-specific reproduction of age-1(hx546) long-lived worms and found that they suffer from substantial reproductive deficiencies in early-life, which are not sufficiently compensated by their improved late-life performance. Mutations such as age-1(hx546), which are beneficial for long life and slow ageing but detrimental for individual fitness, are directly predicted by the AP theory of ageing and support the notion that standing genetic variation approaches may underestimate the importance of AP in the evolution of ageing.
It has been suggested in recent years that energy trade-offs between reproduction and soma can be uncoupled and, therefore, are not necessarily decisive in the evolution of ageing and longevity [18,20,21,29,42,43]. Here, we show that the long-lived age-1 mutant previously reported to lack the fecundity deficit suffers from early-life reduction in fecundity and net individual fitness under standard conditions supporting the hypothesis of the reproduction-longevity trade-off. Future research should focus on understanding the mechanistic basis of the reduced early-life reproduction and improved late-life reproduction and longevity of the age-1(hx546) mutant. Recent studies suggest that there is a trade-off between germline cell maintenance and longevity in C. elegans nematodes [13,44–46], and that germline removal, or arrest of germline development increases protection and repair of the proteome in the somatic cells [45,47]. It is possible that long-lived age-1(hx546) mutant animals have either smaller or slower developing germlines and, therefore, have more resources available for the maintenance of the somatic cells resulting in slower reproductive ageing and increased longevity.
Supplementary Material
Supplementary Material
Supplementary Material
Ethics
All experiments comply with the laws of Sweden.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
A.A.M., A.H., and S.I. conceived, designed, and coordinated the study. A.A.M. and S.I. wrote the initial draft. H.C., P.D., and B.M. conducted the experimental work. M.I.L. and A.A.M. analysed the data. H.C., P.D., M.I.L., B.M., and A.H. worked on the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This work has been supported by the ERC Starting Grant AGINGSEXDIFF 260885 and Swedish Research Council grant no. 2013-04828 to A.A.M. and the ERC Starting Grant HAPSELA 336633 to S.I.
References
- 1.Partridge L, Barton NH. 1993. Optimality, mutation and the evolution of aging. Nature 362, 305–311. ( 10.1038/362305a0) [DOI] [PubMed] [Google Scholar]
- 2.Boonekamp JJ, Salomons M, Bouwhuis S, Dijkstra C, Verhulst S. 2014. Reproductive effort accelerates actuarial senescence in wild birds: an experimental study. Ecol. Lett. 17, 599–605. ( 10.1111/ele.12263) [DOI] [PubMed] [Google Scholar]
- 3.Nussey DH, Froy H, Lemaitre JF, Gaillard JM, Austad SN. 2013. Senescence in natural populations of animals: widespread evidence and its implications for bio-gerontology. Ageing Res. Rev. 12, 214–225. ( 10.1016/j.arr.2012.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre J-F, Berger V, Bonenfant C, Douhard M, Gamelon M, Plard F, Gaillard J-M. 2015. Early–late life trade-offs and the evolution of ageing in the wild. Proc. R. Soc. B 282, 20150209 ( 10.1098/rspb.2015.0209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medawar PB. 1952. An unresolved problem of biology. London, UK: H.K. Lewis. [Google Scholar]
- 6.Wachter KW, Evans SN, Steinsaltz D. 2013. The age-specific force of natural selection and biodemographic walls of death. Proc. Natl Acad. Sci. USA 110, 10 141–10 146. ( 10.1073/pnas.1306656110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wachter KW, Steinsaltz D, Evans SN. 2014. Evolutionary shaping of demographic schedules. Proc. Natl Acad. Sci. USA 111, 10 846–10 853. ( 10.1073/pnas.1400841111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411. ( 10.2307/2406060) [DOI] [Google Scholar]
- 9.Kirkwood TBL. 1977. Evolution of aging. Nature 270, 301–304. ( 10.1038/270301a0) [DOI] [PubMed] [Google Scholar]
- 10.Kirkwood TBL, Holliday R. 1979. Evolution of aging and longevity. Proc. R. Soc. Lond. B 205, 531–546. ( 10.1098/rspb.1979.0083) [DOI] [PubMed] [Google Scholar]
- 11.Kirkwood TBL, Austad SN. 2000. Why do we age? Nature 408, 233–238. ( 10.1038/35041682) [DOI] [PubMed] [Google Scholar]
- 12.Maklakov AA, Rowe L, Friberg U. 2015. Why organisms age: evolution of senescence under positive pleiotropy? Bioessays 37, 802–807. ( 10.1002/bies.201500025) [DOI] [PubMed] [Google Scholar]
- 13.Maklakov AA, Immler S. 2016. The expensive germline and the evolution of ageing. Curr. Biol. 26, R577–R586. ( 10.1016/j.cub.2016.04.012) [DOI] [PubMed] [Google Scholar]
- 14.Jenkins NL, McColl G, Lithgow GJ. 2004. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc. R. Soc. Lond. B 271, 2523–2526. ( 10.1098/rspb.2004.2897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Hekimi S. 2010. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell 9, 433–447. ( 10.1111/j.1474-9726.2010.00571.x) [DOI] [PubMed] [Google Scholar]
- 16.Bartke A, Brown-Borg H. 2004. Life extension in the dwarf mouse. Curr. Top. Dev. Biol. 63, 189–225. ( 10.1016/S0070-2153(04)63006-7) [DOI] [PubMed] [Google Scholar]
- 17.Briga M, Verhulst S. 2015. What can long-lived mutants tell us about mechanisms causing aging and lifespan variation in natural environments? Exp. Gerontol. 71, 21–26. ( 10.1016/j.exger.2015.09.002) [DOI] [PubMed] [Google Scholar]
- 18.Partridge L, Gems D, Withers DJ. 2005. Sex and death: what is the connection? Cell 120, 461–472. ( 10.1016/j.cell.2005.01.026) [DOI] [PubMed] [Google Scholar]
- 19.Kenyon C. 2011. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Phil. Trans. R. Soc. B 366, 9–16. ( 10.1098/rstb.2010.0276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leroi AM. 2001. Molecular signals versus the Loi de Balancement. Trends Ecol. Evol. 16, 24–29. ( 10.1016/S0169-5347(00)02032-2) [DOI] [PubMed] [Google Scholar]
- 21.Leroi AM, et al. 2005. What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech. Ageing Dev. 126, 421–429. ( 10.1016/j.mad.2004.07.012) [DOI] [PubMed] [Google Scholar]
- 22.Friedman DB, Johnson TE. 1988. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson TE, Tedesco PM, Lithgow GJ. 1993. Comparing mutants, selective breeding, and transgenics in the dissection of aging processes of caenorhabditis-elegans. Genetica 91, 65–77. ( 10.1007/BF01435988) [DOI] [PubMed] [Google Scholar]
- 24.Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. 2000. Natural selection—evolution of lifespan in C. elegans. Nature 405, 296–297. ( 10.1038/35012693) [DOI] [PubMed] [Google Scholar]
- 25.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921 ( 10.1126/science.1080147) [DOI] [PubMed] [Google Scholar]
- 26.Savory FR, Sait SM, Hope IA. 2011. DAF-16 and delta(9) desaturase genes promote cold tolerance in long-lived Caenorhabditis elegans age-1 mutants. PLoS ONE 6, e24550 ( 10.1371/journal.pone.0024550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lithgow GJ, White TM, Hinerfeld DA, Johnson TE. 1994. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J. Gerontol. 49, B270–B276. ( 10.1093/geronj/49.6.B270) [DOI] [PubMed] [Google Scholar]
- 28.Savory FR, Benton TG, Varma V, Hope IA, Sait SM. 2014. Stressful environments can indirectly select for increased longevity. Ecol. Evol. 4, 1176–1185. ( 10.1002/ece3.1013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenyon C. 2005. The plasticity of aging: insights from long-lived mutants. Cell 120, 449–460. ( 10.1016/j.cell.2006.02.002) [DOI] [PubMed] [Google Scholar]
- 30.Aprison EZ, Ruvinsky I. 2016. Sexually antagonistic male signals manipulate germline and soma of C. elegans hermaphrodites. Curr. Biol. 26, 2827–2833. ( 10.1016/j.cub.2016.08.024) [DOI] [PubMed] [Google Scholar]
- 31.Ermolaeva MA, Segref A, Dakhovnik A, Ou H-L, Schneider JI, Utermoehlen O, Hoppe T, Schumacher B. 2013. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature 501, 416–420. ( 10.1038/nature12452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson TE, Hartman PS. 1988. Radiation effects on life-span in Caenorhabditis elegans. J. Gerontol. 43, B137–B141. ( 10.1093/geronj/43.5.B137) [DOI] [PubMed] [Google Scholar]
- 33.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. 2000. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5, 435–443. ( 10.1016/s1097-2765(00)80438-4) [DOI] [PubMed] [Google Scholar]
- 34.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lithgow GJ, White TM, Melov S, Johnson TE. 1995. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal-stress. Proc. Natl Acad. Sci. USA 92, 7540–7544. ( 10.1073/pnas.92.16.7540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lionaki E, Tavernarakis N. 2013. High-throughput and longitudinal analysis of aging and senescent decline in Caenorhabditis elegans. In Cell senescence (eds Galluzzi L, Vitale I, Kepp O, Kroemer G), pp. 485–500. Hatfield, UK: Humana Press, Springer; ( 10.1007/978-1-62703-239-1_32) [DOI] [PubMed] [Google Scholar]
- 37.Stiernagle T. 2006. Maintenance of C. elegans. In WormBook (ed. The C. elegans Research Community, WormBook) ( 10.1895/wormbook.1.101.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pletcher SD. 1999. Model fitting and hypothesis testing for age-specific mortality data. J. Evol. Biol. 12, 430–439. ( 10.1046/j.1420-9101.1999.00058.x) [DOI] [Google Scholar]
- 39.Brommer JE, Merila J, Kokko H. 2002. Reproductive timing and individual fitness. Ecol. Lett. 5, 802–810. ( 10.1046/j.1461-0248.2002.00369.x) [DOI] [Google Scholar]
- 40.Voorhies WA Van, Curtsinger JW, Rose MR. 2006. Do longevity mutants always show trade-offs? Exp. Gerontol. 41, 1055–1058. ( 10.1016/j.exger.2006.05.006) [DOI] [PubMed] [Google Scholar]
- 41.Magwire MM, Yamamoto A, Carbone MA, Roshina NV, Symonenko AV, Pasyukova EG, Morozova TV, Mackay TFC. 2010. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 6, e1001037 ( 10.1371/journal.pgen.1001037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenyon CJ. 2010. The genetics of ageing. Nature 464, 504–512. ( 10.1038/nature08980) [DOI] [PubMed] [Google Scholar]
- 43.Gerisch B, Antebi A. 2011. Molecular basis of life history regulation in C. elegans and other organisms. In Mechanisms of life-history evolution: the genetics and physiology of life history traits and trade-offs (eds Flatt T, Heyland A), pp. 284–298. Oxford, UK: Oxford University Press. [Google Scholar]
- 44.Labbadia J, Morimoto RI. 2015. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell 59, 639–650. ( 10.1016/j.molcel.2015.06.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues APC, Manning G, Dillin A. 2012. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268. ( 10.1038/nature11315) [DOI] [PubMed] [Google Scholar]
- 46.Thondamal M, Witting M, Schmitt-Kopplin P, Aguilaniu H. 2014. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat. Commun. 5, 4879 ( 10.1038/ncomms5879) [DOI] [PubMed] [Google Scholar]
- 47.Shemesh N, Shai N, Ben-Zvi A. 2013. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell 12, 814–822. ( 10.1111/acel.12110) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.