Abstract
The synthesis of antimicrobial thymol/carvacrol-loaded polythioether nanoparticles (NPs) via a one-pot, solvent-free miniemulsion thiol-ene photopolymerization process is reported. The active antimicrobial agents, thymol and carvacrol, are employed as “solvents” for the thiol-ene monomer phase in the miniemulsion to enable facile high capacity loading (≈50% w/w), excellent encapsulation efficiencies (>95%), and elimination of all postpolymerization purification processes. The NPs serve as high capacity reservoirs for slow-release and delivery of thymol/carvacrol-combination payloads that exhibit inhibitory and bactericidal activity (>99.9% kill efficiency at 24 h) against gram-positive and gram-negative bacteria, including both saprophytic (Bacillus subtilis ATCC 6633 and Escherichia coli ATCC 25922) and pathogenic species (E. coli ATCC 43895, Staphylococcus aureus RN6390, and Burkholderia cenocepacia K56-2). This report is among the first to demonstrate antimicrobial efficacy of essential oil-loaded nanoparticles against B. cenocepacia – an innately resistant opportunistic pathogen commonly associated with debilitating respiratory infections in cystic fibrosis. Although a model platform, these results point to promising pathways to particle-based delivery of plant-derived extracts for a range of antimicrobial applications, including active packaging materials, topical antiseptics, and innovative therapeutics.
1. Introduction
Microbial threats, particularly multidrug-resistant (MDR) bacterial strains and other emerging pathogens, are greatly impacting public health, burdening healthcare systems, and have potential to disrupt socioeconomic infrastructures in both developing and industrialized nations. In the United States alone, the Center for Disease Control estimates at least two million illnesses and 23 000 deaths are attributed to multidrug-resistant bacterial infections each year. Annually, these cases account for ≈$20 billion in excess health care costs and up to $35 billion in lost productivity due to sick leave and hospitalizations.[1] While Staphylococcus aureus and Pseudomonas aeruginosa tend to dominate the MDR discussion, other microorganisms, such as Burkholderia cepacia complex (Bcc), have emerged as opportunistic pathogens with significant clinical importance in persons with cystic fibrosis (CF).[2] Pulmonary colonization with Burkholderia cenocepacia, the most common isolate of the 18 member Bcc, results in severe respiratory infections in persons with CF and is associated with high morbidity and mortality rates.[3] Effective treatments for B. cenocepacia infections are rare, as these bacteria exhibit high intrinsic resistance to most antibiotics.[4] In general, the MDR microbial threat extends far beyond the effect on humans, and additionally impacts animal agriculture and veterinary medicine. The continuous emergence of MDR pathogens and scarcity of new antimicrobial drug scaffolds in the pharmaceutical discovery pipeline have led to growing interest in natural, plant-derived extracts as alternatives to synthetic antibiotics.[5] In this direction, essential oils (EOs) – typically complex extracts from aromatic plants comprising mixtures of aldehydes, terpenes, and phenols – are well known to exhibit broad spectrum biological and antimicrobial activity.[6,7] With many EOs identified as “Generally Regarded as Safe,” these extracts have been actively explored and continue to garner interest as food preservatives and packaging constituents,[8,9] textile fragrances,[10] pesticides,[11] and other antimicrobial therapeutic applications.[12–14]
Monoterpene phenol isomers, carvacrol and thymol, are major constituents of EOs extracted from oregano, thyme, and other plants belonging to the Lamiaceae family. These isomers show antiviral, antifungal, and broad spectrum antibacterial properties against both gram-negative and gram-positive bacteria, including MDR and biofilm forming microorganisms.[15,16] In one promising example, EO extracts containing carvacrol and thymol as primary constituents were shown to inhibit the growth of several environmental and clinical bacterial strains belonging to the B. cepacia complex.[17] The broad spectrum of activity of these isomers has been attributed to multiple modes of toxicity; however, the primary site of toxicity is the cell membrane.[16] In general, these hydrophobic isomers act by partitioning into the cytoplasmic membrane leading to increased permeability, depletion of proton gradients, and subsequent disruption of adenosine triphosphate (ATP) synthesis. The collapse of the proton motive force and depletion of the ATP pool eventually lead to cell death.
A major challenge for the practical application of carvacrol and thymol as antimicrobial agents stems from the hydrophobicity (poor water solubility), volatility, and instability of these EO constituents. Poor water solubility, in particular, limits the bioavailability of these compounds and significantly lowers their biological and antimicrobial activity, whereas volatility is problematic for achieving sustained release and controlled delivery. To address these challenges, a variety of approaches have been reported to encapsulate hydrophobic EO constituents as colloidal systems, including oil-in-water emulsions, microemulsions, and nanoemulsions; molecular inclusion complexes; lipid-based carriers (e.g., liposomes and solid lipid nanocapsules);[18] and polymer-based carriers (e.g., films, micro/nanocapsules, and nanoparticles).[5,19,20] Polymer nanoparticles (PNPs) are advantageous in that they offer a highly flexible delivery platform for antimicrobial applications, where nanoparticle properties (e.g., morphology, bulk/surface composition, and size) are readily tunable using a variety of synthetic approaches.[21–24] In turn, these tunable PNP properties dictate important features such as particle stability, surface interactions, and EO loading and release kinetics. In this direction, researchers have successfully incorporated carvacrol, thymol, and other phenolic EO constituents into various PNP matrices (chitosan,[25] poly(lactide-co-glycolide),[26,27] methylcellulose,[28] and zein[29]) via emulsification-evaporation,[26,27] emulsification-solvent exchange,[28] and nanoprecipitation[29] methods. While these examples demonstrate facile fabrication, encapsulation, and exhibit antimicrobial activity, many suffer several deficiencies such as low EO loading (i.e., 3% w/w carvacrol in chitosan matrix), poor EO encapsulation efficiencies (e.g., 14%–31% carvacrol-in-water emulsion and ionic gelation of chitosan),[30] use of organic solvents during encapsulation (acetone, ethanol, dichloromethane),[26,29] and burst release profiles of EO.[26,27]
Miniemulsion polymerization is ideally suited for encapsulation of hydrophobic payloads (e.g., lipophilic drugs, pigments, fragrances, inorganic nanomaterials, and polymers) with high loading and encapsulation efficiencies,[31–33] but has received little attention for sequestration and delivery of essential oils – particularly toward antimicrobial applications. Miniemulsion polymerizations are described as aqueous dispersions of small, narrowly distributed monomer droplets stabilized against Ostwald ripening and collisional degradation by addition of an appropriate surfactant and costabilizer. Monomer droplets ranging in size from 50–500 nm are attained via high shear mixing – typically either high-pressure homogenization or ultrasonic processing – and subsequently serve as discrete nanoreactors for the formation of polymer nanoparticles upon polymerization. Since mass transport between monomer droplets is suppressed, encapsulation of hydrophobic materials requires a straightforward addition of the desired material to the organic phase prior to shear mixing. Thus, encapsulation of materials miscible with the monomer phase provides polymer nanoparticles with the payload dispersed throughout the polymer matrix (whereas immiscible materials provide a route to nanocapsules).[33] Recently, we and others have reported thiol-ene[34–37] and thiol-alkyne[38,39] polymerization in miniemulsion as a facile platform for the synthesis of crosslinked polythioether nanoparticles with tunable particle sizes, tailorable network properties, and clickable surface functionality. More importantly, we demonstrated the ability to encapsulate hydrophobic inorganic nanoparticles within the polythioether matrix with high efficiency without sacrificing the rapid polymerization kinetics and high conversions provided by the thiol-mediated photopolymerization process.
Herein, we report a facile, one-pot approach to encapsulate monoterpene phenols (carvacrol/thymol) within polythioether nanoparticles as a model, sustained-release antimicrobial platform. We employ thiol-ene photopolymerization in miniemulsion for rapid nanoparticle synthesis, where carvacrol and thymol serve as miscible compatibilizers in the monomer phase enabling solvent-free encapsulation with high loading capacities (33%–66% w/w, EO relative to the NP) and excellent encapsulation efficiencies (>95%). The absence of solvent and use of a polymerizable surfactant eliminate the need for any postpolymerization purification steps. Release studies reveal the polythioether nanoparticles function as sustained-release reservoirs for carvacrol/thymol – discharging less than 4% of the payload over 24 h. The thymol/carvacrol-loaded (TCNPs) nanoparticles show effective antimicrobial activity (>99.9% kill efficiency at 24 h) against gram-positive (Bacillus subtilis and Staphylococcus aureus) and gram-negative (Escherichia coli and Burkholderia cenocepacia) bacteria. Considering the innate resistance of the bacterial strain, the high efficacy of TCNPs against Burkholderia cenocepacia is particularly significant, and to our knowledge, represents the first demonstration of antibacterial efficacy against B. cenocepacia using thymol/carvacrol-loaded polymer nanoparticles.
2. Results and Discussion
2.1. Synthesis and Characterization of Essential Oil Encapsulated Nanoparticles
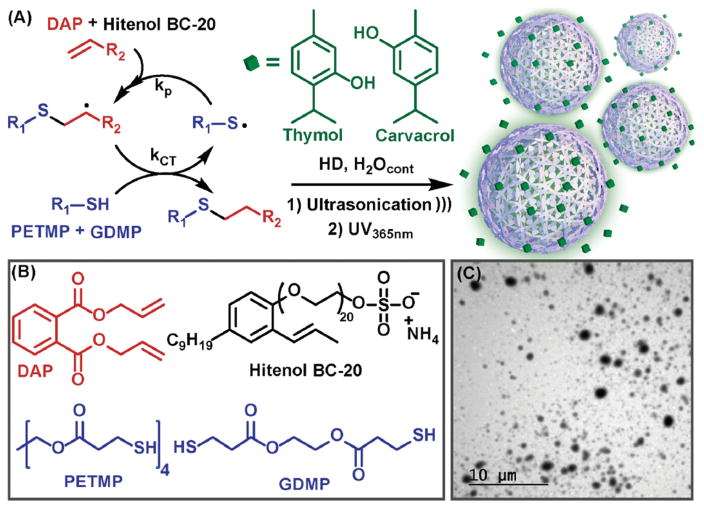
Polythioether NPs loaded with various ratios of carvacrol/thymol were synthesized via thiol-ene photopolymerization in miniemulsion according to Figure 1a. NPs were prepared by dispersing the organic phase, comprising diallyl phthalate (DAP), glycol di(3-mercaptopropionate) (GDMP), pentaerythritol tetra(3-mercaptopropionate) (PETMP), and carvacrol/thymol into deionized water containing surfactant via probe ultrasonication for 25 min (Figure 1b). The full details for the miniemulsion formulation are given in Table S1 (Supporting Information). The monomer droplets were then photopolymerized by exposure to UV light (λmax 365 nm). Control NPs (without carvacrol/thymol) were similarly prepared using butyl acetate as a diluent for the organic phase. After ultrasonication and photopolymerization, aliquots were removed and analyzed via proton nuclear magnetic resonance spectroscopy (1H NMR) (Figure S1, Supporting Information) in D2O. The disappearance of the peaks attributed to the thiol (–CH2CH2–SH, 2.62–2.83 ppm) and alkene (5.07–5.45, 5.87–6.10 ppm) from the monomers, and broadening of other peaks indicated the thiol-ene photopolymerization proceeded to high conversion. Additionally, Raman spectroscopy of freshly emulsified nanoparticles prior to and after UV polymerization showed the disappearance of SH (2580 cm−1) and alkene stretches (1640–1680 cm−1) (Figure S2, Supporting Information). The ability to achieve high thiol-ene monomer conversion in the presence of phenolic functional groups is in good agreement with our previous work.[38,40,41] At near quantitative conversions, crosslink density (0.168 × 10−3 mol cm−3) of the polythioether NPs was estimated from the rubbery plateau storage modulus (1.25 MPa, Tg −12.1 °C, Figure S3, Supporting Information) of the base thiol-ene resin (i.e., GDMP+PETMP+diallyl phthalate prepared at identical stoichiometric ratios used in the miniemulsions) at Tg + 40 °C according to the theory of rubber elasticity.[42] To eliminate surfactant leaching, Hitenol BC-20 was employed as a polymerizable surfactant to covalently incorporate the surfactant into the thiol-ene network (discussed further in the antimicrobial section). A series of exploratory NP syntheses were conducted, in which the concentration of Hitenol BC-20 (Figure S4a, Supporting Information) and organic weight fraction (Figure S4b, Supporting Information) were independently varied, to identify conditions to provide acceptable NP size and size distributions. A 4.5 wt% organic phase and 70 × 10−3 M Hitenol BC-20 were identified as optimum parameters; these conditions provided pure carvacrol-loaded NPs (CNPs), thymol/carvacrol-loaded NPs (TCNPs), and control NPs with mean particle sizes of 148 ± 24 (PDI: 0.560), 147 ± 19 (PDI: 0.387), and 183 ± 19 nm (PDI: 0.473), respectively, as determined by dynamic light scattering (DLS). We note that the reported std. dev. values represent the deviation in the average particle size obtained from a minimum of three separate syntheses. A representative transmission electron microscopy (TEM) image for TCNPs loaded with a 0.75:1 T:C ratio (Figure 1c) showed spherical particles with sizes in good agreement with DLS analysis. TEM for the control NPs and CNPs are shown in Figure S5a,b, respectively. The salient feature of this synthetic approach is that the carvacrol/thymol constituents serve as the “solvent” for the organic phase in the miniemulsion process. This route enabled a high loading capacity of thymol/carvacrol within the NPs (between 33% and 66% w/w, EO/NP) with high encapsulation efficiencies (>95% by GC-MS), eliminated the need for an organic solvent during NP synthesis, and provided a one-pot approach to EO encapsulation without any postpolymerization purification processes (e.g., no freeze drying, evaporation, and extraction processes were required). Thus, the NP dispersions obtained from the previously described synthetic approach were used directly for the antimicrobial studies detailed below.
Figure 1.
a) The thiol-ene reaction involves alternating chain transfer and propagation b) with various multifunctional monomers used to generate polythioether nanoparticles via thiol-alkene photopolymerization in miniemulsion. c) Representative TEM of thymol/carvacrol-loaded nanoparticles.
2.2. Loading and Release of Thymol and Carvacrol
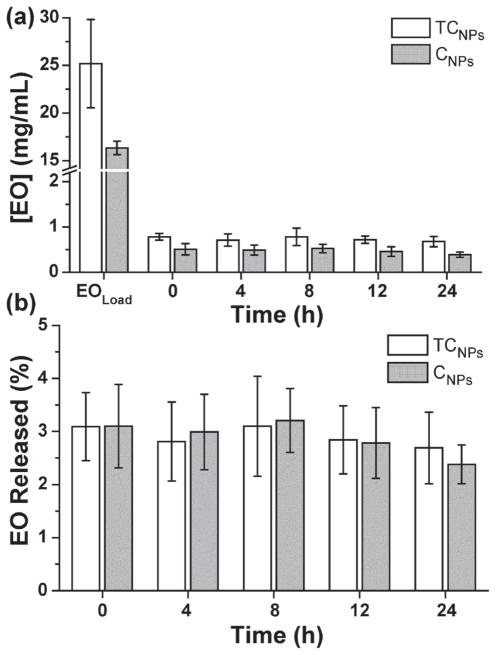
Next, we investigated the kinetics of carvacrol and carvacrol/thymol mixtures released from polythioether NPs into water. The amount of essential oil released was quantified by measuring the concentration of carvacrol/thymol available in water via GC-MS at 0, 4, 8, and 12 h. Figure 2a shows the release data for CNPs (synthesized at 33% w/w C/NP) and TCNPs (synthesized with 47% w/w TC/NP, 0.75:1 T:C). The EOload data points represent the total amount of carvacrol (16 ± 0.7 mg mL−1) and thymol/carvacrol (25 ± 5 mg mL−1) loaded into the NPs during miniemulsion photopolymerization, and are in good agreement with initial formulations for each type of NP. The ratio of the time 0 and EOload concentrations represents the encapsulation efficiency for the miniemulsion process, i.e., ≈96.9% for TCNPs and 96.8% for CNPs. Thus, the concentration of EO within the nanoparticles is up to 20 times higher than the reported solubility limits for thymol (0.85 mg mL−1) and carvacrol (1.25 mg mL−1) at 20 °C.[43] As shown in Figure 2a, the concentration of EO released from the NPs in water was relatively constant over 24 h, where TCNPs and CNPs reached plateau concentrations between 0.7–0.9 mg mL−1 and 0.5–0.6 mg mL−1, respectively. These concentrations correspond to less than 4% release of the total EO payload over 24 h, (Figure 2b) and approach the solubility limit of thymol and carvacrol in water at 20 °C. These results suggest that the polythioether nanoparticles effectively serve as a reservoir for the hydrophobic carvacrol and thymol compounds. This “reservoir effect” enables extended delivery of EO constituents into the bulk aqueous phase, where the solubility limit of the relatively insoluble compounds ultimately dictates the aqueous-phase equilibrium concentration.
Figure 2.
a) Amounts of essential oils extracted from the supernatant after pelleting NPs by ultracentrifugation. b) Calculated release profiles for both CNPs and TCNPs over 24 h.
2.3. Antimicrobial Properties
Antimicrobial activity of the EO-containing NPs was evaluated against a panel of gram-positive and gram-negative bacteria that included both saprophytic (B. subtilis ATCC 6633 and E. coli ATCC 25922) and pathogenic species (E. coli ATCC 43895 [serotype O157:H7], S. aureus RN6390, and B. cenocepacia K56-2). Our preliminary experiments revealed that control NPs prepared using either sodium dodecyl sulfate or cetyltrimethylammonium bromide as surfactant exhibited antimicrobial activity, presumably due to surfactant leaching. Thus, we chose Hitenol BC-20 – a commercially available polymerizable surfactant – for further experiments, as it exhibited no measurable antimicrobial activity in zone of inhibition (ZOI) experiments at the concentration used in NP synthesis (70 × 10−3 M Figure 3a; 140 × 10−3 M Figure S6, Supporting Information). More importantly, control NPs prepared with Hitenol BC-20 showed no measureable ZOI against all five bacteria (Figure 3b). Initially, we evaluated the antimicrobial activity of NPs containing pure carvacrol (33% w/w, carvacrol relative to NP) and various ratios of thymol:carvacrol (Treatment I – 66% w/w, 3:1 T:C; Treatment II – 55% w/w, 1.5:1 T:C; and Treatment III – 47% w/w, 0.75:1) against E. coli ATCC 25922, S. aureus RN6390, and B. subtilis ATCC 6633 (Figure S7, Supporting Information). Since thymol is a solid at room temperature (mp 49–51 °C), thymol-loaded NPs were difficult to synthesize using a solvent-free approach and were not actively pursued in this work. Previous reports have shown that combinations of thymol and carvacrol exhibit higher antimicrobial activity than each individual constituent,[16] thus, carvacrol-loaded NPs were evaluated via ZOI only as a comparison to NPs loaded with combinations of thymol/carvacrol. Interestingly, all treatments with NPs containing thymol/carvacrol showed similar zones of inhibition (Figure S7, Supporting Information), thus treatment III, with the lowest overall thymol/carvacrol loading, was chosen for the experiments discussed hereafter. From this point forward, the acronym TCNP will refer to nanoparticles loaded with 47% w/w, 0.75:1 T:C (treatment III). Figure 3c,d shows the results of well diffusion assays for CNPs and TCNPs against the full panel of bacteria. As shown in Figure 3c, CNPs exhibited moderate activity against all bacteria (ZOI 1–2 mm). As expected, TCNPs were consistently more effective than CNPs, as indicated by the larger ZOIs shown in Figure 3d. TCNPs inhibited the growth of all five bacteria, including S. aureus (2 mm ZOI) and B. cenocepacia (3 mm ZOI) that are known to exhibit high intrinsic resistance to conventional antibiotics. Bacillus subtilis (6 mm ZOI) was the most susceptible species to the TCNP treatment.
Figure 3.
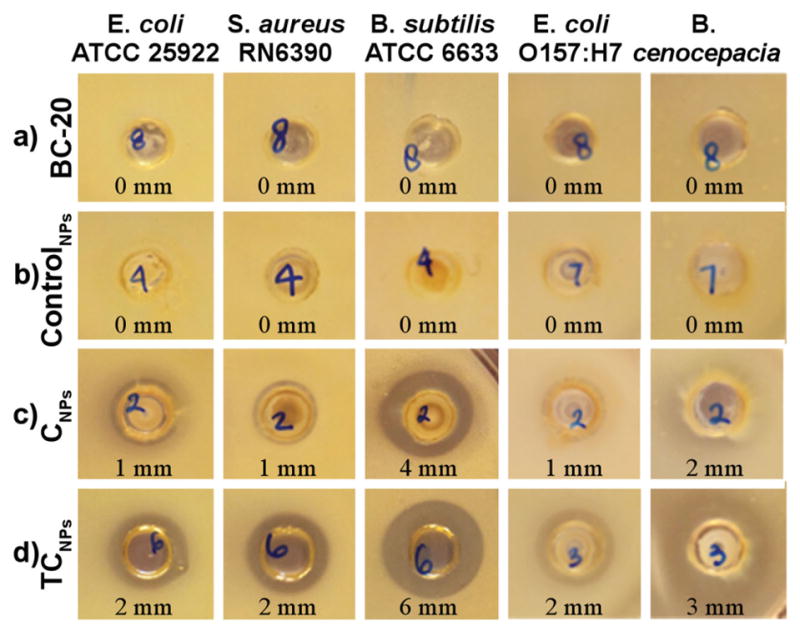
Well diffusion assay identifies treatments with antimicrobial activity. Bacteria were incubated with a) Hitenol BC-20 (70 × 10−3 M), b) ControlNPs, c) CNPs, and d) TCNPs at 1013 NPs mL−1. Zones of inhibition (ZOI, mm) are reported below each image.
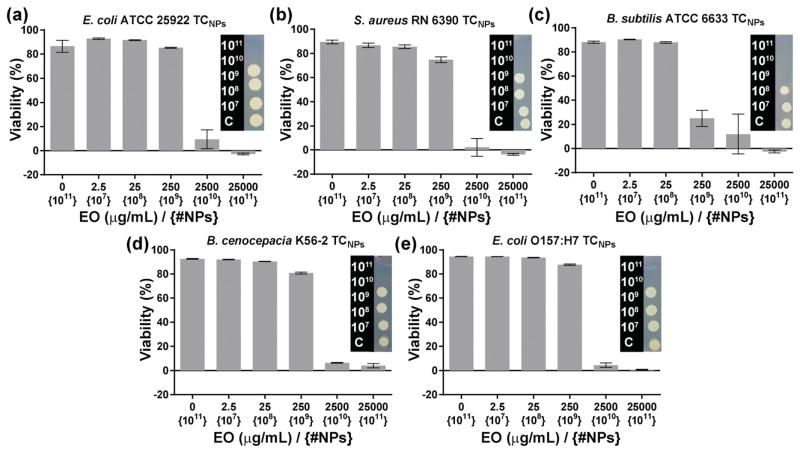
The antimicrobial activity of TCNPs was further evaluated via determination of the minimum inhibitory concentration (MIC). The MIC is defined as the lowest concentration of an antimicrobial agent that inhibits the growth of a microorganism following an overnight incubation period. We adopted a modified broth microdilution method to determine MICs for NP-loaded thymol/carvacrol.[44] We also spot-plated bacteria that were incubated with NPs to check for possible bacteriostatic effects. While MIC assays are reputable for accuracy and standardized comparison across numerous antibiotics, the conventional MIC output presents a challenge for hydrophobic antibiotics encapsulated in polymer nanoparticles – and is particularly problematic for NP delivery systems that function via the “reservoir effect.” As a result of the “reservoir effect,” the concentration loaded in the nanoparticle may be orders of magnitude greater than the concentration delivered to the bacteria during the time frame of the MIC assay. For example, the loaded concentration of thymol/carvacrol in TCNPs was determined to be 25 000 μg mL−1 by GC-MS. These thymol/carvacrol loading levels are up to 100-fold higher than reported MIC values for E. coli ATCC 25922 (MICcarvacrol: 225 μg mL−1, MICthymol: 225 μg mL−1) and S. aureus (MICcarvacrol: 450 μg mL−1, MICthymol: 225 μg mL−1);[45] however, our release studies show that carvacrol and thymol are delivered to the bulk aqueous phase at a relatively constant concentration of 500–900 μg mL−1 over 24 h. Following the precedent of Langer and co-workers,[46] who previously noted the difficulty in defining the MIC for antibiotic-loaded nanoparticles, we report concentrations as the total EO concentration within the NP at the beginning of the treatment to identify the MIC. While the loaded concentrations grossly overestimate the MIC value relative to a conventional definition, we reasoned this route of reporting was the most useful and conservative measure of NP-loaded antimicrobial efficacy. Additionally, we calculated the nanoparticle number density (number of particles per mL) to estimate the number of NPs delivered in each experiment as an alternate MIC output value, with the caveat that the number density is based on the average particle diameter of a polydisperse nanoparticle population. Nonetheless, number density MIC provides a point of comparison among the various bacteria investigated in this paper. Figure 4 shows the results from the viability assays used to determine the MICs for TCNPs. In general, the growth of all five bacteria included in this study was completely inhibited upon exposure to TCNPs loaded at 25 000 μg mL−1 (1010 nanoparticles) in a 50 μL broth volume, which translated to ≈105 NPs per bacterial cell. B. subtilis was the most susceptible species, and was completely inhibited at TCNPs loaded at 2 500 μg mL−1 (109 nanoparticles, ≈104 NPs per cell). The MIC values are in good agreement with the previously described results of agar well diffusion tests.
Figure 4.
Percent viability of various bacteria upon treatment with TCNPs. The 0 μg mL−1 EO {1011} was treated with ControlNPs. Inset images show the corresponding spot tests for the presence of live bacteria.
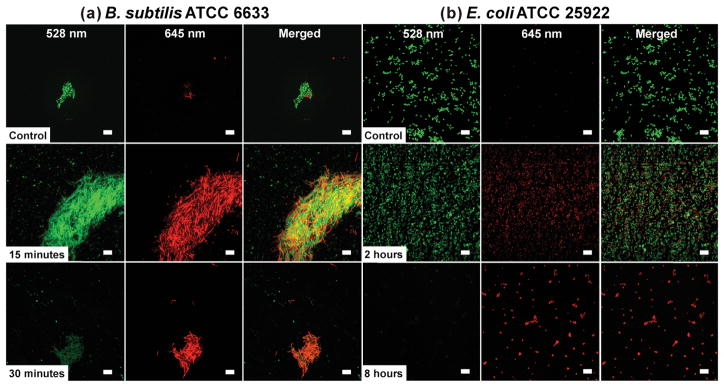
We also investigated the kill kinetics for the panel of bacteria incubated with TCNPs using live/dead and terminal dilution assays. Initially, the kill kinetics of TCNPs against B. subtilis ATCC 6633 and E. coli ATCC 25922 were probed using a live/dead cell viability assay, which stains “dead” bacteria red upon cell membrane damage and uptake of propidium iodide. As shown in Figure 5a,b a large number of viable bacterial cells (stained green) were observed for the B. subtilis and E. coli control samples, respectively. Incubation of TCNPs with B. subtilis for 15 min yielded an apparent 50:50 live/dead ratio, whereas 30 min resulted in mostly dead bacteria (Figure 5a). For E. coli ATCC 25922, the 50:50 live/dead ratio was observed after two hours of incubation with TCNPs, and most bacteria stained dead after 8 h of incubation (Figure 5b). To quantify the kill kinetics for all five bacteria, we used a terminal dilution assay. Figure 6a shows the log reduction in the bacteria count as a function of incubation time with ≈1011 TCNPs. The log CFU reductions at 48 h are reported as % kill values in Figure 6b. In agreement with our previous ZOI data, B. subtilis ATCC 6633 showed the highest susceptibility to the TCNP treatment with 4.3 and 7.1 log reductions observed at 12 and 24 h, respectively. The log reductions for E. coli O157:H7 (ATCC 43895) were 3.5 at 12 h and 7 at 24 h — results that translate into a kill efficacy of >99.99%. The viability of E. coli ATCC 25922 declined at a slower rate than E. coli O157:H7, however, the 48 h exposure to TCNPs ultimately killed >99.99% of the bacteria. Incubation of TCNPs with B. cenocepacia K56-2 produced 3.9 and 6.2 log reductions at 12 and 24 h, respectively. The kill kinetics and high susceptibility (over 99.99% efficacy at 24 h) of B. cenocepacia to the TCNPs is of particular interest, as these bacteria have very high innate resistance to a wide range of antibiotics and biocides.[47] With a 3.6 log reduction, S. aureus RN6390 was the only species that maintained viability after 24 h incubation with TCNPs. However, a 6.3 log reduction, or >99.99 % kill efficacy for S. aureus was ultimately attained at 48 h.
Figure 5.
Effect of TCNPs on the viability of a) B. subtilis ATCC 6633 and b) E. coli ATCC 25922, as monitored by confocal laser scanning microscopy. Representative images of control cultures (top row), and cultures treated with 1011 TCNPs (middle and bottom row) at the indicated time points. The green signal (SYTO 9) indicates viable live cells, whereas red signal (propidium iodide) indicates damaged or dead cells. Scale bars = 10 μm.
Figure 6.
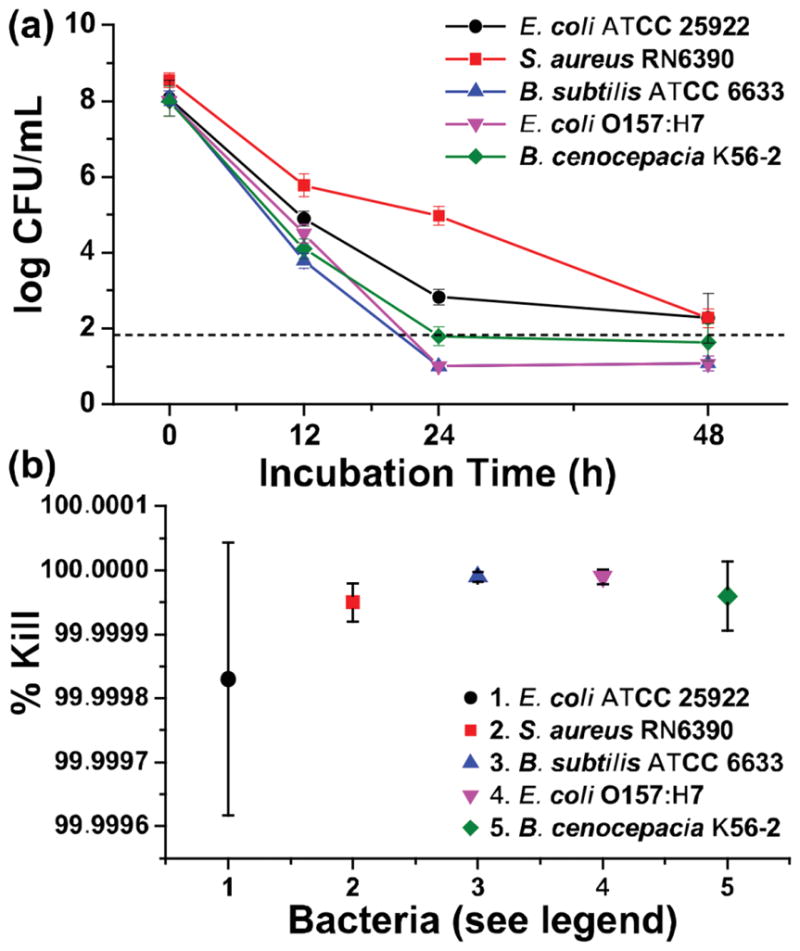
Evaluation of antimicrobial activity for 1011 TCNPs mL–1 on the viability of (●) E. coli ATCC 25922, (■) S. aureus RN6390, (▲) B. subtilis ATCC 6633, (▼) E. coli ATCC 43895 (serotype O157:H7), and (◆) B. cenocepacia K56-2 via a) a kinetic terminal dilution and b) percentage of bacteria killed and over 48 h. *Dashed line represents the limit of quantitation.
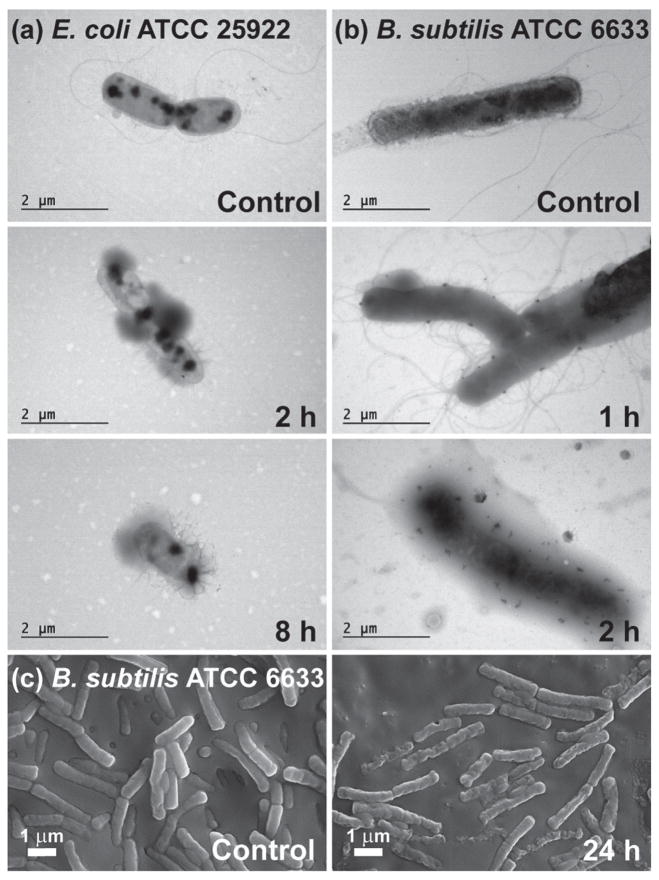
Finally, insight into the antimicrobial mechanism was investigated by visualizing the microbial structure of B. subtilis ATCC 6633 and E. coli ATCC 25922 before and after prolonged contact with TCNPs using electron microscopy. Prior to exposure to TCNPs, TEM of both microbes showed intact cell structure (Figure 7, control). After exposure with TCNPs, distinct indication of damage to the cell envelope (diffuse membrane, cellular debris) and loss of flagella were visible in both E. coli ATCC 25922 (Figure 7a) and B. subtilis ATCC 6633 (Figure 7b). Additionally, the scanning electron microscopy (SEM) image of B. subtilis after 24 h exposure to inhibitory concentrations of TCNPs showed bacteria with crumpled cell envelopes and pore-like lesions, which is consistent with the collapse of the cell structure (Figure 7c). These results are consistent with previously reported postulates that identify the cell membrane as the primary site of toxicity for carvacrol/thymol.[16]
Figure 7.
a) TEM of E. coli ATCC 25922 and b) B. subtilis ATCC 6633 control cultures and cultures that were challenged with 1011 TCNPs for various times. c) High resolution SEM of the control culture of B. subtilis ATCC 6633 and the culture treated for 24 h with 1011 TCNPs.
3. Conclusion
We have reported a one-pot, solvent-free miniemulsion photopolymerization process for the synthesis of thymol/carvacrol-loaded polythioether nanoparticles. Using the thymol/carvacrol payload directly as a diluent for the monomer phase in the presence of a polymerizable surfactant provided the active NPs without any postsynthetic purification. The NPs serve as high capacity reservoirs for slow-release and delivery of thymol/carvacrol-combination payloads that exhibit inhibitory and bactericidal activity (>99.9% kill efficiency at 24 h) against gram-positive (Bacillus subtilis and Staphylococcus aureus) and gram-negative (Escherichia coli and Burkholderia cenocepacia) bacteria. The simplicity, modularity, and efficacy of the essential oil encapsulation platform may combat bacteria with intrinsic resistance to conventional antibiotics, and is potentially adaptable for delivery of EOs as active packaging materials and topical antiseptics. The antimicrobial activity of TCNPs against inherently resistant Burkholderia cenocepacia may provide a route to innovative pulmonary therapeutics by appropriately engineering the nanoparticle properties; investigations of thymol/carvacrol-loaded NPs against B. cepacia complex biofilms — a challenging form of the bacteria commonly associated with cystic fibrosis — are currently underway. In this direction, we are particularly interested to exploit the simple synthetic modularity of this process for the design of EO-loaded biodegradable nanoparticles with specific surface chemistries to serve as multimode (e.g., contact biocidal, targeted NP-bacteria interactions) antimicrobial platforms.
4. Experimental Section
Materials
GDMP and PETMP were provided by Bruno Bock. Hexadecane, ethyl acetate, sodium chloride, and thymol were obtained from Fisher Scientific. DAP, 4-p-methoxy phenol (MEHQ), and butyl acetate were acquired from Sigma-Aldrich. Other reagents were purchased from the following vendors: 1-hydroxycyclohexyl phenyl ketone (Irgacure 184) from CIBA; Hitenol BC-20 from Montello, Inc.; carvacrol from TCI America. Difco Agar, Mueller Hinton II agar (MHA), Mueller Hinton II broth (MHB), and Bacto Tryptone were from Becton, Dickinson and Company. All the materials were obtained at the highest purity available and used without further purification unless otherwise specified.
Characterization Methods
The size and distribution of the NPs were measured by DLS using a Microtrac Nanotrac Ultra NPA150 particle analyzer. Particle size and distribution were obtained using the Microtrac Flex software (v.10.6.1), which employs non-negatively constrained least-squares and cumulants analysis to obtain the intensity-weighted “z-average” mean particle size as the first cumulant, and the polydispersity index from the second cumulant.[48] Transmission electron micrographs were taken with a Zeiss 900 electron microscope operating at 50 kV and outfitted with a Model 785 Erlangshen ES1000 WCCD camera (Gatan). Samples were applied to 200 mesh copper grids (3.05 mm, 200 lines per inch square mesh, Electron Microscopy Sciences) coated with Formvar (5% polyvinyl formal resin). Proton (1H) NMR was recorded on a Bruker Acend 600 MHz spectrometer at 30 °C in D2O, using 128 scans and a 4.27 s relaxation delay. Optical density (OD) and fluorescence readings were performed in a BioTek Synergy 2 programmable microplate reader. Confocal images were taken using a Zeiss LSM 510 confocal laser scanning microscope. High-resolution field emission SEM (FE-SEM) was performed with a Zeiss SIGMA variable pressure field emission scanning electron Microscope operating at 10 kV in high vacuum mode. Samples were sputter coated with silver at instrument-reported thickness of 5 nm with a Quorum Emitech K550X sputter coater. Raman spectra were acquired using a high-performance portable Raman spectrometer (i-Raman Plus, B&W Tek Inc., Delaware, USA). The samples were analyzed at 100 mW, with a 785 nm diode laser and 150 s accumulation time.
General Nanoparticle Sample Preparation
Each nanoparticle synthesis was prepared in a 20 mL scintillation vial with a total volume of 10 mL. The organic stock solutions shown in Table S1 (Supporting Information) were added into a vial containing a stock solution of Hitenol BC 20 and deionized water. As we previously reported, MEHQ serves as a radical inhibitor to suppress thermal polymerization during ultrasonication.[34] The samples were placed into an ice bath and sonicated using a Qsonica Q700 probe ultrasonicator at 25% amplitude for 25 min. The resultant miniemulsions were cured for 15 min at an intensity of 185 mW cm−2 using an OmniCure S1000 UV light source with a 100 W mercury lamp (λmax = 365 nm, 320–500 nm filter). All samples were made in triplicate to ensure data reproducibility.
Preparation of Essential Oil Encapsulated Nanoparticles
Carvacrol encapsulated nanoparticles were prepared by replacing butyl acetate with 33% w/w carvacrol as the solvent. For nanoparticles containing a combination of thymol and carvacrol, 20% w/w thymol was added along with 27% w/w carvacrol. The total organic fraction evaluated was 4.5% w/w for all samples.
Determination of nanoparticle number density (particles per mL)
| (1) |
Where organic added is a known volume that can be converted into cubic nanometers
| (2) |
Where r is the radius (nm) determined from light scattering
| (3) |
The final number of particles is determined by dividing the total volume of organic by the volume of the average nanoparticle size (see Table S2, Supporting Information). This number results in particles per 10 mL (10 mL is the total volume of the emulsified solution), multiplication of volume added by this number results in the number of particles delivered.
Gas Chromatography–Mass Spectrometry (GC–MS) Release Study of Essentials Oil Nanoparticles
Freshly prepared CNPs and TCNPs were transferred in 100 mL volumetric flasks and diluted 1:10 with deionized water. For each diluted suspension, 10 mL aliquots were removed at 0, 4, 8, 12, and 24 h, and NPs were precipitated for 4 h at 40 000 rpm (4 °C) in a Beckman Coulter Optima XE ultracentrifuge. 200 μL of the supernatant was removed and extracted by vortexing for 30 s with 800 μL of ethyl acetate. The liquid phases were separated by centrifugation for 15 min at 13 000 rpm, and the organic layer was transferred to a capped 1.5 mL GC-MS vial. To determine the amounts of EOs in the pre-emulsified organic monomer mixture, 450 μL of organic stock solution (carvacrol and combo) was diluted 1:100 in ethyl acetate. All samples were placed into capped GC-MS vials and run using the following protocol.
GC-MS analysis was performed on an Agilent 6890 GC/MSD equipped with an auto sampler and a Restek RTx-1 30 meter column. All samples and standards were analyzed from 1 μL injection volumes. Analysis parameters include an inlet temperature of 200 °C in splitless mode and a helium flow rate of 30 mL min−1 in gas saver mode. Thermal ramping conditions used include an initial hold at 45 °C for 4 min followed by an increase in temperature at 25 °C min−1 up to 140 °C with a 3 min hold at 140 °C. The temperature ramp continued at 2.5 °C min−1 up to 150 °C with a 3 min hold followed by a final increase at 50 °C min−1 to 220 °C with a 7 min hold (Figure S8, Supporting Information). The method provided unique separation of thymol and carvacrol isomers eluting at 19.41 and 19.58 min, respectively (Figure S9, Supporting Information). Calibration curves for both isomers were prepared in ethyl acetate at 0, 200, 400, 600, 800, and 1000 ng μL−1 (Figure S10, Supporting Information). A representative chromatogram of extracted nanoparticles depicting the elution of thymol, carvacol, and hexadecane can be seen in Figure S11 (Supporting Information).
Evaluation of Antibacterial Activity of Nanoparticles
The antimicrobial activity of NPs was tested against several species of bacteria using a well diffusion method. The indicator microorganisms included Escherichia coli ATCC 25922 (serotype O6, biotype 1), E. coli ATCC 43895 (serotype O157:H7), Staphylococcus aureus RN6390,[49] Bacillus subtilis subsp. spizizenii ATCC 6633, and Burkholderia cenocepacia K56-2 (clinical isolate from Canada).[50] The testing was done on Mueller Hinton II agar (MHA) plates that had been overlaid with soft agar seeded with individual bacterial strains. The soft agar contained (per liter): 10 g of Bacto Tryptone, 6 g of Difco agar, and 8 g of sodium chloride. To create an overlay, the indicator organisms were grown overnight at 27 °C (B. subtilis) or 37 °C (E. coli, S. aureus, and B. cenocepacia) in MHB. The overnight cultures were diluted 1:5 with fresh MHB, and mixed with molten soft agar to achieve the density of ≈108 CFU mL−1. From this mixture, 4 mL aliquots were overlaid onto MHA base plates and allowed to completely solidify.
After solidifying of soft agar, 8 mm wells were made and their bottoms were sealed with 20 μL of MHA. Then, 70 μL of each sample was added to the wells and allowed to freely diffuse into growth medium. Wells containing pure carvacrol or a mixture of carvacrol (1 g) and thymol (0.6 g) were used as a positive control. Negative controls contained 70 × 10−3 M Hitenol BC 20, or empty NPs suspended in butyl acetate. The plates were incubated at each bacterium’s optimum growth temperature as listed above. The zones of inhibition (ZOI) were measured after 24, 48, and 72 h. Three replicates were carried out for each nanoparticle treatment and bacterial strain. Experiment was repeated three times to ensure data reproducibility.
Determination of Minimum Inhibitory Concentrations (MICs)
MICs of nanoparticles were determined using a modified broth microdilution method.[44] Briefly, overnight bacterial cultures were adjusted to ≈105 CFU mL−1, and nanoparticles were serially diluted with sterile water to achieve a range of concentrations between 1011 to 107 particles μL−1. In a 96-well microplate, 150 μL aliquots of adjusted bacterial cultures were mixed with 50 μL aliquots of different NPs. Bacteria suspended in MHB without addition of nanoparticles served as a positive control, while MHB without bacterial inoculum and nanoparticles served as a negative control. The inoculated microplates were incubated at each bacterium’s optimum growth temperature, and the susceptibility of microorganisms to different NPs was assessed by measuring optical density at 600 nm. The results were expressed as % viability obtained through Equation (4)
| (4) |
Where OD is the optical density at 600 nm, t0 is the initial time 0 h, and t20 is the time after 20 h of incubation.
For further verification of inhibition of bacterial growth, 10 μL of bacterial cultures treated for 20 h with different nanoparticles were plated on MHA and incubated at each bacterium’s optimum growth temperature for additional 20 h. The MIC refers to the concentration of NPs that completely inhibited bacterial growth. Each treatment had four replications and the experiment was repeated twice.
Viability Staining Assays
Bacterial viability was determined by using a LIVE/DEAD BacLight Bacterial Viability Kit staining kit (Life Technologies). Overnight bacterial cultures were centrifuged at 8500 rpm for 3 min, supernatants were removed and the bacterial cells were suspended in 0.9% NaCl at OD600 of 0.1 (≈108 CFU mL−1). Freshly prepared nanoparticles were diluted 1:10 with sterile water. The adjusted bacterial cultures were mixed with the diluted nanoparticles at a ratio of 3:1, and incubated at 20 °C in the dark. At 0, 2, 4, 6, 8, 12, and 24 h, 100 μL aliquots of the bacteria-NP mixtures were transferred into a black flat bottom microplate, and mixed with 100 μL of the LIVE/DEAD BacLight staining reagent. The samples were incubated in the dark for 15 min, and fluorescence was measured using a 485/20 nm excitation filter (for both SYTO9 and PI) and a 528/20 nm (SYTO9 emission wavelength) and a 620/40 nm (PI emission wavelength) emission filter. For confocal imaging, 5 μL of the stained sample was cast on a microscope slide. Each sample was assayed in triplicate, and the experiment was repeated twice. The results of viability staining were confirmed by plating 10 μL of each bacterial suspension on MHA, and incubating the plates at an appropriate growth temperature for 20 h.
Terminal Dilution Assays
To compare the rate of bacterial killing by different NPs, the test organisms were exposed to NPs and the decline in numbers of viable bacteria was followed by a modified terminal dilution method.[51] The bacterial cultures were prepared and adjusted to ≈108 CFU mL−1 as described above. The adjusted bacterial cultures (9 mL) were mixed with nanoparticles (1 mL), and the bacterial populations were determined immediately after the addition of NPs (0 h), and at 2, 4, 8, 12, and 24 h of exposure. At each time point, 100 μL aliquots of the bacteria-NP suspensions were transferred into 96 well microplates prefilled with 200 μL of MHB and serially diluted. The inoculated microplates were incubated for 48 h at an appropriate growth temperature, after which the turbidity in each well was measured with a BioTek Synergy 2 microplate reader. An optical density at 600 nm of ≥0.05 was considered positive for bacterial growth. Populations of viable bacteria were calculated from the final dilution (terminal dilution, or TD), in which bacterial growth was observed using Equation (5)
| (5) |
Where TD is the terminal dilution factor obtained from microplate readings.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the financial support from the National Science Foundation (DMR-1056817 and IIA-1430364). D.N.A. acknowledges fellowship support from the NSF GK-12 program “Molecules to Muscles” (Award #0947944) through the University of Southern Mississippi. D.V.A. acknowledges traineeship support from the NSF NRT program “Interface” (Award #1449999) through the University of Southern Mississippi. Confocal microscopy was supported by MS INBRE funded by NCRR (5P20RR-016476-11) and NIGMS/NIH (8 P20 GM103476-11). D.V.M. acknowledges startup funds from the University of Southern Mississippi. The authors thank Montello Inc. for the kind donations of all polymerizable surfactant samples used in this work, and David Delatte of the Rawlins Research Group for help with DMA.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dahlia N. Amato, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Douglas V. Amato, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Dr. Olga V. Mavrodi, Department of Biological Sciences, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Dr. Dwaine A. Braasch, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Susan E. Walley, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Jessica R. Douglas, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Prof. Dmitri V. Mavrodi, Department of Biological Sciences, University of Southern Mississippi, Hattiesburg, MS 39406, USA
Prof. Derek L. Patton, School of Polymers and High Performance Materials, University of Southern Mississippi, Hattiesburg, MS 39406, USA
References
- 1. [accessed:November 2015]; www.cdc.gov/drugresistance/threat-report-2013/
- 2.Mahenthiralingam E, Baldwin A, Dowson CG. J Appl Microbiol. 2008;104:1539. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 3.Holden M, Seth-Smith H, Crossman L, Sebaihia M, Bentley S, Cerdeño-Tárraga A, Thomson N, Bason N, Quail M, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha E, Fialho A, Baldwin A, Dowson C, Barrell B, Govan J, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. J Bacteriol. 2009;191:261. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drevinek P, Mahenthiralingam E. Clin Microbiol Infect. 2010;16:821. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 5.Majeed H, Bian YY, Ali B, Jamil A, Majeed U, Khan QF, Iqbal KJ, Shoemaker CF, Fang Z. RSC Adv. 2015;5:58449. [Google Scholar]
- 6.Bakkali F, Averbeck S, Averbeck D, Waomar M. Food Chem Toxicol. 2008;46:446. doi: 10.1016/j.fct.2007.09.106. [DOI] [PubMed] [Google Scholar]
- 7.Burt S. Int J Food Microbiol. 2004;94:223. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Hyldgaard M, Mygind T, Meyer RL. Front Microbiol. 2012;3:1. doi: 10.3389/fmicb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacchetti G, Maietti S, Muzzoli M, Scaglianti M, Manfredini S, Radice M, Bruni R. Food Chem. 2005;91:621. [Google Scholar]
- 10.Liu C, Liang B, Shi G, Li Z, Zheng X, Huang Y, Lin L. Flavour Frag J. 2015;30:295. [Google Scholar]
- 11.Batish DR, Singh HP, Kohli SK, Kaur S. Forest Ecol Manag. 2008;256:2166. [Google Scholar]
- 12.Duncan B, Li X, Landis RF, Kim ST, Gupta A, Wang LS, Ramanathan R, Tang R, Boerth JA, Rotello VM. ACS Nano. 2015;9:7775. doi: 10.1021/acsnano.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung ML, Lin PY, Hsieh CW, Tai MH, Wu DC, Kuo CH, Hsieh SL, Chen HT, Hsieh S. ACS Sustain Chem Eng. 2014;2:1769. [Google Scholar]
- 14.Carbone-Howell AL, Stebbins ND, Uhrich KE. Biomacromolecules. 2014;15:1889. doi: 10.1021/bm500303a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helander IM, Alakomi HL, Latva-Kala K, Mattila-Sandholm T, Pol I, Smid EJ, Gorris LGM, von Wright A. J Agric Food Chem. 1998;46:3590. [Google Scholar]
- 16.Lambert RJW, Skandamis PN, Coote PJ, Nychas GJE. J Appl Microbiol. 2001;91:453. doi: 10.1046/j.1365-2672.2001.01428.x. [DOI] [PubMed] [Google Scholar]
- 17.Maida I, Lo Nostro A, Pesavento G, Barnabei M, Calonico C, Perrin E, Chiellini C, Fondi M, Mengoni A, Maggini V, Vannacci A, Gallo E, Bilia AR, Flamini G, Gori L, Firenzuoli F, Fani R. J Evid Based Complementary Altern Med. 2014;2014:10. doi: 10.1155/2014/573518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry M, Charcosset C, Fessi H, Greige-Gerges H. J Liposome Res. 2013;23:268. doi: 10.3109/08982104.2013.819888. [DOI] [PubMed] [Google Scholar]
- 19.Asbahani AE, Miladi K, Badri W, Sala M, Addi EHA, Casabianca H, Mousadik AE, Hartmann D, Jilale A, Renaud FNR, Elaissari A. Int J Pharm. 2015;483:220. doi: 10.1016/j.ijpharm.2014.12.069. [DOI] [PubMed] [Google Scholar]
- 20.McClements DJ. Nanoparticle- and Microparticle-Based Delivery Systems: Encapsulation, Protection and Release of Active Compounds. CRC Press; Boca Raton: 2015. [Google Scholar]
- 21.Lee SM, Nguyen ST. Macromolecules. 2013;46:9169. doi: 10.1021/ma401529w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikoba U, Peng H, Li H, Miller C, Yu C, Wang Q. Nanoscale. 2015;7:4291. doi: 10.1039/c4nr07682f. [DOI] [PubMed] [Google Scholar]
- 23.He W, Gu X, Liu S. Adv Funct Mater. 2012;22:4023. [Google Scholar]
- 24.Noimark S, Weiner J, Noor N, Allan E, Williams CK, Shaffer MSP, Parkin IP. Adv Funct Mater. 2015;25:1367. [Google Scholar]
- 25.Woranuch S, Yoksan R. Carbohyd Polym. 2013;96:586. doi: 10.1016/j.carbpol.2012.09.099. [DOI] [PubMed] [Google Scholar]
- 26.Gomes C, Moreira RG, Castell-Perez E. J Food Sci. 2011;76:N16. doi: 10.1111/j.1750-3841.2010.01985.x. [DOI] [PubMed] [Google Scholar]
- 27.Iannitelli A, Grande R, Di Stefano A, Di Giulio M, Sozio P, Bessa LJ, Laserra S, Paolini C, Protasi F, Cellini L. Int J Nanomed. 2011;12:5039. doi: 10.3390/ijms12085039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wattanasatcha A, Rengpipat S, Wanichwecharungruang S. Int J Pharm. 2012;434:360. doi: 10.1016/j.ijpharm.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 29.da Rosa CG, de Oliveira BrisolaMaciel MV, de Carvalho SM, de Melo APZ, Jummes B, da Silva T, Martelli SM, Villetti MA, Bertoldi FC, Barreto PLM. Colloid Surf A. 2015;481:337. [Google Scholar]
- 30.Keawchaoon L, Yoksan R. Colloid Surf B. 2011;84:163. doi: 10.1016/j.colsurfb.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Asua JM. Prog Polym Sci. 2002;27:1283. [Google Scholar]
- 32.Landfester K. Angew Chem Int Ed. 2009;48:4488. doi: 10.1002/anie.200900723. [DOI] [PubMed] [Google Scholar]
- 33.Landfester K, Musyanovych A, Mailänder V. J Polym Sci Part A: Polym Chem. 2010;48:493. [Google Scholar]
- 34.Amato DV, Amato DN, Flynt AS, Patton DL. Polym Chem. 2015;6:5625. [Google Scholar]
- 35.Durham OZ, Krishnan S, Shipp DA. ACS Macro Lett. 2012;1:1134. doi: 10.1021/mz300358j. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Chatani S, Podgorski M, Bowman CN. Polym Chem. 2015;6:3758. doi: 10.1039/C4PY01552E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasinski F, Lobry E, Tarablsi B, Chemtob A, Croutxé-Barghorn C, Le Nouen D, Criqui A. ACS Macro Lett. 2014;3:958. doi: 10.1021/mz500458s. [DOI] [PubMed] [Google Scholar]
- 38.Amato DN, Amato DV, Narayanan J, Donovan BR, Douglas JR, Walley SE, Flynt AS, Patton DL. Chem Commun. 2015;51:10910. doi: 10.1039/c5cc03319e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durham OZ, Norton HR, Shipp DA. RSC Adv. 2015;5:66757. [Google Scholar]
- 40.Sparks BJ, Hoff EFT, Hayes LP, Patton DL. Chem Mater. 2012;24:3633. [Google Scholar]
- 41.Donovan BR, Cobb JS, Hoff EFT, Patton DL. RSC Adv. 2014;4:61927. [Google Scholar]
- 42.Flory PJ. Polymer. 1979;20:1317. [Google Scholar]
- 43.Chen H, Davidson PM, Zhong Q. Appl Environ Microb. 2014;80:907. doi: 10.1128/AEM.03010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiegand I, Hilpert K, Hancock REW. Nat Protocols. 2008;3:163. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 45.Cosentino S, Tuberoso CIG, Pisano B, Satta M, Mascia V, Arzedi E, Palmas F. Lett Appl Microbiol. 1999;29:130. doi: 10.1046/j.1472-765x.1999.00605.x. [DOI] [PubMed] [Google Scholar]
- 46.Radovic-Moreno AF, Lu TK, Puscasu VA, Yoon CJ, Langer R, Farokhzad OC. ACS Nano. 2012;6:4279. doi: 10.1021/nn3008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose H, Baldwin A, Dowson CG, Mahenthiralingam E. J Antimicrob Chemo. 2009;63:502. doi: 10.1093/jac/dkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown JC, Pusey PN, Dietz R. J Chem Phys. 1975;62:1136. [Google Scholar]
- 49.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. EMBO J. 1993;12:3967. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. J Clin Microbiol. 2000;38:3165. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landa BB, Mavrodi DM, Thomashow LS, Weller DM. Phytopathology. 2003;93:982. doi: 10.1094/PHYTO.2003.93.8.982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.