Barrett's esophagus (BE) is a premalignant lesion for esophageal adenocarcinoma, a rapidly increasing, highly fatal cancer.1 Clinical guidelines recommend screening for BE in those with chronic gastroesophageal reflux disease (GERD) and at least two risk factors (e.g., >50 years, white race, obese, tobacco smoking history).2,3 However, providing clinicians with a tool that allows them to estimate a patients' risk may better aid them in deciding who to screen for BE and make future resource utilization more efficient. The Michigan Barrett's Esophagus pREdiction Tool (M-BERET) predicts risk for BE in men attending primary care using information on their frequency of GERD symptoms, age, waist-to-hip ratio (WHR), and pack-years of cigarette smoking.4 In internal validation, the M-BERET discriminated reasonably well between men with and without BE, with an area under the receiver operating characteristic curve (AUROC) of 0.72. This was significantly better than using GERD symptoms alone (0.72 vs. 0.61, p<0.001).4 However, this prediction tool needs to be validated in an independent population before its use can be recommended in clinical practice.5
Methods
We examined the predictive performance of the M-BERET using data from four separate population-based case-control studies in the Barrett's and Esophageal Adenocarcinoma Consortium (http://beacon.tlvnet.org/): 282 cases and 434 controls in the Houston BE study; 140 cases and 162 controls in the Factors Influencing the Barrett's/Adenocarcinoma Relationship study (based in Ireland; “FINBAR”); 128 cases and 128 controls in the Study of Digestive Health (based in Australia; “SDH”); and 45 cases and 73 controls in the Study of Reflux Disease (based in western Washington State; “SRD”). In all four datasets, cases included persons with BE defined by endoscopic evidence of columnar mucosa in the tubular esophagus, accompanied by the presence of specialized intestinal metaplasia in an esophageal biopsy. Cases were compared with population-based controls, representing the underlying source population from which cases arose. We restricted our analyses to men aged between 50 and 79 years because this was the population in which M-BERET was developed4 and this represents the population at greatest risk for BE.
Statistical analysis
Analyses were performed separately in the four studies and we used the following formula to determine BE risk according to the M-BERET (http://mberet.umms.med.umich.edu/): Logit[Prob(BE)] = -9.1422 + (0.04278 × age) + (0.8453 if weekly GERD) + (3.6682 × WHR) + (0.00835 × pack-years). We quantified the discriminatory ability of the M-BERET using the AUROC. To be useful, the M-BERET's predicted risks must discriminate (separate) well between those participants who do have BE (cases) and those participants who do not have BE (population-based controls). The AUROC gives the probability that for any randomly selected pair of individuals, one case and one control, the model assigns a higher probability to the case. A value of 1 indicates the model has perfect discrimination, while a value of 0.5 indicates the model discriminates no better than chance. A moderately predictive AUROC ranges from 0.7 to 0.8 and indicates acceptable model discrimination.5 All statistical analyses were conducted using SAS 9.4.
Results
Mean age of the study populations ranged from 60 to 65 years. In each study, BE cases had higher WHR, greater pack-years smoked and were more likely to have frequent GERD symptoms, compared with controls. When the M-BERET was applied separately to each study, it achieved similar discrimination accuracy between cases and controls in each study: Houston, AUROC=0.71, 95%CI=0.67-0.75; FINBAR, AUROC=0.70, 95%CI 0.64-0.76; SDH, AUROC=0.72, 95%CI=0.66-0.78; and SRD, AUROC=0.70, 95%CI=0.60-0.79 (Figure 1a-d). These compare favorably with the AUROC in the original study (0.72),4 showing little evidence for over-fitting or optimism. The AUROC was higher for long-segment BE (pooled analysis of 270 long-segment BE vs. 669 controls, AUROC=0.73, 95%CI=0.70-0.77).
Figure 1.
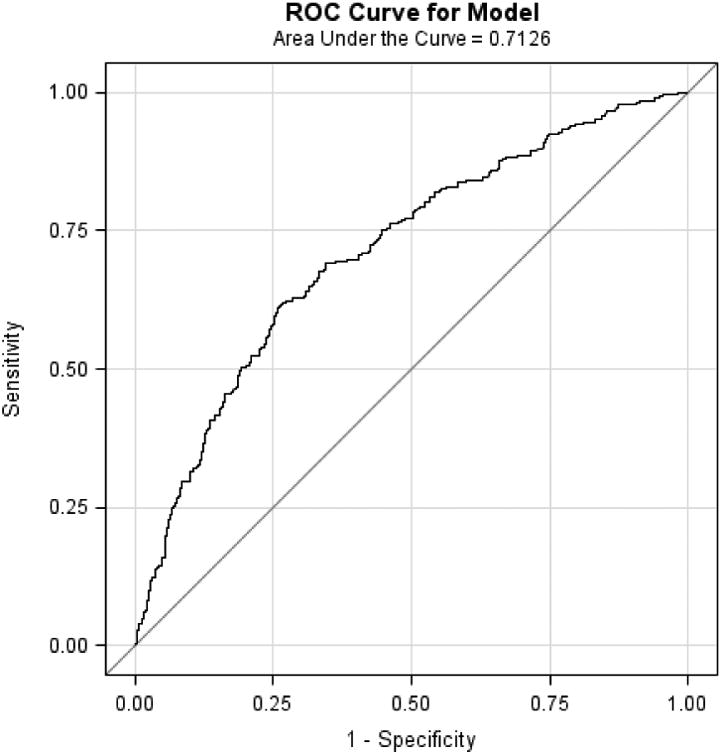
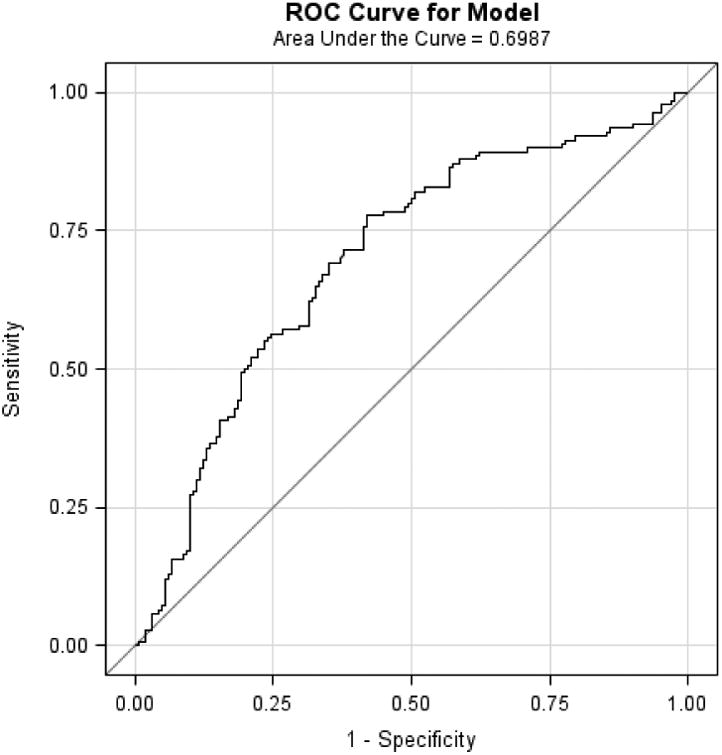
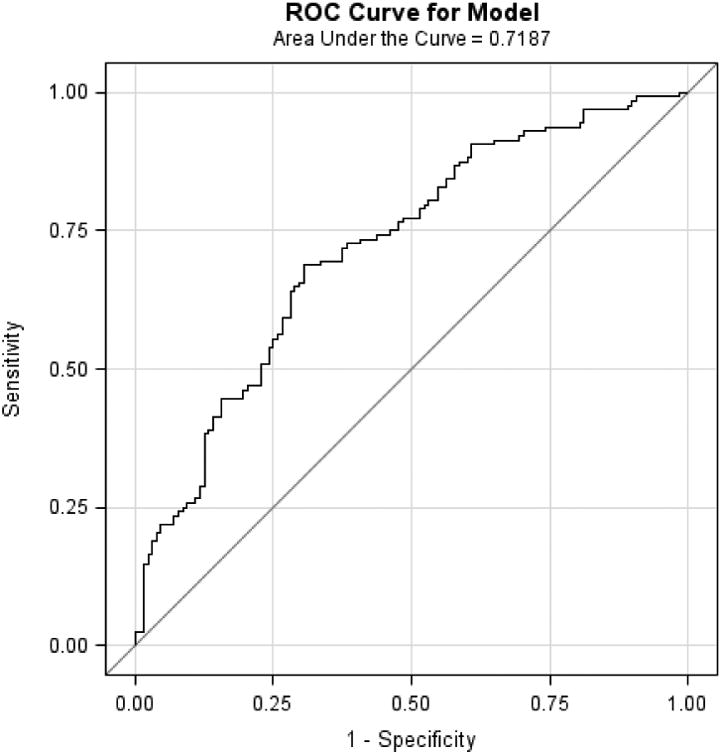
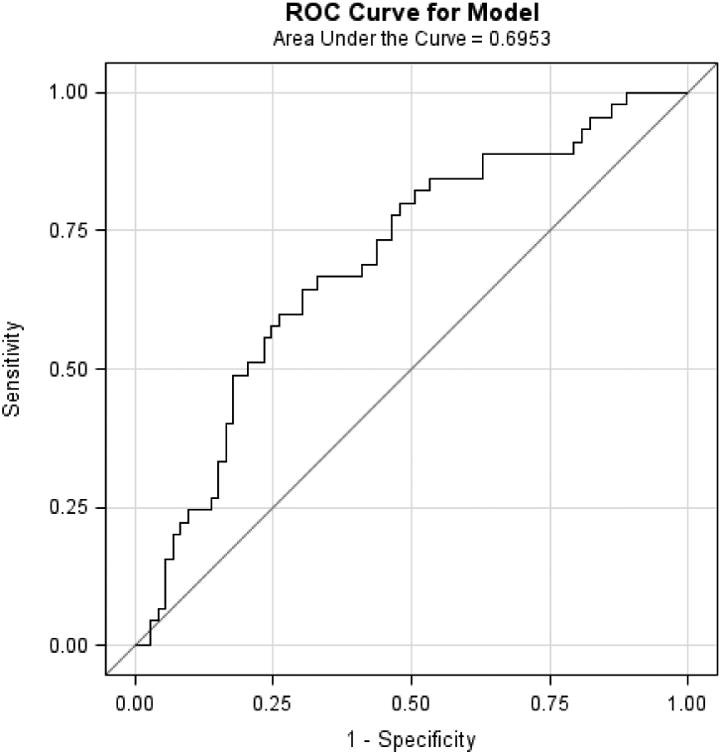
Discussion
Prediction models are increasingly common in the medical literature and may have a role in disease prevention as well as diagnosis and treatment of patients. However, despite the large number of models developed and published, few are externally validated and fewer are used in clinical practice.6 In this study, we externally validated the M-BERET in four independent datasets and confirm that it discriminates patients with BE from population-based controls with reasonable accuracy. However, BE is relatively rare,7 and the number of people who actually have BE as a proportion of those predicted to do so will be low. Thus, even in this setting where M-BERET shows high discriminatory power, most people exposed to the risks of further investigation (i.e., endoscopy) will not see any benefit. While the M-BERET has the potential to allow for more efficient screening for BE, blood based biomarkers may augment or even replace this model.8
Acknowledgments
Funding: This work was supported by the National Institutes of Health R01 CA116845 (HES); K24-04-107 (HES); an Ireland–Northern Ireland cooperation research project grant sponsored by the Northern Ireland Research and Development Office and the Health Research Board, Ireland (for FINBAR; RES/1699/01N/S); the Study of Digestive Health, NCI RO1 CA 001833 (DCW); the Study of Reflux Disease, NCI R01 CA72866 (TLV), and the Established Investigator Award in Cancer Prevention and Control, K05 CA124911 (TLV).
Footnotes
Conflict of interest: The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thrift AP, El-Serag HB. Sex and racial disparity in incidence of esophageal adenocarcinoma: observations and explanations. Clin Gastroenterol Hepatol. 2016;14:330–332. doi: 10.1016/j.cgh.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Spechler SJ, Sharma P, Souza RF, et al. American Gastroenterological Association technical review on the management of Barrett's esophagus. Gastroenterology. 2011;140:e18–52. doi: 10.1053/j.gastro.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaheen NJ, Falk GW, Iyer PG, et al. ACG Clinical Guideline: Diagnosis and Management of Barrett's Esophagus. Am J Gastroenterol. 2016;111:30–50. doi: 10.1038/ajg.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubenstein JH, Morgenstern H, Appelman H, et al. Prediction of Barrett's esophagus among men. Am J Gastroenterol. 2013;108:353–62. doi: 10.1038/ajg.2012.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosmer DW, Lemeshow S. Applied logistic regression. 2nd. New York: Wiley; 2000. [Google Scholar]
- 6.Thrift AP, Kanwal F, El-Serag HB. Prediction models for gastrointestinal and liver diseases: too many developed, too few validated. Clin Gastroenterol Hepatol. 2016;14:1678–680. doi: 10.1016/j.cgh.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett's esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–31. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 8.Thrift AP, Garcia JM, El-Serag HB. A multibiomarker risk score helps predict risk for Barrett's esophagus. Clin Gastroenterol Hepatol. 2014;12:1267–71. doi: 10.1016/j.cgh.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


