Abstract
Background
The development of novel therapeutics and treatment regimens for the management of asthma is hindered by an incomplete understanding of its heterogeneous nature and pathophysiology. Metabolomics can provide an integrated and global profile of a biological system in a dysregulated state, making it a valuable tool to identify biomarkers along the disease development pathway and to understand the biological mechanisms driving that pathway.
Methods
Liquid Chromatography-Mass spectrometry metabolomic profiling was conducted on plasma samples provided at recruitment for 380 children with asthma from the ‘Genetic Epidemiology of Asthma in Costa Rica Cohort’. Metabolites associated with three clinical characteristics of asthma severity (i) airway hyper-responsiveness (AHR) (ii) percent-predicted forced expiratory volume in one second/forced vital capacity ratio (FEV1/FVC), and (iii) FEV1/FVC post-bronchodilator were identified and their discriminatory ability assessed. Metabolite set enrichment analyses was applied to explore the biology underlying these relationships.
Results
AHR was associated (p<0.05) with 91 of 574 metabolites (15.9%), FEV1/FVC pre-bronchodilator with 102(17.8%), and FEV1/FVC post-bronchodilator with 155 (27.0%). The findings suggest these characteristics capture some common and some distinct phenotypic aspects of lung function; glycerophospholipid, linoleic acid and pyrimidine metabolism were common to all three characteristics. The corresponding metabolomic profiles showed moderate but robust discriminatory ability.
Conclusions
The results confirm the existence of an asthma severity metabolome. However, differences in the metabolomic profiles of the three lung function characteristics studied, suggest that refinement of both phenotype classification and metabolite selection should be a priority as the field of asthma metabolomics progresses.
Keywords: Metabolomics, asthma, lung function, FEV1/FVC ratio, Airway hyperresponsiveness
Introduction
The worldwide public health burden of asthma continues to increase [1]. Asthma and related disorders are among the most common chronic diseases in industrialized countries[2–7] affecting 26 million U.S. children and adults[8]. Yet the development of novel therapeutics to treat asthma has not kept pace with the increase in its prevalence. One of the bottlenecks in the progress of such therapeutic advances is an incomplete understanding of the pathophysiology of asthma. Asthma is a heterogeneous syndrome encompassing a number of different subtypes and multiple phenotypic aspects, complicating the interpretation of its underlying biological mechanisms. New approaches are required to address this.
High-throughput omics technologies including epigenetics, genomics, transcriptomics, proteomics and metabolomics represent a novel opportunity to explore the biology of asthma. Metabolomics; the systematic study of all the small molecules in a biological system, reflects the genome, the transcriptome and the proteome as well as their interactions with the environment. Thus it can provide an integrated and global profile of a system in a dysregulated state [9–11]. Metabolomics is increasingly being recognized as a valuable tool to identify biomarkers along a pathogenic pathway and to understand the biological mechanisms driving that pathway. Asthma is a complex disease with both environmental and genetic influences which are as yet not fully understood [12]. As such, it is particularly well suited to metabolomic profiling, both for the development of novel biomarkers and for the improved understanding of pathophysiology. However, to date, metabolomic studies of asthma remain limited and methodologically heterogenous. Despite promising findings there are currently no clinically translatable biomarkers of this disorder [13]. Further work in this area is required.
The aim of this study was to identify metabolites and metabolomic profiles that distinguish children with asthma by their degree of lung function, and to explore the biology underlying these profiles. This study is nested within the ‘Genetic Epidemiology of Costa Rica’ Cohort’. Elucidating the biological mechanisms driving different phenotypic aspects of asthma severity in children will assist the development of strategies for improved management and treatment, helping to ease the global burden of this disorder.
Methods
Study Population
The ‘Genetic Epidemiology of Asthma in Costa Rica’ cohort is based within a Hispanic population isolate from the Central Valley of Costa Rica with one of the highest prevalences of asthma worldwide (24% in children) [14]. From February 2001 to August 2008, screening questionnaires were sent to the parents of children aged 6–14 years who were enrolled in 140 Costa Rican schools. Children were eligible for the study if they had asthma and a high probability of having ≥6 great-grandparents born in the Central Valley of Costa Rica. Asthma was defined by physician-diagnosis and ≥2 respiratory symptoms or asthma attacks in the prior year. A total of 439 unrelated children with asthma and their parents were willing to participate in the study. All participating children completed a protocol including questionnaires, assessment of airway responsiveness to methacholine and spirometry conducted with a Survey Tach Spirometer. The questionnaire was a slightly modified and translated version of the one used in the Collaborative Study on the Genetics of Asthma [15]. A subset of 380 children had metabolomic profiling on blood samples provided at enrolment. Written parental and participating child consent was obtained. The study was approved by the Institutional Review Boards of the Hospital Nacional de Ninos (San Jose, Costa Rica) and Brigham and Women’s Hospital (Boston, Mass, USA).
Metabolomic profiling
Metabolomic profiling methods have been published previously [16] and are described in detail in the supplementary methods. Briefly, profiling was conducted at the Broad Institute (Massachusetts Institute of Technology, Cambridge, MA, USA) using four liquid chromatography-tandem mass spectrometry (LC-MS) methods to measure complementary sets of metabolite classes: (1) HILIC-positive platform: Amines and polar metabolites that ionize in the positive ion mode using hydrophilic interaction liquid chromatography (HILIC) and MS analyses; (2) HILIC-negative platform: Central metabolites (i.e. metabolites central to the normal physiological processes of a biological system) and polar metabolites that ionize in the negative ion mode using HILIC chromatography with an amine column and targeted MS; (3) C8-positive platform: Polar and non-polar lipids using reversed phase chromatography and full scan MS; (4) C18-negative platform: Free fatty acids, bile acids, and metabolites of intermediate polarity using reversed chromatography with a T3 UPLC column (C18 chromatography) and MS analyses in the negative ion mode.
Reference and internal samples were analyzed to confirm LC-MS system sensitivity, chromatography quality, and as a quality control check during the analyses. A pooled reference sample was analyzed throughout the analytical run as an additional quality control measure and to serve as reference for scaling raw LC–MS peak areas across sample batches.
Features with a signal-to-noise ratio <10 were and those which were undetectable/missing for >10% of the samples were excluded All remaining missing values were imputed with the median peak intensity for that feature. Features with a coefficient of variance in the quality-control samples greater than 25% across all batches were excluded to ensure good technical reproducibility. All features were then log transformed to normalize them and pareto-scaled to reduce the variation in fold change differences between them, and to make the effect estimates for each feature comparable. Features were indexed by their mass-to-charge ratio (m/z) and retention time (rt) and metabolite identities were confirmed using known standards. The analyses presented here were restricted to those features that could be annotated to metabolites using these known standards.
Statistical analysis
To assess different aspects of asthma severity, three clinical characteristics were studied (i) airway hyper-responsiveness (AHR, measured as a 20 percent decrement in FEV1 after the administration of <= 16.8mg/ml of methacholine), (ii) pre-bronchodilator lung function, as measured by percent-predicted forced expiratory volume in one second/forced vital capacity ratio (FEV1/FVC), and (iii) FEV1/FVC post-bronchodilator (summarized in Table 1).
Table 1.
Baseline and clinical characteristics of 380 children with asthma
| Variable | Asthmatic Children (n=380) | |
|---|---|---|
| Gender | Male (%) | 226 (59.5%) |
| Age at baseline [yrs] | mean (range) | 9.1 (4.5, 13.3) |
| Age at asthma onset [yrs] | mean (range) | 2.3 (0.0, 10.0) |
| AHR to Methacholine <=16.8 | Yes (%) | 236 (62.1%) |
| % predicted FEV1/FVC Ratio pre-bronchodialator | mean (range) | 97.5 (69.9, 113.7) |
| % predicted FEV1/FVC Ratio post-bronchodialator | mean (range) | 100.3 (73.5, 113.7) |
| Receiving regular treatment for asthma symptom controla | Yes (%) | 346 (91.1%) |
| BMI | mean (range) | 18.2 (11.3, 41.4) |
| Smoke exposure (in utero or during childhood) | Yes (%) | 116 (30.5%) |
| Exercise | Ever (%) | 302 (79.5%) |
defined as regular use of inhaled or oral medication for the control of asthma symptoms
Metabolite features were analyzed as measured LC-MS peak areas, which are proportional to feature concentration. For each endpoint a partial least squares discriminant analysis (PLS-DA) model was utilized to determine the combined ability of the measured metabolites to predict that endpoint using MetaboAnalyst v.2.5 (http://www.metaboanalyst.ca/). FEV1/FCV ratios were dichotomized above and below the median. A seven-fold internal cross-validation procedure was implemented to guard against model over-fitting, and the overall significance of the model’s discriminatory ability was evaluated using permutation testing.
Independent generalized regression models adjusting for age at metabolomic profiling and gender were constructed to identify metabolites associated with the three endpoints of interest; FEV1/FVC ratios were treated as continuous variables and AHR as a binary variable. The ability of the significant metabolites to distinguish children by asthma severity was assessed using a Receiver Operator Characteristic (ROC) curve analysis. For each endpoint a baseline model including age and gender was compared to a model including a summary score for the metabolites identified at a threshold of p<0.05, p<0.01, p<Bonferroni.
Finally, metabolite set enrichment analyses (MetaboAnalyst v.2.5[17]) was applied to the metabolites found to be associated with each endpoint in order to explore the biology underlying these relationships. The hypergeometric test was specified for the over-representation analysis and relative-betweenness centrality for the pathway topology analysis.
Results
The baseline characteristics of the study population are described in Table 1. Metabolomic profiling of plasma samples was available for 380 children. The mean age was 9.1 years (SD: 1.8 years), 59.5% were male and all were Hispanic. The majority of children were defined as mild to moderate asthmatics; 91% were on some form (inhaled or oral) of regular treatment for the control of chronic symptoms.
Metabolomic profiling
Metabolite profiling data were available from four platforms which generated a total of 18,062 metabolite features (Figure 1), after data processing and QC 8185 metabolite features remained. Of these 8185 features, 574 could be annotated to known metabolites using authentic reference standards and were included in these analyses.
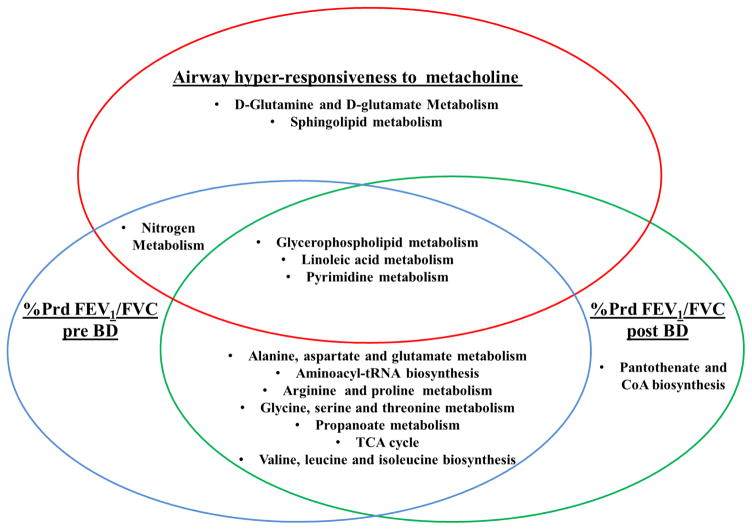
Figure 1. Metabolic pathways that were significantly enriched (p<0.05) among the metabolites significantly associated with three phenotypic aspects of asthma.
For AHR- analysis conducted on 91 metabolites; for %prd FEV1/FVC pre-BD 102 metabolites and for %prd FEV1/FVC post-BD 155 metabolites
When including all metabolites the metabolomic profiles showed poor discriminatory ability under the PLS-DA analysis after seven-fold internal cross-validation (Table 2). The R2 and Q2 metrics were <0.1 for all three endpoints. FEV1/FVC ratios performed slightly better than AHR and permutation testing confirmed that these models were robust. The metabolites with the largest Variable Importance in the Projection (VIP) values for the first component are shown in Table 2
Table 2.
Metabolites significantly associated with three lung function outcomes in 380 asthmatic children at varying significance levels and the ability of these metabolites to distinguish asthmatic children by lung function
| Airway hyper- responsiveness to metacholine | %Predicted FEV1/FVC pre bronchodilatora | %Predicted FEV1/FVC post bronchodilatora | |
|---|---|---|---|
| PLS-DA model (1st component) | |||
| R2 | 0.05 | 0.06 | 0.06 |
| Q2 | 0.004 | 0.01 | 0.02 |
| Permuted p-value | 0.531 | 0.025 | 0.006 |
| Top Ten Metabolites with the highest VIP score | Thiamine; Glycoursodeoxycholate; gamma-linolenic acid; C4-OH carnitine; palmitoleic acid; myristic acid; Taurohyodeoxycholate/Tauroursodeoxycholate; CMP; Taurocholate; oleic acid | 1-Methoxy-1H-indole-3- carboxaldehyde; Calystegine A6; Proline betaine; C50:1 TAG; Uracil; Glycolithocholate; creatinine; C44:0 PG; (ent-2b,4S,9a)- 2,4,9-Trihydroxy- 10(14)-oplopen-3- one 2-(2- methylbutanoate) 9- (3-methyl-2E- pentenoate); Taurocyamine | (2E,6E)-2,6- Nonadienal; Thiamine; C50:1 TAG; 1-Methoxy- 1H-indole-3- carboxaldehyde; TAG source fragment; Taurocyamine; C44:0 PG; C36:4 DAG; Taurocholate; C56:1 TAG |
| Regression modelb (n(%)) | |||
| <0.05 | 91(15.9%) | 102 (17.8%) | 155 (27.0%) |
| <0.01 | 31(5.4%) | 64 (11.1%) | 71 (12.4%) |
| <bonferroni | 1 (0.2%) | 8 (1.4%) | 11 (1.9%) |
| ROC curve analysis (AUC (95% CIc ) | |||
| Baseline <0.05 | 0.585 (0.520, 0.651) | 0.545 (0.487, 0.603) | 0.526 (0.468, 0.585) |
| metabolites <0.01 | 0.652 (0.589, 0.716)* | 0.603 (0.546, 0.660) | 0.590 (0.532, 0.648) |
| metabolites <bonferroni | 0.678 (0.616, 0.740)* | 0.609 (0.552, 0.666)* | 0.602 (0.545, 0.659)* |
| metabolites | 0.666 (0.605, 0.727)* | 0.660 (0.605, 0.714)* | 0.671 (0.617, 0.726)* |
For the regression analyses FEV1/FCV ratios were treated as continuous variables, for the PLS-DA and ROC curve analyses FEV1/FCV ratios were dichotomised at the median
adjusting for age and gender; a logistic model was used for AHR and a linear model for FEV1/FVC ratios
A baseline model including age and gender was compared to a model including a summary score for the metabolites identified at a threshold hold of p<0.05, p<0.01, p<Bonferroni.
indicates whether the models including the metabolite summary score perform significantly better than the baseline model
To refine the ability of the metabolomics profiles to distinguish between children with based on their degree of severity logistic regression models were run adjusting for gender and age at expression profiling. AHR was associated (p<0.05) with 91 metabolites (15.9% of annotated metabolites), FEV1/FVC pre-bronchodilator with 102 (17.8%), and FEV1/FVC post-bronchodilator with 155 (27.0%). At the most stringent Bonferroni corrected significance threshold, FEV1/FVC post-bronchodilator retained the largest number of significant metabolites (n=11, (1.9%)) (Table 2). Overall there was considerable crossover between the three clinical characteristics; AHR shared 24 common metabolites with both FEV1/FVC endpoints (pre- and post-bronchodilator), and pre- and post-bronchodilator FEV1/FVC showed almost identical metabolomic signatures, incorporating 97 common metabolites. The top metabolites for each metabolite included those with the highest VIP scores in the PLS-DA analysis, strengthening the evidence for a relationship with asthma severity.
The inclusion of a summary score based on the first principal component of the significant metabolites for each endpoint resulted in an area under the curve (AUC) for that endpoint that outperformed a baseline model including only age and gender (Table 2). Discriminatory ability varied depending on the threshold used to classify a metabolite as ‘significant’ and therefore the number of metabolites included in the profile. For AHR the metabolite model significantly outperformed baseline when using a classification of p<0.05 (AUC [95%CI] 0.652 [0.589, 0.716], p=0.034) of p<0.01 (0.678 [0.616, 0.740] p=0.006) and of metabolites that were robust to Bonferroni adjustment (0.666 [0.05, 0.727], p=0.014). For FEV1/FVC although the AUC increased with significance level the model only significantly outperformed the baseline when using the p<0.01 (pre: 0.609 [0.552, 0.666], p=0.048, post: 0.660 [0.605, 0.714], p=0.0004) and Bonferroni adjusted (pre: 0.590 [0.532, 0.648], p=0.048, post: 0.671 [0.617, 0.726], p=0.0004) metabolites.
To assess the influence of potential confounders that may be related to lung function in children a fully adjusted logistic regression model additionally including BMI, smoke exposure (in utero or during childhood), exercise (ever/never) and asthma treatment regime (regular use to control chronic symptoms versus sporadic use for acute symptoms) was run. Adjustment for these variables did not alter the findings; the majority of metabolites and metabolomics pathways retained significance. This was particularly evident for those with metabolites with the strongest associations (p<0.01). The results of the ROC curve analysis were also comparable (supplementary table S1).
Metabolite-set enrichment analysis using MetaboAnalyst [17], which uses a combination of over-representation and pathway topology analysis, further supported the notion that FEV1/FVC ratio and AHR capture some common and some distinct phenotypic aspects of lung function (Figure 1). Among the significant (p<0.05) metabolites of all three variables, three pathways were enriched (p<0.05); glycerophospholipid, linoleic acid and pyrimidine metabolism. The two measures of FEV1/FVC ratios shared a total of seven common pathways; four of these pathways are involved in amino acid metabolism, two in carbohydrate metabolism and one pathway; aminoacyl tRNA biosynthesis, in genetic information processing. Two pathways; sphingolipid metabolism and D-glutamine/glutamate metabolism were specific to AHR, while Pantothenate and CoA biosynthesis was enriched only among the FEV1/FVC post BD metabolites.
Discussion
Asthma has doubled in prevalence during recent decades and is among the most common chronic diseases in industrialized countries [1]. New approaches for the identification of novel biomarkers and therapeutic targets that reflect the heterogeneity of asthma are necessary. This requires a greater understanding of the biology underpinning this heterogeneity. One such approach is metabolomics. Asthma represents a particularly interesting disorder for the application of metabolomics, given the interplay between genetics and environment in its etiology, and the somewhat limited understanding of its pathophysiology. In this study a comprehensive analysis of the association between the metabolome and three different measures of lung function was conducted in a large well characterized cohort of Costa-Rican children with asthma. The pathways and metabolites highlighted in this study will serve as a critical foundation for the improved understanding of asthma biology and the identification of potential biomarkers of severity and therapeutic targets.
There was considerable crossover in terms of the significantly associated metabolites between the three investigated clinical characteristics of asthma Pre- and post- bronchodilator FEV1/FVC and AHR. Pre- and post-bronchodilator FEV1/FVC showed almost identical metabolomic signatures; the majority of the constituent metabolites were amines, a metabolite class consistently linked with asthma [18–20]. Thiamine, which can trigger allergic reactions [21], and creatinine, a break down product of creatine which exacerbates lung inflammation [22], were among the top metabolites for all three outcomes. The results also support the reported role of myristic acid, oleic acid and carnitines in asthma [23, 24]. Yet there was also evidence that the metabolomic profile for AHR, a composite variable incorporating airflow obstruction, hyper-responsiveness, and inflammation, included aspects distinct from the FEV1/FVC signatures, which reflect mainly airflow obstruction. The majority of the AHR-associated metabolites were polar and non-polar lipids, in agreement with previous evidence demonstrating altered lipid profiles in asthmatics [25]. Succinate, Alanine and Phenyalanine which have been reported to be associated with asthma and asthma related outcomes in multiple studies and multiple biological media [13] were only significantly associated with FEV1/FVC.
Metabolite-set enrichment analysis using MetaboAnalyst, which uses a combination of over-representation and pathway topology analysis, further supported the notion that FEV1/FVC ratio and AHR capture some common and some distinct phenotypic aspects of lung function. Three pathways, relating to lipid and nucleotide metabolism, were enriched among the metabolites identified as significantly associated with all three asthma endpoints. A number of these pathways and their constituent metabolites have been previously associated with asthma endpoints [20, 26–28]. It is hypothesized they may be involved in the overall asthmatic phenotype through broad mechanisms such as the mediation of the inflammatory response, oxidative stress and alterations in the cellular energy requirements of activated inflammatory cells. Conversely other pathways were only associated with single clinical characteristics, such as sphingolipid metabolism and AHR. This is in agreement with previous evidence specifically linking sphingolipids and sphingolipid metabolism to airway hyperactivity [29]. FEV1/FVC pre- and post- bronchodilator shared eight pathways, however there were still some differences both on a metabolite and pathway level, suggesting that responsiveness to bronchodilators may also be reflected in the metabolome.
The ability of metabolites to classify asthmatic children by lung function was assessed using both PLS-DA models and ROC curve analysis. When all metabolites were included, the PLS-DA model indicated that discriminatory ability was poor, reflecting the broad range of the metabolome that can be captured by metabolomic profiling. This metabolome is sensitive to both internal and external influences that may be unrelated to asthma status and therefore any asthma-severity signal is weak. Conversely when a metabolite summary score based on metabolites shown to be associated with asthma status was created, ROC curve analysis demonstrated that this had moderate to good discriminatory ability using, which outperformed a baseline model. With further refinement such profiles may support the development of asthma biomarkers as the field progresses. Crucially, sensitivity analyses demonstrated these results were not confounded by any of the environmental factors studied.
Asthma metabolomics studies to date report encouraging results including biologically plausible metabolites and metabolomic pathways associated with the development and manifestation of asthma. However, validation of most of these findings is lacking. This may in part be due to the heterogeneous nature of asthma which could lead to incomparable study populations when a single definition of asthma is utilized. These findings suggest that different phenotypic aspects of asthma and asthma severity may be reflected by different aspects of the metabolome. Consequently asthma metabolomics studies must consider the heterogeneity of asthma, it must include detailed phenotypic information on the asthma population studied and it must include a broad range of the metabolome. This study is among the first studies to utilize metabolomics to explore different measures of asthma severity, and adds to the growing body of literature on asthma metabolomics. This study was conducted within the ‘Genetic Epidemiology of Asthma in Costa Rica’ study; a highly homogeneous cohort with consistent and controllable environmental exposures, detailed clinical information, and minimal population stratification. Four metabolomic profiling platforms were utilized providing broad coverage of the metabolome.
However there were some limitations to these analyses. Only a percentage of the metabolite ion features could be annotated and studied. It is likely that there are additional metabolites and therefore pathways involved in the clinical manifestations of asthma which could not be identified here. Given the highly dimensional nature of metabolomics false positives are also possible and a number of the findings would not be robust to correction for multiple testing. However, many false discovery rate methods are considered too stringent for metabolomics analysis, due to the high correlation and redundancy between metabolite features, and there is a lack of agreed standards in the field to deal with this issue [30].
Conclusions
These results further confirm the existence of an asthma severity metabolome in children that can be measured and interrogated to provide further information on the biology and mechanistic pathways underlying lung function in asthma. However, further refinement and validation of this metabolome is still required. The differences in the metabolomic profiles of the three lung function characteristics studied emphasizes the need to carefully consider the phenotypic measures of asthma severity used to characterize asthmatics and asthma subgroups, in order to best capture the phenotypic heterogeneity of this disorder. The fact that the models for FEV1/FVC ratio displayed differing discriminatory ability that those for AHR, may again reflect the diverse nature of the AHR outcome, which encompasses multiple aspects of airway disease that may be associated with different dysregulated metabolomic pathways. The variation in the discriminatory ability of the models when varying p-value thresholds were used to determine a ‘significant’ association further emphasizes the importance of determining the correct metabolites with which to build metabolomic models.
The findings suggest that novel methods to explore both phenotype and metabolite selection should be a priority as the field progresses, in order to develop novel and better targeted management and treatment strategies that can address the significant and growing public health burden of asthma.
Supplementary Material
Highlights.
The metabolome can be interrogated to inform on the biology underlying lung function in asthma
Different aspects of lung function relate to different metabolic pathways
Developments in the field are necessary before clinical translation is possible
Acknowledgments
The authors wish to thank the participants of the Genetic Epidemiology of Asthma in Costa Rica Study, and all those involved in conducting the study. The Genetics of Asthma in Costa Rica Study was supported by grants HL066289 and HL04370 from the U.S. NIH. Dr. Celedón’s contribution was supported by grants HL117191 and HL119952 from the U.S. NIH.
Funding
The Genetics of Asthma in Costa Rica Study was supported by grants HL066289 and HL04370 from the U.S. NIH. Dr. Celedón’s contribution was supported by grants HL117191 and HL119952 from the U.S. NIH. The funding bodies played no role in the study design in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Abbreviations
- AHR
Airway hyper-responsiveness
- AUC
Area under the curve
- FEV1/FVC
Percent-predicted forced expiratory volume in one second/forced vital capacity ratio
- LC-MS
Liquid chromatography tandem mass spectrometry
- PLS-DA
Partial Least Squares-Discriminant Analysis
- QC
Quality Control
- ROC
Receiver operator characteristic
- TCA cycle
tricarboxylic acid
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braman SS. THe global burden of asthma*. Chest. 2006;130(1_suppl):4S–12S. doi: 10.1378/chest.130.1_suppl.4S. [DOI] [PubMed] [Google Scholar]
- 2.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59(5):469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 3.Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma--United States, 1980–1999. MMWR Surveill Summ. 2002;51(1):1–13. [PubMed] [Google Scholar]
- 4.Devereux G. The increase in allergic disease: environment and susceptibility. Clin Exp Allergy; Proceedings of a symposium held at the Royal Society of Edinburgh; 4th June 2002; 2003. pp. 394–406. [DOI] [PubMed] [Google Scholar]
- 5.Mannino DM, Homa DM, Akinbami LJ, Ford ES, Redd SC. Chronic obstructive pulmonary disease surveillance--United States, 1971–2000. MMWR Surveill Summ. 2002;51(6):1–16. [PubMed] [Google Scholar]
- 6.Global Initiative for Asthma Management and Prevention. U.S. Department of Health and Human Services, National Institutes of Health. NHLBI/WHO Workshop Report. 1995:95–3659. [Google Scholar]
- 7.Burney P. Epidemiological trends. In: Barnes P, Grunstein M, Leff A, Woolcock AJ, editors. Asthma. Lippincott-Raven; Philadelphia: 1997. [Google Scholar]
- 8.Akinbami Lara J, Moorman Jeanne E, CBHSZMKCAJaXL . Trends in Asthma Prevalence, Health Care Use, and Mortality in the United States, 2001–2010. Hyattsville, MD: National Center for Health Statistics; 2012. [Google Scholar]
- 9.Nicholson JK, Wilson ID. Opinion: understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 10.Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19(1):45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 11.Holmes E, Loo RL, Stamler J, Bictash M, Yap IKS, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453(7193):396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ober C, Yao T-C. The Genetics of Asthma and Allergic Disease: A 21(st) Century Perspective. Immunological reviews. 2011;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly RS, Dahlin A, McGeachie MJ, Qiu W, Sordillo J, Wan ES, Wu AC, Lasky-Su J. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest. 2016 doi: 10.1016/j.chest.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce N, Aït-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, Robertson C the IPTSG. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62(9):758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blumenthal MN, Banks-Schlegel S, Bleecker ER, Marsh DG, Ober C. Collaborative studies on the genetics of asthma—National Heart, Lung and Blood Institute. Clinical & Experimental Allergy. 1995;25:29–32. doi: 10.1111/j.1365-2222.1995.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 16.Kelly RS, Croteau-Chonka DC, Dahlin A, Mirzakhani H, Wu AC, Wan ES, McGeachie MJ, Qiu W, Sordillo JE, Al-Garawi A, Gray KJ, McElrath TF, Carey VJ, Clish CB, Litonjua AA, Weiss ST, Lasky-Su JA. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 2016;13(1):7. doi: 10.1007/s11306-016-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzpatrick AM, Park Y, Brown LA, Jones DP. Children with severe asthma have unique oxidative stress-associated metabolomic profiles. The Journal of allergy and clinical immunology. 2014;133(1):258–261. e258. doi: 10.1016/j.jaci.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loureiro CC, Duarte IF, Gomes J, Carrola J, Barros AS, Gil AM, Bousquet J, Bom AT, Rocha SM. Urinary metabolomic changes as a predictive biomarker of asthma exacerbation. Journal of Allergy and Clinical Immunology. 2014;133(1):261–263. e265. doi: 10.1016/j.jaci.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Chang C, Guo Z-g, He B, Yao W-z. Metabolic alterations in the sera of Chinese patients with mild persistent asthma: a GC-MS-based metabolomics analysis. Acta Pharmacologica Sinica. 2015;36(11):1356–1366. doi: 10.1038/aps.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drought VJ, Francis HC, Mc L, Niven R, Burge PS. Occupational asthma induced by thiamine in a vitamin supplement for breakfast cereals. Allergy. 2005;60(9):1213–1214. doi: 10.1111/j.1398-9995.2005.00848.x. [DOI] [PubMed] [Google Scholar]
- 22.Vieira RP, Duarte ACS, Claudino RC, Perini A, Santos ÂBG, Moriya HT, Arantes-Costa FM, Martins MA, Carvalho CRF, Dolhnikoff M. Creatine Supplementation Exacerbates Allergic Lung Inflammation and Airway Remodeling in Mice. American Journal of Respiratory Cell and Molecular Biology. 2007;37(6):660–667. doi: 10.1165/rcmb.2007-0108OC. [DOI] [PubMed] [Google Scholar]
- 23.McGeachie MJ, Dahlin A, Qiu W, Croteau-Chonka DC, Savage J, Wu AC, Wan ES, Sordillo JE, Al-Garawi A, Martinez FD, Strunk RC, Lemanske RF, Jr, Liu AH, Raby BA, Weiss S, Clish CB, Lasky-Su JA. The metabolomics of asthma control: a promising link between genetics and disease. Immunity, inflammation and disease. 2015;3(3):224–238. doi: 10.1002/iid3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Biltagi M, Isa M, Bediwy AS, Helaly N, El Lebedy DD. L-carnitine improves the asthma control in children with moderate persistent asthma. J Allergy (Cairo) 2012;2012:509730. doi: 10.1155/2012/509730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinding RK, Stokholm J, Chawes BLK, Bisgaard H. Blood lipid levels associate with childhood asthma, airway obstruction, bronchial hyperresponsiveness, and aeroallergen sensitization. Journal of Allergy and Clinical Immunology. 2016;137(1):68–74. e64. doi: 10.1016/j.jaci.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 26.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, Moqbel R, Sykes BD, Rowe BH, Adamko DJ. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. The Journal of allergy and clinical immunology. 2011;127(3):757–764. e751–756. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 27.Motta A, Paris D, D’Amato M, Melck D, Calabrese C, Vitale C, Stanziola AA, Corso G, Sofia M, Maniscalco M. NMR Metabolomic Analysis of Exhaled Breath Condensate of Asthmatic Patients at Two Different Temperatures. Journal of Proteome Research. 2014;13(12):6107–6120. doi: 10.1021/pr5010407. [DOI] [PubMed] [Google Scholar]
- 28.Jung J, Kim SH, Lee HS, Choi GS, Jung YS, Ryu DH, Park HS, Hwang GS. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin Exp Allergy. 2013;43(4):425–433. doi: 10.1111/cea.12089. [DOI] [PubMed] [Google Scholar]
- 29.Ono JG, Worgall TS, Worgall S. Airway reactivity and sphingolipids–implications for childhood asthma. Molecular and Cellular Pediatrics. 2015;2(1):1–6. doi: 10.1186/s40348-015-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chadeau-Hyam M, Ebbels TMD, Brown IJ, Chan Q, Stamler J, Huang CC, Daviglus ML, Ueshima H, Zhao L, Holmes E, Nicholson JK, Elliott P, De Iorio M. Metabolic Profiling And The Metabolome-Wide Association Study: Significance Level For Biomarker Identification. Journal of proteome research. 2010;9(9):4620–4627. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.