Abstract
Living organisms are continuously exposed to a myriad of DNA damaging agents that can impact health and modulate disease-states. However, robust DNA repair and damage-bypass mechanisms faithfully protect the DNA by either removing or tolerating the damage to ensure an overall survival. Deviations in this fine-tuning are known to destabilize cellular metabolic homeostasis, as exemplified in diverse cancers where disruption or deregulation of DNA repair pathways results in genome instability. Because routinely used biological, physical and chemical agents impact human health, testing their genotoxicity and regulating their use have become important. In this introductory review, we will delineate mechanisms of DNA damage and the counteracting repair/tolerance pathways to provide insights into the molecular basis of genotoxicity in cells that lays the foundation for subsequent articles in this issue.
Keywords: Base excision repair, mismatch repair, nucleotide excision repair, single and double strand break repair, translesion synthesis, telomeres
Introduction
Preserving genomic sequence information in living organisms is important for the perpetuation of life. At the same time, mutagenesis plays an indispensible part in its maintenance and evolution, while also contributing to cancer, certain human diseases and aging. It is known that DNA, the basic unit of inheritance, is an intrinsically reactive molecule and is highly susceptible to chemical modifications by endogenous and exogenous agents. Furthermore, the DNA polymerases engaged in DNA replication and repair make mistakes, thereby burdening cells with potentially disadvantageous mutations. However, cells are equipped with intricate and sophisticated systems—DNA repair, damage tolerance, cell cycle checkpoints and cell death pathways—that collectively function to reduce the deleterious consequences of DNA damage.
Cells respond to DNA damage by instigating robust DNA damage response (DDR) pathways, which allow sufficient time for specified DNA repair pathways to physically remove the damage in a substrate-dependent manner. At least five major DNA repair pathways—base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination (HR) and non-homologous end joining (NHEJ)—are active throughout different stages of the cell cycle, allowing the cells to repair the DNA damage. A few specific lesions can also be removed by direct chemical reversal and interstrand crosslink (ICL) repair. These repair processes are key to maintaining genetic stability in cells. In addition, certain types of DNA damage are substrates for the DNA damage tolerance pathways. In higher eukaryotes, for example, a well-orchestrated group of five main translesion synthesis (TLS) polymerases—REV1, POL ζ, POL η, POL κ and POL ι—bypass the damage to enable the continuation of replication, but with the possibility of a concurrent introduction of an incorrect base that can be fixed into a mutation in the subsequent round of replication. Under the circumstances, when the damaged DNA persists, programmed cell death or apoptosis, a regulatory response to DNA damage, is activated to get rid of cells with extensive genome instability.
Not surprisingly, in many cancers, DNA repair, DNA damage tolerance and DDR pathways are disrupted or deregulated, which increases mutagenesis and genomic instability, thereby promoting cancer progression [Bouwman and Jonkers, 2012; Ghosal and Chen, 2013; Wolters and Schumacher, 2013]. Likewise, aging is attributed to attrition of chromosomal ends and failing capacities of a combination of these pathways. Other diseases, such as neurodegenerative disorders, result from a combinatorial failure of more than one of these processes. The 2015 Nobel Prize in Chemistry to Drs. Lindahl, Modrich and Sancar highlights the importance of mechanisms of DNA damage and repair and their implications for human health. In this review we will discuss the details of various types and mechanisms of DNA damage and the compensatory repair and tolerance pathways.
Types of DNA damage
DNA damage can be categorized into two main classes based on its origin: endogenous and exogenous. The majority of the endogenous DNA damage arises from the chemically active DNA engaging in hydrolytic and oxidative reactions with water and reactive oxygen species (ROS), respectively, that are naturally present within cells. Such inherently predisposed reactions of DNA with molecules from its immediate surroundings fuel the development of hereditary diseases and sporadic cancers [Visconti and Grieco, 2009; Reuter et al., 2010; Perrone et al., 2016]. Exogenous DNA damage, on the other hand, occurs when environmental, physical and chemical agents damage the DNA. Examples include UV and ionizing radiation, alkylating agents, and crosslinking agents. We offer here a brief summary of the main endogenous and environmental agents that produce the different classes of DNA damage that then become substrates for the specific DNA repair pathways discussed in the subsequent section.
Endogenous DNA damage
Replication errors, DNA base mismatches and topoisomerase-DNA complexes
Every time a human cell replicates, approximately 3 X 109 bases are copied over by high fidelity replicative polymerases (δ and ε). However, a battery of other DNA polymerases (α, β, σ, γ, λ, REV1, ζ, η, ι, κ, θ, ν, μ, Tdt and PrimPol) can carry out lower fidelity DNA synthesis during DNA replication or repair (Table 1) [Loeb and Monnat (2008)]. High fidelity DNA synthesis is a consequence of structural and biochemical attributes of replicative DNA polymerases, which ensure the insertion of a correct complementary deoxynucleotide opposite the template base. This is accomplished, for instance, by: 1) the thermodynamic stability and base-pair energetics of the incoming dNTP and template base, 2) the geometric selection of a correctly shaped and sized dNTP in the polymerase’s active site, and 3) removing an incorrectly inserted deoxynucleotide by a 3′-5′ deoxynucleotide exonuclease. In addition, the mismatch repair (MMR) pathway contributes to replication fidelity by more than 100-fold by correcting the rare errors that have escaped proofreading by replicative polymerases [Kunkel, 2004; Kunkel, 2009; Kunkel, 2011].
Table 1.
Shows representative human DNA polymerases. BER (base excision repair), MMR (mismatch repair), NER (nucleotide excision repair), DSBR (double strand break repair), dCTP (deoxycytidine phosphate), FA (fanconi anemia), TLS (translesion synthesis), SHM (somatin hypermutation), ICL (interstrand cross link), dRP (deoxyribosephosphate), TdT (Terminal deoxynucleotidyl transferase), RT (reverse transcriptase), AEP (archaeo-eukaryotic primases).
| Polymerase | Family | Error Rate | Function |
|---|---|---|---|
| α (POLA) | B | 10−4 – 10−5 | An RNA primase during replication; role in S-phase checkpoint |
| β (POLB) | X | 5 X 10−4 | A dRP and AP lyase; role in BER |
| δ (POLD) | B | 10−5 – 10−6 | Has a 3′-5′ exonuclease activity; role in replication; additional roles in BER, MMR, DSBR, NER |
| ε (POLE) | B | 10−6 – 10−7 | Has a 3′-5′ exonuclease activity; role in replication; additional roles BER, MMR, DSBR, NER and S-phase checkpoint |
| REV1 (REV1) | Y | Incorporates only dCTPs | Incorporates only dCTPs and mediate protein-protein interactions during TLS; role in FA, HR |
| ζ (REV3) | B | 10−3 | Roles in TLS, DSBR, FA and SHM |
| η (POLH) | Y | 3.5 X 10−2 | Roles in TLS, SHM, BER |
| ι (POLI) | Y | 10−1 – 10−4 | Roles in TLS, SHM, BER |
| κ (POLK) | Y | 10−2 – 10−3 | Roles in TLS and NER |
| θ (POLQ) | A | 2.4 X 10−3 | Has a helicase motif; ICL repair |
| γ (POLG) | A | 10−5 | Has a 3′-5′ exonuclease activity; role in mitochondrial replication; BER |
| λ (POLL) | X | 1.5 X 10−4 | A dRP lyase; roles in V(D)J recombination, NHEJ and BER |
| μ (POLM) | X | 10−3 – 10−5 | A terminal transferase; roles in V(D)J recombination and NHEJ |
| ν (POLN) | A | 3.5 X 10−3 | Possibly TLS |
| σ (POLS) | X | unknown | Has a 3′-5′ exonuclease activity; role in sister chromatid exchange |
| Tdt | X | unknown | V(D)J recombination, template independent synthesis |
| Telomerase | RT | 2 X 10−3 | Replicates the ends of chromosomes |
| PrimPol | AEP | unknown | Translesion polymerase with high efficiency |
Nevertheless, base substitutions and single base insertion and deletion errors still accumulate at a frequency of 10−6 to 10−8 per cell per generation [Kunkel, 2004; Kunkel, 2009]. Additional replication errors accumulate from strand slippage events at repetitive sequences causing insertions and deletions of nucleotides that can potentially change the reading frame [Viguera et al., 2001; Chatterjee N., 2013]. Other times, the replicative polymerases incorrectly incorporate uracil in the DNA or end up with a compromised fidelity because of the alterations of the relative and absolute concentrations of dNTPs and rNTPs within the cell’s environment [Andersen et al., 2005; Vertessy and Toth, 2009; Kumar et al., 2011; Clausen et al., 2013; Buckland et al., 2014; Potenski and Klein, 2014]. These incorrectly paired/incorporated nucleotides that escape proofreading and MMR become mutations in the next round of replication and are a major source of spontaneous mutagenesis.
Another source of endogenous DNA damage results from the action of topoisomerase enzymes (for example: TOP I, TOP II, TOP III; 7 TOP genes are found in the human genome), which primarily remove superhelical tension on DNA during replication and transcription [Wang, 2002; Pommier et al., 2006]. TOP1, for example, transiently nicks the supercoiled DNA and facilitates rotation of the broken strand around the TOP1-bound DNA strand to relax the DNA. Thereafter, TOP1 religates the breaks by aligning the 5′-OH group of the DNA with the tyrosine-DNA phosphodiester bond to resolve the complex [Stewart et al., 1998; Carey et al., 2003]. Misalignment of the 5′-OH DNA end stabilizes the cleavage complex to form a DNA lesion [Pommier and Cherfils, 2005; Pommier and Marchand, 2005]. Interestingly, anticancer drugs such as camptothecin and many naturopathic compounds are known to stabilize the TOP1-DNA cleavage complexes [Staker et al., 2002; Han et al., 2008]. Additionally, DNA adducts (from UV and benzene derivatives) and aberrant DNA structures (nicks, mismatches, abasic sites) can also irreversibly trap the TOP1-DNA cleavage complex into DNA lesions called suicidal complexes [Burgin et al., 1995; Pourquier and Pommier, 2001; Meng et al., 2003]. TOP1-associated DNA damage is usually repaired by reversal of these complexes or is excised by TDP1 (tyrosyl DNA phosphodiesterase) and endonucleases [Pommier et al., 2006].
Spontaneous base deamination
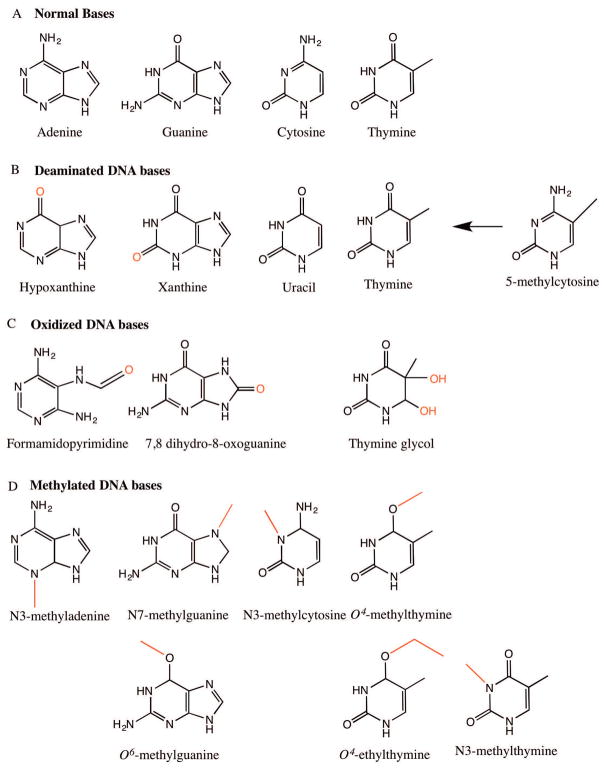
Base deamination is a major source of spontaneous mutagenesis in human cells, where cytosine (C), adenine (A), guanine (G), and 5-methyl cytosine (5mC) in DNA lose their exocyclic amine to become uracil (U), hypoxanthine, xanthine and thymine (T), respectively (Figure 1B). Interestingly, these base deamination events occur at a much higher frequency in single-stranded versus double-stranded DNA and are often exacerbated by transient single strandedness during active replication, transcription and recombination [Lindahl, 1993; Yonekura et al., 2009]. In the case of deamination of cytosine, for instance, the native C:G base pairing alters to a U:A base pair in the first round of replication, which in the next round of replication results in a CG→TA mutation. Cytosine and 5-methyl cytosine are the most frequently deaminated, but 5-methyl cytosine is deaminated three to four times more frequently than cytosine [Lindahl, 1979]. While deaminated cytosine is rapidly removed from DNA by uracil-DNA glycosylase, the G:T base pair resulting from deamination of 5-methylcytosine is instead a substrate for the thymine DNA glycosylase (TDG) and the relatively slow MMR process [Lindahl, 1979; Wiebauer and Jiricny, 1990; Waters and Swann, 1998]. Consequently, the GC→AT transition at the CpG sequences accounts for one-third of the single site mutations responsible for hereditary diseases in humans [Cooper and Youssoufian, 1988; De Bont and van Larebeke, 2004].
Figure 1.
Common DNA base lesions. A) Normal structures of DNA bases: adenine (A), guanine (G), cytosine (C) and thymine (T). B) Deaminated bases: hypoxanthine, xanthine, uracil and thymine arising from deamination of exocyclic bases of adenine, guanine, cytosine and 5-methylcytosine (5-mC) respectively. C) Oxidized DNA bases: formamidopyrimidine derivative of adenine (Fapy-A), 7,8 dihydro-8-oxoguanine (8-oxo-G) and thymine glycol. D) Methylated DNA bases: N3-methyladenine, N7-methylguanine, O6-methylguanine, N3-methylcytosine, O4-methylthymine, O4-ethylthymine and N3-methylthymine.
Paradoxically, cytosine deamination is also a normal route for somatic hypermutagenesis during antibody development due to the action of the deaminase enzymes AID (activation-induced deaminase) and APOBEC1 (Apolipoprotein B mRNA editing enzyme catalytic polypeptide 1), which mediate host defense against reteroviruses [Goff, 2003; Blanc and Davidson, 2010; Chandra et al., 2015]. In addition to the endogenous deamination sources, environmental exposure to UV radiation, intercalating agents, nitrous acid and sodium bisulfite can in general enhance base deamination rates in the DNA [Chen and Shaw, 1993; Moyer et al., 1993; Pfeifer et al., 2005; d’Ischia et al., 2011]. From an evolutionary standpoint, cytosine deamination from endogenous and exogenous sources may serves as a source for genetic diversity [Fryxell and Zuckerkandl, 2000; Nabel et al., 2012].
Abasic sites
Abasic or AP (apurinic/apyrimidic) sites are continuously created in the DNA when the N-glycosyl bond, which links the nitrogenous base and the sugar phosphate backbone, either hydrolyzes spontaneously or gets cleaved by a DNA glycosylase to generate an intermediate in the BER pathway. For example, AP sites are formed when uracil is removed from the DNA by uracil-DNA glycosylase [Lindahl and Barnes, 2000]. In a human cell, about 10,000 abasic sites are created per day; both extreme pH conditions and high temperatures positively impact their generation [Lindahl, 1993; Tropp, 2011]. Abasic sites are inherently unstable and readily convert into single strand breaks (SSBs) from a β-elimination reaction that targets the 3′ phosphodiester bond of the leftover deoxyribose [Bailly and Verly, 1988; Waters and Walker, 2006; Tropp, 2011; Chan et al., 2013]. Most AP sites are effectively removed by AP endonucleases that cleave at their 5′ end and allow the BER pathway to repair them. Alternatively, AP sites can be bypassed by TLS polymerases [Chan et al., 2013]. It is not known whether other exogenous stresses could also directly propel the formation of AP sites in the genome.
Oxidative DNA damage
Reactive oxygen species (ROS) are the typical byproducts of the electron transport chain (ETC) during cellular respiration in aerobic organisms, and are additionally derived from catabolic oxidases, anabolic processes and peroxisomal metabolism [Henle and Linn, 1997]. At low levels, ROS species perform important cellular functions such as serving as cellular messengers in redox signaling reactions and effecting important defense responses to invading pathogens by the immune system [Errol C. Friedberg, 2005; Segal, 2005; Malle et al., 2007]. However, in excess, ROS species can cause a total of approximately 100 different oxidative base lesions and 2-deoxyribose modifications [Bjelland and Seeberg, 2003; Cadet et al., 2010; Cadet et al., 2011; Cadet et al., 2012; Ravanat et al., 2012; Cadet and Wagner, 2014]. Ordinarily, the deleterious consequences of ROS are regulated in cells by 1) restricting respiration in the mitochondrial compartment, thereby protecting other cellular components, 2) protecting DNA by complexing it with histones, and 3) quenching of surplus ROS species by the anti-oxidant enzymes superoxide dismutase, catalase, and peroxiredoxin [Riley, 1994; Mates et al., 1999; Mates and Sanchez-Jimenez, 1999]. Despite this, an overabundance of ROS species is notably associated with the development of human diseases, such as cancer, Alzheimer’s disease, Parkinson’s disease, diabetes, and heart failure [Giacco and Brownlee, 2010; Liou and Storz, 2010; Mohsenzadegan and Mirshafiey, 2012; Dias et al., 2013; Hafstad et al., 2013].
The most conspicuous of the ROS species are the superoxide radicals (•O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (•OH) [Tropp, 2011]. Amongst these ROS species, the •OH radical, produced as a byproduct of a Fenton’s reaction of H2O2 with Fe2+, is by far the most reactive, and is capable of damaging DNA, proteins and lipids [Imlay et al., 1988; Dizdaroglu et al., 1991]. These electrophilic •OH radicals react with DNA bases by 1) adding to their double bonds, 2) abstracting hydrogen atoms from their methyl groups, and 3) attacking the sugar residue in their immediate vicinity [Breen and Murphy, 1995; Winterbourn, 2008]. For example, thymine glycol residues are generated from a •OH attack on the C5/C6 double bonds of thymine (Figure 1C). Similarly, the •OH radical produced as a byproduct of the Fenton reaction of H2O2 and Fe2+ induces an imidazole ring opening in guanine and adenine to form the fragmented purine structure formamidopyrimidine (Figure 1C) [Chetsanga et al., 1981; Errol C. Friedberg, 2005; C., 2006]. Another biologically significant and major oxidative base lesion formed from hydroxylation of the C-8 residue of guanine is the saturated imidazole ring 7,8 dihydro-8-oxoguanine (8-oxo-G) (Figure 1C). 8-oxo-guanine pairs incorrectly with adenine instead of cytosine, thereby adding to the overall mutational load, and is further oxidized to other deleterious secondary DNA lesions because of its low oxidation potential [Kasai and Nishimura, 1984; Cheng et al., 1992; Cadet et al., 1999; Cadet et al., 2010].
Other than attacking DNA bases, ROS radicals can also compromise the DNA backbone causing an estimated 2300 single strand breaks per cell per hour in mammalian cells [Giloni et al., 1981; R, 1981; Henner et al., 1983a; Henner et al., 1983b]. While the BER pathway repairs the oxidized bases, the breaks in the DNA backbone are repaired by the single strand break repair (SSBR) pathways or the double strand break repair (DSBR) pathways [Henner et al., 1983a; Demple and Harrison, 1994]. Finally, lipid peroxidation, the oxidation of lipid molecules by hydroxyl radicals, generates aldehyde products such as malondialdehyde and 4-hydroxynonenal, which can react with adenine, guanine and cytosine to form mutagenic adducts [Marnett, 2000; Plastaras et al., 2000; VanderVeen et al., 2001]. About 1 adduct per 106 – 107 parent DNA bases results from lipid peroxidation events and the number of mutagenic adducts are expected to be even higher for metal storage diseases such as Wilson’s disease and hemochromatosis [Carmichael et al., 1995; Luczaj and Skrzydlewska, 2003; Broedbaek et al., 2009].
DNA methylation
S-adenosylmethionine (SAM), which is used as a methyl donor by methyl transferases during normal methylation reactions, can also spontaneously generate up to 4000 N7-methylguanine, 600 N3-methyladenine and 10–30 O6-methylguanine residues per cell per day in mammals (Figure 1d) [Rydberg and Lindahl, 1982; Holliday and Ho, 1998; De Bont and van Larebeke, 2004]. Other methylating agents include endogenous nitrosated bile salts, betaine, choline, and environmental agents such as tobacco smoke, diet, pollution or derivatives of N-nitroso compounds [O’Driscoll et al., 1999; Zhao et al., 1999]. O6-methylguanine and the related residues O4-methylthymine and O4-ethylthymine are highly mutagenic, producing G:C→A:T and T:A→C:G transition mutations, respectively. In contrast, N3-methyladenine is only partly cytotoxic due to inhibition of DNA synthesis, while the N7-methylguanine residue is essentially harmless unless it undergoes a spontaneous cleavage to generate an AP site or opens the imidazole ring to form formamidopyrimidine [Loveless, 1969; Loechler et al., 1984; Larson et al., 1985; Preston et al., 1986; Preston et al., 1987; O’Connor et al., 1988; Singer et al., 1989; Tudek et al., 1992]. Other minor methyl lesions produced by SAM are the mutagenic N3-methylthymine and N3-methylcytosine (Figure 1D) [Boiteux and Laval, 1982; Saffhill, 1985; Huff and Topal, 1987].
Methylated bases are removed from DNA by two main pathways: 1) direct reversal of the DNA damage by O6-methylguanine DNA methyltransferase or by oxidation by an α-ketoglutarate-dependent dioxygenase AlkB homolog, and 2) BER, which is initiated by DNA glycosylases to remove the methylated bases by catalyzing the cleavage of their glycosidic bonds [Sakumi and Sekiguchi, 1990; Tudek et al., 1992; Huang et al., 1994; Zak et al., 1994; Ye et al., 1998]. In addition, the O6-methylguanine DNA lesion interestingly triggers a cytotoxic and futile cycle of MMR, by means of its abnormal base pairing with other residues [Branch et al., 1993; Kat et al., 1993]. If left unrepaired, methylated DNA bases are a major source of spontaneous DNA damage. Alkylated DNA damage from exogenous compounds will be discussed later in the article.
Exogenous DNA damage
Ionizing Radiation (IR)
Ionizing radiation, composed of alpha, beta, gamma, neutrons and X-rays, is abundant in our environment, being produced from diverse sources ranging from rocks, soil, and radon, to cosmic radiation and medical devices. Each type of radiation can be classified to describe its effect (direct or indirect) and ionization density (linear energy transfer (LET)). Depending on the amount of energy transferred to matter, radiations are classified as either high LET (alpha rays) or low LET (beta and gamma). Cumulatively, IR can damage the DNA either directly, or by indirect means, such as by radiolysis of the surrounding water to generate a cluster of highly reactive hydroxyl radicals (•OH) [Errol C. Friedberg, 2005; Omar Desoukya, 2015]. The presence of oxygen and other reactive species in the surrounding also potentiates the formation of other DNA-reactive free radicals by IR [Wardman, 2009]. In fact, indirect DNA damage from (•OH) radicals accounts to about 65% of the radiation-induced DNA damage [Vignard et al., 2013]. Because of this, IR produces a spectrum of base lesions that is similar to that generated by ROS species (see previous section). Major lesions include 8-oxo-guanaine, thymine glycol and formamidopyrimidines (Figure 1C).
Apart from causing base lesions, ionizing radiation also causes single strand breaks with a unique signature, where the DNA breaks have 3′ phosphate or 3′-phosphoglycolate ends rather than 3′-OH ends. In addition, fragmented sugar derivatives and loss of terminal base residues culminate into clustered damage or single stranded gaps [Henner et al., 1982; Henner et al., 1983b; Obe et al., 1992]. AP endonucleases, polynucleotide kinase/phosphatase (PNKP) and tyrosyl DNA phosphodiesterase 1 (TDP1) can efficiently process the modified ends and enable repair of the IR-induced single strand beaks [Price, 1993; Jilani et al., 1999; Zhou et al., 2005; El-Khamisy et al., 2007]. A particularly important radiation-induced lesion is the double strand break, formed from multiple damaged sites closely positioned on both DNA strands [Hutchinson, 1985; Iliakis, 1991]. Although toxic, IR-induced double strand breaks can be repaired by the HR pathway [Lomax et al., 2013].
Ultraviolet (UV) radiation
UV radiation emanating from the sun is the leading cause of skin cancers in humans [Davies, 1995; KIEFER, 2007]. Typically, UV radiation is categorized into three classes based on the range of wavelength: UV-C (190–290 nm), UV-B (290–320 nm) and UV-A (320–400nm). DNA absorbs maximal UV radiation at 260 nm, beyond which the photo-absorption drops dramatically. Sunlight is composed of 5.1% UV-A, 0.3% UV-B, 62.7% visible light and 31.9% infrared, as the hazardous UV-C is mostly filtered out by the ozone layer [Davies, 1995]. The effects of UV on matter are disseminated in two ways. First, if the UV is absorbable, molecules in matter are excited leading to their photochemical alteration. Second, if UV cannot be directly absorbed, energy transfer from nearby molecules called photosensitizers indirectly affects matter. UV damage DNA by both pathways.
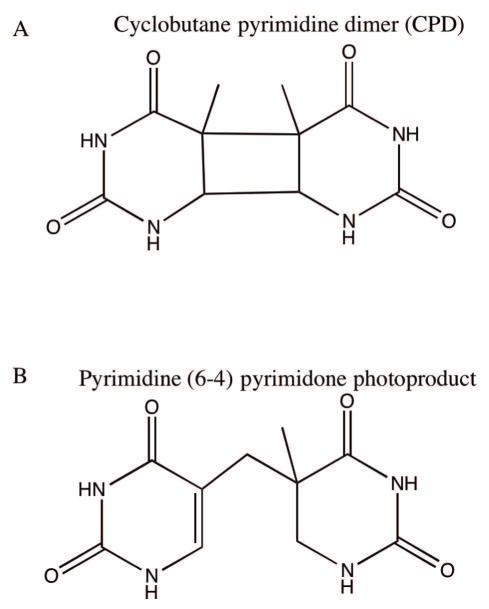
Laboratory studies have shown that UV-C damages DNA primarily by causing covalent linkages between two adjacent pyrimidines. Here the two major photoproducts are the cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6 – 4) pyrimidone photoproducts ((6 – 4) PPs) (Figure 2). Their relative formation frequency depends on wavelength and dose of light [Varghese, 1972; Mitchell and Nairn, 1989; Davies, 1995], although, the yield of (6 – 4) PP is slightly less than CPDs [Mitchell and Nairn, 1989]. Other minor photoproducts are also generated, such as pyrimidine hydrate, thymine glycols, and dipurine adducts [Demple and Linn, 1982; Bose et al., 1983; Kumar et al., 1991; Mitchell et al., 1991]. In CPDs, a cyclobutane ring covalently links the two adjacent pyrimidines, whereas in (6 – 4) PP, the C6 position of one pyrimidine is covalently linked to C4 position of the adjacent pyrimidine. These bulky dimers distort the helix, requiring TLS polymerases for replication past them, thereby contributing to mutagenicity. For example, C:G→T:A, T:A→C:G and characteristic tandem CC→TT transition mutations result from pyrimidine dimers [Chan et al., 1985; Dumaz et al., 1993; Gentil et al., 1996; Naegeli, 1997]. An interesting attribute of the (6 – 4) PP is that it undergoes photoisomerization to a Dewar valence isomer in the presence of UV-B, while reverting to the conventional (6 – 4) PP structure when exposed to UV-C light [Mitchell and Nairn, 1989; Davies, 1995]. If these lesions are left unrepaired or are not bypassed, they result in cytotoxicity.
Figure 2.
Main UV radiation-induced DNA base lesions. A) Representative cyclobutane pyrimidine dimers (CPD). Shown here are cyclobutane thymine dimers. B) Representative pyrimidine (6 – 4) pyrimidone photoproduct [(6 – 4)PP]. Shown here are derivatives of two thymine bases linked via C6 of one thymine base and C4 of the other thymine base.
UV-C is widely used in laboratory investigations because of its maximal absorption by DNA, producing more photoproducts than the UV-A and UV-B radiations, which are also physiologically relevant UV wavelengths that also cause DNA damage [KIEFER, 2007]. UV-B for instance causes the formation of pyrimidine dimers, but does so less efficiently than UV-C [You et al., 2000; Errol C. Friedberg, 2005; Rastogi et al., 2010]. UV-A damages DNA by inducing DNA adduct formation by photooxidation reactions and by the excitation of endogenous (porphyrins and flavins) and exogenous (psoralens, tetracycline, promazine and methylene blue) photosensitizers [Epe, 1991; Kvam and Tyrrell, 1997; Douki et al., 1999]. In addition, UV-A-mediated photosensitization can induce 8-oxoG formation or an excess accumulation of cyclobutane dimers [Epe, 1991; Rochette et al., 2003]. In mammalian cells, near and far UV radiations are known to cause DNA protein crosslinks, while UV-A radiation results in DNA strand breakages [Peak and Peak, 1986; Errol C. Friedberg, 2005]. UV lesions are repaired by direct reversal of UV-damaged bases, NER, interstrand crosslink (ICL) repair, translesion synthesis, or homologous recombinations (HR), all of which either repair the lesions or enable cells to tolerate their presence [Sancar, 1996; Errol C. Friedberg, 2005; Waters and Walker, 2006; Eppink et al., 2011].
Exogenous chemical agents
Alkylating agents
Exogenous alkylating agents are primarily produced from dietary components, tobacco smoke, biomass burning, industrial processing and chemotherapeutic agents [Lawley, 1966; AE, 1990; Crutzen and Andreae, 1990]. The electrophilic alkylating agents react with increased affinity to the highly nucleophilic base ring nitrogens, especially the N7 of guanine and N3 of adenine, and slightly less so with the oxygens. Examples of adducted DNA bases include modified adenine (at N1, N3, N6 and N7), guanine (N1, N2, N3, N7 and O6), cytosine (N3, N4 and O2), thymine (N3, O2 and O4), and alkyl phosphates in the DNA backbone (exocyclic positions on DNA bases are in italicized superscripts) [Singer and Kusmierek, 1982; Singer, 1986; Errol C. Friedberg, 2005]. Mechanistically, alkylating agents add the alkyl group by either 1) an SN1 substitution reaction that progresses via the first order kinetics and involves a carbonium ion intermediate, or, 2) an SN2 substitution reaction that follows the second order kinetics, and in general produces adducts that are less mutagenic and carcinogenic than those of the SN1 pathway [Naegeli, 1997], although evidence has been presented that some SN1 alkylating agents may not proceed via the carbonium ion intermediate [Loechler, 1994].
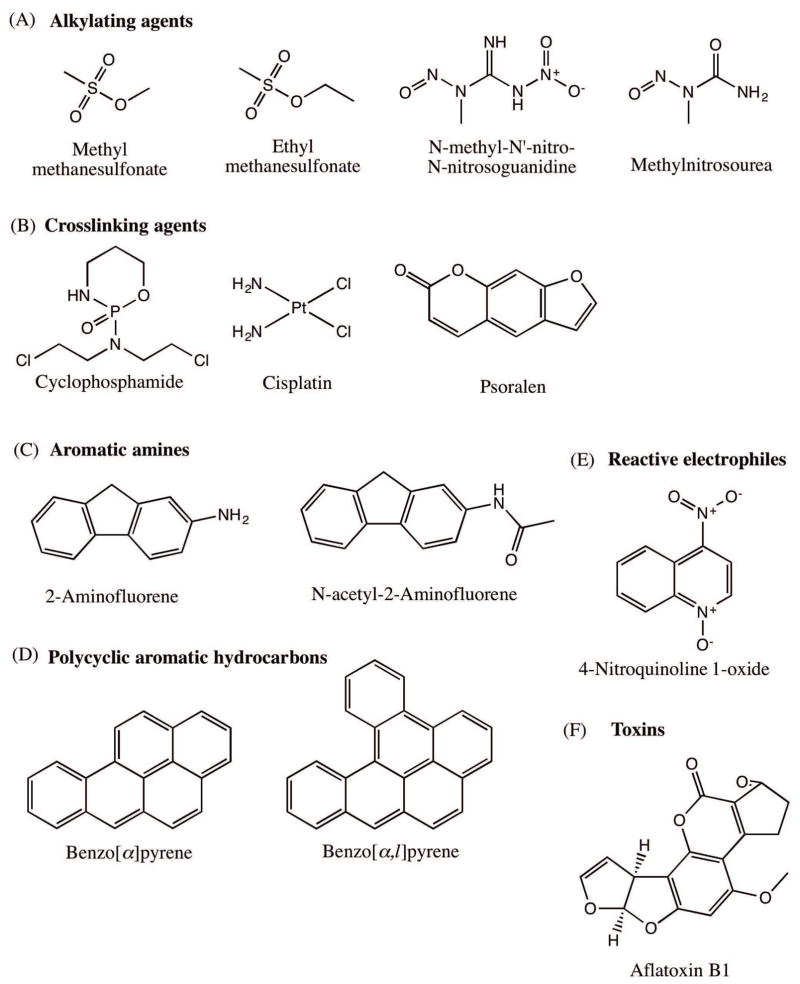
Most common alkylating agents that are regularly used in labs, including methyl methanesulfonate (MMS), ethyl methanesulfonate (EMS), N-methyl -N′ –nitro-N-nitrosoguanidine (MNNG) and methylnitrosourea (MNU) (Figure 3A), react with DNA to generate mutagenic and carcinogenic lesions. For example, MMS produces the mutagenic N7-methylguanine and N3-methyladenine, both of which are susceptible to cleavage of the N-glycosidic bond, thereby generating AP sites, while MNNG and MNU produce O6-methylguanine, which mispairs with T and induces G:C→A:T mutations [Loechler et al., 1984; Beranek, 1990; Wyatt and Pittman, 2006].
Figure 3.
Structures of representative DNA damaging agents. A) Alkylating agents: methyl methanesulfonate (MMS), ethyl methanesulfonate (EMS), N-methyl -N′ –nitro-N-nitrosoguanidine (MNNG) and methylnitrosourea (MNU). B) Crosslinking agents: Cyclophosphamide, cisplatin and psoralen. C) Aromatic amines: 2-aminofluorene (AF) and N-acetyl-2-aminofluorene (AAF). D) Polycyclic aromatic hydrocarbons: benzo(a)pyrene and dibenzo[a,l]pyrene. E) Reactive electrophiles: 4-nitroquinoline 1-oxide (4-NQO). F) Toxins: Afaltoxin B1.
Other classical examples of alkylating agents are sulfur and the nitrogen mustards, first used in World War I, and in many other conflicts since including the present day Syria. The mustards drive SN1 reactions, and are bifunctional in that they carry two reactive groups, instead of one as in monofunctional alkylating agents, and thus have the potential to react with two different sites on the DNA. Such bifunctional reactions result in intra- and interstrand crosslinks, along with the DNA-protein crosslinks, which block DNA metabolic activity [Lawley, 1966; AE, 1990]. These properties of the mustards have been exploited in their use as chemotherapeutic alkylating agents [DeVita and Chu, 2008]. One clinically relevant alkylating agent for chemotherapy is cyclophosphamide (Figure 3B) used in the treatment of lymphomas, leukemias and solid tumors [Emadi et al., 2009]. Another class of crosslinking agents that are used in chemotherapy includes cisplatin (Figure 3B), the first FDA approved platinum compound that is used to treat a wide variety of cancers [Kelland, 2007; Dasari and Tchounwou, 2014]. A crosslinking agent that is not an alkylating agent, psoralen (a furocoumarin) (Figure 3B), intercalates into DNA and cause both interstrand crosslinks and pyrimidine adducts upon photoactivation by UV-A [Yurkow and Laskin, 1991]. The combined psoralen+UV-A or PUVA has been effectively used for treating skin conditions such as psoriasis, eczema and cutaneous T-cell lymphomas. Direct damage reversal, BER and ICL repair are the putative repair pathways that respond to alkylated base damage [Wyatt and Pittman, 2006].
Aromatic amines
Aromatic amines are principally produced from cigarette smoke, fuel, coal, industrial dyes, pesticides and everyday high temperature cooking [Sugimura, 1986; Skipper et al., 2010]. Upon activation by the P450 monooxygenase system, aromatic amines are converted into the carcinogenic (ester and sulfate) alkylating agents that attack the C8 position of guanine [Hammons et al., 1997; Naegeli, 1997]. The most intensively studied examples of aromatic amines are 2-aminofluorene (AF) and its acetylated derivative N-acetyl-2-aminofluorene (AAF) (Figure 3C), which were originally used as insecticides until they were recalled due to their carcinogenic properties [Kriek, 1992]. C8-guanine lesions formed from aminofluorenes are known to form persistent lesions that ultimately give rise to base substitutions and frameshift mutations [Mah et al., 1989; Heflich and Neft, 1994; Shibutani et al., 2001]. The mutagenic properties of the C8-guanine lesion come from its characteristic ability to adopt two conformations while on the DNA [Eckel and Krugh, 1994a]. In the external conformation where the fluorene moiety protrudes out, there is minimal disturbance to Watson-Crick base pairing, which allows these isomers to be effectively bypassed by TLS polymerases [Vooradi and Romano, 2009]. While in internal conformation, the C8-guanine lesion and its partner cytosine are displaced into the minor groove, completely altering the geometry and acting as a very mutagenic substrate on the DNA [Kriek, 1992; Eckel and Krugh, 1994a; Eckel and Krugh, 1994b]. The NER pathway is known to repair C8-guanine adducts in human cells [Mu et al., 2012]
Polycyclic aromatic hydrocarbon (PAH)
Polycyclic aromatic hydrocarbons are carbon compounds with two or more aromatic rings and are generally known to be inert, nonpolar and widely distributed carcinogens in the environment [Harvey, 1991]. Common sources include tobacco smoke, automobile exhaust, charred food and incomplete combustion of organic matter and fossil fuels [Schoket, 1999; Yu, 2002]. The carcinogenicity of these compounds was first documented in 1775, followed by their isolation from coal tar and the later elucidation of their mechanism of action [Butlin, 1892; Phillips, 1983; Fujiki, 2014]. PAHs depend on the P-450 system of the liver to generate reactive intermediates that react with DNA [Phillips, 1983]. Photo-oxidation, one electron oxidation, multiple ring-oxidation and nitrogen-reduction pathways are also known to activate the PAHs [Strniste et al., 1980; Fu, 1990; RamaKrishna et al., 1992; Rogan et al., 1993; Flowers et al., 1997; Penning et al., 1999; Yu, 2002].
Prominent examples of PAHs are naphthalene, anthracene, pyrene, 1-hydroxypyrene, 1-nitropyrene, benzo(a)pyrene and dibenzo[a,l]pyrene. Of these, the most well studied is benzo(a)pyrene (Figure 3D). Upon P-450 activation, benzo(a)pyrene generates the ultimate carcinogen (+)-anti-BPDE [(+)- 7,8-hydroxy-9α, 10α-epoxy-7,8,9,10-tetrahydrobenzo(α)pyrene], along with the (+)-anti-BPDE and the (−)-anti-BPDE intermediates. These intermediates first intercalate into DNA, then the C10 position of the BPDE binds to the N2 exocyclic position of guanine to form DNA adducts [Geacintov, 1986; Graslund and Jernstrom, 1989; Cosman et al., 1992]. In terms of carcinogenicity, dibenzo[a,l]pyrene (Figure 3D) is the most potent PAH and poses a major cancer risk to humans [Luch, 2009]. Normally, the excision repair pathways such as NER and BER repair the PAH DNA lesions if they are not bypassed by TLS polymerases [Braithwaite et al., 1998; Jha et al., 2016].
Other reactive electrophiles
Given the space constraint and scope of this manuscript, we will only briefly touch upon a few other relevant reactive electrophiles that damage the DNA. N-nitrosamines, which are potent carcinogens, are byproducts of tobacco smoke and are also encountered by humans in preserved meats. N-nitrosamines have been implicated in the development of esophagus, stomach and nasopharynx cancers [Bartsch and Montesano, 1984; Tricker and Preussmann, 1991; Hecht, 1999; Herrmann et al., 2015]. Another reactive electrophile, 4-nitroquinoline 1-oxide, has both carcinogenic and mutagenic properties (Figure 3E). Upon its metabolic activation to 4-acetoxyaminoquinoline 1-oxide (Ac-4HAQO), 4NQO1 forms covalent adducts with C8 or N2 of guanine and N6 of adenine, as well as causing oxidative stress that results in 8-hydroxyguanine lesion, all of which significantly adds to the strand breakage events and oral carcinogenesis [Galiegue-Zouitina et al., 1985; Galiegue-Zouitina et al., 1986; Kohda et al., 1986; Hawkins et al., 1994; Kanojia and Vaidya, 2006].
Our final notable compound is the hormone estrogen, frequently used in hormonal replacement therapy, which poses a cumulative increased cancer risk after its prolonged use [Cavalieri et al., 2000; Yager and Davidson, 2006]. Epidemiological and clinical trial studies indicate an increased breast cancer risk and other health issues from a combinatorial use of estrogen and progesterone compared to estrogen alone [Yager and Davidson, 2006]. The P-450 1BI enzyme complex, constitutively expressed in breast and other tissues, hydroxylates estrogen at position 4 to produce reactive catechol estrogens, which are either oxidized to semiquinones and quinones that react with N3 and N7 position of purines, or generate ROS species [Nutter et al., 1991; Nutter et al., 1994; Errol C. Friedberg, 2005]. Both of these unstable bulky adducts and oxidants produce AP sites and strand breakages [Errol C. Friedberg, 2005]. Estrogen is also implicated in the development of prostate cancer, where strand breaks and lipid peroxidation were the phenotypic readouts in a prostate rat model [Ho and Roy, 1994; Nelles et al., 2011].
Toxins
Natural toxins constitute a class of genotoxic and carcinogenic compounds, which are normally used by microorganisms or fungi in defense responses [Ames et al., 1990]. Human and animal exposures result from contaminated cereals, oilseeds, spices, tree nuts, milk and milk products [Lopez et al., 2002]. Aflatoxins are naturally occurring toxins from Aspergillus flavus and Aspergillus parasiticus, of which aflatoxin B1 is the most potent liver carcinogen [Bennett and Klich, 2003]. After passively diffusing into cells, aflatoxin B1 (Figure 3F) is metabolized by the P-450 complex into the active form, aflatoxin B1-8,9-epoxide. This reactive electrophile then adducts with N7 of guanine to form a positively charged product, 8,9-dihydro-8-(N7-guanyl)-9-hydroaflatoxin B1, which weakens the glycosidic bond resulting in depurination [Essigmann et al., 1977; Smela et al., 2001].
Environmental stresses
Environmental sources of stress such as extreme heat or cold, hypoxia, and oxidative stress have been shown to cause DNA damage in human cells [Gregory and Milner, 1994; Gafter-Gvili et al., 2013; Luoto et al., 2013; Neutelings et al., 2013; Kantidze et al., 2016]. These stresses have also been shown to cause mutagenesis at trinucleotide repeats, which are implicated in the development of neurodegenerative disorders via the alt-NHEJ DNA repair pathway [Chatterjee et al., 2015; Chatterjee et al., 2016b]. Equally compelling is the observation that the pathway of environmental stress-induced mutagenesis is akin to the physiological genome instability program operational in many cancer cells [Chatterjee et al., 2015; Chatterjee et al., 2016b]. It is of interest to know whether similar environmental stress-induced phenotypes can be recapitulated in mouse studies.
Other everyday use biological products have increasingly been associated with DNA damage. For example, butyl paraben (BP) and bisphenol A (BPA), found in cosmetics, pharmaceuticals, food-products and beverage processing, are linked to DNA damage in sperm cells [Oishi, 2002; Meeker et al., 2010a; Meeker et al., 2010b; Meeker et al., 2011]. Food preservatives [(sodium benzoate (SB), potassium benzoate (PB) and potassium sorbate (PS)] and food additives [(citric acid (CA), phosphoric acid (PA), brilliant blue (BB) and sunset yellow (SY)] are all known to cause DNA damage [Mamur et al., 2010; Zengin et al., 2011; Yilmaz et al., 2014; Pandir, 2016]. Additionally, plant protection products (PPPs) regularly used by orchard workers have also been associated with DNA damage [Kasiotis et al., 2012]. Such instances stress the importance of global regulatory requirements on the use of chemicals that risk human health, as there may be yet unknown chemicals that have health risks.
DNA damage response (DDR)
After the DNA is damaged, lesion-specific sensor proteins initiate a DNA damage response. The DDR is a collection of mechanisms that sense DNA damage, signal its presence and promote subsequent repair [Harper and Elledge, 2007]. Recruitment of DDR factors is a spatiotemporally regulated process, in which the DDR factors are assembled at the site of damage in a sequential and coordinated manner, as verified by time-lapse microscopy of discrete foci [Harper and Elledge, 2007; Ciccia and Elledge, 2010; Polo and Jackson, 2011]. In addition, chromatin remodeling is an important modulator of DDR response, whereby key post-translational modifications allow assembly of specific DDR and repair factors [Bekker-Jensen et al., 2006; Harper and Elledge, 2007; Misteli and Soutoglou, 2009; Polo and Jackson, 2011; Altmeyer and Lukas, 2013a; Altmeyer and Lukas, 2013b; House et al., 2014]. Mutations affecting DDR network components are the cause of several cancer predisposition syndromes, reflecting their overall importance in avoiding DNA damage-induced human diseases [Ciccia and Elledge, 2010]. However, DNA repair pathways (Figure 4) effectively remove most DNA lesions, which could otherwise result in the formation of mutations or block metabolic processes such as replication and transcription thereby causing senescence and cell death as we discuss below. Readers are directed to excellent reviews on the role of histone modifications during the DNA damage response [van Attikum and Gasser, 2005; Altaf et al., 2007; Zhu and Wani, 2010].
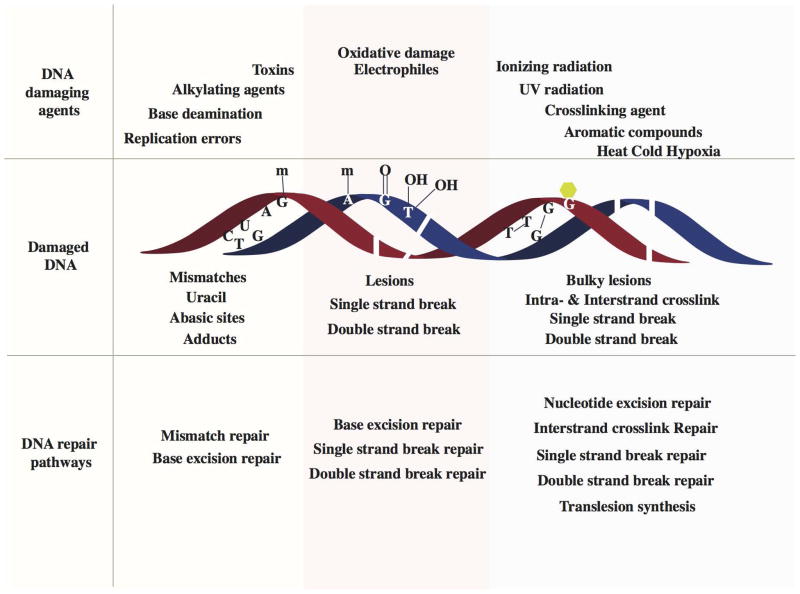
Figure 4.
Schematic of various DNA damage-induced DNA repair pathways. A variety of DNA damaging agents can induce DNA damage, which becomes substrate for specific DNA repair pathways. Upper panel shows representative DNA damaging agents: errors from replication, spontaneous base deamination, alkylating agents, toxins, oxidative agents, ionizing radiation, UV radiation, crosslinking agents, aromatic compounds and environmental agents such as heat, cold and hypoxia. Middle panel represents different kinds of damaged DNA: base mismatches (C:T), uracil from deamination of cytosine, an abasic site from the loss of a base from one DNA strand, methylated guanine, methylated adenine, 8-oxo-G lesion, thymine glycols, single strand breaks, double strand breaks, intrastrand cyclobutane thymine dimers and interstrand guanine crosslinks. The lower panel lists the specific DNA repair pathways that are instigated to repair DNA damages: mismatch repair corrects replication errors and other base mismatches; base excision repair removes base adducts, uracil, abasic sites and oxidative lesions; single strand break repair pathways repairs single stranded breaks in the DNA backbone; double strand break repair pathway repair double strand breaks; nucleotide excision repair removes bulky lesions and intrastrand crosslinks; interstrand crosslink repair removes interstrand linkages and translesion synthesis bypasses intrastrand crosslinks and bulky lesions.
Repair of base DNA damage
Reversal of DNA damage
Small subsets of DNA lesions—UV photolesions and alkylated bases—are simply reversed in an error-free manner. Readers are directed to excellent literature sources on the photolyase-mediated photoreactivation of UV lesions found in lower organisms and marsupials [Kato et al., 1994; Errol C. Friedberg, 2005; Yi and He, 2013]. Here we will briefly discuss the reversal of alkylated DNA damage.
Two different classes of enzymes reverse alkylated bases in humans and mammals. First, the O6- alkylguanine-DNA alkyltransferase (AGT/MGMT) enzyme reverses O-alkylated DNA lesions, such as the O6-methyl, ethyl, 2-chloroethyl, benzyl and aliphatic groups, the pyridyloxobutyl adducts of guanine, and even repair the O6–G-alkyl-O6- G interstrand cross-links [Tubbs et al., 2007; Fang et al., 2008; Pegg, 2011]. A single AGT molecule removes the alkylation adduct in a one step reaction by transferring the alkyl group from the oxygen of the DNA base to the cysteine residue in its catalytic pocket [Kaina et al., 2007]. AGT has a special and complex significance in the cancer field. On one hand, AGT’s potential to target a diverse set of substrates is exploited to synthesize pseudosubstrates that can be used in combination with therapeutic alkylating agents to circumvent resistance to cancer chemotherapy [Tubbs et al., 2007]. On the other hand, lack of AGT expression is associated with certain group of cancers [Lee et al., 2011; Mokarram et al., 2013]. In addition, alkyltransferase-like proteins (ATLs), a family of AGT homologs, inhibit the AGT enzyme by directing the repair of bulky alkyl damage to the NER pathway [Margison et al., 2007; Tubbs et al., 2009].
The second class of direct reversal enzymes, the AlkB-related α-ketoglutarate-dependent dioxygenases (AlkB), reverse N-alkylated base adducts. There are 9 human homologs of E. coli AlkB, which are designated as ALKBH1-8 (Alkylation Repair Homologs) and FTO (Fat Mass and Obesity associated) in human cells [Kurowski et al., 2003; Gerken et al., 2007; Sanchez-Pulido and Andrade-Navarro, 2007; Yi and He, 2013]. For demethylation, the AlkB family proteins hydroxylate the alkyl group in a α-ketoglutarate and iron(II) dependent manner. The oxidized alkyl group is released as formaldehyde, leaving behind the original base [Drablos et al., 2004; Falnes et al., 2007].
Base excision repair (BER)
BER corrects those forms of oxidative, deamination, alkylation and abasic single base damage that are not perceived as significant distortions to the DNA helix. In the nucleus, this repair process is mainly active in the G1 phase of the cell cycle [Dianov and Hubscher, 2013]. For BER transactions, chromatin remodeling at the DNA damage site is followed by lesion recognition by a DNA glycosylase [Odell et al., 2013]. At least 11 different DNA glycosylases can recognize and excise a damaged base from undistorted helices, as well as ones flipped out from the major groove [Huffman et al., 2005; Krokan and Bjoras, 2013]. In terms of function, DNA glycosylases are either monofunctional, with only a glycosylase activity, such as the uracil glycosylases, N-methylpurine DNA Glycosylase (MPG), and MutY Homolog (MUTYH), or are bifunctional with a glycosylase and an additional β-lyase activity. Examples of the latter include the Nth-like DNA glycosylase 1 (NTHL1), Nei-like DNA glycosylase 1 (NEIL1) and Nei-like DNA glycosylase 2 (NEIL2) [Jacobs and Schar, 2012]. It should be noted that 8-oxoguanine DNA glycosylase (OGG1) and NEIL3 function as both mono- and bifunctional glycosylases [Svilar et al., 2011]. An abasic site created from the monofunctional glycosylases gets committed to the short-patch-repair pathway, while the bifunctional glycosylases initiate the long-patch repair pathway of BER [Dianov and Hubscher, 2013].
In short patch repair, the abasic site is the substrate for the AP endonuclease (APE1 in human cells), which cleaves the phosphodiester bond 5′ to the abasic site and generates a hydroxyl residue at the 3′-end while leaving a deoxyribose phosphate (dRP) at the 5′-end. This repair gap is tailored by the 5′-dRP lyase activity of POL β (gap tailoring), followed by filling the single nucleotide gap by POL β and ligation by either LIG1 (DNA ligase 1) or a complex of LIG3 (DNA ligase 3) and XRCC1 (X-ray repair cross-complementing protein 1) [Almeida and Sobol, 2007]. In long patch repair, the repair gap left behind from the bifunctional glycosylase is tailored by the 3′ phosphodiesterase activity of APE1. Thereafter, POL β (in non-proliferating cells) or POL δ/ε (in proliferating cells) synthesize in a strand-displacement manner, which is then followed by flap removal by the flap endonuclease and a LIG1-mediated ligation [Akbari et al., 2009; Svilar et al., 2011].
While BER of 8-oxo-G lesions at CAG repeats is implicated in triplet repeat instability, downregulation of OGG1 is associated with aging, neurodegenerative disorders and cancer [Kovtun et al., 2007; Tian et al., 2009; Curtin, 2012; Mollersen et al., 2012; Chatterjee N., 2013; Krokan and Bjoras, 2013; Chatterjee et al., 2015]. Specifically, mutations in POL β are found in solid cancers and POL β variants can act as dominant negative and sequence specific mutators [Wang et al., 1992; Starcevic et al., 2004; Lang et al., 2007; Murphy et al., 2012]. In addition, PARP1 (Poly [ADP-ribose] polymerase 1) has also been shown to be required for the repair of single strand breaks and damaged purine bases by a sub-pathway of BER [Krokan and Bjoras, 2013; Reynolds et al., 2015]. Finally, mitochondria are also known to carry out both short and long patch BER, where the synthesis step is carried out by POL γ; all of which adds to the significance of this repair pathway in the maintenance of global genome stability [Akbari et al., 2008; Liu and Demple, 2010]. Readers are directed to these reviews for an overview of mitochondrial BER [Bauer et al., 2015; Prakash and Doublie, 2015].
Repair of multiple and bulky base damage
Nucleotide excision repair (NER)
Nucleotide excision repair is the pathway of choice to remove bulky lesions such as CPDs and (6 – 4)PP from UV radiation, benzo[a]pyrene adducts, or damage from chemotherapeutic agents. NER deficiency results in a number of different human syndromes: Xeroderma Pigmentosum (XP), which is associated with a predisposition to skin cancers; Cockayne Syndrome (CS); rare UV-Sensitive Syndrome (UVSS); and Cerebro-Oculo-Facio-Skeletal syndrome (COFS) [Errol C. Friedberg, 2005; Vermeulen and Fousteri, 2013]. However, like the BER pathway, NER contributes to the instability mechanisms in triplet repeat disorders [Lin et al., 2006; Hubert et al., 2011; Dion, 2014]. To begin NER, chromatin remodeling mediated both by chromatin and NER components makes way for the NER machinery on the specified DNA lesions [Scharer, 2013]. There are two major branches of NER: global genome NER (GG−NER) and transcription−coupled NER (TC−NER).
In GG-NER, the main DNA damage sensor is the XPC (Xeroderma Pigmentosum, complementation group C) protein, complexed with RAD23B (UV excision repair protein Radiation sensitive 23B) protein and CETN2 (Centrin 2). This complex scans for the presence of transient single−stranded DNA (ssDNA) caused by disrupted base pairing due to the lesion [Masutani et al., 1994; Nishi et al., 2005]. For repair of UV-induced CPDs, the ultraviolet-damaged DNA damage−binding protein (UV–DDB) complex, consisting of DDB1 (XPE−binding factor) and the GG−NER−specific protein DDB2, directly binds to UV−radiation−induced lesions and then stimulates the binding of XPC [Chu and Chang, 1988; Wakasugi et al., 2002; Scrima et al., 2008]. XPC bound to the lesion becomes the substrate for the transcription initiation factor II H (TFIIH) complex, a transcription initiation and repair factor composed of ten protein subunits that can switch functions in both transcription initiation and in NER [Yokoi et al., 2000; Volker et al., 2001; Compe and Egly, 2012]. The final step of dual excision and gap filling is coordinated to prevent the ssDNA gap formation that can potentially trigger DDR signaling [Marini et al., 2006; Marti et al., 2006; Mocquet et al., 2008].
The incision step of GG-NER commits the assemblage of all the proteins to NER. It involves the use of structure specific endonucleases XPF–ERCC1 and XPG (also known as ERCC5), which cuts the damaged strand short distances away from the 5′ and 3′ end of the lesion respectively [Fagbemi et al., 2011]. The replication proteins PCNA (proliferating cell nuclear antigen), RFC (replication factor C), POL δ, POL ε or POL κ, and LIG1 or XRCC1–LIG3 carry out the final step of gap−filling synthesis and ligation. Proliferative status of the cell determines the choice of polymerase used. For example, POL ε−dependent repair predominates in non−replicating cells, while POL δ and POL κ are the main NER polymerases in replicating cells. LIG1-dependent ligation occurs in replicating cells. However, XRCC1–LIG3 complex seals the gap in non-proliferating cells because of low expression of dNTPs and LIG1 in these cells, [Moser et al., 2007; Ogi et al., 2010].
The second NER pathway, TC-NER, is initiated by a lesion-stalled RNA polymerase II, which begins with the recruitment of TC−NER−specific proteins CSA (Cockayne syndrome WD repeat protein A; also known as ERCC8) and CSB (Cockayne syndrome protein B; also known as ERCC6), which are essential for additional assembly of other TC−NER components [Fousteri et al., 2006]. These include the core NER factors (except for the GG−NER− specific UV–DDB and XPC complexes) and TC−NER−specific proteins, such as UVSSA (UV−stimulated scaffold protein A), USP7 (ubiquitin−specific−processing protease 7; also known as ubiquitin C−terminal hydrolase 7), XAB2 (XPA−binding protein 2; also known as pre−mRNA−splicing factor SYF1) and HMGN1 (high mobility group nucleosome−binding domain−containing protein 1; also known as non−histone chromosomal protein HMG14) [Fousteri et al., 2006; Schwertman et al., 2012]. Once localized at the lesion site, the CSA-CSB complex backtracks (or reverse translocates) RNA polymerase II, exposing the lesion site. TFIIH is recruited to the lesion. The subsequent sequence of events is predicted to be the same as in GG-NER as the lesion is removed from the transcribed strand [Marteijn et al., 2014].
Mismatch repair (MMR)
MMR is an evolutionarily conserved, post replicative repair pathway that contributes to replication fidelity by at least 100-fold [Kunkel, 2009; Arana and Kunkel, 2010]. Typical substrates for the MMR pathway are base mismatches that have arisen during replication and the insertion-deletion loops (IDLs) within repetitive DNA sequences that have resulted from strand slippage events [Errol C. Friedberg, 2005; Jiricny, 2006]. MMR is also implicated in a variety of cellular processes including microsatellite stability, meiotic and mitotic recombination, DNA-damage signaling, apoptosis, class-switch recombination, somatic hypermutation and triplet-repeat expansion [Jiricny, 2006; Jiricny, 2013; Chatterjee et al., 2016a]. Germline mutations in the MMR genes result in Lynch syndrome (also known as hereditary nonpolyposis colorectal cancer or HNPCC), which presents as a familial susceptibility to colon and ovarian cancers as well to a number of other cancers [Peltomaki, 2001]. Chromatin modifications have recently been shown to pave the way for the MMR proteins to gain access to the DNA lesion and initiate repair [Li et al., 2013; Li, 2014].
Of the eight known MSH (MutS homolog) polypeptides in eukaryotes, humans employ the MutSα heterodimer (MSH2/MSH6) to recognize base mismatches and one-to-two nucleotide IDLs, and the MutSβ heterodimer (MSH2/MSH3) to recognize large IDLs [Kunkel and Erie, 2005; Sachadyn, 2010]. The previously accepted model was that after the lesion recognition step, the MutS complex translocates along the DNA in an ATP-dependent manner to make way for the downstream MMR components [Jiricny, 2013]. Recently, the Modrich lab has shown that MutL can trap MutS at the mismatch before it forms a sliding clamp [Qiu et al., 2015]. Next, the MutL complexes are recruited on to DNA and among the 4 known human MutL homologs; the MutLα heterodimer (MLH1/PMS2 heterodimer) plays a major role in MMR [Nicolaides et al., 1994; Papadopoulos et al., 1994; Li and Modrich, 1995; Lipkin et al., 2000]. MutLα regulates termination of mismatch-provoked excision, and its endonuclease activity plays a role in the 3′ nick-directed digestion by EXO1 (Exonuclease 1) in a PCNA/RFC dependent manner [Zhang et al., 2005; Kadyrov et al., 2006]. EXO1 also carries out the 5′ directed mismatch excision creating a gap that is stabilized by RPA [Genschel and Modrich, 2003; Zhang et al., 2005]. POL δ, RFC, HMGB1 (high mobility group box 1 protein) and LIG1 orchestrate the final steps of new DNA synthesis and ligation [Genschel and Modrich, 2003; Yuan et al., 2004; Guo et al., 2006]. PCNA plays an important role in both the initiation step of MMR and in the subsequent DNA synthesis by interacting and localizing MutSα/β and MutLα complexes at the lesion site [Umar et al., 1996; Lau and Kolodner, 2003; Jiricny, 2006].
In addition to mismatch repair and other cellular functions, the mismatch repair genes have recently been shown to be repressed in response to environmental stresses, such as hypoxia, benzo[a]pyrene, inflammation and even tumor microenvironment [Mihaylova et al., 2003; Bindra and Glazer, 2007; Nakamura et al., 2008; Edwards et al., 2009; Chen et al., 2013]. It remains to be seen whether other exogenous stresses can also suppress the expression of MMR genes.
Interstrand cross-link repair (ICL)
Interstrand crosslinks are lesions in which two bases from complementary strands are covalently linked due to damage to the DNA from crosslinking agents such as platinum compounds, nitrogen mustards, MMC, psoralens and alkylating agents [Clauson et al., 2013]. Additional modifications from these crosslinking agents include bases monoadducts, intrastrand crosslinks, and DNA-protein crosslinks. These lesions are recognized and repaired by the Fanconi anemia (FA) proteins. Mutations in the FA genes are the cause of the autosomal recessive FA disorder. FA disorder is a heterogeneous and rare genetic disorder characterized with a high frequency of hematological abnormalities, congenital anomalies and a general predisposition to cancers [Kee and D’Andrea, 2012]. Classically, FA is diagnosed by assessing cellular hypersensitivity—chromosomal breaks and chromosomal radial formations—to DNA ICL agents such as diepoxybutane (DEB) and MMC [D’Andrea, 2010]. In addition, DEB-induced chromosome breakage assay is widely used for the primary diagnosis of FA [Auerbach, 1993].
Interstrand crosslink repair is initiated by chromatin loading of the FA proteins in a cell cycle-dependent manner [Mi and Kupfer, 2005; Kim et al., 2008]. The FA family consists of 21 different functional complementation groups (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P, Q, R, S, T, U, V), which are known to suppress ICL sensitivity [Bluteau et al., 2016; Michl et al., 2016]. Upon ICL damage, FANCM is recruited to the damaged site along with FAAP24 (Fanconi Anemia associated protein of 24 kDa) and MFH (histone fold protein complex) [Ciccia et al., 2007; Niedernhofer, 2007; Yan et al., 2010]. Replication fork remodeling stimulated by MFH and FANCM promotes Holliday junction migration and the creation of ssDNA gaps [Gari et al., 2008a; Gari et al., 2008b; Huang et al., 2010]. RPA bound ssDNA signals ATR activation [Zou and Elledge, 2003; Ben-Yehoyada et al., 2009]. ATR phosphorylates downstream target CHK1, which in turn phosphorylates FANCE, FANCD2, FANCI and MRN [Andreassen et al., 2004; Smogorzewska et al., 2007; Wang et al., 2007a; Cui et al., 2009; Duquette et al., 2012]. In a yet unknown way, other FA core complex components (FANCA/B/C/E/F/G/L/T) assemble at the damaged site and activate the phosphorylated FANCI–FANCD2 heterodimer through FANCL-mediated monoubiquitination [Smogorzewska et al., 2007]. This activation of FANCI-D2 marks as the major activation switch for the FA pathway [Wang, 2008; Tomida et al., 2013]. Subsequently, excision of the DNA strand (5′ and 3′) of the lesion is coordinated by structure specific endonucleases—XPF-ERCC1, MUS8-EME1, SLX4-SLX1, FAN1, SNM1A/SNM1B—in an as-yet unclear fashion [Clauson et al., 2013]. Next, depending upon the proliferation state of the cells, ICL repair bifurcates into either of the two pathways below.
In replicating cells, the presence of ICL stalls the ongoing replication on the leading strand, as well as on the 5′ end of the lagging strand at some distance from the lesion [Raschle et al., 2008; Knipscheer et al., 2009]. Next, XPF-ERCC1 and SNM1A induce incisions on either side of the lesion that unhook the ICL from the lagging strand, thereby producing a gap [Wang et al., 2011]. The leading strand with the ICL becomes the template for new DNA synthesis—by TLS polymerases POL ι, POL κ, POL ν and REV1—that proceeds up to the lesion, bypasses it, and extends beyond the lesion until it reaches the first downstream Okazaki fragment [Minko et al., 2008; Raschle et al., 2008; Yamanaka et al., 2010; Ho et al., 2011; Klug et al., 2012]. After this step, the 3′ overhang of the leftover lagging strand invades the newly synthesized strand in a RAD51-dependent manner in a tightly coordinated manner [Long et al., 2011]. Interestingly, resolution of this HR intermediate depends on the FANCD2-FANCI complex. NER pathway eventually removes the ICL hook that was still hanging on to the leading strand.
In non-replicating cells, ICL repair of psoralen, MMC, cisplatin, and alkyl ICLs depend on NER and TLS polymerases such as REV1 and POL ζ [Clauson et al., 2013]. Helix-distorting lesions are recognized by both the GG-NER and the TC-NER pathways to initiate repair, although some lesions such as cisplatin may escape recognition [Enoiu et al., 2012]. After lesion recognition, the components of the NER pathway are known to cut only the 5′ side of the lesion, with further incisions possibly aided by the MutSβ complex create a ssDNA-gap [Bessho et al., 1997; Mu et al., 2000; Smeaton et al., 2008; Zhao et al., 2009]. Next, the error-prone TLS polymerase synthesize across the gap and finally a second round of NER removes the ICL hook on the other strand [Clauson et al., 2013].
Recent studies provide a striking evidence of crosstalk between the FA and other repair pathways. For example, the FA pathway suppresses non-homologous end joining (NHEJ) by interacting with CtIP, and recruiting the NHEJ-inhibiting molecules—PARP1 and RAD18 to the DNA [Saberi et al., 2007; Ceccaldi et al., 2016a]. Similarly, inhibition of NHEJ components alleviates the sensitivity of FA deficient cells to crosslinking agents, while the same FA deficient cells show enrichment of 53BP1, RIF1 and RAP80 components at damaged chromatin [Adamo et al., 2010; Pace et al., 2010; Ceccaldi et al., 2016b; Renaud et al., 2016]. A second intriguing example of the FA pathway’s crosstalk is its undefined role in promoting alternate end joining events, as seen in patients with FANCA mutations who lack immunoglobulin class switch recombination. In addition, loss of FANCD2 confers a synthetic lethal phenotype in POL θ null mice [Nguyen et al., 2014; Ceccaldi et al., 2015; Howard et al., 2015]. Finally, the FA pathway has now been implicated in trinucleotide repeat instability [Chatterjee N., 2016].
Translesion Synthesis (TLS)
Translesion synthesis is carried out by highly conserved TLS polymerases. TLS polymerases are specialized DNA polymerases that can replicate opposite and past aberrant DNA lesions in a relatively lower fidelity manner than replicative DNA polymerases [Sale, 2013]. If incorrect nucleotides were incorporated by TLS polymerases, they would become mutations in the next round of replication, which propel tumorigenesis and disease, but can also contribute to the overall fitness and evolution of organisms. A total of eleven TLS polymerases are known (REV1, POL η, POL ι, POL κ, POL ζ, POL μ, POL λ, POL β, POL ν, POL θ), which are distributed in four families (Y, B, X and A) and PrimPol (Table 1). Although all TLS polymerases are less accurate than replicative polymerases, certain TLS polymerases are able to copy relatively accurately over certain cognate lesions. For example, cyclobutane thymine-thymine CPDs are cognate lesions for POL η (Table 1). The frequency of DNA synthesis errors during translesion synthesis depends on several factors, such as whether the lesion is a cognate for the particular TLS DNA polymerase, the biochemical characteristics of the particular TLS polymerase and the DNA sequence context [Pages and Fuchs, 2002; McCulloch et al., 2004; Waters and Walker, 2006]. XPV patients, who exhibit a photosensitive phenotype with a high incidence of skin cancer, highlight the physiological significance of certain TLS DNA polymerases bypassing particular lesions. These patients lack the POL η enzyme and are highly susceptible to UV radiation because alternate TLS polymerases (POL ι and POL κ) instead bypass the UV-induced cyclobutane dimers (CPD) in an error-prone fashion [Yamada et al., 2000; Sweasy et al., 2006; Wang et al., 2007b; Ziv et al., 2009].
The TLS polymerases’ fascinating ability to help cells tolerate DNA damage arises from their structural and biochemical features [Rothwell and Waksman, 2005; Pavlov et al., 2006; Waters and Walker, 2006]. Unique functional attributes of TLS polymerases that distinguishes them from the classical replicative polymerases, stems from their discrete physical features. Notable features include the very limited sequence homology to replicative DNA polymerases, the absence of a 3′-5′ exonuclease domain to proofread incoming nucleotides, and their smaller thumb and finger domains, which make fewer contacts with the DNA than those found in replicative DNA polymerases [Rothwell and Waksman, 2005; Waters and Walker, 2006; Sale, 2013]. These structural differences orient the thumb, fingers, palm, and little finger catalytic domains into a relatively larger open active site, while being aided by other physical features such as the polymerase-associated domain (PAD), wrist, and N-clasp region found in Y Family DNA polymerases that facilitate additional DNA binding. Together, these structural features provide the TLS polymerases with a unique architecture that enable them to bypass DNA damage or fill ssDNA gaps [Trincao et al., 2001; Ling et al., 2003; Lone et al., 2007; Jansen et al., 2009a; Jansen et al., 2009b].
Two models have been proposed to explain the DNA lesion bypass process via translesion synthesis. In the polymerase switch model, TLS polymerases come together sequentially in a two-step process to replicate pass the DNA lesion at a stalled replication fork. First, an ‘inserter’ TLS enzyme, usually a POL η, POL ι, or POL κ, and less often REV1 or POL ζ, incorporates a nucleotide opposite the DNA lesion [Korzhnev and Hadden, 2016]. In the second step, an extender TLS enzyme, a role usually fulfilled by POL ζ exclusively but in some cases by POL κ, replaces the inserter polymerase and extends the primer-template termini [Washington et al., 2002; Yuji Masuda, 2016]. This two-step model is proposed to direct both the error-free and error-prone translesion DNA synthesis across the damage [Shachar et al., 2009]. The central molecule that orchestrates both the insertion and extension step is REV1, and by way of its unique scaffolding function facilitates an assemblage of the TLS polymerases by binding to an RIR-containing polymerase—POL η, POL ι, or POL κ—via one interface, and also to POL ζ4 (REV3-REV7-POLD2-POLD3), via a second interface, a central step in this model’s execution [Wojtaszek et al., 2012a; Wojtaszek et al., 2012b]. Moreover, POLD3, which is part of the POL ζ4 complex, interacts with REV1 via its RIR, thereby assisting the switch from RIR directed insertion to POL ζ4-mediated extension during damage bypass [Pustovalova et al., 2016].
In the gap-filling model, single strand gaps left behind by replicative polymerases during replication or via an incomplete DNA repair process, such as during immunoglobulin gene hypermutation, are the targets of TLS synthesis [Sale et al., 2009]. Usually, these type of TLS events are expected to fall outside of the S phase, but based on the type of DNA lesion, a cell-cycle independence is sometimes conferred [Quinet et al., 2016]. Using a gapped plasmid assay, it has been shown that TLS is as high or higher in G2 compared to S phase in human cells, with slightly higher amounts of POL η in G2 compared to S phase of the cell cycle [Diamant et al., 2012]. An exact order of events for a gap-TLS is still unknown, with only a few isolated studies implying the role of TLS polymerases in gap filling. For example, REV1 is very important in mouse cells for synthesis across post-replicative gaps where REV1 gets recruited to the gaps by the 5′-end, unlike the gap-filling step of NER, where POL κ is the polymerase of choice [Ogi and Lehmann, 2006; de Groote et al., 2011; Sale, 2013]. Likewise, REV3 engages in TLS across gaps opposite 6-4 photoproducts [Quinet et al., 2016], all suggestive of a role of TLS at replicating across ssDNA gaps.
In addition to their traditional DNA damage bypass functions, TLS polymerases are now known to play a role in other cellular pathways. For instance, as previously discussed, TLS polymerases are required for ICL repair and can also play role in the BER and NER pathways to synthesize new DNA after the excision step. Exogenous stressors—for example UV-C, MNNG, and BPDE—regulate the transcriptional expression of POL η, POL ι, POL κ and POL ζ [Zhu et al., 2003; Yu et al., 2004; Liu and Chen, 2006; Zhu et al., 2010; Zhu et al., 2012]. Likewise, an HSP90 inhibitor reduces expression of REV1 and POL η in human cells, indicating an evolutionary regulation of these polymerases [Sekimoto et al., 2010; Pozo et al., 2011]. Interestingly, the TLS polymerases, REV1 and REV3, were also implicated in the development of chemoresistance in human cells and mice models, opening the possibility for a whole new class of promising chemotherapeutic drugs [Doles et al., 2010; Xie et al., 2010; Xu et al., 2013].
Repair of DNA breaks
Single stranded break repair (SSBR)
Single strand breaks (SSBs) are often generated from oxidative damage to the DNA, from abasic sites, or from erroneous activity of the DNA topoisomerase 1 (TOP1) enzyme [Wang, 2002; Hegde et al., 2008]. Unresolved SSBs often collapse DNA replication, stall ongoing transcription, and effect PARP1 activation, which releases cellular NAD+, ATP and apoptosis inducing factor (AIF) in cells [Zhou and Doetsch, 1993; Heeres and Hergenrother, 2007] At least two human genetic disorders, spinocerebellar ataxia with axonal neuropathy 1 (SCAN1) and ataxia-oculomotor apraxia 1 (AOA1), are associated with an abortive SSBR. These patients often manifest genetic instability and high incidence of cancers [El-Khamisy et al., 2005; Reynolds et al., 2009]. SSBR is predicted to occur through three different pathways depending on the source of SSB.
In the long patch SSBR pathway, SSBs are transiently detected by PARP1, which undergoes a rapid cycle of poly(ADP) ribosylation and dissociates to detect the next SSB [D’Amours et al., 1999; Davidovic et al., 2001]. After this, the ends undergo end processing by the apurinic-apyrimidic endonuclease 1 APE1, PNKP (polynuceotide kinase 3′-phosphate) and aprataxin (APTX) [McKinnon and Caldecott, 2007]. Next, FEN1 removes the damaged 5′ termini aided by PARP1 and PCNA, leaving behind a ssDNA gap, which is filled by POL β, in combination with POL δ/ε. The final step of ligation is carried out by the LIG1, which is dependent on the presence of PCNA and XRCC1 [Lan et al., 2004; Mortusewicz et al., 2006; McKinnon and Caldecott, 2007]. In the short patch SSBR pathway, SSBs generated during the BER are recognized by APE1 followed by a similar end-processing pathway as the long patch repair. The gap-filling step, however, is only carried out by POL β enzyme, followed by LIG3-catalyzed ligation [McKinnon and Caldecott, 2007]. Finally, the TOP1-SSB pathway is a variant of the PARP1-dependent long patch repair in which the end-processing is carried out by the TDP1 (tyrosyl-DNA phosphodiesterase 1) enzyme that removes the TOP1 from the 3′-end [Caldecott, 2008].
Double strand break repair (DSBR)
Highly toxic DSBs are induced by various chemical and physical DNA damaging agents [Pfeiffer et al., 2000]. Unresolved DSBs are implicated in various human disorders and cancers [Jackson and Bartek, 2009]. We will briefly discuss the two major pathways—homologous recombination (HR) and non-homologous end joining (NHEJ)—that organisms have evolved to resolve the DSBs. Chromatin modification is the first event that registers the presence of a DSB and triggers a cascade of events including ATM activation, targeted phosphorylation of H2AX, chromatin PARylation, MDC1 recruitment and finally recruitment of 53BP1 and BRCA1 [Rogakou et al., 1998; Rothkamm et al., 2003; Gottschalk et al., 2009; Chou et al., 2010; Lukas et al., 2011; Price and D’Andrea, 2013; Liu et al., 2014]. Interestingly, both 53BP1 and BRCA1 exhibit an antagonistic influence on each other and 53BP1 depletion rescues embryonic lethality of BRCA1 null mice [Xie et al., 2007; Cao et al., 2009; Bunting et al., 2010].
In the NHEJ pathway of DSBR, 53BP1 plays an important regulatory role by recruiting the NHEJ components to the break site, activating checkpoint signaling and facilitating synapsis of the two ends [Panier and Boulton, 2014]. The Ku (Ku70 and Ku80) heterodimer is the first to recognize and bind the DSBs within seconds to prevent end resection and serves as a scaffold to recruit other NHEJ components [Pang et al., 1997; Mari et al., 2006; Soutoglou et al., 2007; Mimitou and Symington, 2010]. Other recruited components include DNA-PKcs, XRCC4, LIG4 and XLF (XRCC4-like factor), APLF (Aprataxin-and-PNK-like factor) [and also TdT (terminal deoxynucleotidyl transferase) in lymphocytes] [Gottlieb and Jackson, 1993; Nick McElhinny et al., 2000; Costantini et al., 2007; Yano et al., 2008; Grundy et al., 2013]. Recent studies indicate that the order of recruitment of these components may depend on the complexity of DNA damage; for instance, DNA-PKcs recruitment depends on the nature of the break [Mari et al., 2006; Yano and Chen, 2008]. However, once DNA-PKcs gets recruited, it is activated in a DNA dependent manner; it pushes Ku inwardly on the DNA and then phosphorylates other nearby components, including autoautophosphorylating itself [Gottlieb and Jackson, 1993; Yoo and Dynan, 1999; Weterings and Chen, 2008]. At the same time, XRCC4 is believed to help stabilize the NHEJ complex by tethering the ends and acting as an additional scaffold with Ku to recruit other components [Malivert et al., 2010; Hammel et al., 2011; Andres et al., 2012]. Once the ends are bridged and stabilized, Artemis, PNKP, APLF, WRN, Aprataxin and Ku initiate DNA end processing, which involves removing groups that are blocking the ends and resecting the resultant naked strands [Ma et al., 2002; Bernstein et al., 2005; Ahel et al., 2006; Perry et al., 2006; Roberts et al., 2010; Li et al., 2011]. The gaps left behind after resection are filled by family X polymerases in a template-dependent (POL μ) or template-independent (POL λ) manner [Ramadan et al., 2004; Roberts et al., 2010]. LIG4 joins the ends and completes the NHEJ process [Grawunder et al., 1997].
The HR pathway consist of a set of related sub-pathways that utilize DNA strand invasion and template-directed DNA repair synthesis to effect a high-fidelity repair [Li and Heyer, 2008]. In addition to the traditional DSBR-induced HR pathway, synthesis-dependent strand annealing (SDSA) and break-induced repair (BIR) are two other variations following the HR premise [Li and Heyer, 2008]. Here, we will very briefly summarize the HR pathway of DSBR.
The MRN (MRE11-RAD50-NBS1) complex initiates HR at a DSB, where it recognizes and binds the DSB and then recruits ATM and TIP60 to the DNA [Sun et al., 2005; Stracker and Petrini, 2011]. Activated ATM (from TIP60) phosphorylates H2AX, which then serves as an anchor for MDC1 [Bhatti et al., 2011]. Next, MDC1 is phosphorylated by ATM, and the phosphorylated MDC1 functions as a scaffold to bring in the ubiquitin E3 ligases RNF8 and RNF168 [Altmeyer and Lukas, 2013b]. Both of these E3 ligases ubiquitinate H2AX, which then serves as a docking site for 53BP1 and BRCA1. In the S/G2 phase where HR is predominant, BRCA1 (recruited by ubiquitinated chromatin) successfully counteracts 53BP1 and initiates ubiquitination of the downstream component, CtIP [Yu et al., 2006; Chapman et al., 2012]. At this time, the other HR components, RPA, and RAD51 proteins make their way on to the DNA.
The next step of end resection involves a 5′-to-3′ nucleolytic degradation to generate 3′ overhangs, committing cells to the HR pathway. Initial resection occurs by the endonuclease activity of MRN, with the help of CtIP, followed by long-range resection by EXO1 or BLM together with DNA2 [Chen et al., 2008; Nimonkar et al., 2011]. Next, RPA coats the 3′ overhang, which is then displaced by RAD51, generating a nucleoprotein filament. BRCA2 and PALB2 aid in the formation of the nucleoprotein filament formation that invades a nearby duplex DNA forming a D-loop [Zhang et al., 2009; Holloman, 2011]. Several other proteins function together at this step. For the strand to invade the template DNA, RAD54 and RAD54B remove RAD51 and allow the 3′-OH group to prime synthesis by Polymerases δ, κ and ν [Mazin et al., 2010; Sebesta et al., 2013]. If the new DNA synthesis stops after a limited distance, as is the case in SDSA, the RTEL1 enzyme dissolves the D-loop [Barber et al., 2008]. Otherwise, the Holliday junction is collectively processed by the BLM-TOPOIII-RMI1-RMI2 complex, GEN1 endonuclease, the MUS81-EME1 complex and the SLX1-SLX4 complex [Chang et al., 2008; Ciccia et al., 2008; Xu et al., 2008; Fekairi et al., 2009; Rass et al., 2010].
DNA damage and telomeres
Telomeres are well-conserved nucleoprotein structures found at the end of linear chromosomes that help differentiate normal chromosomal ends from DSBs [Longhese, 2008; Shammas, 2011]. Telomeric DNA consists of tandem repetitive DNA (TTAGGG in humans), where the G-rich strand (also called the G-tail), bound by sheltrin protein POT1 (protection of telomeres 1), extends beyond the complementary C-rich strand and invades into the double-stranded telomeric DNA. The t-loop thus generated complexes with other sheltrin proteins such as TRF1 (telomeric-repeat binding factor 1), TRF2, TIN2 (TRF-interacting protein 2), the transcriptional repressor/activator protein RAP1, and the TPP1 (POT1- and TIN2- organizing protein), which together prevent the chromosomal ends from being recognized as DNA damage [Takai et al., 2003; d’Adda di Fagagna et al., 2004; Liu et al., 2004; de Lange, 2005]. In addition, telomeric DNA is replicated and maintained by a specialized ribonucleoprotein complex called telomerase [composed of a telomere RNA component (TERC) and a telomere reverse transcriptase (TERT)], which is the only positive regulator of telomere length [Bachand et al., 2001; Blasco, 2003]. A decline in telomerase activity contributes to telomere attrition, which is associated with aging, cancer and several inherited bone marrow failure (IBMF) disorders [Chang and Harley, 1995; Shay et al., 2001; Alter et al., 2015].
Deprotected telomeres elicit a DNA damage response, recruiting DSBR components that attempt to repair the exposed ends, causing deleterious nucleolytic degradation, recombination, and chromosomal fusions [Longhese, 2008]. For example, short telomeres often assemble DDR factors such as 53BP1, ATM, γH2AX, and MRE11 as foci, called Telomere Dysfunction-Induced Foci (TIF), which are highly prone to NHEJ-mediated end-to-end fusion [Takai et al., 2003; Hewitt et al., 2012; Marcand, 2014]. Surprisingly, telomere maintenance requires the presence of some of the same components of DSBR/NHEJ components—for example, Ku and the MRN complex—complicating our understanding of the exact mechanism of telomere stability [Maser and DePinho, 2004; Marcand, 2014]. Establishing the dynamics of this telomere biology is an active area of research.
Recently, several environmental toxins have been implicated in telomere shortening. For example, tobacco smoke causes telomere shortening via oxidative stress [Valdes et al., 2005; McGrath et al., 2007; Song et al., 2010; Babizhayev and Yegorov, 2011]. Obesity has also been associated with accelerated shortening of telomeres in adipose tissue of mice, where levels of ROS were high [Song et al., 2010]. Likewise, white blood cells of obese women harbor shorter telomeres than lean women [Valdes et al., 2005]. Genotoxic stressors such as certain pollutants (e.g. toluene and benzene, and PAHs) are also associated with telomere shortening [Hoxha et al., 2009; Pavanello et al., 2010; Trusina, 2014]. Finally, everyday stresses, including psychological stress, is known to shorten telomeres, whereas meditation and mindfulness may bring about an opposite effect [Epel et al., 2004; Cherkas et al., 2006; Simon et al., 2006; Epel et al., 2009; Mathur et al., 2016]. Critically shortened telomeres are associated with aging and cancer [Shammas, 2011].
Conclusion
In conclusion, DNA is continually being exposed to both endogenous and exogenous DNA damaging agents that chemically modify the DNA constituents. Unresolved DNA damages are implicated in human diseases and cancers. However, robust DNA repair and damage tolerance pathways help remove or tolerate the lesions to allow survival (Figure 4). An understanding of these pathways helps evaluate possible toxic exposures and design strategies to control deleterious consequences on human health.
Acknowledgments
The writing of this review was supported by National Institute of Environmental Health Sciences grants ES-015818 to G.C.W. and P30 ES-002109 to the MIT Center for Environmental Health Sciences. G.C.W. is an American Cancer Society Professor. Nimrat Chatterjee and Graham Walker wrote the manuscript.
Footnotes
There are no conflicts of interests.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39(1):25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- AEP . In: DNA repair and carcinogenesis by alkylating agents. CCaG PL, editor. Berlin: Springer; 1990. pp. 103–131. [Google Scholar]
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443(7112):713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- Akbari M, Pena-Diaz J, Andersen S, Liabakk NB, Otterlei M, Krokan HE. Extracts of proliferating and non-proliferating human cells display different base excision pathways and repair fidelity. DNA Repair (Amst) 2009;8(7):834–843. doi: 10.1016/j.dnarep.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7(4):605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6(6):695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat Res. 2007;618(1–2):81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Rosenberg PS. Telomere length in inherited bone marrow failure syndromes. Haematologica. 2015;100(1):49–54. doi: 10.3324/haematol.2014.114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Lukas J. Guarding against collateral damage during chromatin transactions. Cell. 2013a;153(7):1431–1434. doi: 10.1016/j.cell.2013.05.044. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Lukas J. To spread or not to spread--chromatin modifications in response to DNA damage. Curr Opin Genet Dev. 2013b;23(2):156–165. doi: 10.1016/j.gde.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Ames BN, Profet M, Gold LS. Dietary pesticides (99.99% all natural) Proc Natl Acad Sci U S A. 1990;87(19):7777–7781. doi: 10.1073/pnas.87.19.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S, Heine T, Sneve R, Konig I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26(3):547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18(16):1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012;40(4):1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Kunkel TA. Mutator phenotypes due to DNA replication infidelity. Semin Cancer Biol. 2010;20(5):304–311. doi: 10.1016/j.semcancer.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21(6):731–733. [PubMed] [Google Scholar]
- Babizhayev MA, Yegorov YE. Smoking and health: association between telomere length and factors impacting on human disease, quality of life and life span in a large population-based cohort under the effect of smoking duration. Fundam Clin Pharmacol. 2011;25(4):425–442. doi: 10.1111/j.1472-8206.2010.00866.x. [DOI] [PubMed] [Google Scholar]
- Bachand F, Triki I, Autexier C. Human telomerase RNA-protein interactions. Nucleic Acids Res. 2001;29(16):3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V, Verly WG. Possible roles of beta-elimination and delta-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem J. 1988;253(2):553–559. doi: 10.1042/bj2530553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135(2):261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch H, Montesano R. Relevance of nitrosamines to human cancer. Carcinogenesis. 1984;5(11):1381–1393. doi: 10.1093/carcin/5.11.1381. [DOI] [PubMed] [Google Scholar]
- Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43(21):10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35(5):704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16(3):497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231(1):11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, Durocher D, Weinfeld M, Glover JN. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17(5):657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol Cell Biol. 1997;17(12):6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti S, Kozlov S, Farooqi AA, Naqi A, Lavin M, Khanna KK. ATM protein kinase: the linchpin of cellular defenses to stress. Cell Mol Life Sci. 2011;68(18):2977–3006. doi: 10.1007/s00018-011-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett. 2007;252(1):93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531(1–2):37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2(5):594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA. Telomeres in cancer and aging: lessons from the mouse. Cancer Lett. 2003;194(2):183–188. doi: 10.1016/s0304-3835(02)00705-x. [DOI] [PubMed] [Google Scholar]
- Bluteau D, Masliah-Planchon J, Clairmont C, Rousseau A, Ceccaldi R, Dubois d’Enghien C, Bluteau O, Cuccuini W, Gachet S, Peffault de Latour R, Leblanc T, Socie G, Baruchel A, Stoppa-Lyonnet D, D’Andrea AD, Soulier J. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126(9):3580–3584. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Laval J. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982;64(8–9):637–641. doi: 10.1016/s0300-9084(82)80103-x. [DOI] [PubMed] [Google Scholar]
- Bose SN, Davies RJ, Sethi SK, McCloskey JA. Formation of an adenine-thymine photoadduct in the deoxydinucleoside monophosphate d(TpA) and in DNA. Science. 1983;220(4598):723–725. doi: 10.1126/science.6836308. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12(9):587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- Braithwaite E, Wu X, Wang Z. Repair of DNA lesions induced by polycyclic aromatic hydrocarbons in human cell-free extracts: involvement of two excision repair mechanisms in vitro. Carcinogenesis. 1998;19(7):1239–1246. doi: 10.1093/carcin/19.7.1239. [DOI] [PubMed] [Google Scholar]
- Branch P, Aquilina G, Bignami M, Karran P. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993;362(6421):652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18(6):1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- Broedbaek K, Poulsen HE, Weimann A, Kom GD, Schwedhelm E, Nielsen P, Boger RH. Urinary excretion of biomarkers of oxidatively damaged DNA and RNA in hereditary hemochromatosis. Free Radic Biol Med. 2009;47(8):1230–1233. doi: 10.1016/j.freeradbiomed.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Buckland RJ, Watt DL, Chittoor B, Nilsson AK, Kunkel TA, Chabes A. Increased and imbalanced dNTP pools symmetrically promote both leading and lagging strand replication infidelity. PLoS Genet. 2014;10(12):e1004846. doi: 10.1371/journal.pgen.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin AB, Jr, Huizenga BN, Nash HA. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 1995;23(15):2973–2979. doi: 10.1093/nar/23.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin HT. THREE LECTURES on CANCER of the SCROTUM in CHIMNEY-SWEEPS and OTHERS: Delivered at the Royal College of Surgeons of England. Br Med J. 1892;1(1643):1341–1346. doi: 10.1136/bmj.1.1643.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VSC . Free-radical induced DNA damage and its repair. Berlin/Heidelberg: Springer-Verlag; 2006. [Google Scholar]
- Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl radicals and DNA base damage. Mutat Res. 1999;424(1–2):9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49(1):9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Cadet J, Douki T, Ravanat JL. Measurement of oxidatively generated base damage in cellular DNA. Mutat Res. 2011;711(1–2):3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D. Oxidatively generated complex DNA damage: tandem and clustered lesions. Cancer Lett. 2012;327(1–2):5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Cadet J, Wagner JR. Oxidatively generated base damage to cellular DNA by hydroxyl radical and one-electron oxidants: similarities and differences. Arch Biochem Biophys. 2014;557:47–54. doi: 10.1016/j.abb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35(4):534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey JF, Schultz SJ, Sisson L, Fazzio TG, Champoux JJ. DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc Natl Acad Sci U S A. 2003;100(10):5640–5645. doi: 10.1073/pnas.1031537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael PL, Hewer A, Osborne MR, Strain AJ, Phillips DH. Detection of bulky DNA lesions in the liver of patients with Wilson’s disease and primary haemochromatosis. Mutat Res. 1995;326(2):235–243. doi: 10.1016/0027-5107(94)00177-7. [DOI] [PubMed] [Google Scholar]
- Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents--DNA adducts and mutations. J Natl Cancer Inst Monogr. 2000;(27):75–93. doi: 10.1093/oxfordjournals.jncimonographs.a024247. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016a;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016b;17(6):337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Chan GL, Doetsch PW, Haseltine WA. Cyclobutane pyrimidine dimers and (6-4) photoproducts block polymerization by DNA polymerase I. Biochemistry. 1985;24(21):5723–5728. doi: 10.1021/bi00342a006. [DOI] [PubMed] [Google Scholar]
- Chan K, Resnick MA, Gordenin DA. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst) 2013;12(11):878–889. doi: 10.1016/j.dnarep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Bortnick A, Murre C. AID targeting: old mysteries and new challenges. Trends Immunol. 2015;36(9):527–535. doi: 10.1016/j.it.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Harley CB. Telomere length and replicative aging in human vascular tissues. Proc Natl Acad Sci U S A. 1995;92(24):11190–11194. doi: 10.1073/pnas.92.24.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y. Crystal structure of the Mus81-Eme1 complex. Genes Dev. 2008;22(8):1093–1106. doi: 10.1101/gad.1618708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Lin Y, Santillan BA, Yotnda P, Wilson JH. Environmental stress induces trinucleotide repeat mutagenesis in human cells. Proc Natl Acad Sci U S A. 2015;112(12):3764–3769. doi: 10.1073/pnas.1421917112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Lin Y, Wilson JH. Mismatch repair enhances convergent transcription-induced cell death at trinucleotide repeats by activating ATR. DNA Repair (Amst) 2016a;42:26–32. doi: 10.1016/j.dnarep.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Lin Y, Yotnda P, Wilson JH. Environmental Stress Induces Trinucleotide Repeat Mutagenesis in Human Cells by Alt-Nonhomologous End Joining Repair. J Mol Biol. 2016b;428(15):2978–2980. doi: 10.1016/j.jmb.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, LY, Wilson JH. Fanconi anemia pathway regulates convergent transcription-induced cell death at trinucleotide repeats in human cells. PostDoc Journal. 2016;4(5):46–54. doi: 10.14304/surya.jpr.v4n5.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, SBA, Wilson JH. Microsatellite Repeats: Canaries in the Coalmine. New York: 2013. [Google Scholar]
- Chen H, Shaw BR. Kinetics of bisulfite-induced cytosine deamination in single-stranded DNA. Biochemistry. 1993;32(14):3535–3539. doi: 10.1021/bi00065a003. [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283(12):7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Huang C, Bai C, Gao H, Ma R, Liu X, Dong Q. Benzo[alpha]pyrene repressed DNA mismatch repair in human breast cancer cells. Toxicology. 2013;304:167–172. doi: 10.1016/j.tox.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992;267(1):166–172. [PubMed] [Google Scholar]
- Cherkas LF, Aviv A, Valdes AM, Hunkin JL, Gardner JP, Surdulescu GL, Kimura M, Spector TD. The effects of social status on biological aging as measured by white-blood-cell telomere length. Aging Cell. 2006;5(5):361–365. doi: 10.1111/j.1474-9726.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- Chetsanga CJ, Lozon M, Makaroff C, Savage L. Purification and characterization of Escherichia coli formamidopyrimidine-DNA glycosylase that excises damaged 7-methylguanine from deoxyribonucleic acid. Biochemistry. 1981;20(18):5201–5207. doi: 10.1021/bi00521a016. [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242(4878):564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, Wang W, West SC. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25(3):331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12(2):121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5(10):a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13(6):343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89(5):1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 2007;6(6):712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Crutzen PJ, Andreae MO. Biomass burning in the tropics: impact on atmospheric chemistry and biogeochemical cycles. Science. 1990;250(4988):1669–1678. doi: 10.1126/science.250.4988.1669. [DOI] [PubMed] [Google Scholar]
- Cui B, Johnson SP, Bullock N, Ali-Osman F, Bigner DD, Friedman HS. Bifunctional DNA alkylator 1,3-bis(2-chloroethyl)-1-nitrosourea activates the ATR-Chk1 pathway independently of the mismatch repair pathway. Mol Pharmacol. 2009;75(6):1356–1363. doi: 10.1124/mol.108.053124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12(12):801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18(15):1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362(20):1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Ischia M, Napolitano A, Manini P, Panzella L. Secondary targets of nitrite-derived reactive nitrogen species: nitrosation/nitration pathways, antioxidant defense mechanisms and toxicological implications. Chem Res Toxicol. 2011;24(12):2071–2092. doi: 10.1021/tx2003118. [DOI] [PubMed] [Google Scholar]
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268(1):7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- Davies RJ. Royal Irish Academy Medal Lecture. Ultraviolet radiation damage in DNA. Biochem Soc Trans. 1995;23(2):407–418. doi: 10.1042/bst0230407. [DOI] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19(3):169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- de Groote FH, Jansen JG, Masuda Y, Shah DM, Kamiya K, de Wind N, Siegal G. The Rev1 translesion synthesis polymerase has multiple distinct DNA binding modes. DNA Repair (Amst) 2011;10(9):915–925. doi: 10.1016/j.dnarep.2011.04.033. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Demple B, Linn S. 5,6-Saturated thymine lesions in DNA: production by ultraviolet light or hydrogen peroxide. Nucleic Acids Res. 1982;10(12):3781–3789. doi: 10.1093/nar/10.12.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita VT, Jr, Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68(21):8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- Diamant N, Hendel A, Vered I, Carell T, Reissner T, de Wind N, Geacinov N, Livneh Z. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012;40(1):170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41(6):3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias V, Junn E, Mouradian MM. The role of oxidative stress in Parkinson’s disease. J Parkinsons Dis. 2013;3(4):461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V. Tissue specificity in DNA repair: lessons from trinucleotide repeat instability. Trends Genet. 2014;30(6):220–229. doi: 10.1016/j.tig.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M, Rao G, Halliwell B, Gajewski E. Damage to the DNA bases in mammalian chromatin by hydrogen peroxide in the presence of ferric and cupric ions. Arch Biochem Biophys. 1991;285(2):317–324. doi: 10.1016/0003-9861(91)90366-q. [DOI] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010;107(48):20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douki T, Perdiz D, Grof P, Kuluncsics Z, Moustacchi E, Cadet J, Sage E. Oxidation of guanine in cellular DNA by solar UV radiation: biological role. Photochem Photobiol. 1999;70(2):184–190. [PubMed] [Google Scholar]
- Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3(11):1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Drougard C, Sarasin A, Daya-Grosjean L. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1993;90(22):10529–10533. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Zhu Q, Taylor ER, Tsay AJ, Shi LZ, Berns MW, McGowan CH. CtIP is required to initiate replication-dependent interstrand crosslink repair. PLoS Genet. 2012;8(11):e1003050. doi: 10.1371/journal.pgen.1003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel LM, Krugh TR. 2-Aminofluorene modified DNA duplex exists in two interchangeable conformations. Nat Struct Biol. 1994a;1(2):89–94. doi: 10.1038/nsb0294-89. [DOI] [PubMed] [Google Scholar]
- Eckel LM, Krugh TR. Structural characterization of two interchangeable conformations of a 2-aminofluorene-modified DNA oligomer by NMR and energy minimization. Biochemistry. 1994b;33(46):13611–13624. doi: 10.1021/bi00250a012. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Witherspoon M, Wang K, Afrasiabi K, Pham T, Birnbaumer L, Lipkin SM. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69(16):6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6(10):1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434(7029):108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6(11):638–647. doi: 10.1038/nrclinonc.2009.146. [DOI] [PubMed] [Google Scholar]
- Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40(18):8953–8964. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epe B. Genotoxicity of singlet oxygen. Chem Biol Interact. 1991;80(3):239–260. doi: 10.1016/0009-2797(91)90086-m. [DOI] [PubMed] [Google Scholar]
- Epel E, Daubenmier J, Moskowitz JT, Folkman S, Blackburn E. Can meditation slow rate of cellular aging? Cognitive stress, mindfulness, and telomeres. Ann N Y Acad Sci. 2009;1172:34–53. doi: 10.1111/j.1749-6632.2009.04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci U S A. 2004;101(49):17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppink B, Tafel AA, Hanada K, van Drunen E, Hickson ID, Essers J, Kanaar R. The response of mammalian cells to UV-light reveals Rad54-dependent and independent pathways of homologous recombination. DNA Repair (Amst) 2011;10(11):1095–1105. doi: 10.1016/j.dnarep.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Friedberg Errol C, GCW, Siede Wolfram, Wood Richard D, Schultz Roger A, Ellenberger Tom. DNA Repair and Mutagenesis. 2. ASM Press; 2005. p. 1118. [Google Scholar]
- Essigmann JM, Croy RG, Nadzan AM, Busby WF, Jr, Reinhold VN, Buchi G, Wogan GN. Structural identification of the major DNA adduct formed by aflatoxin B1 in vitro. Proc Natl Acad Sci U S A. 1977;74(5):1870–1874. doi: 10.1073/pnas.74.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10(7):722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Klungland A, Alseth I. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience. 2007;145(4):1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Fang Q, Noronha AM, Murphy SP, Wilds CJ, Tubbs JL, Tainer JA, Chowdhury G, Guengerich FP, Pegg AE. Repair of O6-G-alkyl-O6-G interstrand cross-links by human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2008;47(41):10892–10903. doi: 10.1021/bi8008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138(1):78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers L, Ohnishi ST, Penning TM. DNA strand scission by polycyclic aromatic hydrocarbon o-quinones: role of reactive oxygen species, Cu(II)/Cu(I) redox cycling, and o-semiquinone anion radicals. Biochemistry. 1997;36(28):8640–8648. doi: 10.1021/bi970367p. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23(4):471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol Biol Evol. 2000;17(9):1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- Fu PP. Metabolism of nitro-polycyclic aromatic hydrocarbons. Drug Metab Rev. 1990;22(2–3):209–268. doi: 10.3109/03602539009041085. [DOI] [PubMed] [Google Scholar]
- Fujiki H. Gist of Dr. Katsusaburo Yamagiwa’s papers entitled “Experimental study on the pathogenesis of epithelial tumors” (I to VI reports) Cancer Sci. 2014;105(2):143–149. doi: 10.1111/cas.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafter-Gvili A, Zingerman B, Rozen-Zvi B, Ori Y, Green H, Lubin I, Malachi T, Gafter U, Herman-Edelstein M. Oxidative stress-induced DNA damage and repair in human peripheral blood mononuclear cells: protective role of hemoglobin. PLoS One. 2013;8(7):e68341. doi: 10.1371/journal.pone.0068341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiegue-Zouitina S, Bailleul B, Ginot YM, Perly B, Vigny P, Loucheux-Lefebvre MH. N2-guanyl and N6-adenyl arylation of chicken erythrocyte DNA by the ultimate carcinogen of 4-nitroquinoline 1-oxide. Cancer Res. 1986;46(4 Pt 1):1858–1863. [PubMed] [Google Scholar]
- Galiegue-Zouitina S, Bailleul B, Loucheux-Lefebvre MH. Adducts from in vivo action of the carcinogen 4-hydroxyaminoquinoline 1-oxide in rats and from in vitro reaction of 4-acetoxyaminoquinoline 1-oxide with DNA and polynucleotides. Cancer Res. 1985;45(2):520–525. [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008a;105(42):16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008b;29(1):141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Geacintov NE. Is intercalation a critical factor in the covalent binding of mutagenic and tumorigenic polycyclic aromatic diol epoxides to DNA? Carcinogenesis. 1986;7(5):759–766. doi: 10.1093/carcin/7.5.759. [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12(5):1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Gentil A, Le Page F, Margot A, Lawrence CW, Borden A, Sarasin A. Mutagenicity of a unique thymine-thymine dimer or thymine-thymine pyrimidine pyrimidone (6-4) photoproduct in mammalian cells. Nucleic Acids Res. 1996;24(10):1837–1840. doi: 10.1093/nar/24.10.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O’Rahilly S, Schofield CJ. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2(3):107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107(9):1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloni L, Takeshita M, Johnson F, Iden C, Grollman AP. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981;256(16):8608–8615. [PubMed] [Google Scholar]
- Goff SP. Death by deamination: a novel host restriction system for HIV-1. Cell. 2003;114(3):281–283. doi: 10.1016/s0092-8674(03)00602-0. [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72(1):131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106(33):13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graslund A, Jernstrom B. DNA-carcinogen interaction: covalent DNA-adducts of benzo(a)pyrene 7,8-dihydrodiol 9,10-epoxides studied by biochemical and biophysical techniques. Q Rev Biophys. 1989;22(1):1–37. [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388(6641):492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Gregory CD, Milner AE. Regulation of cell survival in Burkitt lymphoma: implications from studies of apoptosis following cold-shock treatment. Int J Cancer. 1994;57(3):419–426. doi: 10.1002/ijc.2910570321. [DOI] [PubMed] [Google Scholar]
- Grundy GJ, Rulten SL, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, Oliver A, Caldecott KW. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32(1):112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zhang Y, Yuan F, Gao Y, Gu L, Wong I, Li GM. Regulation of replication protein A functions in DNA mismatch repair by phosphorylation. J Biol Chem. 2006;281(31):21607–21616. doi: 10.1074/jbc.M603504200. [DOI] [PubMed] [Google Scholar]
- Hafstad AD, Nabeebaccus AA, Shah AM. Novel aspects of ROS signalling in heart failure. Basic Res Cardiol. 2013;108(4):359. doi: 10.1007/s00395-013-0359-8. [DOI] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem. 2011;286(37):32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammons GJ, Milton D, Stepps K, Guengerich FP, Tukey RH, Kadlubar FF. Metabolism of carcinogenic heterocyclic and aromatic amines by recombinant human cytochrome P450 enzymes. Carcinogenesis. 1997;18(4):851–854. doi: 10.1093/carcin/18.4.851. [DOI] [PubMed] [Google Scholar]
- Han HJ, Tan NH, Zeng GZ, Fan JT, Huang HQ, Ji CJ, Jia RR, Zhao QS, Zhang YJ, Hao XJ, Wang LQ. Natural inhibitors of DNA topoisomerase I with cytotoxicities. Chem Biodivers. 2008;5(7):1364–1368. doi: 10.1002/cbdv.200890124. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Harvey RG. Polycyclic Aromatic Hydrocarbons: Chemistry and Carcinogenicity. Cambridge University Press; 1991. [Google Scholar]
- Hawkins BL, Heniford BW, Ackermann DM, Leonberger M, Martinez SA, Hendler FJ. 4NQO carcinogenesis: a mouse model of oral cavity squamous cell carcinoma. Head Neck. 1994;16(5):424–432. doi: 10.1002/hed.2880160506. [DOI] [PubMed] [Google Scholar]
- Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424(1–2):127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11(6):644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Heflich RH, Neft RE. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat Res. 1994;318(2):73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle ES, Linn S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem. 1997;272(31):19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- Henner WD, Grunberg SM, Haseltine WA. Sites and structure of gamma radiation-induced DNA strand breaks. J Biol Chem. 1982;257(19):11750–11754. [PubMed] [Google Scholar]
- Henner WD, Grunberg SM, Haseltine WA. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983a;258(24):15198–15205. [PubMed] [Google Scholar]
- Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma Ray induced deoxyribonucleic acid strand breaks. 3′ Glycolate termini. J Biol Chem. 1983b;258(2):711–713. [PubMed] [Google Scholar]
- Herrmann SS, Granby K, Duedahl-Olesen L. Formation and mitigation of N-nitrosamines in nitrite preserved cooked sausages. Food Chem. 2015;174:516–526. doi: 10.1016/j.foodchem.2014.11.101. [DOI] [PubMed] [Google Scholar]
- Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun. 2012;3:708. doi: 10.1038/ncomms1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM, Roy D. Sex hormone-induced nuclear DNA damage and lipid peroxidation in the dorsolateral prostates of Noble rats. Cancer Lett. 1994;84(2):155–162. doi: 10.1016/0304-3835(94)90370-0. [DOI] [PubMed] [Google Scholar]
- Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Scharer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39(17):7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Ho T. Gene silencing and endogenous DNA methylation in mammalian cells. Mutat Res. 1998;400(1–2):361–368. doi: 10.1016/s0027-5107(98)00034-7. [DOI] [PubMed] [Google Scholar]
- Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18(7):748–754. doi: 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House NC, Koch MR, Freudenreich CH. Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet. 2014;5:296. doi: 10.3389/fgene.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SM, Yanez DA, Stark JM. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11(1):e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxha M, Dioni L, Bonzini M, Pesatori AC, Fustinoni S, Cavallo D, Carugno M, Albetti B, Marinelli B, Schwartz J, Bertazzi PA, Baccarelli A. Association between leukocyte telomere shortening and exposure to traffic pollution: a cross-sectional study on traffic officers and indoor office workers. Environ Health. 2009;8:41. doi: 10.1186/1476-069X-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Hsu DS, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Natl Acad Sci U S A. 1994;91(25):12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39(2):259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L, Jr, Lin Y, Dion V, Wilson JH. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum Mol Genet. 2011;20(24):4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff AC, Topal MD. DNA damage at thymine N-3 abolishes base-pairing capacity during DNA synthesis. J Biol Chem. 1987;262(26):12843–12850. [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577(1–2):55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Acid Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- Iliakis G. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. Bioessays. 1991;13(12):641–648. doi: 10.1002/bies.950131204. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240(4852):640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LH, Haracska L, de Wind N. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol. 2009a;29(11):3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst) 2009b;8(12):1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jha V, Bian C, Xing G, Ling H. Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase kappa. Nucleic Acids Res. 2016;44(10):4957–4967. doi: 10.1093/nar/gkw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem. 1999;274(34):24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6(8):1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kanojia D, Vaidya MM. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. 2006;42(7):655–667. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Kantidze OL, Velichko AK, Luzhin AV, Razin SV. Heat Stress-Induced DNA Damage. Acta Naturae. 2016;8(2):75–78. [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12(4):2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasiotis KM, Kyriakopoulou K, Emmanouil C, Tsantila N, Liesivuori J, Souki H, Manakis S, Machera K. Monitoring of systemic exposure to plant protection products and DNA damage in orchard workers. Toxicol Lett. 2012;210(2):182–188. doi: 10.1016/j.toxlet.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci U S A. 1993;90(14):6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Jr, Todo T, Ayaki H, Ishizaki K, Morita T, Mitra S, Ikenaga M. Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental mammals. Nucleic Acids Res. 1994;22(20):4119–4124. doi: 10.1093/nar/22.20.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012;122(11):3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- KIEFER JR. In: Effects of Ultraviolet Radiation on DNA. Günter Obe V, editor. Berlin; New York: Springer; 2007. [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111(10):5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases eta and iota. Chem Res Toxicol. 2012;25(3):755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K, Tada M, Kasai H, Nishimura S, Kawazoe Y. Formation of 8-hydroxyguanine residues in cellular DNA exposed to the carcinogen 4-nitroquinoline 1-oxide. Biochem Biophys Res Commun. 1986;139(2):626–632. doi: 10.1016/s0006-291x(86)80036-5. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Hadden MK. Targeting the Translesion Synthesis Pathway for the Development of Anti-Cancer Chemotherapeutics. J Med Chem. 2016 doi: 10.1021/acs.jmedchem.6b00596. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447(7143):447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek E. Fifty years of research on N-acetyl-2-aminofluorene, one of the most versatile compounds in experimental cancer research. J Cancer Res Clin Oncol. 1992;118(7):481–489. doi: 10.1007/BF01225261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39(4):1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Joshi PC, Sharma ND, Bose SN, Jeremy R, Davies H, Takeda N, McCloskey JA. Adenine photodimerization in deoxyadenylate sequences: elucidation of the mechanism through structural studies of a major d(ApA) photoproduct. Nucleic Acids Res. 1991;19(11):2841–2847. doi: 10.1093/nar/19.11.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279(17):16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Balancing eukaryotic replication asymmetry with replication fidelity. Curr Opin Chem Biol. 2011;15(5):620–626. doi: 10.1016/j.cbpa.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4(1):48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E, Tyrrell RM. Artificial background and induced levels of oxidative base damage in DNA from human cells. Carcinogenesis. 1997;18(11):2281–2283. doi: 10.1093/carcin/18.11.2281. [DOI] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(38):13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol Cell Biol. 2007;27(15):5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150(1–2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J Biol Chem. 2003;278(1):14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- Lawley PD. Effects of some chemical mutagens and carcinogens on nucleic acids. Prog Nucleic Acid Res Mol Biol. 1966;5:89–131. doi: 10.1016/s0079-6603(08)60232-9. [DOI] [PubMed] [Google Scholar]
- Lee KH, Lee JS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Lee JH. Promoter methylation status of hMLH1, hMSH2, and MGMT genes in colorectal cancer associated with adenoma-carcinoma sequence. Langenbecks Arch Surg. 2011;396(7):1017–1026. doi: 10.1007/s00423-011-0812-9. [DOI] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153(3):590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM. New insights and challenges in mismatch repair: getting over the chromatin hurdle. DNA Repair (Amst) 2014;19:48–54. doi: 10.1016/j.dnarep.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci U S A. 1995;92(6):1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, Lieber MR. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem. 2011;286(42):36368–36377. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13(2):179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424(6952):1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44(5):479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, Collins FS. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24(1):27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- Liu C, Srihari S, Cao KA, Chenevix-Trench G, Simpson PT, Ragan MA, Khanna KK. A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy. Nucleic Acids Res. 2014;42(10):6106–6127. doi: 10.1093/nar/gku284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O’Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279(49):51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26(4):1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51(5):417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9(8):594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Loechler EL. A violation of the Swain-Scott principle, and not SN1 versus SN2 reaction mechanisms, explains why carcinogenic alkylating agents can form different proportions of adducts at oxygen versus nitrogen in DNA. Chem Res Toxicol. 1994;7(3):277–280. doi: 10.1021/tx00039a001. [DOI] [PubMed] [Google Scholar]
- Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol (R Coll Radiol) 2013;25(10):578–585. doi: 10.1016/j.clon.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25(4):601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333(6038):84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22(2):125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C, Ramos L, Bulacio L, Ramadan S, Rodriguez F. Aflatoxin B1 content in patients with hepatic diseases. Medicina (B Aires) 2002;62(4):313–316. [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Luch A. On the impact of the molecule structure in chemical carcinogenesis. EXS. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- Luczaj W, Skrzydlewska E. DNA damage caused by lipid peroxidation products. Cell Mol Biol Lett. 2003;8(2):391–413. [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13(10):1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Luoto KR, Kumareswaran R, Bristow RG. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013;4(1):5. doi: 10.1186/2041-9414-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108(6):781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Mah MC, Maher VM, Thomas H, Reid TM, King CM, McCormick JJ. Mutations induced by aminofluorene-DNA adducts during replication in human cells. Carcinogenesis. 1989;10(12):2321–2328. doi: 10.1093/carcin/10.12.2321. [DOI] [PubMed] [Google Scholar]
- Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, Callebaut I, de Villartay JP. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J Biol Chem. 2010;285(34):26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malle E, Furtmuller PG, Sattler W, Obinger C. Myeloperoxidase: a target for new drug development? Br J Pharmacol. 2007;152(6):838–854. doi: 10.1038/sj.bjp.0707358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamur S, Yuzbasioglu D, Unal F, Yilmaz S. Does potassium sorbate induce genotoxic or mutagenic effects in lymphocytes? Toxicol In Vitro. 2010;24(3):790–794. doi: 10.1016/j.tiv.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Marcand S. How do telomeres and NHEJ coexist? Mol Cell Oncol. 2014;1(3):e963438. doi: 10.4161/23723548.2014.963438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison GP, Butt A, Pearson SJ, Wharton S, Watson AJ, Marriott A, Caetano CM, Hollins JJ, Rukazenkova N, Begum G, Santibanez-Koref MF. Alkyltransferase-like proteins. DNA Repair (Amst) 2007;6(8):1222–1228. doi: 10.1016/j.dnarep.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103(49):18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci U S A. 2006;103(46):17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21(3):361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15(7):465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci U S A. 2006;103(26):9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, DePinho RA. Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amst) 2004;3(8–9):979–988. doi: 10.1016/j.dnarep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13(8):1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates JM, Perez-Gomez C, Nunez de Castro I. Antioxidant enzymes and human diseases. Clin Biochem. 1999;32(8):595–603. doi: 10.1016/s0009-9120(99)00075-2. [DOI] [PubMed] [Google Scholar]
- Mates JM, Sanchez-Jimenez F. Antioxidant enzymes and their implications in pathophysiologic processes. Front Biosci. 1999;4:D339–345. doi: 10.2741/mates. [DOI] [PubMed] [Google Scholar]
- Mathur MB, Epel E, Kind S, Desai M, Parks CG, Sandler DP, Khazeni N. Perceived stress and telomere length: A systematic review, meta-analysis, and methodologic considerations for advancing the field. Brain Behav Immun. 2016;54:158–169. doi: 10.1016/j.bbi.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. Rad54, the motor of homologous recombination. DNA Repair (Amst) 2010;9(3):286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Kunkel TA. Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle. 2004;3(5):580–583. [PubMed] [Google Scholar]
- McGrath M, Wong JY, Michaud D, Hunter DJ, De Vivo I. Telomere length, cigarette smoking, and bladder cancer risk in men and women. Cancer Epidemiol Biomarkers Prev. 2007;16(4):815–819. doi: 10.1158/1055-9965.EPI-06-0961. [DOI] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Meeker JD, Calafat AM, Hauser R. Urinary bisphenol A concentrations in relation to serum thyroid and reproductive hormone levels in men from an infertility clinic. Environ Sci Technol. 2010a;44(4):1458–1463. doi: 10.1021/es9028292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker JD, Ehrlich S, Toth TL, Wright DL, Calafat AM, Trisini AT, Ye X, Hauser R. Semen quality and sperm DNA damage in relation to urinary bisphenol A among men from an infertility clinic. Reprod Toxicol. 2010b;30(4):532–539. doi: 10.1016/j.reprotox.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker JD, Yang T, Ye X, Calafat AM, Hauser R. Urinary concentrations of parabens and serum hormone levels, semen quality parameters, and sperm DNA damage. Environ Health Perspect. 2011;119(2):252–257. doi: 10.1289/ehp.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng LH, Liao ZY, Pommier Y. Non-camptothecin DNA topoisomerase I inhibitors in cancer therapy. Curr Top Med Chem. 2003;3(3):305–320. doi: 10.2174/1568026033452546. [DOI] [PubMed] [Google Scholar]
- Mi J, Kupfer GM. The Fanconi anemia core complex associates with chromatin during S phase. Blood. 2005;105(2):759–766. doi: 10.1182/blood-2004-01-0001. [DOI] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016;35(9):909–923. doi: 10.15252/embj.201693860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23(9):3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29(19):3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283(25):17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10(4):243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DL, Jen J, Cleaver JE. Relative induction of cyclobutane dimers and cytosine photohydrates in DNA irradiated in vitro and in vivo with ultraviolet-C and ultraviolet-B light. Photochem Photobiol. 1991;54(5):741–746. doi: 10.1111/j.1751-1097.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49(6):805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27(1):155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsenzadegan M, Mirshafiey A. The immunopathogenic role of reactive oxygen species in Alzheimer disease. Iran J Allergy Asthma Immunol. 2012;11(3):203–216. [PubMed] [Google Scholar]
- Mokarram P, Zamani M, Kavousipour S, Naghibalhossaini F, Irajie C, Moradi Sarabi M, Hosseini SV. Different patterns of DNA methylation of the two distinct O6-methylguanine-DNA methyltransferase (O6-MGMT) promoter regions in colorectal cancer. Mol Biol Rep. 2013;40(5):3851–3857. doi: 10.1007/s11033-012-2465-3. [DOI] [PubMed] [Google Scholar]
- Mollersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveteras L, Bjolgerud A, Wilson DM, 3rd, Bjoras M, Klungland A. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet. 2012;21(22):4939–4947. doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34(12):3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27(2):311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Moyer R, Briley D, Johnsen A, Stewart U, Shaw BR. Echinomycin, a bis-intercalating agent, induces C-->T mutations via cytosine deamination. Mutat Res. 1993;288(2):291–300. doi: 10.1016/0027-5107(93)90097-y. [DOI] [PubMed] [Google Scholar]
- Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20(7):2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;40(19):9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DL, Donigan KA, Jaeger J, Sweasy JB. The E288K colon tumor variant of DNA polymerase beta is a sequence specific mutator. Biochemistry. 2012;51(26):5269–5275. doi: 10.1021/bi3003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel CS, Manning SA, Kohli RM. The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem Biol. 2012;7(1):20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli H. DNA structure: Inherent instability and genomic reactions. Springer; 1997. [Google Scholar]
- Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E, Nishiyama M. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene. 2008;27(30):4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- Nelles JL, Hu WY, Prins GS. Estrogen action and prostate cancer. Expert Rev Endocrinol Metab. 2011;6(3):437–451. doi: 10.1586/eem.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutelings T, Lambert CA, Nusgens BV, Colige AC. Effects of mild cold shock (25 degrees C) followed by warming up at 37 degrees C on the cellular stress response. PLoS One. 2013;8(7):e69687. doi: 10.1371/journal.pone.0069687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Riou L, Aoufouchi S, Rosselli F. Fanca deficiency reduces A/T transitions in somatic hypermutation and alters class switch recombination junctions in mouse B cells. J Exp Med. 2014;211(6):1011–1018. doi: 10.1084/jem.20131637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20(9):2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371(6492):75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ. The Fanconi anemia signalosome anchor. Mol Cell. 2007;25(4):487–490. doi: 10.1016/j.molcel.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol. 2005;25(13):5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter LM, Ngo EO, Abul-Hajj YJ. Characterization of DNA damage induced by 3,4-estrone-o-quinone in human cells. J Biol Chem. 1991;266(25):16380–16386. [PubMed] [Google Scholar]
- Nutter LM, Wu YY, Ngo EO, Sierra EE, Gutierrez PL, Abul-Hajj YJ. An o-quinone form of estrogen produces free radicals in human breast cancer cells: correlation with DNA damage. Chem Res Toxicol. 1994;7(1):23–28. doi: 10.1021/tx00037a004. [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Boiteux S, Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988;16(13):5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, Macpherson P, Xu YZ, Karran P. The cytotoxicity of DNA carboxymethylation and methylation by the model carboxymethylating agent azaserine in human cells. Carcinogenesis. 1999;20(9):1855–1862. doi: 10.1093/carcin/20.9.1855. [DOI] [PubMed] [Google Scholar]
- Obe G, Johannes C, Schulte-Frohlinde D. DNA double-strand breaks induced by sparsely ionizing radiation and endonucleases as critical lesions for cell death, chromosomal aberrations, mutations and oncogenic transformation. Mutagenesis. 1992;7(1):3–12. doi: 10.1093/mutage/7.1.3. [DOI] [PubMed] [Google Scholar]
- Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J Cell Physiol. 2013;228(2):258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8(6):640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Oishi S. Effects of butyl paraben on the male reproductive system in mice. Arch Toxicol. 2002;76(7):423–429. doi: 10.1007/s00204-002-0360-8. [DOI] [PubMed] [Google Scholar]
- Omar Desoukya ND, Zhoub Guangming. Targeted and non-targeted effects of ionizing radiation. Journal of Radiation Research and Applied Sciences. 2015;8(2):247–254. [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21(58):8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- Pandir D. DNA damage in human germ cell exposed to the some food additives in vitro. Cytotechnology. 2016;68(4):725–733. doi: 10.1007/s10616-014-9824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang D, Yoo S, Dynan WS, Jung M, Dritschilo A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57(8):1412–1415. [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263(5153):1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Pavanello S, Pesatori AC, Dioni L, Hoxha M, Bollati V, Siwinska E, Mielzynska D, Bolognesi C, Bertazzi PA, Baccarelli A. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis. 2010;31(2):216–221. doi: 10.1093/carcin/bgp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- Peak MJ, Peak JG. DNA-to-protein crosslinks and backbone breaks caused by far- and near-ultraviolet, and visible light radiations in mammalian cells. Basic Life Sci. 1986;38:193–202. doi: 10.1007/978-1-4615-9462-8_20. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem Res Toxicol. 2011;24(5):618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P. DNA mismatch repair and cancer. Mutat Res. 2001;488(1):77–85. doi: 10.1016/s1383-5742(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Penning TM, Burczynski ME, Hung CF, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem Res Toxicol. 1999;12(1):1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- Perrone S, Lotti F, Geronzi U, Guidoni E, Longini M, Buonocore G. Oxidative Stress in Cancer-Prone Genetic Diseases in Pediatric Age: The Role of Mitochondrial Dysfunction. Oxid Med Cell Longev. 2016;2016:4782426. doi: 10.1155/2016/4782426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13(5):414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571(1–2):19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, Goedecke W, Obe G. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis. 2000;15(4):289–302. doi: 10.1093/mutage/15.4.289. [DOI] [PubMed] [Google Scholar]
- Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303(5917):468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- Plastaras JP, Riggins JN, Otteneder M, Marnett LJ. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: base propenals, malondialdehyde, and N(epsilon)-oxopropenyllysine. Chem Res Toxicol. 2000;13(12):1235–1242. doi: 10.1021/tx0001631. [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Cherfils J. Interfacial inhibition of macromolecular interactions: nature’s paradigm for drug discovery. Trends Pharmacol Sci. 2005;26(3):138–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr Med Chem Anticancer Agents. 2005;5(4):421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- Potenski CJ, Klein HL. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42(16):10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- Pozo FM, Oda T, Sekimoto T, Murakumo Y, Masutani C, Hanaoka F, Yamashita T. Molecular chaperone Hsp90 regulates REV1-mediated mutagenesis. Mol Cell Biol. 2011;31(16):3396–3409. doi: 10.1128/MCB.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Doublie S. Base Excision Repair in the Mitochondria. J Cell Biochem. 2015;116(8):1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Singer B, Loeb LA. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986;83(22):8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Singer B, Loeb LA. Comparison of the relative mutagenicities of O-alkylthymines site-specifically incorporated into phi X174 DNA. J Biol Chem. 1987;262(28):13821–13827. [PubMed] [Google Scholar]
- Price A. The repair of ionising radiation-induced damage to DNA. Semin Cancer Biol. 1993;4(2):61–71. [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152(6):1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustovalova Y, Magalhaes MT, D’Souza S, Rizzo AA, Korza G, Walker GC, Korzhnev DM. Interaction between the Rev1 C-Terminal Domain and the PolD3 Subunit of Polzeta Suggests a Mechanism of Polymerase Exchange upon Rev1/Polzeta-Dependent Translesion Synthesis. Biochemistry. 2016;55(13):2043–2053. doi: 10.1021/acs.biochem.5b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, Sakato M, Sacho EJ, Wilkins H, Zhang X, Modrich P, Hingorani MM, Erie DA, Weninger KR. MutL traps MutS at a DNA mismatch. Proc Natl Acad Sci U S A. 2015;112(35):10914–10919. doi: 10.1073/pnas.1505655112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A, Martins DJ, Vessoni AT, Biard D, Sarasin A, Stary A, Menck CF. Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res. 2016;44(12):5717–5731. doi: 10.1093/nar/gkw280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RS . Chromosome damage and repair. New York: Plenum press; 1981. [Google Scholar]
- Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu and terminal deoxyribonucleotidyl transferase. J Mol Biol. 2004;339(2):395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- RamaKrishna NV, Devanesan PD, Rogan EG, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Mechanism of metabolic activation of the potent carcinogen 7,12-dimethylbenz[a]anthracene. Chem Res Toxicol. 1992;5(2):220–226. doi: 10.1021/tx00026a011. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Compton SA, Matos J, Singleton MR, Ip SC, Blanco MG, Griffith JD, West SC. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010;24(14):1559–1569. doi: 10.1101/gad.585310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids. 2010;2010:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanat JL, Cadet J, Douki T. Oxidatively generated DNA lesions as potential biomarkers of in vivo oxidative stress. Curr Mol Med. 2012;12(6):655–671. doi: 10.2174/156652412800792651. [DOI] [PubMed] [Google Scholar]
- Renaud E, Barascu A, Rosselli F. Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. Nucleic Acids Res. 2016;44(2):648–656. doi: 10.1093/nar/gkv1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29(5):1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P, Cooper S, Lomax M, O’Neill P. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 2015;43(8):4028–4038. doi: 10.1093/nar/gkv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65(1):27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464(7292):1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31(11):2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogan EG, Devanesan PD, RamaKrishna NV, Higginbotham S, Padmavathi NS, Chapman K, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Identification and quantitation of benzo[a]pyrene-DNA adducts formed in mouse skin. Chem Res Toxicol. 1993;6(3):356–363. doi: 10.1021/tx00033a017. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23(16):5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, Helleday T, Teraoka H, Arakawa H, Buerstedde JM, Takeda S. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol Cell Biol. 2007;27(7):2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachadyn P. Conservation and diversity of MutS proteins. Mutat Res. 2010;694(1–2):20–30. doi: 10.1016/j.mrfmmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Saffhill R. In vitro miscoding of alkylthymines with DNA and RNA polymerases. Chem Biol Interact. 1985;53(1–2):121–130. doi: 10.1016/s0009-2797(85)80090-9. [DOI] [PubMed] [Google Scholar]
- Sakumi K, Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990;236(2–3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(3):a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Batters C, Edmunds CE, Phillips LG, Simpson LJ, Szuts D. Timing matters: error-prone gap filling and translesion synthesis in immunoglobulin gene hypermutation. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):595–603. doi: 10.1098/rstb.2008.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi: 10.1186/1471-2091-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(10):a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoket B. DNA damage in humans exposed to environmental and dietary polycyclic aromatic hydrocarbons. Mutat Res. 1999;424(1–2):143–153. doi: 10.1016/s0027-5107(99)00015-9. [DOI] [PubMed] [Google Scholar]
- Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, Marteijn JA. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44(5):598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135(7):1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, Lee MY, Haracska L, Krejci L. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair (Amst) 2013;12(9):691–698. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Oda T, Pozo FM, Murakumo Y, Masutani C, Hanaoka F, Yamashita T. The molecular chaperone Hsp90 regulates accumulation of DNA polymerase eta at replication stalling sites in UV-irradiated cells. Mol Cell. 2010;37(1):79–89. doi: 10.1016/j.molcel.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28(4):383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA. Telomeres, lifestyle, cancer, and aging. Curr Opin Clin Nutr Metab Care. 2011;14(1):28–34. doi: 10.1097/MCO.0b013e32834121b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Hum Mol Genet. 2001;10(7):677–685. doi: 10.1093/hmg/10.7.677. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40(12):3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- Simon NM, Smoller JW, McNamara KL, Maser RS, Zalta AK, Pollack MH, Nierenberg AA, Fava M, Wong KK. Telomere shortening and mood disorders: preliminary support for a chronic stress model of accelerated aging. Biol Psychiatry. 2006;60(5):432–435. doi: 10.1016/j.biopsych.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Singer B. O-alkyl pyrimidines in mutagenesis and carcinogenesis: occurrence and significance. Cancer Res. 1986;46(10):4879–4885. [PubMed] [Google Scholar]
- Singer B, Chavez F, Goodman MF, Essigmann JM, Dosanjh MK. Effect of 3′ flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc Natl Acad Sci U S A. 1989;86(21):8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Kusmierek JT. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Skipper PL, Kim MY, Sun HL, Wogan GN, Tannenbaum SR. Monocyclic aromatic amines as potential human carcinogens: old is new again. Carcinogenesis. 2010;31(1):50–58. doi: 10.1093/carcin/bgp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, McGregor Mason T, Noronha AM, Wilds CJ, Miller PS. Distortion-dependent unhooking of interstrand cross-links in mammalian cell extracts. Biochemistry. 2008;47(37):9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22(4):535–545. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, von Figura G, Liu Y, Kraus JM, Torrice C, Dillon P, Rudolph-Watabe M, Ju Z, Kestler HA, Sanoff H, Lenhard Rudolph K. Lifestyle impacts on the aging-associated expression of biomarkers of DNA damage and telomere dysfunction in human blood. Aging Cell. 2010;9(4):607–615. doi: 10.1111/j.1474-9726.2010.00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9(6):675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc Natl Acad Sci U S A. 2002;99(24):15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3(8):998–1001. [PubMed] [Google Scholar]
- Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279(5356):1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12(2):90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strniste GF, Martinez E, Martinez AM, Brake RJ. Photo-induced reactions of benzo(a)pyrene with DNA in vitro. Cancer Res. 1980;40(2):245–252. [PubMed] [Google Scholar]
- Sugimura T. Past, present, and future of mutagens in cooked foods. Environ Health Perspect. 1986;67:5–10. doi: 10.1289/ehp.86675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102(37):13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14(12):2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166(5):693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tian F, Tong TJ, Zhang ZY, McNutt MA, Liu XW. Age-dependent down-regulation of mitochondrial 8-oxoguanine DNA glycosylase in SAM-P/8 mouse brain and its effect on brain aging. Rejuvenation Res. 2009;12(3):209–215. doi: 10.1089/rej.2009.0849. [DOI] [PubMed] [Google Scholar]
- Tomida J, Itaya A, Shigechi T, Unno J, Uchida E, Ikura M, Masuda Y, Matsuda S, Adachi J, Kobayashi M, Meetei AR, Maehara Y, Yamamoto K, Kamiya K, Matsuura A, Matsuda T, Ikura T, Ishiai M, Takata M. A novel interplay between the Fanconi anemia core complex and ATR-ATRIP kinase during DNA cross-link repair. Nucleic Acids Res. 2013;41(14):6930–6941. doi: 10.1093/nar/gkt467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker AR, Preussmann R. Carcinogenic N-nitrosamines in the diet: occurrence, formation, mechanisms and carcinogenic potential. Mutat Res. 1991;259(3–4):277–289. doi: 10.1016/0165-1218(91)90123-4. [DOI] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8(2):417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- Tropp BE. Molecular biology, From gene to protein. 4. Jones & Bartlett Learning; 2011. p. 1100. [Google Scholar]
- Trusina A. Stress induced telomere shortening: longer life with less mutations? BMC Syst Biol. 2014;8:27. doi: 10.1186/1752-0509-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459(7248):808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6(8):1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudek B, Boiteux S, Laval J. Biological properties of imidazole ring-opened N7-methylguanine in M13mp18 phage DNA. Nucleic Acids Res. 1992;20(12):3079–3084. doi: 10.1093/nar/20.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87(1):65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, Aviv A, Spector TD. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366(9486):662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6(10):757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J Biol Chem. 2001;276(12):9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- Varghese AJ. Photochemistry of nucleic acids and their constituents. Photophysiology. 1972;(7):207–274. [PubMed] [Google Scholar]
- Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol. 2013;5(8):a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertessy BG, Toth J. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc Chem Res. 2009;42(1):97–106. doi: 10.1021/ar800114w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignard J, Mirey G, Salles B. Ionizing-radiation induced DNA double-strand breaks: a direct and indirect lighting up. Radiother Oncol. 2013;108(3):362–369. doi: 10.1016/j.radonc.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20(10):2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti R, Grieco D. New insights on oxidative stress in cancer. Curr Opin Drug Discov Devel. 2009;12(2):240–245. [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8(1):213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Vooradi V, Romano LJ. Effect of N-2-acetylaminofluorene and 2-aminofluorene adducts on DNA binding and synthesis by yeast DNA polymerase eta. Biochemistry. 2009;48(19):4209–4216. doi: 10.1021/bi9000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J Biol Chem. 2002;277(3):1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- Wang AT, Sengerova B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, Gileadi O, Hartley JA, McHugh PJ. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25(17):1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang L, Patel U, Ghosh L, Banerjee S. DNA polymerase beta mutations in human colorectal cancer. Cancer Res. 1992;52(17):4824–4827. [PubMed] [Google Scholar]
- Wang W. A major switch for the Fanconi anemia DNA damage-response pathway. Nat Struct Mol Biol. 2008;15(11):1128–1130. doi: 10.1038/nsmb1108-1128. [DOI] [PubMed] [Google Scholar]
- Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007a;27(8):3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007b;67(7):3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- Wardman P. The importance of radiation chemistry to radiation and free radical biology (The 2008 Silvanus Thompson Memorial Lecture) Br J Radiol. 2009;82(974):89–104. doi: 10.1259/bjr/60186130. [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci U S A. 2002;99(4):1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci U S A. 2006;103(24):8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J Biol Chem. 1998;273(32):20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18(1):114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Wiebauer K, Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4(5):278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Wojtaszek J, Lee CJ, D’Souza S, Minesinger B, Kim H, D’Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012a;287(40):33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J, Liu J, D’Souza S, Wang S, Xue Y, Walker GC, Zhou P. Multifaceted recognition of vertebrate Rev1 by translesion polymerases zeta and kappa. J Biol Chem. 2012b;287(31):26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters S, Schumacher B. Genome maintenance and transcription integrity in aging and disease. Front Genet. 2013;4:19. doi: 10.3389/fgene.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19(12):1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28(6):1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci U S A. 2010;107(48):20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22(20):2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xie K, Zhang XQ, Pridgen EM, Park GY, Cui DS, Shi J, Wu J, Kantoff PW, Lippard SJ, Langer R, Walker GC, Farokhzad OC. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc Natl Acad Sci U S A. 2013;110(46):18638–18643. doi: 10.1073/pnas.1303958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Yamada A, Masutani C, Iwai S, Hanaoka F. Complementation of defective translesion synthesis and UV light sensitivity in xeroderma pigmentosum variant cells by human and mouse DNA polymerase eta. Nucleic Acids Res. 2000;28(13):2473–2480. doi: 10.1093/nar/28.13.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase nu in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol. 2010;23(3):689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, Lin T, Schuster B, Decaillet C, Stasiak A, Stasiak AZ, Stone S, Hoatlin ME, Schindler D, Woodcock CL, Joenje H, Sen R, de Winter JP, Li L, Seidman MM, Whitby MC, Myung K, Constantinou A, Wang W. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37(6):865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle. 2008;7(10):1321–1325. doi: 10.4161/cc.7.10.5898. [DOI] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9(1):91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Holmquist GP, O’Connor TR. Heterogeneous repair of N-methylpurines at the nucleotide level in normal human cells. J Mol Biol. 1998;284(2):269–285. doi: 10.1006/jmbi.1998.2138. [DOI] [PubMed] [Google Scholar]
- Yi C, He C. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol. 2013;5(1):a012575. doi: 10.1101/cshperspect.a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Unal F, Yuzbasioglu D, Celik M. DNA damage in human lymphocytes exposed to four food additives in vitro. Toxicol Ind Health. 2014;30(10):926–937. doi: 10.1177/0748233712466132. [DOI] [PubMed] [Google Scholar]
- Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem. 2000;275(13):9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- Yonekura S, Nakamura N, Yonei S, Zhang-Akiyama QM. Generation, biological consequences and repair mechanisms of cytosine deamination in DNA. J Radiat Res. 2009;50(1):19–26. doi: 10.1269/jrr.08080. [DOI] [PubMed] [Google Scholar]
- Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27(24):4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You YH, Szabo PE, Pfeifer GP. Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis. 2000;21(11):2113–2117. doi: 10.1093/carcin/21.11.2113. [DOI] [PubMed] [Google Scholar]
- Yu H. Environmental carcinogenic polycyclic aromatic hydrocarbons: photochemistry and phototoxicity. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2002;20(2):149–183. doi: 10.1081/GNC-120016203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20(13):1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yang J, Zhu F, Xu F. Response of REV3 promoter to N-methyl-N′-nitro-N-nitrosoguanidine. Mutat Res. 2004;550(1–2):49–58. doi: 10.1016/j.mrfmmm.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem. 2004;279(20):20935–20940. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- Yuji Masuda FH, Masutani Chikahide. In: Translesion DNA Synthesis and Damage Tolerance Pathways. Hanaoka FKS, editor. Japan: Springer; 2016. [Google Scholar]
- Yurkow EJ, Laskin JD. Mechanism of action of psoralens: isobologram analysis reveals that ultraviolet light potentiation of psoralen action is not additive but synergistic. Cancer Chemother Pharmacol. 1991;27(4):315–319. doi: 10.1007/BF00685118. [DOI] [PubMed] [Google Scholar]
- Zak P, Kleibl K, Laval F. Repair of O6-methylguanine and O4-methylthymine by the human and rat O6-methylguanine-DNA methyltransferases. J Biol Chem. 1994;269(1):730–733. [PubMed] [Google Scholar]
- Zengin N, Yuzbasioglu D, Unal F, Yilmaz S, Aksoy H. The evaluation of the genotoxicity of two food preservatives: sodium benzoate and potassium benzoate. Food Chem Toxicol. 2011;49(4):763–769. doi: 10.1016/j.fct.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7(7):1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122(5):693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Zhao C, Tyndyk M, Eide I, Hemminki K. Endogenous and background DNA adducts by methylating and 2-hydroxyethylating agents. Mutat Res. 1999;424(1–2):117–125. doi: 10.1016/s0027-5107(99)00013-5. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jain A, Iyer RR, Modrich PL, Vasquez KM. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37(13):4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33(1):289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc Natl Acad Sci U S A. 1993;90(14):6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Yang J, Xu F, Yu YN. Cloning and bioinformatics of human REV3 gene promoter region and its response to carcinogen N-methyl-N′-nitro-N-nitrosoguanidine. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2003;32(5):393–397. doi: 10.3785/j.issn.1008-9292.2003.05.006. [DOI] [PubMed] [Google Scholar]
- Zhu H, Fan Y, Jiang H, Shen J, Qi H, Mei R, Shao J. Response of human DNA polymerase iota promoter to N-methyl-N′-nitro-N-nitrosoguanidine. Environ Toxicol Pharmacol. 2010;29(1):79–86. doi: 10.1016/j.etap.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Zhu H, Fan Y, Shen J, Qi H, Shao J. Characterization of human DNA polymerase kappa promoter in response to benzo[a]pyrene diol epoxide. Environ Toxicol Pharmacol. 2012;33(2):205–211. doi: 10.1016/j.etap.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Wani AA. Histone modifications: crucial elements for damage response and chromatin restoration. J Cell Physiol. 2010;223(2):283–288. doi: 10.1002/jcp.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci U S A. 2009;106(28):11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]