Abstract
Purpose
To explore factors associated with response and resistance to anti-PD-1 therapy, we analyzed multiple disease sites at autopsy in a patient with widely metastatic melanoma who had a heterogeneous response.
Materials and Methods
Twenty-six melanoma specimens (four pre-mortem, 22 post-mortem) were subjected to whole-exome sequencing. Candidate immunologic markers and gene expression were assessed in ten cutaneous metastases showing response or progression during therapy.
Results
The melanoma was driven by biallelic inactivation of NF1. All lesions had highly concordant mutational profiles and copy number alterations, indicating linear clonal evolution. Expression of candidate immunologic markers was similar in responding and progressing lesions. However, progressing cutaneous metastases were associated with over-expression of genes associated with extracellular matrix and neutrophil function.
Conclusions
Although mutational and immunologic differences have been proposed as the primary determinants of heterogeneous response/resistance to targeted therapies and immunotherapies, respectively, differential lesional gene expression profiles may also dictate anti-PD-1 outcomes.
Keywords: melanoma, cancer genetics, immunotherapy, anti-PD-1
INTRODUCTION
Complex interactions between the immune system and melanoma influence tumor invasion and metastasis, and ultimately define patient survival. Immunologic and genetic factors may contribute independently to prognosis and treatment resistance in melanoma, however, a deeper knowledge of their interactions is central to understanding disease evolution and developing synergistic combination treatment regimens. Melanoma has the capacity to metastasize to virtually any organ in the body; often, the full extent of metastasis is discovered only on postmortem examination. To date, studies of melanoma heterogeneity and metastatic evolution have been hampered by the limited availability of tissues from readily accessible anatomic sites in living patients. This limitation can be overcome by a rapid autopsy approach.1 This approach has been applied to pancreatic and other cancer types, revealing diverse patterns of clonal evolution from the primary lesion to metastases, in which intratumoral subclones and metastatic tumors accumulate private mutations superimposed on shared or ubiquitous mutations.2–4 In some cases, an unexpectedly long chronology from primary lesion to the development of metastasis has been observed.
Immune checkpoint blockade with PD-1/PD-L1 antibodies, recently approved for treating metastatic melanoma, Hodgkin lymphoma, and cancers of the lung, kidney, bladder, and head and neck, unleashes endogenous antitumor immunity to eliminate cancer cells.5 To date, most research into anti-PD-1 treatment resistance has focused on immunologic factors such as intratumoral PD-L1 expression and infiltration of cytolytic CD8+ T cells.6–8 However, the extent to which genetic and other factors may play equally important roles is unknown. We therefore combined genetic and immunologic analyses of tumor evolution within one patient’s cutaneous melanoma over a 5-year clinical course from diagnosis to metastasis and death. This patient received anti-PD-1 therapy up to the time of death, at which time some metastases were responding to therapy while others were progressing. Surprisingly, genetic and immunologic factors were relatively homogenous among responding and progressing lesions, while a non-genomic program characterized the progressing metastases.
RESULTS
Clinical history and tumor specimens
A 60-year-old man presented with a primary cutaneous melanoma of the left upper extremity, 7 mm in Breslow thickness. Clinical testing indicated that no mutations were present in the most frequently mutated regions of BRAF, KIT, NRAS, and PIK3CA. Over the ensuing 2 years, the patient underwent surgical resection of locoregional recurrences, including lymph node and in-transit cutaneous metastases. Distant metastases then appeared, for which he received two 4-dose courses of ipilimumab (anti-CTLA-4) over 15 months. The patient’s disease was initially stable for several months, but then progressed. The patient subsequently began a 6-month course of nivolumab (anti-PD-1), during which he died suddenly of an unrelated cause. A rapid autopsy was performed within 6 hours of death, under consent on an IRB-approved protocol.
Twenty-six tumor specimens were selected for study, including 4 pre-mortem specimens (the primary melanoma, biopsies of two locoregional recurrences, and a distant cutaneous metastasis) (Table 1). Twenty-two metastases were sampled at autopsy. Post-mortem specimens were derived from 5 organ sites including skin, stomach, small bowel, lung and brain. Ten cutaneous metastases were specifically selected as they showed unequivocally divergent clinical behavior on anti-PD-1 therapy, including 6 responding and 4 progressing lesions (Figure 1), thereby facilitating the study anti-PD-1 response/resistance in multiple lesions derived from a single organ in an individual patient.
Table 1.
Tumor and normal tissue specimens used for whole exome sequencing
| ID | Metastatic site | Anatomic Location | Date collected | Tissue Preservation | Source | Notes |
|---|---|---|---|---|---|---|
| M26 | Primary melanoma | Left upper arm | 12/31/08 | FFPE | Surgery, no prior therapy | |
| M23 | Axillary lymph node | Left axilla | 9/27/10 | FFPE | Surgery, no prior therapy | |
| M24 | Skin | Left axilla | 5/25/11 | FFPE | Surgery, no prior therapy | |
| M25 | Skin | Left chest wall | 11/5/12 | FFPE | Surgery, post ipilimumab | |
| M1 | Lymph node | Left axilla | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M2 | Lymph node | Left axilla | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M3 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M4 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M5 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M6 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M7 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M8 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Regressing on anti-PD-1 |
| M9 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M10 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M11 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M12 | Skin | Left chest wall | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 |
| M13 | Local recurrence | Left arm surgical site | 11/5/13 | Snap frozen and FFPE | Autopsy | |
| M14 | Lung | 11/5/13 | Snap frozen and FFPE | Autopsy | Progressing on anti-PD-1 | |
| M15 | Small Bowel | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M16 | Small Bowel | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M17 | Small Bowel | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M18 | Small Bowel | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M19 | Stomach | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M20 | Stomach | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M21 | Stomach | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| M22 | Brain | 11/5/13 | FFPE | Autopsy | Discovered at autopsy | |
| Germline | Normal kidney | 11/5/13 | Snap frozen and FFPE | Autopsy |
Abbreviations: FFPE, formalin-fixed, paraffin-embedded specimens.
Figure 1.
Cutaneous melanoma metastases demonstrating progression (white arrows) or regression (black arrows) after 25 and 19 weeks of nivolumab anti-PD-1 therapy, respectively.
Genomic analyses
Overall, at the time of autopsy, this patient had a classical high mutational burden cutaneous melanoma. Mutational signature decomposition analysis revealed the expected abundance of C>T/G>A mutations associated with prior ultraviolet light exposure.9 Whole exome sequencing of all 26 specimens confirmed that this tumor was wildtype for canonical activating mutations in RAS, RAF, and KIT, and was NF1-driven with clonal biallelic inactivation of the gene present in all primary, recurrent, and metastatic specimens collected pre- and post-mortem.10–11 All tumors harbored a truncating NF1 Q519* mutation that was accompanied by a focal heterozygous loss of the single remaining wildtype allele (Figure 2).
Figure 2.
Mutational profiling of a metastatic melanoma of cutaneous origin. (A) The abundance of mutations in every lesion is typical of cutaneous melanoma. Most mutations are clonal (n=618) and present in every specimen. Each specimen, including the primary lesion, 3 pre-mortem and 22 post-mortem specimens, harbors the NF1 Q519* driver mutation. Excluding lesions M13 (local recurrence left arm skin) and M14 (lung metastasis), which have the greatest number of private mutations (281 and 622, respectively), 50% of all somatic nonsynonymous mutations are present in all tumors. Melanoma lesions (described in Table 1) are ordered from top to bottom by increasing numbers of private mutations. Intensity of blue shading indicates fraction of mutant tumor cells. (B) Phylogenetic tree of mutational evolution in this patient. The variants in Fig. 2A were filtered for phylogenetic analysis (see Methods). The branches colored in light green, clustered with the 10 regressing/progressing cutaneous metastases on nivolumab therapy in 2013, represent cutaneous lesions M24 and M25 obtained in 2011 and 2012, respectively. The primary melanoma lesion from 2008 (M26) is highly similar to these cutaneous metastases.
We sought to explore the evolutionary trajectory of this melanoma from diagnosis until the patient’s death and throughout immunotherapy with anti-CTLA-4 followed by anti-PD-1. At the genomic level, the tumors did not display a significant number of genetic subclones with respect to cancer cell fraction (Figure 2A). Indeed, the majority of mutations were either present in all samples or private to one. The homogeneity of the genomic profile observed at the somatic mutational level was also found in DNA copy number alteration (CNA) analysis. All tumors possessed a high burden, but very stable pattern, of copy number changes, with few changes acquired after diagnosis (Supplementary Figure S1). Consequently, phylogenetic analysis indicated that while all tumors harbored a small number of private mutations, there was only modest ongoing genomic evolution after the initial branch point defined by 586 shared truncal somatic mutations (Figure 2B). Review of the somatic mutations that defined the first major branch point did not reveal evidence of additional driver genes or copy number alterations as the underlying basis of these two distinct subclonal populations.
As most late-arising branching in the phylogeny was supported by few mutations, we can infer only major patterns in this patient. The local recurrence in the skin of the left arm after primary tumor resection (lesion M13) was the closest in evolutionary relationship to the primary tumor (M26), though the local recurrence had evidence of ongoing genomic evolution in the form of additional private mutations. Moreover, the distant cutaneous metastasis resected four years after diagnosis, as well as those present post-mortem, were highly similar to each other and appear to have emerged through linear clonal evolution from the primary tumor. The cutaneous metastases were relatively distant evolutionarily from most but not all visceral metastases, some of which branched off at an evolutionary point prior to diagnosis (Figure 2B). Notably, no somatic mutations cleanly divided the two groups of cutaneous metastases that were either responding or progressing on anti-PD-1 therapy. Furthermore, none of the mutated genes were differentially expressed at the transcriptional level between the two groups (see gene expression profiling, below). Collectively these findings indicate this patient’s melanoma underwent a singular branching event prior to diagnosis, and the two major branches each proceeded by linear evolution with nearly all samples acquiring private mutations. Importantly, both the responding and progressing cutaneous metastases were derived from a single subclone, whereas visceral metastases arose from one of two distinct subclones that branched early in the phylogenetic history of this neoplasm. Similar to the mutational data, neoantigen prediction analysis did not indicate any neoepitopes common to the regressing or progressing cutaneous metastases, although many putative neoantigens were detected across the tumor samples (Supplementary Table S1).
Assessment of candidate immunologic correlative markers in responding vs. progressing cutaneous metastases
The occurrence of ten metastases in a single organ site (skin), that had clearly dichotomous clinical behavior in response to anti-PD-1 therapy but highly similar genomic profiles, led us to explore candidate immunological factors as the basis for differential response. Tissue integrity in all post-mortem specimens was intact without significant histologic evidence of autolysis. We found no significant differences between regressing and progressing cutaneous lesions with regard to the extent of tumor necrosis, presence of lymphoid aggregates, or geographic pattern of immune cell infiltration. We then assessed candidate immune markers with immunonhistochemical (IHC) labeling on formalin-fixed, paraffin-embedded (FFPE) specimens. When present, the major PD-1 ligand, PD-L1, was displayed in association with immune cell infiltrates, suggesting “adaptive” expression in response to intratumoral inflammation.12 There were no significant differences in the prevalence of PD-L1 expression on either tumor or infiltrating immune cells, or on immune cell subsets, when comparing cutaneous metastases that were responding or progressing during anti-PD-1 therapy (Supplementary Figure S2A). Furthermore, although others have correlated the density and geography of tumor infiltrating CD8 T cells with response to anti-PD-1,7 in a quantitative analysis of these parameters we found no significant differences in CD8 T cell densities in intratumoral or peritumoral locations in progressing vs. regressing cutaneous metastases (Figure 3, Supplementary Figure S2B). The tumor microenvironments of both the progressing and regressing lesions showed maximum CD8 T cell densities in the peritumoral stroma, particularly within 100 μm of the tumor border (Figure 3B, Supplementary Figure S2B). Similar geographic findings were observed in the primary melanoma and three metastatic lesions biopsied before death, although the density of CD8 cells was much lower in these lesions compared to those collected on anti-PD-1 therapy (Figure 3C). Moreover, there were no significant differences between responding and progressing lesions among additional immune regulatory molecules and cellular subsets, including LAG-3 and TIM-3 (Supplementary Figure S2A), and PD-1, CD20 and the CD4:CD8 ratio (not shown).
Figure 3.
Geography of CD8 infiltrates does not distinguish regressing from progressing cutaneous melanoma metastases. (A) Representative regressing and progressing cutaneous melanoma metastases, stained with hematoxylin/eosin (H&E) or anti-CD8. The distribution of intratumoral and peritumoral CD8+T cells does not differ between these lesions. (B) Density of CD8 T cells in 6 regressing and 4 progressing melanoma metastases, mapped according to intratumoral and peripheral zones. (C) Density mapping of CD8 T cells in the primary melanoma lesion and 3 metastases biopsied before death. Note the difference in Y-axis scale between (B) and (C). Two specimens, the primary melanoma and metastasis M25, did not contain sufficient peritumoral tissue to perform a complete analysis. See also Supplementary Figure S2B.
In melanoma, coordinate gene expression has been observed for PD-L1 (CD274), an immunosuppressive ligand found on tumor and stromal cells in the melanoma microenvironment, and genes whose expression is associated with CD8 T cell activation (CD8A, IFNG, PRF1, CCL5), antigen presentation (CD163, TLR3, CXCL1, LYZ), and immunosuppression [PD1(PDCD1), LAG3, IL10].13 To assess whether these or other candidate immune-related genes were differentially expressed between regressing and progressing cutaneous melanoma metastases, we employed the same 60-gene multiplex qRT-PCR array as in our prior melanoma study. Gene expression was normalized to the pan-immune cell marker PTPRC (CD45). We found that none of the screened molecules was significantly differentially expressed according to clinical response characteristics (data not shown), consistent with the IHC results described above for selected immune markers. Thus, neither major genomic events, differences in immune cell infiltration, or candidate immune-related gene expression correlated with the divergent clinical behavior of progressing versus regressing cutaneous metastases on anti-PD-1 therapy.
Unbiased examination of gene expression in regressing vs. progressing metastases
Given the lack of a genomic or immunologic profile to explain the differential clinical response patterns in the ten cutaneous metastases on anti-PD-1 therapy, we next turned to transcriptome-wide profiling. RNA was isolated from tumor-enriched areas in 6 regressing and 4 progressing lesions, by manually scraping unstained tissue sections guided by neighboring sections stained with H&E. By comparing the profiles of regressing vs. progressing metastases, we identified 827 differentially expressed genes between the two groups, based on fold change magnitude ≥1.5. Principal component analysis illustrated a segregation of gene expression profiles between regressing and progressing lesions (Supplementary Figure S3A). Enrichment analyses of the over-expressed genes in responding lesions yielded only 4 DAVID gene sets (terms), including Signal and Immunoglobulin domain [DAVID Benjamini false discovery rate (FDR) ≤0.05]. In contrast, a similar analysis of genes over-expressed in progressing lesions yielded 42 gene sets involving epithelial, adhesion and cell differentiation functions (DAVID Benjamini FDR ≤0.010) (Supplementary Table S2).
Next, in order to better differentiate regressing vs. progressing metastases, more stringent criteria were applied for gene selection (p-value ≤0.05 and expression fold change magnitude ≥1.7), resulting in 140 probes corresponding to 138 distinct genes differentially expressed between the two groups (Supplementary Table S3). Inspection showed up-regulation in progressing lesions of keratin genes such as KRT1, KRT5, KRT10, KRT15, KRT78; cellular adhesion genes such as loricrin (LOR), filaggrin (FLG2), desmocollin (DSC1, DSC3), galectin 7 (LGALS7), laminin (LAMA3), and kallikrein (KLK7); and the Wnt pathway genes WNT3 and WNT5A. Among 138 unique genes that were differentially expressed between regressing and progressing cutaneous melanoma metastases after anti-PD-1 therapy, 13 were also found in TCGA “keratin” subclass associated with poor prognosis in patients with melanoma, representing a substantial enrichment -- KRT1, KRT5, KRT15, KRT78, KRTDAP, EVPL, KLK7, KLK11, LGALS7, LOR, SCEL, SPINK5, and SPRR1A.11
Analysis of differential gene expression with qRT-PCR
Following global gene expression profiling, PCR-based custom arrays were designed to further characterize the differential expression of unique gene targets in regressing vs. progressing cutaneous metastases. First, a multiplex array was assembled with 61 genes selected from among the 138 genes that were differentially expressed in microarray analysis (Supplementary Table S3). Criteria employed for gene selection included the following: expression fold-change magnitude ≥2 and p-value ≤0.01, comparing regressing vs. progressing tumors; biological associations; and overlap with genes included in the TCGA “keratin” profile (11). Twenty-eight among the 61 genes tested were confirmed to be significantly differentially expressed (FC magnitude ≥2 and p-value ≤0.05) in cutaneous metastases with divergent clinical outcomes (not shown). In particular, this assay confirmed up-regulation of genes associated with keratin/epithelial pathways in progressing cutaneous metastases. Among them, KRT1 showed the greatest differential expression, being up-regulated >200,000-fold in progressing lesions. To further investigate this finding, IHC for the cytokeratin 1 protein was performed on the 10 cutaneous metastases with clinically divergent behavior. Notably, we failed to observe tumor cell expression of KRT1 protein, but instead saw robust expression in normal overlying squamous epithelium and eccrine sweat gland ducts (not shown). Similarly, IHC with an antibody cocktail directed against low and high molecular weight keratins, including the protein products of KRT1, KRT10 and KRT15, failed to detect cytokeratin expression in tumor cells among 21 different melanoma specimens collected from this patient. Retrospective H&E staining of FFPE slides that were manually scraped to isolate RNAs revealed that normal overlying skin had been inadvertently and selectively captured from the progressing skin lesions, generating an artifactual “keratin” tumor gene expression profile.
Next, to insure RNA isolation from pure tumor areas free from normal epithelium or necrosis, laser capture microdissection under direct visualization was performed on tissue sections from the 10 cutaneous metastases.12 Multiplex qRT-PCR arrays were employed to assess expression of 122 among 138 genes that were previously found to be differentially expressed in whole genome microarray analysis, excluding non-coding RNAs and those for which commercial probes were unavailable (Supplementary Table S3). Thirteen among 122 genes were confirmed to be differentially expressed in these highly tumor-enriched RNA specimens from progressing vs. regressing cutaneous metastases (FC magnitude ≥2 and p-value ≤0.1), when normalized to either 18S or GUSB (Table 2). Genes up-regulated in the progressing metastases were associated with cellular adhesion and extracellular matrix formation (LAMA3, CCM2L, CST2, DACT1) and with neutrophil function (FAM183B, PTPRC, CXCR2). Among them, LAMA3 encoding the laminin subunit alpha 3 found in laminins-5, -6, and -7 showed the greatest degree of differential expression, being over-expressed ~2000-fold in progressing subcutaneous metastases (p ≤0.005). This gene was also well-expressed in cultured melanoma cell lines (Figure 4), supporting tumor cell-intrinsic expression.
Table 2.
Genes differentially expressed in regressing vs. progressing metastases, assessed by qRT-PCR
| Normalizing gene | |||||||
|---|---|---|---|---|---|---|---|
| 18S | GUSB | ||||||
| Gene symbola | Alternative gene namea | Gene descriptiona | Protein functiona | FC, R/P b | p-value | FC, R/P b | p-value |
| LAMA3 | BM600; E170; LAMNA; LOCS | Laminin subunit alpha 3 | The protein encoded by this gene belongs to the laminin family of secreted molecules. Laminins are essential for formation and function of the basement membrane and have additional functions in regulating cell migration, wound healing and mechanical signal transduction. Their activity is coordinated by the presence of metalloproteineses (MMP). This gene encodes an alpha subunit and is responsive to several epithelial-mesenchymal regulators including keratinocyte growth factor, epidermal growth factor and insulin-like growth factor. | −2624.7 | 0.003 | −1904.8 | 0.005 |
| CCM2L | C20orf160 | CCM2 like scaffolding protein | The cerebral cevernous malformation (CCM) signaling pathway utilizes the adaptor protein CCM2 to strength the endothelial cell junctions and stabilize vessels. The CCM2 paralog, CCM2L, functions as a molecular mechanism through which CCM signaling converts endothelial cells from a stable to an angiogenic phenotype thus promoting tumor growth, endothelial-mesenchymal transition (EMT) and wound healing. | −267.3 | 0.054 | −155.3 | 0.071 |
| FAM183B | FAM183BP; THEG6 | Family with sequence similarity 183 member B, pseudogene | Acyloxyacyl hydrolase (neutrophil) | −69.4 | 0.051 | −40.3 | 0.073 |
| CST2 | N/Ac | Cystatin SA | The type 2 cystatin proteins are a class of cysteine proteinase inhibitors found in a variety of human fluids and secretions, where they appear to provide protective functions. Proteases and their interactions with their inhibitors play a role in remodeling extracellular matrix (ECM) and contribute to ECM composition. | −19.2 | 0.002 | −13.9 | 0.002 |
| PTPRC (expt. 1)d | B220; CD45; CD45R; GP180; LCA; L-CA; LY5; T200 | Protein tyrosine phosphatase, receptor type C | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitosis, and oncogenic transformation. This PTP has been shown to be an essential regulator of T- and B-cell antigen receptor signaling but also to induce a neutrophil migration. | −4.2 | 0.010 | −3.1 | 0.003 |
| PTPRC (expt. 2)d | −3.1 | 0.077 | −1.8 | 0.171 | |||
| CXCR2 | CD182; CDw128b; CMKAR2; IL8R2; IL8RA; IL8RB | C-X-C motif chemokine receptor 2 | This protein is a receptor for interleukin 8 (IL8). This receptor also binds to chemokine (C-X-C motif) ligand 1 (CXCL1/MGSA), a protein with melanoma growth stimulating activity, and has been shown to be a major component required for serum-dependent melanoma cell growth. This receptor mediates neutrophil migration to sites of inflammation and has angiogenic effects. | −4.2 | 0.102 | −2.5 | 0.238 |
| NDUFA4L2 | N/A | NDUFA4, mitochondrial complex associated like 2 | NADH dehydrogenase (ubiquinone) 1α subcomplex subunit 4-like 2 (NDUFA4L2) is considered as an inhibitory component of Complex I and a direct transcriptional target of HIF. HIF induces expression of NDUFA4L2, which in turn inhibits ETC Complex I activity, thereby attenuating oxygen consumption and mitochondrial ROS production. In several cancer types, it is likely that NDUFA4L2 has a role in mediating the Warburg effect. | −3.4 | 0.099 | −2.0 | 0.235 |
| DACT1 | DAPPER; DAPPER1; DPR1; FRODO; HDPR1; THYEX3 | Dishevelled binding antagonist of beta catenin 1 | DACT1 has been identified as a modulator of Wnt signaling through its interaction with Dishevelled (Dvl), a central mediator of both the canonical and noncanonical Wnt pathways. DACT1 induces cancer cell proliferation and stability of β-catenin. Its expression has been also shown to support the proliferation of human epidermal keratinocytes. | −2.9 | 0.061 | −1.7 | 0.096 |
| EXO1 | HEX1; hExoI | Exonuclease 1 | This gene encodes a protein with 5′ to 3′ exonuclease activity as well as an RNase H activity. Exo-1 is a developmental antigen of human epithelial cells. Exo-1 is a marker for an early embryonic differentiation pathway of human keratinocytes and in adult tissue reveals abnormal differentiation associated with certain stages of hyperproliferation | −2.3 | 0.099 | −1.7 | 0.195 |
| TSLP | N/A | Thymic stromal lymphopoietin | This gene encodes a hemopoietic cytokine proposed to signal through a heterodimeric receptor complex composed of the thymic stromal lymphopoietin receptor and the IL-7R alpha chain. It mainly impacts myeloid cells and induces the release of T cell-attracting chemokines from monocytes and enhances the maturation of CD11c(+) dendritic cells. The protein promotes T helper type 2 (TH2) cell responses that are associated with immunity in various inflammatory diseases. | 1.4 | 0.370 | 2.0 | 0.019 |
| LOXL4 | LOXC | Lysyl oxidase like 4 | This gene encodes a member of the lysyl oxidase gene family. The prototypic member of the family is essential to the biogenesis of connective tissue, encoding an extracellular copper-dependent amine oxidase that catalyzes the first step in the formation of crosslinks in collagens and elastin | 2.3 | 0.170 | 3.2 | 0.008 |
| ARNT2 | bHLHe1; WEDAS | Hydrocarbon receptor nuclear translocator 2 | This gene encodes a member of the basic-helix-loop-helix-Per-Arnt-Sim (bHLH-PAS) superfamily of transcription factors. Under hypoxic conditions, the encoded protein complexes with HIF1 in the nucleus and this complex binds to hypoxia-responsive elements in enhancers and promoters of oxygen-responsive genes. | 2.9 | 0.089 | 5.0 | 0.015 |
| SBSN | UNQ698 | Suprabasin | Suprabasin has been identified as an epidermal differentiation marker and has been detected in the suprabasal layers of the epithelium in the epidermis. | 51.7 | 0.125 | 71.3 | 0.100 |
Listed are genes with expression fold change (FC) ≥2 and p-value ≤0.1 (Welch’s two-tailed t-test) when normalized to either 18S or GUSB expression in qRT-PCR. Genes are ordered according to ascending FC when results are normalized using 18S. Positive FC indicates genes over-expressed in tumors from regressing (R) metastases; negative FC indicates over-expression in tumors from progressing (P) metastases. All specimens were obtained using laser capture microdissection.
Obtained from (http://www.ncbi.nlm.nih.gov/gene/)
Data were analyzed using the 2−ΔΔCt method.
N/A, not applicable.
The expression of PTPRC (CD45) was evaluated twice. Comparable results were observed between the two experiments.
Figure 4.
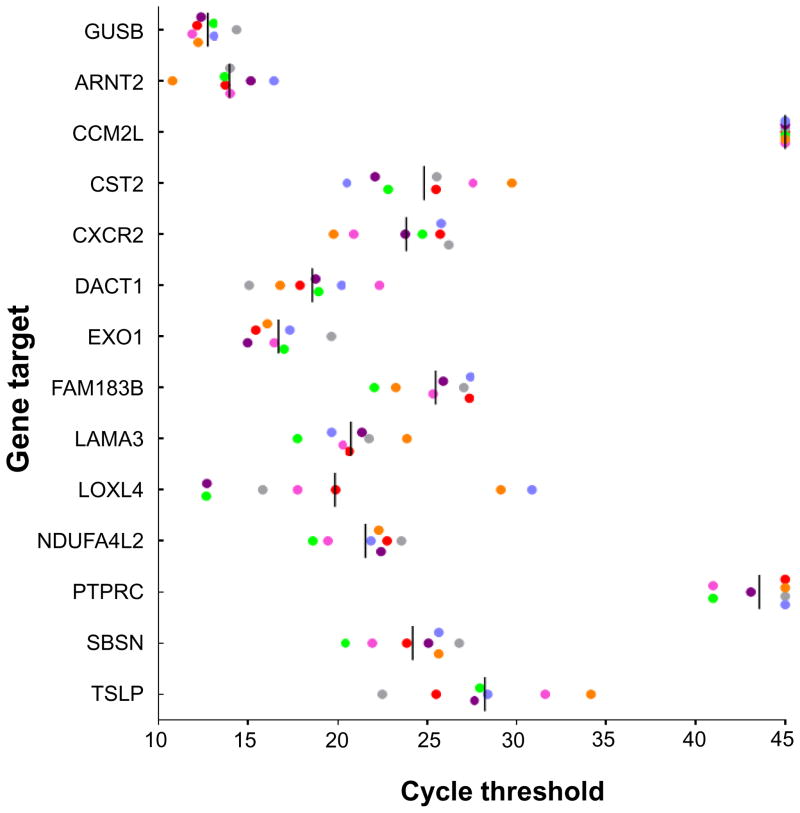
Expression of genes associated with progressing cutaneous metastases, in melanoma cell lines. The expression of 13 genes significantly differentially expressed in progressing compared to regressing cutaneous metastases (Table 2) was assessed in seven established human melanoma cell lines with qRT-PCR. Each dot represents a single cell line. Vertical bars, mean values. GUSB expression is shown as an internal control. Undetermined values are depicted as having a cycle threshold of 45, the maximum number of PCR cycles in this assay.
Expression of LAMA3 protein in progressing cutaneous melanoma metastases
Immunohistochemistry was utilized to assess LAMA3 protein expression in the melanoma primary lesion and metastases. As shown in Figure 5, tumor cells in progressing cutaneous metastases expressed higher levels of LAMA3 protein than did those in regressing metastases. Furthermore, progressing metastases in other organs such as lung, lymph node and bowel also expressed the LAMA3 protein. Interestingly, LAMA3 was not expressed in the primary melanoma (Figure 5). Mutations and copy number alterations in LAMA3 were not observed in the primary or metastatic tumors from this patient, suggesting that LAMA3 expression was acquired during tumor evolution through non-genomic, transcriptional or epigenetic events.
Figure 5.
LAMA3 protein expression in melanoma lesions assessed by IHC. (A) LAMA3 is expressed in a progressing cutaneous melanoma metastasis, and in three non-cutaneous metastases, but not in the primary melanoma lesion nor in a regressing melanoma cutaneous metastasis. Brown staining detects LAMA3 protein. Specimen numbers are found in Table 1. Met, metastasis. (B) LAMA3 protein expression quantified by histoscore (H-score), the product of the percentage of positively staining cells with IHC, by the staining intensity (graded as 0, no stain; 1, weak; 2, medium; 3, strong staining). Horizontal bars, mean values. P-value derived from a one-sided Wilcoxon rank sum test.
In silico analyses of gene expression profile associated with response/resistance to anti-PD-1
Melanoma gene expression classification in TCGA, based on mRNA sequencing of treatment-naïve tumors from 281 patients, identified a “keratin” subclass associated with decreased survival, an “immune” subclass associated with prolonged survival, and an intermediate “MITF-low” subclass.11 In our study of multiple melanoma metastases from one patient, a 13-gene profile was identified that differentiated lesional clinical behavior (progressing vs. regressing) after anti-PD-1 therapy. To investigate whether this gene expression profile might correlate with overall survival in the melanoma patient population, we downloaded and analyzed TCGA melanoma gene expression data. An unsupervised cluster analysis based on these 13 genes did not provide any interpretable picture on TCGA expression data, and there was no significant separation of overall survival curves based on this analysis when evaluated by the Wilcoxon test (Supplementary Figure S4A). This suggests that our 13-gene expression profile, which specifically discriminates melanoma lesional behavior following anti-PD-1 therapy, is not a strong prognostic indicator for patients with melanoma in general. Furthermore, a similar in silico analysis was applied to gene expression data from pretreatment melanoma biopsies in 27 patients who did or did not respond to anti-PD-1 therapy.14 Although there was an apparent association of gene expression with response status that was seen in an unsupervised clustering analysis of these samples, this was not robust to small changes in the gene signature (Supplementary Figure S4B). Therefore, our 13-gene profile appeared to be specifically relevant to the on-treatment behavior of individual metastatic melanoma lesions during anti-PD-1 therapy.
DISCUSSION
Melanoma is a disease often characterized by a prolonged “disease-free” interval between diagnosis and clinically apparent metastasis, which can exceed five years. Melanoma is also one of the most immunogenic human cancers, and anti-melanoma immune responses are readily detected in vitro.15 Thus, primary melanoma lesions likely metastasize early in the course of tumor evolution, and a dynamic balance between melanoma and the immune system in some patients may durably maintain the tumor in a microscopic state.
The findings in the patient described here are consistent with these ideas. This patient’s melanoma was apparently driven by a functional loss of NF1 (neurofibromin), previously shown to induce RAS activation in melanoma cells10 and to confer resistance to therapy with selective BRAF inhibitors via mitogen-activated protein kinase (MAPK) pathway reactivation.16 NF1-altered tumors were also recently defined by TCGA as a major genomic class in melanoma.11 Although the clinical history of this patient’s melanoma spanned 5 years, and distant metastases were not detected until 3 years after initial diagnosis, relatively little genetic heterogeneity was observed in the multiple metastases sequenced. Instead, the phylogeny of this tumor’s evolution revealed a singular early branching event, each branch then proceeding by linear evolution. Under the selection pressure of anti-PD-1 therapy, several distant cutaneous metastases regressed while others progressed. In a similar scenario with molecularly targeted drugs such as MAPK inhibitors, most studies have focused on genetic alterations as the basis for treatment response/resistance, although more recently epigenetic, tumor microenvironmental and immunologic events have also been invoked.17,18 In the current study, we found remarkable genetic similarities among regressing/progressing cutaneous metastases, indicating that all were derived from a single subclone that had not diverged significantly. There were also immunological similarities among these metastases, and no defining characteristics in candidate immune molecules were associated with treatment response or resistance. However, unbiased global gene expression profiling revealed a distinct signature differentiating progressing from regressing tumors. In particular, the most highly differentially expressed gene, LAMA3, was also found to be overexpressed at the protein level in progressing metastases. LAMA3 is a subunit of laminin-5, which has been associated with epithelial-to-mesenchymal transition and poor prognosis in lung cancer,19 and which promotes the aggressive behavior of melanoma cells in three-dimensional culture.20 Laminin-5 is a secreted molecule that plays an important role in tissue architecture and adhesion of normal epithelial cells to the basement membrane.21 The normal adhesive role of laminin presents one possible explanation for the selective contamination of progressing tumor specimens with overlying normal epidermis in the manually scraped tissue sections used for RNA isolation in our preliminary experiments. It also raises the possibility that over-expression of laminins might create a barrier excluding immune cells from penetrating the tumor, and thus impairing immunotherapy, similar to a recent report of aberrant beta-catenin over-expression in melanoma.22 Although we did not observe differential exclusion of CD8 effector cells from progressing vs. regressing metastases in the current study, it is possible that dynamic exclusion effects were not captured at the time of tissue procurement, or that other cell types were involved. Interestingly, PTPRC, a pan-leukocyte marker, was over-expressed in progressing metastases. Because we did not detect differences in T and B lymphocyte and macrophage markers with IHC or qRT-PCR in progressing vs. regressing cutaneous metastases, this raises the possibility that PTPRC reflects neutrophil infiltration or activation in progressing tumors. This is consistent with the coordinate over-expression of FAM183B (neutrophil-associated acyloxyacyl hydrolase) and CXCR2 (IL-8 receptor mediating neutrophil migration) in these specimens. CXCR2-mediated accumulation of tumor infiltrating neutrophils has been associated with intratumoral immunosuppression in other studies.23
A recent report of a group of 27 patients with melanoma receiving anti-PD-1 therapy correlated innate treatment resistance with a complex transcriptional signature marked by up-regulation of genes regulating mesenchymal transition, cell adhesion, extracellular matrix remodeling, T cell suppressive inflammation, angiogenesis and wound healing.14 Our report of a single patient receiving anti-PD-1, which focuses on the post-treatment dichotomy of progressing vs. responding metastases, illustrates a more highly focused and potentially adaptive transcriptional profile that characterizes the progressing lesions. Transcriptional tumor profiling offers a powerful dimension in our understanding of response and resistance to cancer therapies. However, it also poses serious challenges stemming from the instability and fragmentation of RNA, especially in formalin-fixed specimens, and the possible contamination of tumor with normal tissues. Especially in melanoma, with many immediate pretreatment and on-treatment biopsies obtained from biopsies of cutaneous and superficial lymph node metastases, contamination with normal epithelium is a concern and indeed contributed to an artifactual “keratin” profile found in our preliminary analysis. This pitfall can be avoided only by tumor microdissection under direct visualization.
New therapies designed to address the complexities of cancer should be based on an understanding of how genetic and immunological diversity intersect. While studies of tumor biology in living patients with melanoma are facilitated by easy access to cutaneous and lymph nodal metastases which often occur, they are also limited by the typically small volumes of these disease sites and the inaccessibility of most visceral metastases. The current study based on the autopsy of an individual with widespread melanoma offers a window on how cancer evolution can be characterized concordantly at the genetic, transcriptional and immunological levels, and how we can begin to understand the basis of resistance to anti-PD-1 therapy.
MATERIALS AND METHODS
Tissue processing
At autopsy, entire cutaneous metastases, or subsections in the case of very large lesions, were collected from the skin prior to opening the body. The body cavity was subsequently opened with standard autopsy techniques and each grossly identified metastasis was removed. All tumor and normal tissues were immediately flash-frozen in liquid nitrogen and stored at minus 80°C. For each tumor, one-half of the tissue was fixed in 10% buffered-formalin, and the remaining tissue was stored at −80°C. Each metastasis was embedded and frozen in Tissue-Tek OCT for sectioning using a Leica Cryostat. For the FFPE samples, slides were cut for tissue macrodissection using a Leica Microtome. Once visualized, each tumor was macrodissected to remove surrounding non-neoplastic tissue. A 5-micron thick section was used to create a hematoxylin and eosin (H&E) slide for estimating neoplastic cellularity with a standard microscope. We estimated that the neoplastic cellularity was >80% for all samples collected.
Cell lines
The established melanoma cell lines 397-, 537-, 624-, 1011-, 1088-, 1558-, and 2048-mel were generated from metastatic melanoma lesions, each from a unique patient, in our laboratory and maintained as previously described;15 they were characterized by morphology, expression of melanoma antigens, and HLA typing to confirm identity with the donor of origin. They were confirmed to be mycoplasma-free. Growing cell cultures were harvested from flasks with trypsin and washed with centrifugation, and cell pellets were snap-frozen on dry ice for subsequent RNA isolation.
Genomic analysis
DNA extraction and quantification
Genomic DNA (gDNA) was extracted from each tissue piece using phenol and chloroform, followed by ethanol precipitation in ethanol, then quantified by LINE assay [i.e., long interspersed elements (LINE)] using real-time PCR. This method is highly sensitive for calculating the concentration of gDNA. The LINE forward primer used was 5′-AAAGCCGCTCAACTACATGG-3′ and the reverse primer used was 5′-TGCTTTGAATGCGTCCCAGAG-3′. The real-time PCR protocol was 50°C for 2 min, 95°C for 2 min, 40 cycles of 94°C for 10 s, 58°C for 15 s, and 70°C for 30 s, 95°C for 15 s, and 60°C for 30 s. All PCR reactions utilized Platinum SYBR Green qPCR mastermix (Invitrogen). Only high quality tissue samples with adequate concentrations were used for further study. For FFPE samples, gDNA was extracted using standard protocols and quantified with a Qbit prior to the generation of sequencing libraries.
Whole exome sequencing and analysis
Frequently mutated regions of BRAF, KIT, NRAS, and PIK3CA were assessed in a diagnostic tumor biopsy (lesion M26, Table 1) by next-generation sequencing (Ion AmpliSeq Cancer HotSpot Panel v2, Life Technologies).
Exome sequencing was performed on 4 pre-mortem FFPE tumor specimens, 22 autopsy tumor specimens (FFPE or fresh frozen) and two matched normal samples acquired at autopsy (FFPE and fresh frozen normal kidney) using between 500 ng and 1 ug of gDNA captured by hybridization using the SureSelect XT Human All Exon V4 (Agilent Technologies) (Table 1). Samples were prepared according to the manufacturer’s instructions. PCR amplification of the libraries was carried out for 6 cycles in the pre-capture step, and either 8 or 10–12 cycles post-capture for frozen and FFPE materials, respectively. Samples were barcoded and run on a Hiseq 2500 in a 75 bp paired end run using the TruSeq SBS Kit v3 (Illumina). The average depth of coverage in target regions was 180x and 86x, respectively, for tumor and matched normal samples with an average duplication rate of ~14% across all specimens, and 96.7% of the targeted regions were sequenced to 30-fold coverage or higher. Raw sequencing reads were aligned and mutations detected as previously described. Briefly, paired-end sequencing data from exome capture libraries were aligned to the reference human genome (hg19) using BWA.24 Read de-duplication, base quality recalibration, and multiple sequence realignment were performed using the Picard Suite and GATK.25,26 Coordinate sorted BAM files were further processed for quality control and point mutations and small insertions and deletions were detected with MuTect and HaplotypeCaller respectively.25,27 In total, 21,550 candidate somatic mutations were detected, of which 2207 were unique variants across the tumor samples.
Total, allele-specific, and integer DNA copy number genome-wide were determined with the FACETS algorithm from exome data in all cases.28 The FACETS framework segments total and allele-specific DNA copy number simultaneously from the coverage and genotypes of polymorphic SNPs genome-wide. Allele-specific segmentation is based on the log odds-ratio of allele fractions at SNPs identified as heterozygous in the matched normal sample. A fit is applied to the resulting segments, identifying in each sample the 1) log ratio corresponding to diploidy, 2) purity, and 3) average ploidy. Major and minor integer copy number is then assigned to each segment by maximum likelihood. The median purity and ploidy across all pre- and post-mortem specimens was estimated from sequencing data to be 0.75 and 2.88 respectively. The presence of whole-genome duplication was determined from a major integer copy number of 4 for the majority of the autosomal genome not affected by CNAs with the presence of a large-scale (chromosome/arm-length) heterozygous loss in total copy number that appeared to be in allelic balance with a major and minor copy number of 4 and 2. Finally, the purity and integer copy number results from FACETS analysis, along with coverage levels and allele frequencies, were used to estimate the fraction of cancer cells harboring each mutation (cancer cell fraction, CCF) in all specimens. For mutations in regions of genomic gain, two CCFs were calculated, assuming the minimum and maximum possible number of copies.29
To identify driver gene mutations, we utilized multiple methods to cross-reference the somatic variants relying on two pan-cancer studies.30,31 that leveraged ratiometric and statistical approaches to identify driver genes and hotspot mutations. We also analyzed significantly altered genes from three recent sequencing studies of cutaneous melanoma.11,32,33 as well as anti-PD-1 therapy sequencing studies.14,34–36 The somatic mutations were queried and manually reviewed for quality and biochemical effects. Sequence data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002195. Further information about EGA can be found at https://ega-archive.org and “The European Genome-phenome Archive of human data consented for biomedical research” (http://www.nature.com/ng/journal/v47/n7/full/ng.3312.html).
Evolutionary analysis was performed from a subset of the highest confidence somatic mutations. Phylogeny construction was based on the subset of somatic mutations (n=795) with greater than 20-fold coverage in every tumor and normal specimen, with a minimum allelic fraction of 10% in at least one tumor specimen, and no more than a single variant read or 2% allelic fraction in both the frozen and FFPE matched normal samples. The phylogenetic tree was rooted using the patient’s matched normal sample and the leaves comprised the primary tumor, local and distant pre-mortem recurrences, and post-mortem metastatic samples. We generated a binary classification to describe the presence or absence of each somatic mutation in each sample based on an allelic fraction of >5% in a given sample. We then utilized the discrete character parsimony algorithm to construct the most parsimonious tree of this patient’s disease utilizing PHYLIP.37 The resulting phylogeny indicated that some mutations were either lost during evolution or were undetected during sequencing, possibly due to sample cellularity and sequencing sensitivity of chromosome-level genetic mutations.
The patient samples were HLA typed and analyzed as described previously.38 Neoantigen analysis discovered 812 unique nonamers across the tumors predicted to have ≤500 nm HLA binding affinity.
Histologic review and immunohistochemistry
Ten post-treatment cutaneous metastases harvested at autopsy were submitted for routine H&E staining as well as IHC. Cases were reviewed by a board-certified dermatopathologist (JMT) for the presence of post-autopsy autolysis. Percent tumor necrosis and the presence of lymphoid aggregates were scored for each specimen on H&E. Additionally, the architectural pattern of immune cell infiltration was scored as mild, moderate, or severe, as previously described.12 IHC for CD3, CD4, CD8, CD68, CD20, and FoxP3 was performed according to standard automated methods. IHC stains for PD-L1, PD-1 and LAG-3 were performed as previously described.12,13,39 TIM-3 IHC was performed using clone F38-2E2 (BioLegend, San Diego, CA) at a concentration of 1.5 ug/ml. Antigen retrieval was performed in citrate buffer, pH 6.0 using a decloaking chamber (Biocare Medical, Concord, CA) set to 120°C for 10 minutes. The primary antibody was incubated at 4°C overnight. An anti-mouse IgG1-biotin secondary antibody (BD Biosciences, San Jose, CA) was used at 1.0 ug/ml. The signal was amplified (Dako K1500) followed by addition of streptavidin-HRP, and visualization with DAB. Membranous PD-L1 expression on tumor or infiltrating immune cells was scored separately as none, <5%, 5–10% and at increasing 10% intervals. The geographic association of PD-L1 expression with the presence or absence of immune infiltrates was also noted. Lymphocyte subset markers (PD-1, CD20, FoxP3, LAG-3, TIM-3) were scored on a 0–3 scale, representing the proportion of lymphocytes present expressing the marker of interest (0, none; 1, <5%; 2, 5–50%; 3, >50%).39 The CD4:CD8 ratio of cells was also determined for each lesion.
Quantitative analysis was performed for CD8+ T-cell infiltration. Slides stained for CD8 were scanned using the Aperio ImageScope (Leica Biosystems). The slide images were then annotated and quantitatively analyzed using Halo Image Analysis Software (Indica labs). After the tumor border was delineated and necrotic regions were excluded, 25 μM zones on each sides of the tumor border were generated. The intratumoral zones were designated as “T” zones, and the extratumoral (i.e., peripheral) zones were designated as “P” zones (Supplementary Figure S2B), with zones T1 and P1 representing the 25 um zone closest to the tumor border on the respective sides. The density of CD8+ T-cells in each zone was quantified using the Immune Cell Module of the Halo software. CD8+ T cell densities were compared between progressing and regressing lesions using the Wilcoxon rank-sum test.
LAMA3 protein expression was assessed by IHC per standard methods using Dako Target Retrieval Solution, a mouse monoclonal antibody (mAb) raised against human LAMA3 protein (Atlas Antibodies, cat. #AMAb91123), and a Dako Envision+ Dual Link Kit for detection of immunolabeled protein. Sections were incubated with a 1:100 dilution of anti-LAMA3 overnight at room temperature. Both the intensity of labeling (0–3) and the percent of melanoma cells labeling (0–100%) were recorded and used to generate a Histo score (intensity x percent positive cells) that ranged from 0–300. Cytokeratin 1 (KRT-1) was detected with the mAb LHK1 (ThermoFisher/Invitrogen, cat. #MA5-16032). Low and high molecular weight cytokeratins were detected with the mAb cocktail AE1/AE3 (Agilent/Dako, cat. #M351529-2).
Gene expression analysis with quantitative (q)RT-PCR
For FFPE tumor specimens, areas of tumor were identified with H&E staining on neighboring tissue sections, and tissue was manually dissected by scraping with a scalpel for RNA isolation with the High Pure RNA Paraffin Kit (Roche, Indianapolis, IN), as previously described.12 Alternatively, areas of tumor that were free from necrotic or normal tissues were isolated with laser capture microdissection as described.12 For cultured melanoma cells, total RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. One hundred fifty ng of total RNA was reverse-transcribed in a 10 ul reaction volume using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg MD) per protocol. From each RT reaction, 7.5 ul was pre-amplified in a total volume of 30 ul using a 14-cycle PCR reaction per PreAmp protocol (Applied Biosystems, Foster City CA). Fourteen ul of each pre-amplification reaction was expanded into a 440 ul total volume reaction mix and added to custom-made TaqMan Low-Density Array Micro Fluidic Cards per protocol (TLDA; Applied Biosystems, Waltham, MA). One TLDA Card contained 60 candidate immune genes of interest and 4 endogenous control genes as previously described.13 Other TLDA Cards contained genes derived from whole genome microarray results (Supplementary Table S3). Results from FFPE tissue specimens were analyzed using the 2−ΔΔCt method,40 with the Welch t-test and visualized with TIBCO Spotfire Software (Somerville, MA). Results from melanoma cell lines were visualized by using GraphPad software (La Jolla, CA).
Gene expression analysis with whole genome microarray
Global gene expression in tumor specimens from melanoma cutaneous metastases [regressing on therapy (R), n=6; or progressing on anti-PD-1 therapy (P), n=4] was measured by DASL (cDNA-mediated Annealing, Selection, extension, and Ligation) assays arrayed on the Illumina Human HT-12 WG-DASL V4.0 R2 expression BeadChip, per the manufacturer’s specifications (Illumina, San Diego, CA). This platform detects 29,377 annotated transcripts and is designed to detect partially degraded mRNAs such as typically found in FFPE tissue specimens. Details are provided in Supplementary Methods. For each Illumina probe on the DASL array, the expression levels in R and P lesions were compared. Principal component analysis based on a list of 866 Illumina probes corresponding to 827 transcripts with R vs. P fold change (FC) magnitude ≥1.5 was performed using Partek Genomic Suite (St Louis, MI). For functional analysis, two lists of genes (i.e., Illumina Probe_IDs) for which the fold change (FC) was ≥1.5 (up in R), or at or below −1.5 (up in P),were examined for significant enrichment in gene ontology (GO) categories and in functional categories including KEGG pathways, using the DAVID 6.7 web tool (http://david.abcc.ncifcrf.gov/).41,42 Specifically, for each list of Illumina Probe IDs submitted to DAVID, the DAVID Functional Annotation Tool was run on the resulting Homo sapiens genes recognized in the DAVID database, followed by Functional Annotation Clustering. The Illumina Probe IDs from the submitted list that were in each Term (gene set) in the DAVID output file were then replaced by the corresponding currently available (as of January 2016) NCBI/HUGO gene symbol.
To generate a heat map, genes derived from the R vs. P class comparison using the criteria of Welch t test p-value ≤ 0.05 and fold change magnitude ≥1.7 (calculated using the geometric mean of the expression levels in each group) were visualized using the Stanford Cluster 3.0 and TreeView software as elsewhere described.43 The gene expression data have been deposited in NCBI’s Gene Expression Omnibus44 and will be accessible through GEO Series accession number GSE79691. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79691). The GEO platform for this dataset is GPL14951.
In silico analysis of gene expression profile
Briefly, 140 probes were mapped to genes according to IlluminaHumanWGDASLv4.db45 (Supplementary Table S3), resulting in 133 unique gene annotations among which 124 were also represented in TCGA RNAseq dataset. An additional seven genes were mapped between platforms manually, through updated probe annotations, and/or gene aliases, to reach a total of 131 genes. The same mapping was used for the 13 genes confirmed as differentially expressed by RT-PCR. Logarithmic signal values and Manhattan distance were used for the clustering procedure. Kaplan–Meier survival curves were performed, in R, as elsewhere described, by applying the Peto & Peto modification of the Gehan-Wilcoxon test.46,47 We also ran the same analysis on [Hugo, 2016/GSE78220].14
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This rapid autopsy study of a patient with widespread melanoma experiencing a mixed response to anti-PD-1 therapy revealed highly homogeneous genomic and immunologic characteristics in responding and non-responding lesions. However, a distinct gene expression profile was associated with non-response, suggesting that intratumoral transcriptional programs can determine response/resistance to anti-PD-1 therapy.
Acknowledgments
Financial Support: This study was supported by the Melanoma Research Alliance (to SLT and CI-D), the Bloomberg~Kimmel Institute for Cancer Immunotherapy (to JMT, DMP, and SLT), the Barney Family Foundation (to SLT), Moving for Melanoma of Delaware (to SLT), the Laverna Hahn Charitable Trust (to SLT), the National Cancer Institute (R01 CA142779 to JMT, DMP and SLT; 5T32 CA193145 to TRC; R01 CA179991 to CI-D; P30-CA008748 to Memorial Sloan Kettering Cancer Center), the MSKCC TROT Fellowship (to AM-M), the Starr Cancer Consortium (to TAC), the Pershing Square Sohn Cancer Research Foundation (to TAC), the Immunogenomics and Precision Oncology Platform (to TAC), and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology (to DBS).
We thank Haiying Xu, Aleksandra Ogurtsova, and George A. Crabill (Johns Hopkins University School of Medicine) and Rajya Kappagantula (Memorial Sloan-Kettering Cancer Center) for technical assistance, Steven Hashagen (Indica Labs, Corrales, NM) for assistance with CD8 T cell mapping, Annamalai Selvakumar (Memorial Sloan-Kettering Cancer Center) for assisting with HLA genotyping, and Ludmila Danilova (Johns Hopkins) for advice on statistical analyses.
Footnotes
The following authors have declared relevant financial relationships: EJL receives research funding from Genentech and Merck, and consults for Amgen, Bristol-Myers Squibb and Merck. JMT receives research funding from Bristol-Myers Squibb and serves on advisory boards for Bristol-Myers Squibb, Merck, and AstraZeneca. RHH receives royalty payments through his institution, from Myriad Genetics, and serves on the board of directors of MiDiagnostics. TAC receives research funding from Bristol-Myers Squibb, and is a co-founder of and holds equity in Gritstone Oncology. DMP receives research grants from Bristol-Myers Squibb; consults for Five Prime Therapeutics, GlaxoSmithKline, Jounce Therapeutics, MedImmune, Pfizer, Potenza Therapeutics, and Sanofi; holds stock options in Jounce and Potenza; and is entitled to receive patent royalties through his institution, from Bristol-Myers Squibb and Potenza. DBS consults for Pfizer and Loxo Oncology. SLT receives research funding from Bristol-Myers Squibb. MLA, AM-M, TLM, AEB, JF, GJK, TRC, ZAK, AF, VM, NR, LC, BST, and CI-D do not declare any conflicts.
References
- 1.Embuscado EE, Laheru D, Ricci F, Yuni KJ, Witzell SD, Seigel A, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–54. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlinger M. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–72. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu BJ, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–87. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nissan MH, Pratilas CA, Jones AM, Ramirez R, Won HL, Liu CL, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–50. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taube JM, Anders RA, Young GD, Xu HY, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127–37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taube JM, Young GD, McMiller TL, Chen SM, Salas JT, Pritchard TS, et al. Differential expression of immune-regulatory genes associated with PD-L1 display in melanoma: implications for PD-1 pathway blockade. Clin Cancer Res. 2015;21:3969–76. doi: 10.1158/1078-0432.CCR-15-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–25. [PubMed] [Google Scholar]
- 16.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugo W, Shi HB, Sun L, Piva M, Song CY, Kong XJ, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532:250–4. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Liu C-C, Lin J-H, Hsu T-W, Hsu J-W, Chang J-W, Su K, et al. Collagen XVII/laminin-5 activates epithelial-to-mesenchymal transitionand is assocaited with poor prognosis in lung cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11208. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seftor REB, Seftor EA, Kirschmann DA, Hendrix MJC. Targeting the tumor microenvironment with chemically modified tetracyclines: inhibition of laminin 5 γ2 chain promigratory fragments and vaculogenic mimicry. Mol Cancer Ther. 2002;1:1173–9. [PubMed] [Google Scholar]
- 21.Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–23. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spranger S, Bao RY, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–35. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 23.Chao T, Furth EE, Vonderheide RH. CXCR2-dependent accumulation of tumor-associated neutrophils regulates T-cell immunity in pancreatic ductal adenocarcinoma. Cancer Immunol Res. 2016;4:968–82. doi: 10.1158/2326-6066.CIR-16-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mose LE, Wilkerson MD, Hayes DN, Perou CM, Parker JS. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics. 2014;30:2813–5. doi: 10.1093/bioinformatics/btu376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen R, Seshan V. FACETS: Fraction and allele-specific copy number estimates from tumor sequencing. Meml Sloan-Kettering Cancer Center, Dept Epidemiol Biostat Work Pap Ser. 2015 doi: 10.1007/978-1-0716-2293-3_7. [DOI] [PubMed] [Google Scholar]
- 29.Greenman CD, Pleasance ED, Newman S, Yang F, Fu B, Nik-Zainal S, et al. Estimation of rearrangment phylogeny for cancer genomes. Genome Res. 2012;22:346–61. doi: 10.1101/gr.118414.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen P-L, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–37. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsenstein J. PHYLIP- Phylogeny Inference Package (version 3. 2) Cladistics. 1989;5:164–6. [Google Scholar]
- 38.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim J, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. Bmc Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunning M, Lynch A, Eldridge M. R package version 1.26.0. IlluminaHuman WGDASLv4.db: Illumina Human WDASLv4 annotation data. [Google Scholar]
- 46.Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J R Stat Soc Ser a-G. 1972;135:185–7. [Google Scholar]
- 47.Kalbfleisch JD, Prentice RL. The statistical analysis of failure time data. 2. 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.