Abstract
The ATP-binding cassette subfamily C member 4 gene encodes a transmembrane protein involved in the export of proinflammatory molecules, including leukotriene, prostaglandin, and sphingosine-1-phosphate across the plasma membrane. Those metabolites play important roles in asthma. We investigated the potential associations between ABCC4 gene polymorphisms and asthma phenotype. In total, 270 asthma patients and 120 normal healthy controls were enrolled for a genetic association study. Two polymorphisms (−1508A>G and −642C>G) in the ABCC4 promoter were genotyped. The functional variability of the promoter polymorphisms was analyzed by luciferase reporter assay. Inflammatory cytokine levels were measured by enzyme-linked immunosorbent assay. Serum and urinary eicosanoid metabolites, sphingosine-1-phosphate, were evaluated by quadrupole time-of-flight mass spectrometry. Asthma patients carrying the G allele at −1508A>G had significantly higher serum levels of periostin, myeloperoxidase, and urinary levels of 15-hydroxyeicosatetraenoic acid and sphingosine-1-phosphate (P = 0.016, P = 0.027, P = 0.032, and P = 0.010, resp.) compared with noncarrier asthma patients. Luciferase activity was significantly enhanced in human epithelial A549 cells harboring a construct containing the −1508G allele (P < 0.01 for each) compared with a construct containing the −1508A allele. A functional polymorphism in the ABCC4 promoter, −1508A>G, may increase extracellular 15-hydroxyeicosatetraenoic acid, sphingosine-1-phosphate, and periostin levels, contributing to airway inflammation in asthmatics.
1. Introduction
Multidrug resistance protein 4 (ABCC4) is a member of the family of ATP-binding cassette transporters required for the active transport of many bioactive substrates across the cell membrane [1]. ABCC4 pumps various substrates, including eicosanoids, cyclic nucleotides, bile salts, steroids, and other drugs, out of the cell to control multiple cellular signaling processes, including inflammation, cancer, cardiovascular homeostasis, platelet function, endothelial barrier function, vascular smooth muscle cell proliferation, and vasodilation [1–6].
Eicosanoids such as prostaglandin E2 (PGE2) and cysteinyl leukotriene (LT) E4, along with cyclic adenosine monophosphate (cAMP), are exported via ABCC4, and these molecules play important roles in airway inflammation [7, 8]. Previous studies have demonstrated that intracellular elevation of cAMP exerts an anti-inflammatory effect and PGE2 induces cAMP [9, 10]. The exposure of eosinophils to LTE4 also induces cAMP production [11]. Additionally, cAMP is involved in the induction and regulation of T helper (Th2) immunity, particularly in allergic asthma via dendritic cells [12]. Pharmacological inhibition of ABCC4 reduces the migration of human dendritic cells, indicating an important role for ABCC4 in human immunology [13]. Therefore, ABCC4 may play an important role in inflammatory diseases, particularly in asthma, by regulating the intracellular concentration of cAMP [12]. In addition, ABCC4 facilitates the transport of sphingosine-1-phosphate (S1P), the main active metabolite of sphingolipids, from the platelets [14].
ABCC4 is ubiquitously expressed, with particularly high expression in hematopoietic stem cells and blood cells [15]. Limited data on the functions of its variants are available despite the fact that ABCC4 is a highly polymorphic gene. ABCC4 variants are associated with various diseases; however, no report has implicated an association between ABCC4 and immunological diseases. Copsel et al. demonstrated that an ABCC4 polymorphism regulates the cellular levels of cAMP and controls human leukemia cell proliferation and differentiation, indicating its role in cellular processes [16]. Therefore, ABCC4 variants may also play an important role in the pathogenesis of asthma.
There is little evidence regarding associations of ABCC4 polymorphisms with asthma in Korean patients. Therefore, we investigated the potential associations between asthma and ABCC4 polymorphisms in a Korean population.
2. Materials and Methods
2.1. Study Subjects
We enrolled 270 asthma patients and 120 normal healthy controls (NCs) from the Department of Allergy and Clinical Immunology, Ajou University Hospital, Suwon, Korea. Written informed consent was obtained from each subject, and the study was approved by the Institutional Review Board of Ajou University Hospital (AJIRB-GEN-SMP-13-108).
Methacholine bronchial challenge tests were performed as described previously [17]. NCs were selected from the general population using a screening questionnaire. Participants with a history of respiratory symptoms or aspirin hypersensitivity were excluded. All NC subjects exhibited a forced expiratory volume 1 (FEV1) > 80% of the predicted value, a provocation concentration (PC20) of methacholine > 25 mg/mL and normal findings on chest radiographs. Atopy was defined as one or more positive reactions on a skin prick test of 12 common aeroallergens (Bencard Co., Brendford, UK); histamine and saline served as controls. Serum total IgE levels were measured using the UniCAP system (Thermo Scientific, Uppsala, Sweden) according to the manufacturer's instructions. The threshold cut-off value for a specific IgE level was 0.35 kU/L, as measured by UniCAP. The presence of rhinosinusitis and nasal polyps was determined using paranasal sinus X-ray and rhinoscopy.
2.2. DNA Extraction, Single Nucleotide Polymorphism (SNP) Identification, and Genotyping
Each of the twenty Korean asthma patients and NCs was used for SNP identification. Total genomic DNA was isolated from peripheral blood samples using the Puregene DNA Purification Kit (Gentra, Minneapolis, MN, USA) according to the manufacturer's protocol. Our objective was to screen for promoter and 5′-untranslated region (UTR) SNPs. Based on previous findings and sequencing results, we chose two SNPs: one in the promoter and one in the 5′UTR of ABCC4 (−1508A>G and −642C>G, resp.). The two SNPs were genotyped using the TaqMan Allelic Discrimination assay with TaqMan probes (rs868853, −1508A>G assay ID c__7461591_10; rs869951, −642C>G, c__7461587_10; Applied Biosystems, Foster City, CA, USA).
2.3. Quantification of Serum and Urinary Metabolites
For the serum and urinary metabolites, we enrolled 60 and 31 asthmatic patients for measurement, respectively. The serum and urinary levels of S1P and five eicosanoid metabolites, LTE4, prostaglandin F2α (PGF2α), thromboxane B2 (TXB2), 15-hydroxyeicosatetraenoic acid (15-HETE), and eoxin C4, were determined using the Agilent 6530 quadrupole time-of-flight (Q-TOF) mass spectrometer. The device settings have been described in detail in a previous study [18].
2.4. Measurement of Serum Inflammatory Cytokines
Several inflammatory biomarkers, including myeloperoxidase (MPO), interleukin- (IL-) 8, IL-18, eotaxin-1, and eotaxin-2, were measured by enzyme-linked immunosorbent assay (ELISA) (Quantikine, R&D Systems, Minneapolis, MN, USA). Serum periostin levels were measured using a proprietary sandwich ELISA kit (Shino-test, Kanagawa, Japan) [19]. Each sample was run in duplicate. Serum samples were stored at −80°C prior to use.
2.5. Activity of the ABCC4 Promoter Constructs
Human mast cells (HMC-1) were cultured in Iscove's Modified Dulbecco's Medium (Gibco, Grand Island, NY, USA). A549, human alveolar type II epithelial-like, and U937, human leukemic monocyte lymphoma cell lines, were cultured in Roswell Park Memorial Institute-1640 Medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate (Gibco) at 37°C in a 5% CO2 incubator.
A 1681 bp fragment of the human ABCC4 gene was amplified from the genomic DNA of −1508GG and −1508AA homozygous subjects by PCR using the following primers: (forward) 5′-TCTATCGATAGGTACGGCCATGCTTAGACATAGGCTTA-3′ and (reverse) 5′-GATCGCAGATCTCGAAGAACACGCGTGAGCAGAGGTT-3′. PCR products were gel purified using an Agarose Gel Purification Kit (GeneAll Biotechnology, Seoul, Korea) and ligated into the pGL3-basic vector (Promega, Madison, WI, USA) after digestion with KpnІ and XhoІ (Takara, Shuzo, Japan) using the In-Fusion® HD Cloning Kit (Clontech Laboratories Inc., Mountain View, CA, USA). All constructs were confirmed by a restriction enzyme analysis and DNA sequencing. Plasmid DNAs were prepared from these constructs using the Endo-Free Plasmid Maxi Kit (Qiagen, Hilden, Germany), and the concentration and purity were assessed by UV spectrophotometry and agarose gel electrophoresis. Before transfection, the constructs were verified by direct sequencing.
The constructs were transfected into A549, HMC-1, and U937 cells using Lipofectamine (Invitrogen) according to the manufacturer's protocol. Briefly, 1 × 105 cells were seeded into 12-well plates and, after reaching 70–80% confluency, were transfected with 1 μg of the reporter construct, 5 ηg Renilla plasmid DNA and 5 μL Lipofectamine. Forty-eight hours after transfection, the cells were lysed and assayed for firefly luciferase activity according to the manufacturer's instructions (Promega). Transfection and luciferase assays were repeated three times according to the method described above.
2.6. Statistical Analyses
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). Differences in clinical characteristics among the groups were examined using the independent t-test for continuous variables and the χ2 test for categorical variables. Genotype frequency was examined between the subject groups using a χ2 test, and differences in clinical characteristics, cytokines and metabolites, according to genotype were examined using a logistic regression analysis with codominant, dominant, and recessive models after accounting for age and sex as covariables. Statistical significance was established at P < 0.05.
3. Results
3.1. Clinical Characteristics of the Study Subjects
The clinical characteristics of the study population are summarized in Table 1. The mean age of the asthma patients was 43.8 (±13.85) years and that of the NCs was 27.04 (±7.17) years (P < 0.001). The percentage of males was significantly higher in the asthma group (58.21%) than in the NC group (38.35%) (P < 0.001). In the asthma group, atopy was observed in 48.50% of patients, rhinosinusitis in 83.75%, and nasal polyps in 39.44%.
Table 1.
Clinical characteristics of study subjects.
| Asthmatics (n = 270) |
NC (n = 120) |
P value | |
|---|---|---|---|
| Age (years)† | 43.80 ± 13.85 | 27.04 ± 7.17/98 | <0.001 |
| Male (%)‡ | 58.21 | 38.35 | <0.001 |
| Atopy (%)† | 48.50 | NA | NA |
| Total IgE (kU/L)† | 0.431 ± 0.9 | NA | NA |
| Rhinosinusitis (%)‡ | 83.75 | NA | NA |
| Nasal polyp (%)‡ | 39.44 | NA | NA |
| FEV1 (%)† | 85.69 ± 20.30 | NA | NA |
| PC20 methacholine (mg/mL) |
9.17 ± 15.99 | NA | NA |
| Asthma duration (years)† |
7.69 ± 15.97 | NA | NA |
NC: normal control; n: number of subjects; NA: not applicable; FEV1: forced expiratory volume in 1 s; PC20 methacholine: provocative concentration of methacholine producing a 20% fall in FEV1. †This value was presented as mean ± SD, whereas ‡ value was shown as percentage. Values in bold indicate significant P value. Each P value <0.05 was considered to be significant. P value was obtained by t-test for continuous variables and the χ2 test for categorical variables.
3.2. No Association of the ABCC4 Promoter Polymorphisms with Asthma
Two promoter polymorphisms of ABCC4 gene (−1508A>G and −642C>G) were examined in this study. Linkage disequilibrium analysis was performed between the two ABCC4 SNPs. Three common haplotypes, ht1 [AC], ht2 [AG], and ht3 [GG], were constructed using the EM algorithm (Table 2), which revealed the genotype and haplotype frequencies of SNPs in the study subjects. There were no significant differences with respect to genotype or haplotype between the study groups.
Table 2.
Distribution of genotypes and haplotypes of ABCC4 gene polymorphisms.
| SNP | Genotype | Asthmatics (n = 270) |
NC (n = 120) |
P value |
|---|---|---|---|---|
| −1508A>G (rs868853) |
AA | 224 (83.3) | 102 (86.4) | 0.288 |
| AG | 47 (17.5) | 16 (13.6) | 0.417 | |
| GG | 2 (0.74) | 2 (1.69) | 0.648 | |
| −642C>G (rs869951) |
CC | 94 (34.9) | 52 (44.1) | 0.61 |
| CG | 139 (51.7) | 47 (39.8) | 0.101 | |
| GG | 36 (13.4) | 19 (16.1) | 0.068 | |
| ht1 [AC] | +/+ | 94 (34.9) | 52 (44.0) | 0.171 |
| +/− | 139 (51.7) | 47 (39.8) | 0.705 | |
| −/− | 36 (13.4) | 19 (16.1) | 0.068 | |
| ht2 [AG] | +/+ | 17 (6.3) | 14 (11.9) | 0.077 |
| +/− | 156 (57.9) | 50 (42.3) | 0.094 | |
| −/− | 96 (35.7) | 54 (45.7) | 0.366 | |
| ht3 [GG] | +/+ | 2 (0.74) | 2 (1.7) | 0.404 |
| +/− | 17 (6.3) | 3 (2.5) | 0.75 | |
| −/− | 250 (92.9) | 113 (95.7) | 0.454 |
NC: normal healthy control; n: number of subjects; ht: haplotype. P value was obtained by logistic regression analysis with age and sex as covariates.
3.3. Associations between the ABCC4 −1508A>G Polymorphism and Urinary Levels of Metabolites
We next examined potential associations between ABCC4 gene polymorphisms and serum and urinary eicosanoid metabolites and S1P (Table 3).
Table 3.
Association of ABCC4 −1508A>G polymorphism with clinical features, metabolite, and cytokine profiles in asthmatic patients.
| Clinical features | AA (n = 305) | AG/GG (n = 48) | P value |
|
| |||
| Age (years)† | 39.01 ± 14.34 | 40.92 ± 15.09 | 0.735 |
| Sex, male (%)‡ | 32.14 | 28.57 | 0.335 |
| Atopy (%)‡ | 57.46 | 51.18 | 0.928 |
| Total IgE (IU/mL)† | 451.48 ± 965.78 | 278.32 ± 319.68 | 0.240 |
| Rhinosinusitis (%)‡ | 81.21 | 18.78 | 0.143 |
| Nasal polyp (%)‡ | 81.11 | 18.88 | 0.889 |
| FEV1 (%)† | 86.45 ± 19.66 | 87.54 ± 14.57 | 0.638 |
| PC20 methacholine (mg/mL) | 10.19 ± 17.07 | 5.82 ± 8.04 | 0.119 |
| Total IgE | 451.48 ± 965.78 | 278.32 ± 319.69 | 0.24 |
| Asthma duration (years)† | 6.30 ± 6.03 | 12.81 ± 32.36 | 0.237 |
|
| |||
| Inflammatory cytokines | AA (n = 121) | AG/GG (n = 26) | P value |
|
| |||
| MPO (μg/L)† | 108.26 ± 79.5 | 150.91 ± 94.13 | 0.027 |
| IL-8 (pg/mL)† | 16.82 ± 14.45 | 14.47 ± 7.5 | 0.616 |
| IL-18 (pg/mL)† | 247.77 ± 166.06 | 281.92 ± 197.10 | 0.560 |
| Eotaxin-1 (ng/mL)† | 85.67 ± 63.67 | 85.22 ± 65.11 | 0.841 |
| Eotaxin-2 (ng/mL)† | 1146.92 ± 802.14 | 1104.95 ± 641.09 | 0.650 |
| Periostin (ng/mL)† | 71.07 ± 33.62 | 91.83 ± 50.85 | 0.016 |
|
| |||
| Serum metabolites (ng/mL) | AA (n = 43) | AG/GG (n = 17) | P value |
|
| |||
| 15-HETE | 265.98 ± 185.69 | 334.34 ± 132.47 | 0.195 |
| LTE4 | 18.22 ± 17 | 18.8 ± 14.71 | 0.731 |
| PGF2α | 23.55 ± 9.3 | 21.96 ± 8.69 | 0.653 |
| TXB2 | 0.41 ± 0.42 | 0.39 ± 0.17 | 0.952 |
| Eoxin C4 | 4.82 ± 9.5 | 8.14 ± 11.95 | 0.224 |
| S1P | 111.75 ± 42.16 | 120.91 ± 42.88 | 0.419 |
|
| |||
| Urinary metabolites (pmol/mg Cr) | AA (n = 23) | AG/GG (n = 8) | P value |
|
| |||
| 15- HETE | 271.91 ± 89.87 | 332.5 ± 99.31 | 0.032 |
| LTE4 | 7648.29 ± 14378.31 | 2601.73 ± 3043.59 | 0.616 |
| PGF2α | 8618.51 ± 22942.8 | 1678.71 ± 978.76 | 0.65 |
| TXB2 | 5686.61 ± 4305.7 | 4798.25 ± 3957.21 | 0.765 |
| Eoxin C4 | 144.78 ± 94.07 | 124.75 ± 46.65 | 0.602 |
| S1P | 32.56 ± 8.25 | 41.5 ± 9.35 | 0.010 |
†Values were presented as mean ± SD, whereas ‡ values were shown as percentage. Cr: creatinine; LTE4: leukotriene E4; PGF2α: prostaglandin F2α; TXB2: thromboxane B2; 15-HETE: 15-hydroxyeicosatetraenoic acid; MPO: myeloperoxidase; S1P: sphingosine-1-phosphate. P value was obtained by logistic regression analysis with age and sex as covariates.
Among five eicosanoid metabolites (LTE4, PGF2α, TXB2, 15-HETE, and eoxin C4), the urinary 15-HETE level was significantly associated with the ABCC4 −1508A>G polymorphism; asthma patients carrying the −1508G allele showed a significantly higher level than that of noncarriers (332 ± 99.31 versus 271.91 ± 89.87 pmol/mg creatinine [pmol/mg Cr], P = 0.032; Figure 1(a)). Serum levels of LTE4, PGF2α, TXB2, and eoxin C4 did not differ significantly between −1508G carriers and noncarriers among asthma patients (Table 3).
Figure 1.
Asssociation of the baseline levels of (a) urinary 15-HETE, (b) urinary S1P, (c) serum MPO, and (d) serum periostin with the ABCC4 −1508A>G polymorphisms in asthma patients. ABCC4: ATP-binding cassette subfamily C member 4; 15-HETE: 15-hydroxyeicosatetraenoic acid; MPO: myeloperoxidase; S1P: sphingosine-1-phosphate. P values were obtained by logistic regression with age and sex as covariates. The data represented as mean values ± SD.
Regarding to S1P, asthma patients carrying the −1508G allele showed a significantly higher level of the urinary level of S1P than that of noncarriers (41.5 ± 9.35 versus 32.56 ± 8.25 pmol/mg Cr, P = 0.010; Figure 1(b)).
3.4. Associations between the ABCC4 −1508A>G Polymorphism and Clinical Characteristics, Serum Periostin and MPO
Several inflammatory cytokines that are important biomarkers for asthma were also measured (Table 3). Among the cytokines examined, asthma patients carrying the −1508G allele showed significantly higher serum MPO levels (150.91 ± 94.13 versus 108.26 ± 79.5 mg/L, P = 0.027) and serum periostin level than those of noncarrier asthma patients (91.83 ± 50.85 versus 71.07 ± 33.62 ng/mL, P = 0.016), (Figures 1(c) and 1(d)).
3.5. Effects of the ABCC4 Polymorphisms on Transcriptional Activity
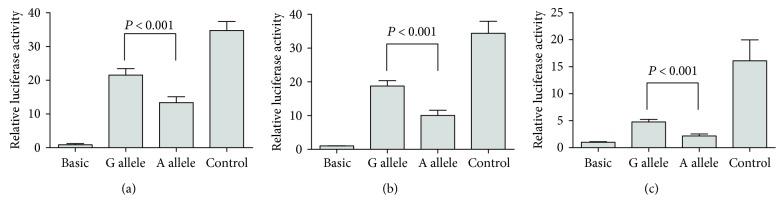
The luciferase reporter assay was performed using constructs containing two different ABCC4 alleles, −1508A and −1508G, to determine the transcriptional effects of the ABCC4 –1508A>G polymorphism. The constructs comprised of the ABCC4 sequence and a luciferase reporter gene were transfected into A549, U937, and HMC-1 cells. The reporter activities of the −1508G allele and −1508A allele constructs were compared. Luciferase activity was significantly enhanced in the construct with the −1508G allele compared with the −1508A allele in all cell lines (P < 0.01 for each, Figure 2).
Figure 2.
Effect of the ABCC4 −1508A>G polymorphism on the promoter activity. Transfection of plasmid constructs carrying ABCC4 −1508A or G allele into (a) A549, (b) U937, and (c) HMC-1 cells. All P values were obtained by Mann–Whitney U test. Data represent the mean values of three independent experiments ± SD. Each experiment was conducted in triplicate.
4. Discussion
Asthma is a complex, chronic respiratory disease with a wide clinical spectrum, with contributions from several environmental and genetic factors [20, 21]. Recent studies have shown that various gene polymorphisms influence the onset and progression of asthma [22, 23]. In the present study, we selected two SNPs within the ABCC4 gene to examine their potential roles in asthma pathogenesis based on their relationships with eicosanoid, sphingolipid metabolites, and proinflammatory cytokines. To date, the roles of these genes in asthma have not been determined.
Most studies on ABCC4 have focused on its role in cancer chemotherapy, particularly its ability to confer clinical drug resistance [24]. Diverse studies have shown that ABCC4 induces the extrusion of cyclic nucleotides in various cell types; however, it has emerged as the main transporter of cAMP [25]. van de Ven et al. reported that ABCC4 plays an important role in dendritic cell migration in humans and that inhibition of ABCC4 activity decreases dendritic cell migration in the skin [13]. However, no association study to date has examined the relationships between this gene and immunological diseases such as asthma, although this gene is associated with immunological processes.
We first identified a significant association between the ABCC4 −1508A>G polymorphism and the urinary levels of metabolites, including 15-HETE and S1P. 15-HETE is the major metabolite of arachidonic acid in the 15-lipoxygenase pathway [26]. 15-HETE was recently proposed as a biomarker for asthma severity, as its levels were 5-fold higher in eosinophils from severe asthmatics than from mild asthmatics [27]. 15-HETE undergoes reaction to produce 14,15-epoxides, designated eoxins A4, C4, D4, and E4 in eosinophils, mast cells, and nasal polyps from allergic subjects [28]. Similar to cysteinyl leukotrienes, eoxins are potent proinflammatory agents [28]. We found a tendency toward increased eoxin C4 levels in asthmatics with the −1508G allele, although no significance was observed, suggesting inflammation of the airways in eosinophilic asthma. Beside, S1P, a major metabolite of sphingolipid pathway, has been identified as a biomarker for asthma in our previous study [29]. S1P is suggested to contribute to airway hyperreactivity and release of IL-4 and IL-13, thereby involving in asthma pathogenesis [30, 31]. Inhibitors of ABCC4 block the release of S1P from platelet granules [14]. Therefore, the −1508G allele of ABCC4 polymorphism may be associated with the transport of metabolites from immune cells.
Secondly, we found enhanced serum periostin levels in asthma patients carrying the −1508G allele [32]. Periostin is a multifunctional protein expressed in many types of inflammatory cells, such as epithelial cells, mast cells, and so forth [33]. Recent studies have suggested that periostin modulates Th2-mediated asthma pathogenesis by assisting in the recruitment of inflammatory cells, particularly eosinophils, to the lungs [34]. Moreover, periostin is known to facilitate the activation of dendritic cells, thereby rendering airway hyperresponsiveness and airway inflammation in mice [35, 36]. Beside, we discovered an increased serum level of MPO in the ABCC4 −1508G allele carriers comparing to that of noncarriers. Activated neutrophils secrete MPO which then induces oxidative stress, thereby resulting in oxidative damage of respiratory cells, lung inflammation, cytotoxicity, airway obstruction, and decrease of lung function [37, 38]. A polymorphism of the MPO gene was proposed to be associated with asthma susceptibility [38]. Taken together, the ABCC4 –1508G allele may also interfere the release of periostin and MPO from inflammatory cells.
Although we did not find any genetic association in asthma patients in the present study, we found that a functional polymorphism of the ABCC4 gene (−1508A>G) may affect its promoter activity, thereby affecting release of 15-HETE, S1P, periostin, and MPO from innate immune cells in asthma.
5. Conclusions
To our knowledge, this is the first study to provide evidence of associations between ABCC4 and 15-HETE, S1P, periostin, and MPO in asthma patients. The present findings further suggest that ABCC4 represents a new potential target of asthma therapy. However, further studies are required to understand the functional mechanism of ABCC4 polymorphisms on airway inflammation in asthmatics.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korean government (MSIP) (Grant no. 2015R1C1A2A01053492) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) grant, and funded by the Ministry of Health and Welfare, Republic of Korea (Grant no. HI16C0992).
Conflicts of Interest
The authors have no conflict of interest to declare.
Authors' Contributions
Sailesh Palikhe and Eun-Mi Yang performed the experiments. Udval Uuganbayar and Hoang Kim Tu Trinh performed the statistical analysis and participated in writing the manuscript. Ga-Young Ban recruited patients and normal healthy controls. Hae-Sim Park and Seung-Hyun Kim interpreted the data, revised the manuscript, and supervised all the steps of this study.
References
- 1.Borst P., de Wolf C., van de Wetering K. Multidrug resistance-associated proteins 3, 4, and 5. Pflügers Archiv. 2007;453(5):661–673. doi: 10.1007/s00424-006-0054-9. [DOI] [PubMed] [Google Scholar]
- 2.Gelhaus S. L., Gilad O., Hwang W. T., Penning T. M., Blair I. A. Multidrug resistance protein (MRP) 4 attenuates benzo[a]pyrene-mediated DNA-adduct formation in human bronchoalveolar H358 cells. Toxicology Letters. 2012;209(1):58–66. doi: 10.1016/j.toxlet.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Z. P., Zhu Y. L., Johnson D. R., et al. Disruption of cAMP and prostaglandin E2 transport by multidrug resistance protein 4 deficiency alters cAMP-mediated signaling and nociceptive response. Molecular Pharmacology. 2008;73(1):243–251. doi: 10.1124/mol.107.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rees D. C., Johnson E., Lewinson O. ABC transporters: the power to change. Nature Reviews. Molecular Cell Biology. 2009;10(3):218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid G., Wielinga P., Zelcer N., et al. The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9244–9249. doi: 10.1073/pnas.1033060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao X., Guo Y., Yue W., Zhang L., Gu M., Wang Y. ABCC4 is required for cell proliferation and tumorigenesis in non-small cell lung cancer. Onco Targets Therapy. 2014;7:343–351. doi: 10.2147/OTT.S56029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billington C. K., Ojo O. O., Penn R. B., Ito S. cAMP regulation of airway smooth muscle function. Pulmonary Pharmacology & Therapeutics. 2013;26(1):112–120. doi: 10.1016/j.pupt.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rius M., Hummel-Eisenbeiss J., Keppler D. ATP-dependent transport of leukotrienes B4 and C4 by the multidrug resistance protein ABCC4 (MRP4) The Journal of Pharmacology and Experimental Therapeutics. 2008;324(1):86–94. doi: 10.1124/jpet.107.131342. [DOI] [PubMed] [Google Scholar]
- 9.Janciauskiene S. M., Nita I. M., Stevens T. α1-Antitrypsin, old dog, new tricks. α1-antitrypsin exerts in vitro anti-inflammatory activity in human monocytes by elevating cAMP. The Journal of Biological Chemistry. 2007;282(12):8573–8582. doi: 10.1074/jbc.M607976200. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Klein R. D. Prostaglandin E2 induces vascular endothelial growth factor secretion in prostate cancer cells through EP2 receptor-mediated cAMP pathway. Molecular Carcinogenesis. 2007;46(11):912–923. doi: 10.1002/mc.20320. [DOI] [PubMed] [Google Scholar]
- 11.Steinke J. W., Negri J., Payne S. C., Borish L. Biological effects of leukotriene E_{4} on eosinophils. Prostaglandins, Leukotrienes, and Essential Fatty Acids. 2014;91(3):105–110. doi: 10.1016/j.plefa.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J., Kim T. H., Murray F., et al. Cyclic AMP concentrations in dendritic cells induce and regulate Th2 immunity and allergic asthma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(5):1529–1534. doi: 10.1073/pnas.1417972112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Ven R., Scheffer G. L., Reurs A. W., et al. A role for multidrug resistance protein 4 (MRP4; ABCC4) in human dendritic cell migration. Blood. 2008;112(6):2353–2359. doi: 10.1182/blood-2008-03-147850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrych T., Böhm A., Polzin A., et al. Release of sphingosine-1-phosphate from human platelets is dependent on thromboxane formation. Journal of Thrombosis and Haemostasis. 2011;9(4):790–798. doi: 10.1111/j.1538-7836.2011.04194.x. [DOI] [PubMed] [Google Scholar]
- 15.Abla N., Chinn L. W., Nakamura T., et al. The human multidrug resistance protein 4 (MRP4, ABCC4): functional analysis of a highly polymorphic gene. The Journal of Pharmacology and Experimental Therapeutics. 2008;325(3):859–868. doi: 10.1124/jpet.108.136523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copsel S., Garcia C., Diez F., et al. Multidrug resistance protein 4 (MRP4/ABCC4) regulates cAMP cellular levels and controls human leukemia cell proliferation and differentiation. The Journal of Biological Chemistry. 2011;286(9):6979–6988. doi: 10.1074/jbc.M110.166868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S. H., Bae J. S., Holloway J. W., et al. A polymorphism of MS4A2 (−109T>C) encoding the beta-chain of the high-affinity immunoglobulin E receptor (FcepsilonR1beta) is associated with a susceptibility to aspirin-intolerant asthma. Clinical and Experimental Allergy. 2006;36(7):877–883. doi: 10.1111/j.1365-2222.2006.02443.x. [DOI] [PubMed] [Google Scholar]
- 18.Ban G. Y., Cho K., Kim S. H., et al. Metabolomic analysis identifies potential diagnostic biomarkers for aspirin-exacerbated respiratory disease. Clinical and Experimental Allergy. 2017;47(1):37–47. doi: 10.1111/cea.12797. [DOI] [PubMed] [Google Scholar]
- 19.Kim M. A., Yoon M. K., Lee Y. S., et al. Clinical implication of the serum periostin level for differentiating phenotypes of NSAID hypersensitivity. Allergology International. 2016;65(4):492–494. doi: 10.1016/j.alit.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Kauffmann F., Demenais F. Gene-environment interactions in asthma and allergic diseases: challenges and perspectives. The Journal of Allergy and Clinical Immunology. 2012;130(6):1229–1240. doi: 10.1016/j.jaci.2012.10.038. quiz 1241-2. [DOI] [PubMed] [Google Scholar]
- 21.Baos S., Calzada D., Cremades L., et al. Biomarkers associated with disease severity in allergic and nonallergic asthma. Molecular Immunology. 2017;82:34–45. doi: 10.1016/j.molimm.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Tizaoui K., Kaabachi W., Hamzaoui K., Hamzaoui A. Association of single nucleotide polymorphisms in toll-like receptor genes with asthma risk: a systematic review and meta-analysis. Allergy, Asthma & Immunology Research. 2015;7(2):130–140. doi: 10.4168/aair.2015.7.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vercelli D. Genetic polymorphism in allergy and asthma. Current Opinion in Immunology. 2003;15(6):609–613. doi: 10.1016/j.coi.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Sampath J., Adachi M., Hatse S., et al. Role of MRP4 and MRP5 in biology and chemotherapy. AAPS PharmSci. 2002;4(3):p. E14. doi: 10.1208/ps040314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassi Y., Abi-Gerges A., Fauconnier J., et al. Regulation of cAMP homeostasis by the efflux protein MRP4 in cardiac myocytes. The FASEB Journal. 2012;26(3):1009–1017. doi: 10.1096/fj.11-194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamberg M., Hedqvist P., Radegran K. Identification of 15-hydroxy-5,8,11,13-eicosatetraenoic acid (15-HETE) as a major metabolite of arachidonic acid in human lung. Acta Physiologica Scandinavica. 1980;110(2):219–221. doi: 10.1111/j.1748-1716.1980.tb06656.x. [DOI] [PubMed] [Google Scholar]
- 27.James A., Daham K., Backman L., et al. The influence of aspirin on release of eoxin C4, leukotriene C4 and 15-HETE, in eosinophilic granulocytes isolated from patients with asthma. International Archives of Allergy and Immunology. 2013;162(2):135–142. doi: 10.1159/000351422. [DOI] [PubMed] [Google Scholar]
- 28.Feltenmark S., Gautam N., Brunnström A., et al. Eoxins are proinflammatory arachidonic acid metabolites produced via the 15-lipoxygenase-1 pathway in human eosinophils and mast cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(2):680–685. doi: 10.1073/pnas.0710127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trinh H. K., Kim S. C., Cho K., et al. Exploration of the sphingolipid metabolite, sphingosine-1-phosphate and sphingosine, as novel biomarkers for aspirin-exacerbated respiratory disease. Scientific Reports. 2016;6 doi: 10.1038/srep36599.36599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roviezzo F., D’Agostino B., Brancaleone V., et al. Systemic administration of sphingosine-1-phosphate increases bronchial hyperresponsiveness in the mouse. American Journal of Respiratory Cell and Molecular Biology. 2010;42(5):572–577. doi: 10.1165/rcmb.2009-0108OC. [DOI] [PubMed] [Google Scholar]
- 31.Idzko M., Panther E., Corinti S., et al. Sphingosine 1-phosphate induces chemotaxis of immature and modulates cytokine-release in mature human dendritic cells for emergence of Th2 immune responses. The FASEB Journal. 2002;16(6):625–627. doi: 10.1096/fj.01-0625fje. [DOI] [PubMed] [Google Scholar]
- 32.Palikhe S., Kim S. H., Kim R. Y., Yang E. M., Park H. S. XXIV World Allergy Congress: Seoul, Korea. The World Allergy Organization Journal. 2016;9(Supplement 1):14–17. doi: 10.1186/s40413-016-0096-1. [DOI] [Google Scholar]
- 33.Liu A. Y., Zheng H., Ouyang G. Periostin, a multifunctional matricellular protein in inflammatory and tumor microenvironments. Matrix Biology. 2014;37:150–156. doi: 10.1016/j.matbio.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Li W., Gao P., Zhi Y., et al. Periostin: its role in asthma and its potential as a diagnostic or therapeutic target. Respiratory Research. 2015;16(1):p. 57. doi: 10.1186/s12931-015-0218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentley J. K., Chen Q., Hong J. Y., et al. Periostin is required for maximal airways inflammation and hyperresponsiveness in mice. The Journal of Allergy and Clinical Immunology. 2014;134(6):1433–1442. doi: 10.1016/j.jaci.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal S., Townley R. G. Role of periostin, FENO, IL-13, lebrikzumab, other IL-13 antagonist and dual IL-4/IL-13 antagonist in asthma. Expert Opinion on Biological Therapy. 2014;14(2):165–181. doi: 10.1517/14712598.2014.859673. [DOI] [PubMed] [Google Scholar]
- 37.Monteseirin J., Bonilla I., Camacho J., Conde J., Sobrino F. Elevated secretion of myeloperoxidase by neutrophils from asthmatic patients: the effect of immunotherapy. The Journal of Allergy and Clinical Immunology. 2001;107(4):623–626. doi: 10.1067/mai.2001.113566. [DOI] [PubMed] [Google Scholar]
- 38.Polonikov A., Solodilova M., Ivanov V. Genetic variation of myeloperoxidase gene contributes to atopic asthma susceptibility: a preliminary association study in Russian population. The Journal of Asthma. 2009;46(5):523–528. doi: 10.1080/02770900902818389. [DOI] [PubMed] [Google Scholar]