Abstract
Infective endocarditis (IE) is a septic inflammation of the endocardium. Recognition of microbial patterns, cytokine and acute phase responses, hemostasis features, and alterations in plasma lipid and calcium profile all have been reported to affect pathogenesis and clinical course of IE. Having recruited 123 patients with IE and 300 age-, sex-, and ethnicity-matched healthy blood donors, we profiled their genomic DNA for 35 functionally significant polymorphisms within the 22 selected genes involved in the abovementioned pathways, with the further genetic association analysis. We found that the G/A genotype of the rs1143634 polymorphism within the IL1B gene, the G/T genotype of the rs3212227 polymorphism within the IL12B gene, the A/G genotype of the rs1130864 polymorphism within the CRP gene, and the G allele of the rs1801197 polymorphism within the CALCR gene were associated with a decreased risk of IE whereas the T/T genotype of the rs1205 polymorphism within the CRP gene was associated with a higher risk of IE. Furthermore, heterozygous genotypes of the rs1143634 and rs3212227 polymorphisms were associated with the higher plasma levels of IL-1β and IL-12, respectively. Our results indicate that inherited variation in the cytokine, acute phase response, and calcium metabolism pathways may be linked to IE.
1. Introduction
Infective endocarditis (IE) is defined as an infection of the endocardial surface, which may involve native or prosthetic heart valves, the mural endocardium, interventricular septum, chordae tendineae, or surfaces of intracardiac devices [1]. In general, IE is caused by bacteria, in particular Staphylococcus, Streptococcus, and Enterococcus spp. [2], but fungi have also been reported as a culprit [3]. The most common signs and symptoms of IE are malaise, fatigue, coughing, chills, fever, weight loss, and heart murmur [1]. Incidence of IE significantly varies in different countries (from 1.5/100,000 in Netherlands to 11.6/100,000 in United States), with an average case fatality rate of 25% [4]. However, case fatality rates considerably depend on the etiological agent: from 10% in patients infected with Streptococcus spp. to 40% in those infected with Staphylococcus aureus [5–9]. The development and course of IE may depend on (1) the recognition of pathogen-associated molecular patterns as well as cytokine and acute phase response to causative agents [10–12], (2) hemostasis [13–15], (3) plasma lipid profile [16, 17], and (4) plasma calcium profile [18, 19].
Progress in genotyping technologies reasoned the studies on the association of single nucleotide polymorphisms (SNPs) with human diseases [20]. SNPs located in different genomic regions may cause distinct consequences, including altered (1) transcription initiation; (2) mRNA splicing; (3) protein folding, stability, and expression; and (4) posttranslational modifications [21]. Previously, we reported that the C/C genotype of the rs3775073 polymorphism within the TLR6 gene is associated with a twofold decreased risk of IE; other polymorphisms within the genes encoding pattern recognition receptors did not show a predictive value. Here, we investigated whether the SNPs within the cytokine, acute phase response, lipid metabolism, and calcium metabolism genes can alter individual susceptibility to IE.
2. Materials and Methods
2.1. Population
Inclusion criteria were as follows: (1) living in Kemerovo region for ≥2 generations; (2) Russian ethnicity; (3) clinically (modified Duke criteria, at least 1 major and 1 minor criteria or 3 minor criteria being fulfilled [22]) and histologically verified diagnosis of IE; and (4) written informed consent. Exclusion criteria were as follows: (1) belonging to the immigrant or aboriginal populations, (2) previous cancer diagnosis, (3) concomitant mental disorders and/or autoimmune diseases, (4) drug addiction, and (5) refusal to sign a written informed consent. All the patients underwent antibiotic therapy in the acute phase during the first admission to their district hospital according to the European guidelines [22]. Antibiotic therapy and treatment of all concomitant diseases were further performed during the preoperative period at the Kemerovo Cardiology Centre.
In total, we recruited 161 patients with IE admitted to our Research Institute during 2009–2016. After exclusion of 38 patients due to the abovementioned criteria, the study group finally included 123 patients (Table 1). The control group for this study was formed from 300 age- (±6 years), sex-, and ethnicity-matched asymptomatic blood donors with no history of drug addiction, cardiovascular disease, cancer, and autoimmune and mental disorders (Table 1). Data on the clinicopathological features and hospital complications/case fatality rate of patients with IE are presented in Table 2. The local ethical committee of the Research Institute for Complex Issues of Cardiovascular Diseases approved the study protocol. All the participants provided written informed consent after the study was fully explained.
Table 1.
Characteristics of the study population.
| Controls | Cases | Total | p value | |
|---|---|---|---|---|
| Number of subjects | 300 (70.92%) | 123 (29.07%) | 423 (100.00%) | |
| Median age with 95% CI | 55.00 (53.00–56.00) | 50.00 (48.00–53.00) | 53.00 (52.00–55.00) | 0.12 |
| Interquartile range | 44–62 | 37–59 | 42–61 | |
| Males | 190 (63.00%) | 77 (63.00%) | 267 (63.12%) | 0.99 |
| Females | 110 (37.00%) | 46 (37.00%) | 156 (36.88%) |
CI: confidence interval.
Table 2.
Characteristics of the patients with infective endocarditis.
| Basic characteristics | ||
| Type of infective endocarditis | Location | Valve involved |
| Native (93/123, 75.61%) | Left-sided (116/123, 94.31%) | Aortic (45/123, 36.58%) |
| Mitral (62/123, 50.41%) | ||
| Aortic and mitral (9/123, 7.31%) | ||
| Prosthetic (30/123, 24.39%) | Right-sided (6/123, 4.88%) | Tricuspid (6/123, 4.88%) |
| Device (0/123, 0.0%) | Double-sided (1/123, 0.81%) | Mitral and tricuspid (1/123, 0.81%) |
| Echocardiography characteristics (mean ± standard deviation) | ||
| Left atrial diameter, cm | 4.78 ± 0.90 | |
| Left ventricular end-diastolic diameter, cm | 6.01 ± 1.17 | |
| Left ventricular end-systolic diameter, cm | 3.92 ± 0.76 | |
| Left ventricular end-diastolic volume, cm3 | 207.80 ± 93.98 | |
| Left ventricular end-systolic volume, cm3 | 72.13 ± 36.05 | |
| Interventricular septal thickness, cm | 1.06 ± 0.18 | |
| Left ventricular posterior wall thickness, cm | 1.07 ± 0.19 | |
| Left ventricular ejection fraction, % | 64.00 ± 7.64 | |
| Right atrial diameter, cm | 5.20 ± 1.19 | |
| Right ventricular diameter, cm | 2.51 ± 1.08 | |
| Hospital complications | ||
| Hydrothorax | 32/123 (26.02%) | |
| Pneumonia | 21/123 (17.07%) | |
| Multiple organ dysfunction syndrome | 17/123 (13.82%) | |
| Arrhythmia | 33/123 (26.83%) | |
| Heart block | 7/123 (5.69%) | |
| Heart failure | 46/123 (37.40%) | |
| Myocardial infarction | 4/123 (3.25%) | |
| Stroke | 2/123 (1.63%) | |
| Hospital case fatality rate | ||
| Death | 1/123 (0.81%) | |
2.2. SNP Selection and Genotyping
For this study, we defined four main criteria for SNP selection: (1) location within cytokine, acute phase response, hemostasis, lipid metabolism, or calcium metabolism genes; (2) minor allele frequency ≥ 5% for Russian population tested with HapMap; (3) functional alteration of protein expression; and (4) few or no studies investigating the role of an SNP in IE. The National Center for Biotechnology Information dbSNP, SNPinfo, and SNPnexus databases were utilized for the SNP selection [23, 24]. In total, we selected 35 SNPs within 22 genes (Table 3).
Table 3.
Features of the gene polymorphisms used in the study.
| Single nucleotide polymorphism | Nucleotide substitution | Chromosomal position | Amino acid substitution | Forward 5′-3′ and reverse 3′-5′ polymerase chain reaction primers |
|---|---|---|---|---|
| IL1B gene | ||||
| rs1143634 | G > A | 113590390 | Phe105Phe | F: cataagcctcgttatcccatgtgtc R: aagaagataggttctgaaatgtgga |
| IL6 gene | ||||
| rs1554606 | T > G | 22768707 | Intronic | F: ttagttcatcctgggaaaggtactc R: cagggccttttccctctctggctgc |
| rs1800796 | G > C | 22766246 | 5′-upstream | F: atggccaggcagttctacaacagcc R: ctcacagggagagccagaacacaga |
| rs2069827 | G > T | 22765456 | 5′-upstream | F: gcccaacagaggtcactgttttatc R: atcttgaagagatctcttcttagca |
| IL6R gene | ||||
| rs2228145 | A > T/C | 154426970 | Asp358Val/Ala | F: aattttttttttaacctagtgcaag R: ttcttcttcagtaccactgcccaca |
| rs2229238 | T > C | 154437896 | 3′-UTR | F: ccagcagcctggaccctgtggatga R: aaaacacaaacgggctcagcaaaag |
| IL8 gene | ||||
| rs2227306 | C > T | 74607055 | Intronic | F: aactctaactctttatataggaagt R: gttcaatgttgtcagttatgactgt |
| IL10 gene | ||||
| rs1800871 | A > G | 206946634 | 5′-upstream | F: agtgagcaaactgaggcacagagat R: ttacatcacctgtacaagggtacac |
| rs1800872 | T > G | 206946407 | 5′-upstream | F: ttttactttccagagactggcttcctacag R: acaggcggggtcacaggatgtgttccaggc |
| rs1800896 | T > C | 206946897 | 5′-upstream | F: tcctcttacctatccctacttcccc R: tcccaaagaagccttagtagtgttg |
| IL12B gene | ||||
| rs3212227 | T > G | 158742950 | 3′-UTR | F: attgtttcaatgagcatttagcatc R: aactatacaaatacagcaaagatat |
| IL12RB gene | ||||
| rs375947 | A > G | 18180451 | Met365Thr | F: aggctgccattcaatgcaatacgtc R: tgctctgagcccgggctggccaata |
| TNF gene | ||||
| rs361525 | G > A | 31543101 | 5′-upstream | F: ggcccagaagacccccctcggaatc R: gagcagggaggatggggagtgtgag |
| rs1800629 | G > A | 31543031 | 5′-upstream | F: gaggcaataggttttgaggggcatg R: ggacggggttcagcctccagggtcc |
| CRP gene | ||||
| rs3093077 | A > C | 159679636 | Not announced | F: ggaatccaggcaagtacgacaaccc R: tctgagactagtgggcagttgtcct |
| rs1130864 | G > A | 159683091 | 3′-UTR | F: cctcaaattctgattcttttggacc R: tttcccagcatagttaacgagctcc |
| rs1205 | C > T | 159682233 | 3′-UTR | F: acttccagtttggcttctgtcctca R: agtctctctccatgtggcaaacaag |
| APOB gene | ||||
| rs1042031 | C > T | 21225753 | Glu4181Lys | F: caatcagatgcttgactttcatatggaatt R: ttgagtaactcgtaccaagccatcaaacac |
| rs6725189 | G > T | 21219001 | Not announced | F: ttcccagcctcagctcaacagagctatggg R: cagcagtcggccctctctattgttctttcc |
| APOE gene | ||||
| rs7412 | C > T | 45412079 | Arg176Cys | F: ctcctccgcgatgccgatgacctgcagaag R: gcctggcagtgtaccaggccggggcccgcg |
| rs429358 | T > C | 45411941 | Cys130Arg | F: gcccggctgggcgcggacatggaggacgtg R: gcggccgcctggtgcagtaccgcggcgagg |
| LIPC gene | ||||
| rs1800588 | C > T | 58723675 | 5′-upstream | F: tctttgcttcttcgtcagctccttttgaca R: gggggtgaagggttttctgcaccacacttt |
| LPA gene | ||||
| rs10455872 | A > G | 161010118 | Intronic | F: tcagacaccttgttctcagaaccca R: tgtgtttatacaggttagaggagaa |
| NOTCH1 gene | ||||
| rs13290979 | A > G | 139425634 | Intronic | F: ccagcccagcagtgaagaaactgagcccac R: accctcctggcctgacctacactcgggctt |
| VDR gene | ||||
| rs2228570 | A > G | 48272895 | Met1Thr/Lys/Arg | F: ggcagggaagtgctggccgccattgcctcc R: tccctgtaagaacagcaagcaggccacggt |
| CASR gene | ||||
| rs1042636 | A > G | 122003769 | Arg990Gly | F: gatgagcctcagaagaacgccatggcccac R: ggaattctacgcaccagaactccctggagg |
| OPG gene | ||||
| rs3134069 | A > C | 119964988 | 5′-upstream | F: ggagcttcctacgcgctgaacttctggagt R: gcctcctcgaggtctttccactagcctcaa |
| rs2073618 | G > C | 119964052 | Asn3Lys | F: gggacttaccacgagcgcgcagcacagcaa R: ttgttcattgtggtccccggaaacctcagg |
| rs3102735 | T > C | 119965070 | 5′-upstream | F: ctttgctctagggttcgctgtctcccccat R: aattccctggtctagaagttagacttgatg |
| CALCR gene | ||||
| rs1801197 | A > G | 93055753 | Leu481Pro | F: tcgccttggttgttggctggttcattcctc R: gctcctgatggcagatgtaaattgggatgt |
| F2 gene | ||||
| rs1799963 | G > A | 46761055 | 3′-UTR | F: gttcccaataaaagtgactctcagc R: agcctcaatgctcccagtgctattc |
| F5 gene | ||||
| rs6025 | T > C | 169519049 | Gln534Arg | F: ttacttcaaggacaaaatacctgtattcct R: gcctgtccagggatctgctcttacagatta |
| rs6027 | T > C | 169483561 | Asp2222Gly | F: gggtttttgaatgttcaattctagtaaata R: cacagccaaagagttccaggcgaagtgcaa |
| F7 gene | ||||
| rs6046 | G > A | 113773159 | Arg412Gln/Pro/Leu | F: acagtggaggcccacatgccacccactacc R: gggcacgtggtacctgacgggcatcgtcag |
| ITGB3 gene | ||||
| rs5918 | T > C | 45360730 | Leu59Pro | F: tttgggctcctgacttacaggccctgcctc R: gggctcacctcgctgtgacctgaaggagaa |
IL: interleukin; TNF: tumor necrosis factor; CRP: C-reactive protein; APO: apolipoprotein; LIPC: hepatic lipase; LPA: lipoprotein (a); VDR: vitamin D receptor; CASR: calcium-sensing receptor; OPG: osteoprotegerin; CALCR: calcitonin receptor; ITGB: integrin beta.
The procedures of DNA extraction and genotyping were the same as previously described [12, 25–27]. Briefly, 5 mL of venous blood was collected into a tube with ethylenediaminetetraacetic acid. Then, 0.5 mL of blood was mixed with 1 mL of saline-sodium citrate buffer (Promega) following centrifugation at 12,000 rpm for 2 min. The pellet was digested in a mixture of 10% sodium dodecyl sulfate (Sigma) with 100 μg/mL proteinase K (Helicon) for 3 h at 50°C. Upon digestion, we added phenol : chloroform : isoamyl alcohol (25 : 24 : 1) to the lysate, vortexed it for 20 seconds, and centrifuged at 12,000 rpm for 15 min; 70% ethanol was further utilized to precipitate genomic DNA from a viscous interphase layer. The sample was finally centrifuged at 12,000 rpm for 5 min. DNA pellet was incubated overnight in deionized water at room temperature and was further stored at −70°C until use.
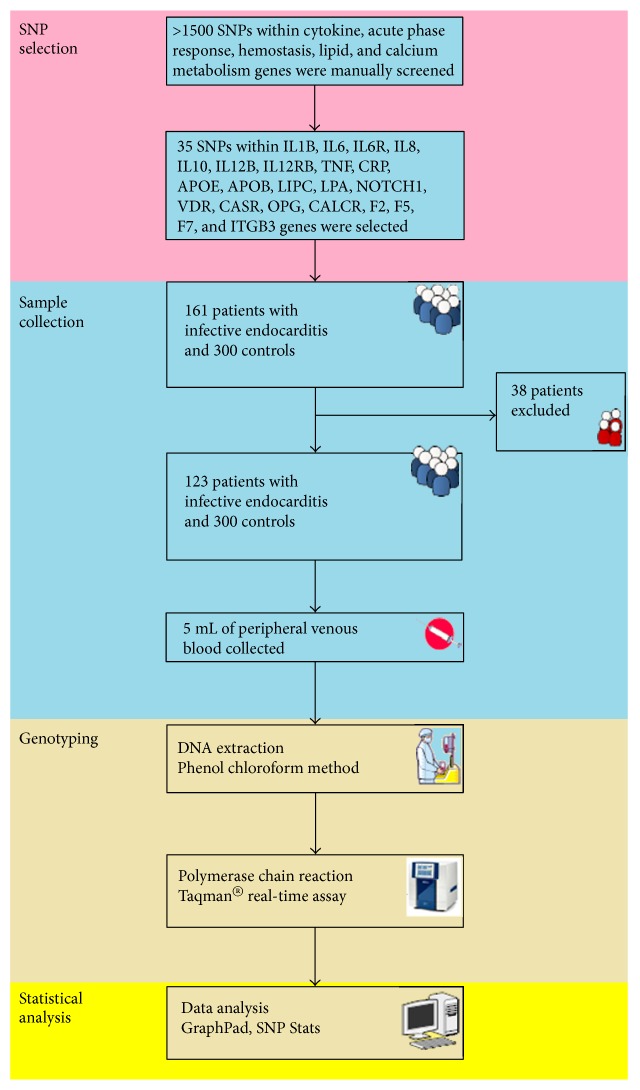
Genotyping was carried out in a 96-well format using the TaqMan SNP assay on the ViiA™ 7 Real-Time PCR System (Life Technologies) according to the manufacturer's instructions. Amplification mixture contained 100 ng of DNA, 1.25 μL of each primer, 2.5 mM of MgCl2, 1 mM of dNTPs, and 1 U of Taq polymerase (Life Technologies), in a total volume of 10 μL. We employed the following polymerase chain reaction (PCR) protocol: hold stage 50°C for 120 s and 95°C for 10 min and PCR stage 95°C for 15 s and 60°C for 1 min repeated in 40 cycles. Table 3 demonstrates the sequence-specific primers for the genotyped SNPs. Laboratory staff was blinded to patient status, and one-tenth of the samples was repeatedly genotyped for quality control purposes. The study workflow is summarized in Figure 1.
Figure 1.
Study workflow.
2.3. Measurement of Plasma Cytokine Level
Venous blood was withdrawn during hospital admission and 7 days postoperation. The plasma was obtained with a centrifugation for 15 min at 1780 ×g and −4°C; 300 μL aliquots have been stored at −80°C until use. The plasma levels of interleukin- (IL-) 1β, IL-6, IL-8, IL-10, IL-12, tumor necrosis factor- (TNF-) α, and C-reactive protein (CRP) were measured by enzyme-linked immunosorbent assay using the kits purchased from eBioscience (BMS224/2, BMS213/2, BMS204/3CE, BMS215/2, BMS238CE, BMS223/4CE, and 88-7502-28, resp.) according to the manufacturer's instructions. All samples were plated in duplicates, with average concentrations used for further analysis.
2.4. Statistical Analysis
The statistical analysis was performed as in [12, 25–27] using GraphPad Prism (GraphPad Software) and SNPStats, a web tool for the analysis of genetic association studies [28].
3. Results
Here, we evaluated the distribution of SNPs within 22 core genes involved in cytokine activity, acute phase response, lipid metabolism, and calcium metabolism genes in a sample of 123 patients with IE and 300 matched asymptomatic control individuals (Table 3). Tables 1 and 2 summarize demographic and clinicopathological characteristics of cases and controls. We did not find statistically significant gender differences between the groups (Table 1). The genotype distributions in both groups are presented in Table 4. All the genotype distributions were in Hardy-Weinberg equilibrium that confirmed a good quality of the genotyping.
Table 4.
Association of the polymorphisms within the cytokine immunity genes, acute phase response genes, hemostasis genes, genes of lipid metabolism, and genes of calcium metabolism with infective endocarditis.
| Model | Genotype | Without infective endocarditis |
With infective endocarditis |
OR (95% CI) | p value | AIC | HWE |
|---|---|---|---|---|---|---|---|
| IL1B rs1143634 | |||||||
| Codominant | G/G | 154 (51.3%) | 82 (67.8%) | 1.00 | 0.0029 | 472.5 | 0.89 |
| G/A | 123 (41%) | 28 (23.1%) | 0.43 (0.26–0.72) | ||||
| A/A | 23 (7.7%) | 11 (9.1%) | 0.97 (0.43–2.18) | ||||
| Dominant | G/G | 154 (51.3%) | 82 (67.8%) | 1.00 | 0.0036 | 473.8 | |
| G/A-A/A | 146 (48.7%) | 39 (32.2%) | 0.51 (0.32–0.81) | ||||
| Recessive | G/G-G/A | 277 (92.3%) | 110 (90.9%) | 1.00 | 0.52 | 481.8 | |
| A/A | 23 (7.7%) | 11 (9.1%) | 1.30 (0.59–2.88) | ||||
| Overdominant | G/G-A/A | 177 (59%) | 93 (76.9%) | 1.00 | 0.0016 | 470.6 | |
| G/A | 123 (41%) | 28 (23.1%) | 0.43 (0.26–0.71) | ||||
| Log-additive | — | — | — | 0.70 (0.48–1.00) | 0.046 | 478.2 | |
| IL6 rs1554606 | |||||||
| Codominant | G/G | 92 (30.7%) | 31 (25.4%) | 1.00 | 0.31 | 483 | 0.99 |
| G/T | 149 (49.7%) | 62 (50.8%) | 1.38 (0.82–2.33) | ||||
| T/T | 59 (19.7%) | 29 (23.8%) | 1.57 (0.84–2.95) | ||||
| Dominant | G/G | 92 (30.7%) | 31 (25.4%) | 1.00 | 0.15 | 481.2 | |
| G/T–T/T | 208 (69.3%) | 91 (74.6%) | 1.44 (0.87–2.36) | ||||
| Recessive | G/G-G/T | 241 (80.3%) | 93 (76.2%) | 1.00 | 0.36 | 482.5 | |
| T/T | 59 (19.7%) | 29 (23.8%) | 1.28 (0.76–2.17) | ||||
| Overdominant | G/G-T/T | 151 (50.3%) | 60 (49.2%) | 1.00 | 0.58 | 483 | |
| G/T | 149 (49.7%) | 62 (50.8%) | 1.13 (0.73–1.76) | ||||
| Log-additive | — | — | — | 1.26 (0.92–1.72) | 0.14 | 481.2 | |
| IL6 rs1800796 | |||||||
| Codominant | G/G | 260 (86.7%) | 104 (85.2%) | 1.00 | 0.98 | 485.3 | 0.64 |
| C/G | 38 (12.7%) | 17 (13.9%) | 1.07 (0.56–2.02) | ||||
| C/C | 2 (0.7%) | 1 (0.8%) | 1.17 (0.08–17.14) | ||||
| Dominant | G/G | 260 (86.7%) | 104 (85.2%) | 1.00 | 0.83 | 483.3 | |
| C/G-C/C | 40 (13.3%) | 18 (14.8%) | 1.07 (0.57–2.00) | ||||
| Recessive | G/G-C/G | 298 (99.3%) | 121 (99.2%) | 1.00 | 0.91 | 483.3 | |
| C/C | 2 (0.7%) | 1 (0.8%) | 1.16 (0.08–16.99) | ||||
| Overdominant | G/G-C/C | 262 (87.3%) | 105 (86.1%) | 1.00 | 0.85 | 483.3 | |
| C/G | 38 (12.7%) | 17 (13.9%) | 1.06 (0.56–2.02) | ||||
| Log-additive | — | — | — | 1.07 (0.60–1.92) | 0.82 | 483.3 | |
| IL6 rs2069827 | |||||||
| Codominant | G/G | 245 (81.7%) | 99 (81.2%) | 1.00 | 0.82 | 484.9 | 0.51 |
| G/T | 51 (17%) | 22 (18%) | 1.20 (0.68–2.14) | ||||
| T/T | 4 (1.3%) | 1 (0.8%) | 0.93 (0.10–8.66) | ||||
| Dominant | G/G | 245 (81.7%) | 99 (81.2%) | 1.00 | 0.55 | 483 | |
| G/T–T/T | 55 (18.3%) | 23 (18.9%) | 1.19 (0.68–2.08) | ||||
| Recessive | G/G-G/T | 296 (98.7%) | 121 (99.2%) | 1.00 | 0.93 | 483.3 | |
| T/T | 4 (1.3%) | 1 (0.8%) | 0.90 (0.10–8.33) | ||||
| Overdominant | G/G-T/T | 249 (83%) | 100 (82%) | 1.00 | 0.53 | 482.9 | |
| G/T | 51 (17%) | 22 (18%) | 1.20 (0.68–2.14) | ||||
| Log-additive | — | — | — | 1.15 (0.69–1.92) | 0.6 | 483.1 | |
| IL6R rs2228145 | |||||||
| Codominant | A/A | 144 (48%) | 62 (50.8%) | 1.00 | 0.46 | 483.8 | 0.89 |
| C/A | 127 (42.3%) | 45 (36.9%) | 0.84 (0.52–1.34) | ||||
| C/C | 29 (9.7%) | 15 (12.3%) | 1.32 (0.64–2.73) | ||||
| Dominant | A/A | 144 (48%) | 62 (50.8%) | 1.00 | 0.71 | 483.2 | |
| C/A-C/C | 156 (52%) | 60 (49.2%) | 0.92 (0.59–1.43) | ||||
| Recessive | A/A-C/A | 271 (90.3%) | 107 (87.7%) | 1.00 | 0.32 | 482.3 | |
| C/C | 29 (9.7%) | 15 (12.3%) | 1.43 (0.72–2.87) | ||||
| Overdominant | A/A-C/C | 173 (57.7%) | 77 (63.1%) | 1.00 | 0.32 | 482.3 | |
| C/A | 127 (42.3%) | 45 (36.9%) | 0.79 (0.51–1.25) | ||||
| Log-additive | — | — | — | 1.03 (0.74–1.43) | 0.85 | 483.3 | |
| IL6R rs2229238 | |||||||
| Codominant | C/C | 158 (52.7%) | 62 (50.8%) | 1.00 | 0.48 | 483.8 | 0.88 |
| C/T | 121 (40.3%) | 47 (38.5%) | 1.00 (0.63–1.60) | ||||
| T/T | 21 (7%) | 13 (10.7%) | 1.63 (0.73–3.61) | ||||
| Dominant | C/C | 158 (52.7%) | 62 (50.8%) | 1.00 | 0.69 | 483.2 | |
| C/T–T/T | 142 (47.3%) | 60 (49.2%) | 1.09 (0.71–1.70) | ||||
| Recessive | C/C-C/T | 279 (93%) | 109 (89.3%) | 1.00 | 0.22 | 481.8 | |
| T/T | 21 (7%) | 13 (10.7%) | 1.63 (0.75–3.51) | ||||
| Overdominant | C/C-T/T | 179 (59.7%) | 75 (61.5%) | 1.00 | 0.78 | 483.3 | |
| C/T | 121 (40.3%) | 47 (38.5%) | 0.94 (0.60–1.47) | ||||
| Log-additive | — | — | — | 1.16 (0.82–1.63) | 0.4 | 482.6 | |
| IL8 rs2227306 | |||||||
| Codominant | C/C | 100 (33.3%) | 49 (40.2%) | 1.00 | 0.42 | 483.6 | 0.99 |
| C/T | 147 (49%) | 59 (48.4%) | 0.83 (0.52–1.33) | ||||
| T/T | 53 (17.7%) | 14 (11.5%) | 0.63 (0.31–1.28) | ||||
| Dominant | C/C | 100 (33.3%) | 49 (40.2%) | 1.00 | 0.29 | 482.2 | |
| C/T–T/T | 200 (66.7%) | 73 (59.8%) | 0.78 (0.50–1.23) | ||||
| Recessive | C/C-C/T | 247 (82.3%) | 108 (88.5%) | 1.00 | 0.28 | 482.2 | |
| T/T | 53 (17.7%) | 14 (11.5%) | 0.70 (0.37–1.35) | ||||
| Overdominant | C/C-T/T | 153 (51%) | 63 (51.6%) | 1.00 | 0.79 | 483.3 | |
| C/T | 147 (49%) | 59 (48.4%) | 0.94 (0.61–1.46) | ||||
| Log-additive | — | — | — | 0.80 (0.58–1.12) | 0.19 | 481.6 | |
| IL10 rs1800871 | |||||||
| Codominant | G/G | 183 (61%) | 69 (56.6%) | 1.00 | 0.88 | 485.1 | 0.61 |
| A/G | 105 (35%) | 47 (38.5%) | 1.12 (0.71–1.78) | ||||
| A/A | 12 (4%) | 6 (4.9%) | 0.98 (0.34–2.81) | ||||
| Dominant | G/G | 183 (61%) | 69 (56.6%) | 1.00 | 0.66 | 483.1 | |
| A/G-A/A | 117 (39%) | 53 (43.4%) | 1.11 (0.71–1.73) | ||||
| Recessive | G/G-A/G | 288 (96%) | 116 (95.1%) | 1.00 | 0.9 | 483.3 | |
| A/A | 12 (4%) | 6 (4.9%) | 0.94 (0.33–2.64) | ||||
| Overdominant | G/G-A/A | 195 (65%) | 75 (61.5%) | 1.00 | 0.61 | 483.1 | |
| A/G | 105 (35%) | 47 (38.5%) | 1.13 (0.71–1.77) | ||||
| Log-additive | — | — | — | 1.06 (0.73–1.55) | 0.74 | 483.2 | |
| IL10 rs1800872 | |||||||
| Codominant | G/G | 185 (62.1%) | 67 (55.4%) | 1.00 | 0.55 | 481.3 | 0.86 |
| T/G | 101 (33.9%) | 49 (40.5%) | 1.27 (0.80–2.01) | ||||
| T/T | 12 (4%) | 5 (4.1%) | 0.86 (0.28–2.61) | ||||
| Dominant | G/G | 185 (62.1%) | 67 (55.4%) | 1.00 | 0.39 | 479.8 | |
| T/G-T/T | 113 (37.9%) | 54 (44.6%) | 1.22 (0.78–1.90) | ||||
| Recessive | G/G-T/G | 286 (96%) | 116 (95.9%) | 1.00 | 0.65 | 480.3 | |
| T/T | 12 (4%) | 5 (4.1%) | 0.78 (0.26–2.34) | ||||
| Overdominant | G/G-T/T | 197 (66.1%) | 72 (59.5%) | 1.00 | 0.29 | 479.4 | |
| T/G | 101 (33.9%) | 49 (40.5%) | 1.28 (0.81–2.01) | ||||
| Log-additive | — | — | — | 1.11 (0.76–1.63) | 0.57 | 480.2 | |
| IL10 rs1800896 | |||||||
| Codominant | T/T | 88 (29.3%) | 34 (28.1%) | 1.00 | 0.82 | 481.7 | 0.35 |
| T/C | 157 (52.3%) | 66 (54.5%) | 1.17 (0.70–1.95) | ||||
| C/C | 55 (18.3%) | 21 (17.4%) | 1.17 (0.59–2.29) | ||||
| Dominant | T/T | 88 (29.3%) | 34 (28.1%) | 1.00 | 0.53 | 479.7 | |
| T/C-C/C | 212 (70.7%) | 87 (71.9%) | 1.17 (0.71–1.91) | ||||
| Recessive | T/T–T/C | 245 (81.7%) | 100 (82.6%) | 1.00 | 0.87 | 480 | |
| C/C | 55 (18.3%) | 21 (17.4%) | 1.05 (0.59–1.88) | ||||
| Overdominant | T/T-C/C | 143 (47.7%) | 55 (45.5%) | 1.00 | 0.66 | 479.9 | |
| T/C | 157 (52.3%) | 66 (54.5%) | 1.10 (0.71–1.72) | ||||
| Log-additive | — | — | — | 1.09 (0.78–1.52) | 0.61 | 479.8 | |
| IL12B rs3212227 | |||||||
| Codominant | T/T | 191 (63.7%) | 86 (70.5%) | 1.00 | 0.076 | 480.2 | 0.86 |
| G/T | 96 (32%) | 29 (23.8%) | 0.57 (0.34–0.96) | ||||
| G/G | 13 (4.3%) | 7 (5.7%) | 1.20 (0.45–3.24) | ||||
| Dominant | T/T | 191 (63.7%) | 86 (70.5%) | 1.00 | 0.067 | 480 | |
| G/T-G/G | 109 (36.3%) | 36 (29.5%) | 0.64 (0.40–1.04) | ||||
| Recessive | T/T-G/T | 287 (95.7%) | 115 (94.3%) | 1.00 | 0.5 | 482.9 | |
| G/G | 13 (4.3%) | 7 (5.7%) | 1.41 (0.53–3.75) | ||||
| Overdominant | T/T-G/G | 204 (68%) | 93 (76.2%) | 1.00 | 0.025 | 478.3 | |
| G/T | 96 (32%) | 29 (23.8%) | 0.57 (0.34–0.94) | ||||
| Log-additive | — | — | — | 0.78 (0.53–1.16) | 0.21 | 481.8 | |
| IL12RB rs375947 | |||||||
| Codominant | A/A | 137 (45.7%) | 53 (43.4%) | 1.00 | 0.58 | 484.3 | 0.60 |
| A/G | 135 (45%) | 55 (45.1%) | 1.18 (0.74–1.89) | ||||
| G/G | 28 (9.3%) | 14 (11.5%) | 1.44 (0.68–3.03) | ||||
| Dominant | A/A | 137 (45.7%) | 53 (43.4%) | 1.00 | 0.37 | 482.5 | |
| A/G-G/G | 163 (54.3%) | 69 (56.6%) | 1.23 (0.79–1.91) | ||||
| Recessive | A/A-A/G | 272 (90.7%) | 108 (88.5%) | 1.00 | 0.45 | 482.7 | |
| G/G | 28 (9.3%) | 14 (11.5%) | 1.32 (0.65–2.68) | ||||
| Overdominant | A/A-G/G | 165 (55%) | 67 (54.9%) | 1.00 | 0.66 | 483.1 | |
| A/G | 135 (45%) | 55 (45.1%) | 1.10 (0.71–1.72) | ||||
| Log-additive | — | — | — | 1.19 (0.85–1.67) | 0.3 | 482.3 | |
| TNF rs361525 | |||||||
| Codominant | G/G | 274 (91.3%) | 112 (91.8%) | 1.00 | 0.81 | 484.9 | 0.46 |
| A/G | 25 (8.3%) | 10 (8.2%) | 0.90 (0.40–1.99) | ||||
| A/A | 1 (0.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Dominant | G/G | 274 (91.3%) | 112 (91.8%) | 1.00 | 0.74 | 483.2 | |
| A/G-A/A | 26 (8.7%) | 10 (8.2%) | 0.88 (0.39–1.94) | ||||
| Recessive | G/G-A/G | 299 (99.7%) | 122 (100%) | 1.00 | 0.55 | 483 | |
| A/A | 1 (0.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Overdominant | G/G-A/A | 275 (91.7%) | 112 (91.8%) | 1.00 | 0.79 | 483.3 | |
| A/G | 25 (8.3%) | 10 (8.2%) | 0.90 (0.40–2.00) | ||||
| Log-additive | — | — | — | 0.86 (0.39–1.87) | 0.7 | 483.2 | |
| TNF rs1800629 | |||||||
| Codominant | G/G | 229 (76.3%) | 97 (79.5%) | 1.00 | 0.66 | 484.5 | 0.99 |
| A/G | 67 (22.3%) | 24 (19.7%) | 0.89 (0.51–1.54) | ||||
| A/A | 4 (1.3%) | 1 (0.8%) | 0.41 (0.04–3.93) | ||||
| Dominant | G/G | 229 (76.3%) | 97 (79.5%) | 1.00 | 0.56 | 483 | |
| A/G-A/A | 71 (23.7%) | 25 (20.5%) | 0.85 (0.50–1.46) | ||||
| Recessive | G/G-A/G | 296 (98.7%) | 121 (99.2%) | 1.00 | 0.42 | 482.7 | |
| A/A | 4 (1.3%) | 1 (0.8%) | 0.42 (0.04–4.02) | ||||
| Overdominant | G/G-A/A | 233 (77.7%) | 98 (80.3%) | 1.00 | 0.71 | 483.2 | |
| A/G | 67 (22.3%) | 24 (19.7%) | 0.90 (0.52–1.55) | ||||
| Log-additive | — | — | — | 0.83 (0.51–1.37) | 0.47 | 482.8 | |
| CRP rs3093077 | |||||||
| Codominant | C/C | 262 (87.3%) | 112 (92.6%) | 1.00 | 0.28 | 481.5 | 0.99 |
| A/C | 37 (12.3%) | 8 (6.6%) | 0.55 (0.24–1.23) | ||||
| A/A | 1 (0.3%) | 1 (0.8%) | 1.99 (0.12–34.12) | ||||
| Dominant | C/C | 262 (87.3%) | 112 (92.6%) | 1.00 | 0.17 | 480.2 | |
| A/C-A/A | 38 (12.7%) | 9 (7.4%) | 0.59 (0.27–1.29) | ||||
| Recessive | C/C-A/C | 299 (99.7%) | 120 (99.2%) | 1.00 | 0.61 | 481.8 | |
| A/A | 1 (0.3%) | 1 (0.8%) | 2.10 (0.12–36.04) | ||||
| Overdominant | C/C-A/A | 263 (87.7%) | 113 (93.4%) | 1.00 | 0.13 | 479.7 | |
| A/C | 37 (12.3%) | 8 (6.6%) | 0.55 (0.24–1.23) | ||||
| Log-additive | — | — | — | 0.67 (0.33–1.36) | 0.25 | 480.7 | |
| CRP rs1130864 | |||||||
| Codominant | G/G | 142 (47.3%) | 68 (55.7%) | 1.00 | 0.018 | 477.9 | 0.41 |
| A/G | 134 (44.7%) | 38 (31.1%) | 0.58 (0.36–0.93) | ||||
| A/A | 24 (8%) | 16 (13.1%) | 1.45 (0.70–3.00) | ||||
| Dominant | G/G | 142 (47.3%) | 68 (55.7%) | 1.00 | 0.12 | 481.4 | |
| A/G-A/A | 158 (52.7%) | 54 (44.3%) | 0.71 (0.45–1.09) | ||||
| Recessive | G/G-A/G | 276 (92%) | 106 (86.9%) | 1.00 | 0.094 | 481.1 | |
| A/A | 24 (8%) | 16 (13.1%) | 1.83 (0.91–3.69) | ||||
| Overdominant | G/G-A/A | 166 (55.3%) | 84 (68.8%) | 1.00 | 0.0083 | 476.9 | |
| A/G | 134 (44.7%) | 38 (31.1%) | 0.54 (0.34–0.86) | ||||
| Log-additive | — | — | — | 0.93 (0.67–1.30) | 0.68 | 483.7 | |
| CRP rs1205 | |||||||
| Codominant | C/C | 112 (37.3%) | 38 (31.9%) | 1.00 | 0.018 | 468.4 | 0.11 |
| C/T | 154 (51.3%) | 56 (47.1%) | 1.06 (0.64–1.74) | ||||
| T/T | 34 (11.3%) | 25 (21%) | 2.50 (1.28–4.90) | ||||
| Dominant | C/C | 112 (37.3%) | 38 (31.9%) | 1.00 | 0.28 | 473.3 | |
| C/T–T/T | 188 (62.7%) | 81 (68.1%) | 1.29 (0.81–2.07) | ||||
| Recessive | C/C-C/T | 266 (88.7%) | 94 (79%) | 1.00 | 0.0047 | 466.5 | |
| T/T | 34 (11.3%) | 25 (21%) | 2.42 (1.32–4.43) | ||||
| Overdominant | C/C-T/T | 146 (48.7%) | 63 (52.9%) | 1.00 | 0.34 | 473.6 | |
| C/T | 154 (51.3%) | 56 (47.1%) | 0.80 (0.52–1.25) | ||||
| Log-additive | — | — | — | 1.47 (1.05–2.05) | 0.023 | 469.3 | |
| APOB rs1042031 | |||||||
| Codominant | C/C | 210 (70.5%) | 83 (69.8%) | 1.00 | 0.86 | 477.7 | 0.40 |
| C/T | 78 (26.2%) | 33 (27.7%) | 1.00 (0.61–1.65) | ||||
| T/T | 10 (3.4%) | 3 (2.5%) | 0.69 (0.18–2.71) | ||||
| Dominant | C/C | 210 (70.5%) | 83 (69.8%) | 1.00 | 0.88 | 476 | |
| C/T–T/T | 88 (29.5%) | 36 (30.2%) | 0.96 (0.59–1.56) | ||||
| Recessive | C/C-C/T | 288 (96.6%) | 116 (97.5%) | 1.00 | 0.59 | 475.7 | |
| T/T | 10 (3.4%) | 3 (2.5%) | 0.69 (0.18–2.69) | ||||
| Overdominant | C/C-T/T | 220 (73.8%) | 86 (72.3%) | 1.00 | 0.95 | 476 | |
| C/T | 78 (26.2%) | 33 (27.7%) | 1.01 (0.62–1.67) | ||||
| Log-additive | — | — | — | 0.94 (0.62–1.42) | 0.76 | 475.9 | |
| APOB rs6725189 | |||||||
| Codominant | G/G | 196 (65.8%) | 72 (59.5%) | 1.00 | 0.53 | 481.2 | 0.46 |
| G/T | 89 (29.9%) | 44 (36.4%) | 1.31 (0.82–2.10) | ||||
| T/T | 13 (4.4%) | 5 (4.1%) | 1.01 (0.33–3.04) | ||||
| Dominant | G/G | 196 (65.8%) | 72 (59.5%) | 1.00 | 0.3 | 479.4 | |
| G/T–T/T | 102 (34.2%) | 49 (40.5%) | 1.27 (0.81–2.00) | ||||
| Recessive | G/G-G/T | 285 (95.6%) | 116 (95.9%) | 1.00 | 0.87 | 480.5 | |
| T/T | 13 (4.4%) | 5 (4.1%) | 0.92 (0.31–2.73) | ||||
| Overdominant | G/G-T/T | 209 (70.1%) | 77 (63.6%) | 1.00 | 0.26 | 479.2 | |
| G/T | 89 (29.9%) | 44 (36.4%) | 1.31 (0.82–2.08) | ||||
| Log-additive | — | — | — | 1.17 (0.80–1.71) | 0.42 | 479.9 | |
| APOE rs7412 | |||||||
| Codominant | C/C | 251 (83.7%) | 104 (86%) | 1.00 | 0.63 | 483.2 | 0.71 |
| C/T | 48 (16%) | 16 (13.2%) | 0.79 (0.42–1.49) | ||||
| T/T | 1 (0.3%) | 1 (0.8%) | 2.37 (0.13–43.65) | ||||
| Dominant | C/C | 251 (83.7%) | 104 (86%) | 1.00 | 0.53 | 481.7 | |
| C/T–T/T | 49 (16.3%) | 17 (14.1%) | 0.82 (0.44–1.53) | ||||
| Recessive | C/C-C/T | 299 (99.7%) | 120 (99.2%) | 1.00 | 0.55 | 481.7 | |
| T/T | 1 (0.3%) | 1 (0.8%) | 2.45 (0.13–45.09) | ||||
| Overdominant | C/C-T/T | 252 (84%) | 105 (86.8%) | 1.00 | 0.44 | 481.5 | |
| C/T | 48 (16%) | 16 (13.2%) | 0.78 (0.42–1.48) | ||||
| Log-additive | — | — | — | 0.86 (0.48–1.56) | 0.63 | 481.8 | |
| APOE rs429358 | |||||||
| Codominant | T/T | 239 (79.7%) | 96 (78.7%) | 1.00 | 0.71 | 485.2 | 0.99 |
| C/T | 58 (19.3%) | 24 (19.7%) | 1.22 (0.70–2.14) | ||||
| C/C | 3 (1%) | 2 (1.6%) | 1.58 (0.24–10.40) | ||||
| Dominant | T/T | 239 (79.7%) | 96 (78.7%) | 1.00 | 0.44 | 483.3 | |
| C/T-C/C | 61 (20.3%) | 26 (21.3%) | 1.24 (0.72–2.14) | ||||
| Recessive | T/T-C/T | 297 (99%) | 120 (98.4%) | 1.00 | 0.67 | 483.7 | |
| C/C | 3 (1%) | 2 (1.6%) | 1.52 (0.23–9.97) | ||||
| Overdominant | T/T-C/C | 242 (80.7%) | 98 (80.3%) | 1.00 | 0.5 | 483.4 | |
| C/T | 58 (19.3%) | 24 (19.7%) | 1.21 (0.69–2.12) | ||||
| Log-additive | — | — | — | 1.23 (0.75–2.01) | 0.41 | 483.2 | |
| LIPC rs1800588 | |||||||
| Codominant | C/C | 173 (57.7%) | 77 (63.6%) | 1.00 | 0.2 | 480.8 | 0.52 |
| C/T | 113 (37.7%) | 41 (33.9%) | 0.69 (0.43–1.11) | ||||
| T/T | 14 (4.7%) | 3 (2.5%) | 0.48 (0.13–1.81) | ||||
| Dominant | C/C | 173 (57.7%) | 77 (63.6%) | 1.00 | 0.084 | 479.1 | |
| C/T–T/T | 127 (42.3%) | 44 (36.4%) | 0.67 (0.43–1.06) | ||||
| Recessive | C/C-C/T | 286 (95.3%) | 118 (97.5%) | 1.00 | 0.35 | 481.2 | |
| T/T | 14 (4.7%) | 3 (2.5%) | 0.55 (0.15–2.03) | ||||
| Overdominant | C/C-T/T | 187 (62.3%) | 80 (66.1%) | 1.00 | 0.16 | 480.1 | |
| C/T | 113 (37.7%) | 41 (33.9%) | 0.72 (0.45–1.15) | ||||
| Log-additive | — | — | — | 0.69 (0.46–1.04) | 0.071 | 478.8 | |
| LPA rs10455872 | |||||||
| — | A/A | 275 (92%) | 108 (91.5%) | 1.00 | 0.86 | 475.8 | 0.99 |
| A/G | 24 (8%) | 10 (8.5%) | 1.08 (0.48–2.39) | ||||
| NOTCH1 rs13290979 | |||||||
| Codominant | A/A | 98 (32.8%) | 53 (43.8%) | 1.00 | 0.024 | 476.2 | 0.48 |
| A/G | 152 (50.8%) | 44 (36.4%) | 0.53 (0.33–0.87) | ||||
| G/G | 49 (16.4%) | 24 (19.8%) | 0.98 (0.53–1.82) | ||||
| Dominant | A/A | 98 (32.8%) | 53 (43.8%) | 1.00 | 0.05 | 477.9 | |
| A/G-G/G | 201 (67.2%) | 68 (56.2%) | 0.64 (0.41–1.00) | ||||
| Recessive | A/A-A/G | 250 (83.6%) | 97 (80.2%) | 1.00 | 0.28 | 480.6 | |
| G/G | 49 (16.4%) | 24 (19.8%) | 1.37 (0.78–2.41) | ||||
| Overdominant | A/A-G/G | 147 (49.2%) | 77 (63.6%) | 1.00 | 0.0062 | 474.2 | |
| A/G | 152 (50.8%) | 44 (36.4%) | 0.54 (0.34–0.84) | ||||
| Log-additive | — | — | — | 0.88 (0.65–1.21) | 0.44 | 481.1 | |
| VDR rs2228570 | |||||||
| Codominant | G/G | 94 (31.5%) | 36 (29.8%) | 1.00 | 0.19 | 479.1 | 0.81 |
| A/G | 145 (48.7%) | 51 (42.1%) | 0.93 (0.55–1.57) | ||||
| A/A | 59 (19.8%) | 34 (28.1%) | 1.54 (0.85–2.80) | ||||
| Dominant | G/G | 94 (31.5%) | 36 (29.8%) | 1.00 | 0.67 | 480.3 | |
| A/G-A/A | 204 (68.5%) | 85 (70.2%) | 1.11 (0.69–1.79) | ||||
| Recessive | G/G-A/G | 239 (80.2%) | 87 (71.9%) | 1.00 | 0.07 | 477.2 | |
| A/A | 59 (19.8%) | 34 (28.1%) | 1.61 (0.97–2.68) | ||||
| Overdominant | G/G-A/A | 153 (51.3%) | 70 (57.9%) | 1.00 | 0.25 | 479.2 | |
| A/G | 145 (48.7%) | 51 (42.1%) | 0.77 (0.50–1.20) | ||||
| Log-additive | — | — | — | 1.23 (0.91–1.66) | 0.19 | 478.8 | |
| CASR rs1042636 | |||||||
| Codominant | A/A | 244 (81.6%) | 103 (85.1%) | 1.00 | 0.2 | 480.5 | 0.75 |
| A/G | 52 (17.4%) | 18 (14.9%) | 0.71 (0.39–1.30) | ||||
| G/G | 3 (1%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Dominant | A/A | 244 (81.6%) | 103 (85.1%) | 1.00 | 0.19 | 480 | |
| A/G-G/G | 55 (18.4%) | 18 (14.9%) | 0.67 (0.37–1.23) | ||||
| Recessive | A/A-A/G | 296 (99%) | 121 (100%) | 1.00 | 0.16 | 479.8 | |
| G/G | 3 (1%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Overdominant | A/A-G/G | 247 (82.6%) | 103 (85.1%) | 1.00 | 0.28 | 480.5 | |
| A/G | 52 (17.4%) | 18 (14.9%) | 0.72 (0.39–1.32) | ||||
| Log-additive | — | — | — | 0.65 (0.37–1.17) | 0.14 | 479.6 | |
| OPG rs3134069 | |||||||
| Codominant | A/A | 248 (83.2%) | 110 (90.9%) | 1.00 | 0.19 | 479.2 | 0.71 |
| A/C | 49 (16.4%) | 10 (8.3%) | 0.53 (0.25–1.11) | ||||
| C/C | 1 (0.3%) | 1 (0.8%) | 1.92 (0.09–41.36) | ||||
| Dominant | A/A | 248 (83.2%) | 110 (90.9%) | 1.00 | 0.1 | 477.9 | |
| A/C-C/C | 50 (16.8%) | 11 (9.1%) | 0.56 (0.28–1.15) | ||||
| Recessive | A/A-A/C | 297 (99.7%) | 120 (99.2%) | 1.00 | 0.65 | 480.3 | |
| C/C | 1 (0.3%) | 1 (0.8%) | 2.06 (0.09–44.96) | ||||
| Overdominant | A/A-C/C | 249 (83.6%) | 111 (91.7%) | 1.00 | 0.078 | 477.4 | |
| A/C | 49 (16.4%) | 10 (8.3%) | 0.53 (0.25–1.11) | ||||
| Log-additive | — | — | — | 0.62 (0.32–1.21) | 0.15 | 478.4 | |
| OPG rs2073618 | |||||||
| Codominant | C/C | 76 (25.4%) | 32 (26.7%) | 1.00 | 0.17 | 478.3 | 0.56 |
| C/G | 155 (51.8%) | 51 (42.5%) | 0.79 (0.46–1.36) | ||||
| G/G | 68 (22.7%) | 37 (30.8%) | 1.32 (0.73–2.41) | ||||
| Dominant | C/C | 76 (25.4%) | 32 (26.7%) | 1.00 | 0.86 | 479.9 | |
| C/G-G/G | 223 (74.6%) | 88 (73.3%) | 0.96 (0.58–1.58) | ||||
| Recessive | C/C-C/G | 231 (77.3%) | 83 (69.2%) | 1.00 | 0.089 | 477 | |
| G/G | 68 (22.7%) | 37 (30.8%) | 1.54 (0.94–2.52) | ||||
| Overdominant | C/C-G/G | 144 (48.2%) | 69 (57.5%) | 1.00 | 0.098 | 477.2 | |
| C/G | 155 (51.8%) | 51 (42.5%) | 0.69 (0.44–1.07) | ||||
| Log-additive | — | — | — | 1.16 (0.85–1.58) | 0.35 | 479 | |
| OPG rs3102735 | |||||||
| Codominant | T/T | 218 (73.2%) | 95 (78.5%) | 1.00 | 0.36 | 480.4 | 0.82 |
| C/T | 73 (24.5%) | 25 (20.7%) | 0.89 (0.52–1.52) | ||||
| C/C | 7 (2.4%) | 1 (0.8%) | 0.25 (0.03–2.27) | ||||
| Dominant | T/T | 218 (73.2%) | 95 (78.5%) | 1.00 | 0.46 | 480 | |
| C/T-C/C | 80 (26.9%) | 26 (21.5%) | 0.82 (0.49–1.39) | ||||
| Recessive | T/T-C/T | 291 (97.7%) | 120 (99.2%) | 1.00 | 0.17 | 478.6 | |
| C/C | 7 (2.4%) | 1 (0.8%) | 0.26 (0.03–2.32) | ||||
| Overdominant | T/T-C/C | 225 (75.5%) | 96 (79.3%) | 1.00 | 0.74 | 480.4 | |
| C/T | 73 (24.5%) | 25 (20.7%) | 0.91 (0.53–1.56) | ||||
| Log-additive | — | — | — | 0.78 (0.49–1.26) | 0.3 | 479.4 | |
| CALCR rs1801197 | |||||||
| Codominant | A/A | 140 (46.8%) | 76 (62.8%) | 1.00 | 0.0077 | 474 | 0.22 |
| A/G | 136 (45.5%) | 40 (33.1%) | 0.52 (0.33–0.84) | ||||
| G/G | 23 (7.7%) | 5 (4.1%) | 0.36 (0.13–1.03) | ||||
| Dominant | A/A | 140 (46.8%) | 76 (62.8%) | 1.00 | 0.0024 | 472.5 | |
| A/G-G/G | 159 (53.2%) | 45 (37.2%) | 0.50 (0.32–0.79) | ||||
| Recessive | A/A-A/G | 276 (92.3%) | 116 (95.9%) | 1.00 | 0.13 | 479.5 | |
| G/G | 23 (7.7%) | 5 (4.1%) | 0.48 (0.17–1.33) | ||||
| Overdominant | A/A-G/G | 163 (54.5%) | 81 (66.9%) | 1.00 | 0.018 | 476.2 | |
| A/G | 136 (45.5%) | 40 (33.1%) | 0.58 (0.36–0.92) | ||||
| Log-additive | — | — | — | 0.56 (0.38–0.82) | 0.002 | 472.2 | |
| F2 rs1799963 | |||||||
| — | G/G | 286 (95.7%) | 118 (98.3%) | 1.00 | 0.10 | 477 | 0.99 |
| A/G | 13 (4.3%) | 2 (1.7%) | 0.32 (0.07–1.49) | ||||
| F5 rs6025 | |||||||
| Codominant | C/C | 286 (95.7%) | 117 (97.5%) | 1.00 | 0.32 | 479.4 | 0.14 |
| C/T | 12 (4%) | 3 (2.5%) | 0.56 (0.15–2.11) | ||||
| T/T | 1 (0.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Dominant | C/C | 286 (95.7%) | 117 (97.5%) | 1.00 | 0.24 | 478.3 | |
| C/T–T/T | 13 (4.3%) | 3 (2.5%) | 0.48 (0.13–1.79) | ||||
| Recessive | C/C-C/T | 298 (99.7%) | 120 (100%) | 1.00 | 0.23 | 478.2 | |
| T/T | 1 (0.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Overdominant | C/C-T/T | 287 (96%) | 117 (97.5%) | 1.00 | 0.38 | 478.9 | |
| C/T | 12 (4%) | 3 (2.5%) | 0.57 (0.15–2.13) | ||||
| Log-additive | — | — | — | 0.47 (0.14–1.58) | 0.19 | 477.9 | |
| F5 rs6027 | |||||||
| Codominant | T/T | 254 (85%) | 104 (86.7%) | 1.00 | 0.35 | 479.5 | 0.12 |
| C/T | 41 (13.7%) | 16 (13.3%) | 1.00 (0.52–1.91) | ||||
| C/C | 4 (1.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Dominant | T/T | 254 (85%) | 104 (86.7%) | 1.00 | 0.79 | 479.6 | |
| C/T-C/C | 45 (15.1%) | 16 (13.3%) | 0.92 (0.48–1.74) | ||||
| Recessive | T/T-C/T | 295 (98.7%) | 120 (100%) | 1.00 | 0.15 | 477.5 | |
| C/C | 4 (1.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Overdominant | T/T-C/C | 258 (86.3%) | 104 (86.7%) | 1.00 | 0.98 | 479.6 | |
| C/T | 41 (13.7%) | 16 (13.3%) | 1.01 (0.53–1.93) | ||||
| Log-additive | — | — | — | 0.86 (0.47–1.56) | 0.61 | 479.4 | |
| F7 rs6046 | |||||||
| Codominant | G/G | 236 (78.9%) | 88 (72.7%) | 1.00 | 0.06 | 477.8 | 0.77 |
| A/G | 59 (19.7%) | 33 (27.3%) | 1.58 (0.94–2.64) | ||||
| A/A | 4 (1.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Dominant | G/G | 236 (78.9%) | 88 (72.7%) | 1.00 | 0.14 | 479.2 | |
| A/G-A/A | 63 (21.1%) | 33 (27.3%) | 1.48 (0.89–2.46) | ||||
| Recessive | G/G-A/G | 295 (98.7%) | 121 (100%) | 1.00 | 0.1 | 478.8 | |
| A/A | 4 (1.3%) | 0 (0%) | 0.00 (0.00–NA) | ||||
| Overdominant | G/G-A/A | 240 (80.3%) | 88 (72.7%) | 1.00 | 0.074 | 478.2 | |
| A/G | 59 (19.7%) | 33 (27.3%) | 1.60 (0.96–2.68) | ||||
| Log-additive | — | — | — | 1.32 (0.82–2.14) | 0.26 | 480.1 | |
| ITGB3 rs5918 | |||||||
| Codominant | T/T | 214 (71.6%) | 88 (73.3%) | 1.00 | 0.5 | 480.2 | 0.66 |
| C/T | 77 (25.8%) | 26 (21.7%) | 0.84 (0.49–1.43) | ||||
| C/C | 8 (2.7%) | 6 (5%) | 1.69 (0.55–5.19) | ||||
| Dominant | T/T | 214 (71.6%) | 88 (73.3%) | 1.00 | 0.76 | 479.5 | |
| C/T-C/C | 85 (28.4%) | 32 (26.7%) | 0.93 (0.56–1.52) | ||||
| Recessive | T/T-C/T | 291 (97.3%) | 114 (95%) | 1.00 | 0.32 | 478.6 | |
| C/C | 8 (2.7%) | 6 (5%) | 1.77 (0.58–5.39) | ||||
| Overdominant | T/T-C/C | 222 (74.2%) | 94 (78.3%) | 1.00 | 0.45 | 479 | |
| C/T | 77 (25.8%) | 26 (21.7%) | 0.82 (0.48–1.38) | ||||
| Log-additive | — | — | — | 1.02 (0.68–1.54) | 0.92 | 479.6 | |
All the ORs and 95% CIs are adjusted for age and gender. IL: interleukin; TNF: tumor necrosis factor; CRP: C-reactive protein; APO: apolipoprotein; LIPC: hepatic lipase; LPA: lipoprotein (a); VDR: vitamin D receptor; CASR: calcium-sensing receptor; OPG: osteoprotegerin; CALCR: calcitonin receptor; ITGB: integrin beta; OR: odds ratio; CI: confidence interval; AIC: Akaike information criterion; HWE: Hardy-Weinberg equilibrium.
We first asked whether an inherited variation within the genes encoding cytokines and acute phase proteins may play a role in the risk of IE development. Having performed the genetic association analysis with the adjustments for age and gender, we found that the G/A genotype of the rs1143634 polymorphism within the IL1B gene was associated with a lower risk of IE (OR = 0.43, 95% CI = 0.26–0.71, p = 0.0016, overdominant model, Table 4). Similarly, the G/T genotype of the rs3212227 polymorphism within the IL12B gene correlated with a decreased risk of IE (OR = 0.57, 95% CI = 0.34–0.94, p = 0.0250, overdominant model, Table 4). Finally, we observed that the A/G genotype of the rs1130864 polymorphism within the CRP gene was also associated with a lower risk of IE (OR = 0.54, 95% CI = 0.34–0.86, p = 0.0083, overdominant model, Table 4). Conversely, the T/T genotype of the rs1205 polymorphism within the CRP gene was associated with a higher risk of IE (OR = 2.42, 95% CI = 1.32–4.43, p = 0.0047 according to a recessive model, Table 4). Other SNPs were not significantly different between cases and controls (Table 4).
We further investigated whether the inherited variation in the pathways of hemostasis, lipid metabolism, and calcium metabolism can be linked to IE. The A/G genotype of the rs13290979 polymorphism within the NOTCH1 gene and the G allele of the rs1801197 polymorphism within the CALCR gene were associated with a lower risk of IE (OR = 0.54, 95% CI = 0.34–0.84, p = 0.0062 according to an overdominant model; OR = 0.56, 95% CI = 0.38–0.82, p = 0.0020 according to a log-additive model, resp., Table 4). Other SNPs were not significantly associated with IE (Table 4).
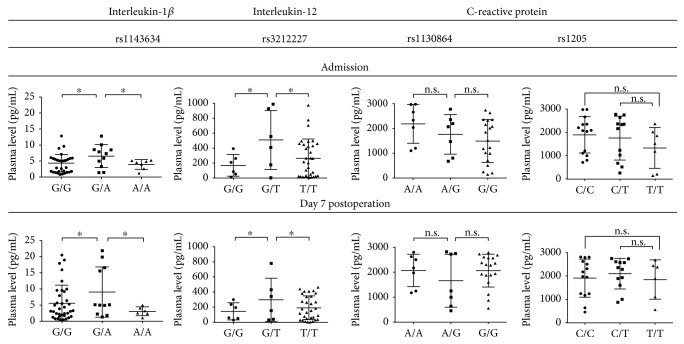
As cytokines are easily detected in blood, we then sought to explore the functional consequences of four SNPs reaching the significance threshold in IL1B, IL12, and CRP genes. We evaluated the plasma levels of these proteins obtained from the patients with IE before admission and 7 days postoperation (Figure 2). The G/A genotype of the rs1143634 polymorphism within the IL1B gene and the G/T genotype of the rs3212227 polymorphism within the IL12B gene were associated with a higher plasma level of IL-1β and IL-12, respectively, at either time point (Figure 2). However, no significant associations were revealed for CRP (Figure 2). To further reinforce the associations between the marker SNPs, plasma cytokine levels, and risk of IE, we hypothesized that those SNPs which did not demonstrate a predictive value would not show associations with altered plasma levels of the corresponding proteins. Hence, we performed the similar analysis for the SNPs within the genes encoding cytokines which are generally elevated in patients with IE, that is, TNF-α, IL-6, IL-8, and IL-10 (Supplementary Figures 1 and 2 available online at https://doi.org/10.1155/2017/7962546). Expectedly, no statistically significant associations were found.
Figure 2.
Measurement of plasma interleukin-1β, interleukin-12, and C-reactive protein levels in patients with infective endocarditis at the hospital admission and 7 days postoperation. Two-tailed Student's t-test with further Tukey's post hoc test to adjust for multiple comparisons; each dot is a measure from one patient, ∗p < 0.05, n.s. is for not significant.
4. Discussion
Despite the recent progress in diagnosis and treatment [1, 2], the basis of genetic susceptibility to IE remains vaguely uncovered. Early investigation by Vollmer et al. revealed the G allele of the rs2232596 polymorphism and the T allele of the rs2232582 polymorphism within the LBP gene being associated with a higher risk of IE [29]. As known, lipopolysaccharide-binding protein is released into the bloodstream as an acute phase response protein during IE [19]. Further studies by Daga et al. [30, 31] and Durante-Mangoni et al. [32] did not find any association between the SNPs within the hemostasis genes (PTH, FV, GPIb, GPIIIa, and FcγRIIa) and IE. Our previous study identified the C/C genotype of the rs3775073 polymorphism within the TLR6 gene as a protective factor [12] while a study by Bustamante et al. suggested the A allele of the rs5743708 polymorphism within the TLR2 gene as a risk factor [33]. In this study, we selected 35 SNPs within 22 genes involved in the development of IE: IL1B, IL6, IL6R, IL8, IL10, IL12B, IL12RB, TNF, CRP, APOB, APOE, LIPC, LPA, NOTCH1, VDR, CASR, OPG, CALCR, F2, F5, F7, and ITGB3. Previous studies demonstrated the elevated serum levels of IL-1β, IL-6, IL-8, IL-10, IL-12, TNF-α, and CRP in patients with IE compared to the healthy blood donors which indicates the possible importance of these cytokines in the clinical course of IE; however, the exact role of these inflammatory molecules in IE remains elusive [11, 34]. Apolipoproteins B and E, lipoprotein (a), and hepatic lipase are involved in the metabolism of lipids that can be important for IE development since patients with IE have lower level of serum high-density lipoprotein cholesterol (HDL) than healthy blood donors; furthermore, low serum HDL level is indicative of a complicated IE course [16]. Another well-established risk factor of IE is heart valve calcification, with NOTCH1, vitamin D receptor, calcium-sensing receptor, osteoprotegerin, and calcitonin receptor being the major regulators of serum calcium and phosphorus levels [27, 35, 36]. Finally, patients with IE are prone to thromboembolism due to increased systemic coagulation, activation of platelets, and impaired fibrinolysis [13, 14]. F2, F5, F7, and integrin beta 3 all are crucial proteins responsible for the maintenance of hemostasis [14, 27].
Here, we found that the G/A genotype of the rs1143634 polymorphism within the IL1B gene, the G/T genotype of the rs3212227 polymorphism within the IL12B gene, the A/G genotype of the rs1130864 polymorphism within the CRP gene, the A/G genotype of the rs13290979 polymorphism within the NOTCH1 gene, and the G allele of the rs1801197 polymorphism within the CALCR gene were associated with a decreased risk of IE whereas the T/T genotype of the rs1205 polymorphism within the CRP gene was associated with a higher risk of IE. Recent studies by Weinstock et al. [37] and Giannitsioti et al. [38] revealed that the SNPs within the IL1B, IL6, and TNF genes can be associated with IE; however, we did not confirm these findings with regard to IL6 and TNF. Small sample sizes, sample differences (e.g., age, gender, ethnicity, and clinical features), and geographical variations in the microbial causes of IE [39, 40] may be responsible for these discrepancies. Since all the SNPs in our study were in Hardy-Weinberg equilibrium, genotyping errors were unlikely to affect the results.
The G/A genotype of the rs1143634 polymorphism within the IL1B gene and the G/T genotype of the rs3212227 polymorphism within the IL12B gene were associated with a higher plasma level of IL-1β and IL-12 that may provide an insight into their possible protective role. It has been reported that both cytokines are abundant in the serum of the patients with IE compared to those with other infections [34]; one can explain this as a specific immune response to the bacterial or fungal infection of the heart valves or chambers. We suggest that IL-1β and IL-12 can limit the infection, preventing further progression of IE. However, we did not find the associations of two SNPs within the CRP gene reaching statistical significance with CRP plasma level, although it is also known to be higher in patients with IE compared to other subjects [11]. Other SNPs within the TNF-α, IL-6, IL-8, and IL-10 genes did not show a predictive value and did not correlate with altered levels of the corresponding cytokines.
Our study has a number of limitations. First, the sample size is relatively small due to the low incidence rate of IE; however, this is a common drawback of genetic association studies on IE. With our sample size (123 cases and 300 controls), we had at least 83% and 99.7% power to detect odds ratio (OR) = 2 and OR = 3, respectively, with 5% alpha risk. Unfortunately, it was still impossible to analyze the genetic associations with the features or severity of IE. Second, technical difficulties made it unable to collect information on the potential confounders, that is, alcohol consumption, smoking status, and so forth. Third, we could not properly assess the microbiological profile due to extensive antibiotic therapy of IE at the district hospitals prior to the admission to our clinic. Finally, this study included only those patients requiring surgical treatment, as other patients with IE are not admitted to our clinic due to peculiarities of Russian healthcare.
5. Conclusions
Inherited variation within the cytokine, acute phase response, and calcium metabolism genes can be linked to IE, providing additional insight into its pathogenesis. In particular, heterozygous genotypes of the rs1143634 and rs3212227 polymorphisms are associated with both decreased risk of IE and higher level of IL-1β and IL-12, respectively, suggesting their possible importance. Further studies are needed to confirm our findings and for the further understanding of the genetic susceptibility to IE.
Supplementary Material
Supplementary Figure 1. Measurement of plasma tumor necrosis factor-α and interleukin-6 in patients with infective endocarditis at the hospital admission and 7 days postoperation. Two-tailed Student's t-test with the further Tukey's post hoc test to adjust for multiple comparisons, each dot is a measure from one patient, n.s. is for not significant. Supplementary Figure 2. Measurement of plasma interleukin-8 and interleukin-10 in patients with infective endocarditis at the hospital admission and 7 days postoperation. Two-tailed Student's t-test with the further Tukey's post hoc test to adjust for multiple comparisons, each dot is a measure from one patient, n.s. is for not significant.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Anastasia V. Ponasenko, Anton G. Kutikhin, Leonid S. Barbarash, and Arseniy E. Yuzhalin designed the research. Anastasia V. Ponasenko, Natalia V. Rutkovskaya, Natalia V. Kondyukova, Yuri N. Odarenko, and Yana V. Kazachek recruited the patients with IE. Arseniy E. Yuzhalin recruited the healthy controls. Anastasia V. Ponasenko, Maria V. Khutornaya, Anna V. Tsepokina, and Arseniy E. Yuzhalin isolated the DNA, performed the genotyping, and conducted the enzyme-linked immunosorbent assay. Anton G. Kutikhin carried out the statistical analysis. Anton G. Kutikhin and Arseniy E. Yuzhalin wrote the paper. All the authors have read and approved the final article.
References
- 1.Habib G., Lancellotti P., Antunes M. J., et al. 2015 ESC guidelines for the management of infective endocarditis: the Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM) European Heart Journal. 2015;36(44):3075–3128. doi: 10.1093/eurheartj/ehv319. [DOI] [PubMed] [Google Scholar]
- 2.Cahill T. J., Prendergast B. D. Infective endocarditis. Lancet. 2016;387(10021):882–893. doi: 10.1016/S0140-6736(15)00067-7. [DOI] [PubMed] [Google Scholar]
- 3.Muñoz P., Kestler M., De Alarcon A., et al. Current epidemiology and outcome of infective endocarditis: a multicenter, prospective, cohort study. Medicine (Baltimore) 2015;94(43, article e1816) doi: 10.1097/MD.0000000000001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bin Abdulhak A. A., Baddour L. M., Erwin P. J., et al. Global and regional burden of infective endocarditis, 1990-2010: a systematic review of the literature. Global Heart. 2014;9(1):131–143. doi: 10.1016/j.gheart.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Chu V. H., Cabell C. H., Benjamin D. K., et al. Early predictors of in-hospital death in infective endocarditis. Circulation. 2004;109(14):1745–1749. doi: 10.1161/01.CIR.0000124719.61827.7F. [DOI] [PubMed] [Google Scholar]
- 6.San Román J. A., López J., Vilacosta I., et al. Prognostic stratification of patients with left-sided endocarditis determined at admission. The American Journal of Medicine. 2007;120(4):369, e1–367. doi: 10.1016/j.amjmed.2006.05.071. [DOI] [PubMed] [Google Scholar]
- 7.Wang A., Athan E., Pappas P. A., et al. Contemporary clinical profile and outcome of prosthetic valve endocarditis. Journal of the American Medical Association. 2007;297(12):1354–1361. doi: 10.1001/jama.297.12.1354. [DOI] [PubMed] [Google Scholar]
- 8.Hill E. E., Herijgers P., Claus P., Vanderschueren S., Herregods M. C., Peetermans W. E. Infective endocarditis: changing epidemiology and predictors of 6-month mortality: a prospective cohort study. European Heart Journal. 2007;28(2):196–203. doi: 10.1093/eurheartj/ehl427. [DOI] [PubMed] [Google Scholar]
- 9.Murdoch D. R., Corey G. R., Hoen B., et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Archives of Internal Medicine. 2009;169(5):463–473. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNicholas S., Talento A. F., O'Gorman J., et al. Cytokine responses to Staphylococcus aureus bloodstream infection differ between patient cohorts that have different clinical courses of infection. BMC Infectious Diseases. 2014;14(1):p. 580. doi: 10.1186/s12879-014-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkin R. W., Harper L. V., Vernallis A. B., et al. Pro-inflammatory cytokines IL6, TNF-alpha, IL1beta, procalcitonin, lipopolysaccharide binding protein and C-reactive protein in infective endocarditis. The Journal of Infection. 2007;55(3):220–225. doi: 10.1016/j.jinf.2007.05.174. [DOI] [PubMed] [Google Scholar]
- 12.Golovkin A. S., Ponasenko A. V., Yuzhalin A. E., et al. An association between single nucleotide polymorphisms within TLR and TREM-1 genes and infective endocarditis. Cytokine. 2015;71(1):16–21. doi: 10.1016/j.cyto.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Buyukasýk N. S., Ileri M., Alper A., et al. Increased blood coagulation and platelet activation in patients with infective endocarditis and embolic events. Clinical Cardiology. 2004;27(3):154–158. doi: 10.1002/clc.4960270312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durante-Mangoni E., Molaro R., Iossa D. The role of hemostasis in infective endocarditis. Current Infectious Disease Reports. 2014;16(11):p. 435. doi: 10.1007/s11908-014-0435-8. [DOI] [PubMed] [Google Scholar]
- 15.İleri M., Kanat S., Orhan G., et al. Increased mean platelet volume in patients with infective endocarditis and embolic events. Cardiology Journal. 2015;22(1):37–43. doi: 10.5603/CJ.a2014.0021. [DOI] [PubMed] [Google Scholar]
- 16.Kahveci G., Bayrak F., Mutlu B., et al. Clinical significance of high-density lipoprotein cholesterol in left-sided infective endocarditis. The American Journal of Cardiology. 2008;101(8):1170–1173. doi: 10.1016/j.amjcard.2007.11.075. [DOI] [PubMed] [Google Scholar]
- 17.Elkhatib W. F., Hair P. S., Nyalwidhe J. O., Cunnion K. M. New potential role of serum apolipoprotein E mediated by its binding to clumping factor a during Staphylococcus aureus invasive infections to humans. Journal of Medical Microbiology. 2015;64(Part 4):335–343. doi: 10.1099/jmm.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thuny F., Textoris J., Amara A. B., et al. The gene expression analysis of blood reveals S100A11 and AQP9 as potential biomarkers of infective endocarditis. PloS One. 2012;7(2, article e31490) doi: 10.1371/journal.pone.0031490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snipsøyr M. G., Ludvigsen M., Petersen E., Wiggers H., Honoré B. A systematic review of biomarkers in the diagnosis of infective endocarditis. International Journal of Cardiology. 2016;202:564–570. doi: 10.1016/j.ijcard.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Yuzhalin A. E., Kutikhin A. G. Integrative systems of genomic risk markers for cancer and other diseases: future of predictive medicine. Cancer Management and Research. 2012;4:131–135. doi: 10.2147/CMAR.S30855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakhtiar S. M., Ali A., Baig S. M., Barh D., Miyoshi A., Azevedo V. Identifying human disease genes: advances in molecular genetics and computational approaches. Genetics and Molecular Research. 2014;13(3):5073–5087. doi: 10.4238/2014.July.4.23. [DOI] [PubMed] [Google Scholar]
- 22.Habib G., Hoen B., Tornos P., et al. Guidelines on the prevention, diagnosis, and treatment of infective endocarditis (new version 2009): the Task Force on the Prevention, Diagnosis, and Treatment of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the International Society of Chemotherapy (ISC) for Infection and Cancer. European Heart Journal. 2009;30(19):2369–2413. doi: 10.1093/eurheartj/ehp285. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z., Taylor J. A. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Research. 2009;37(Supplement 2):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dayem Ullah A. Z., Lemoine N. R., Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Research. 2012;40(Web Server issue):W65–W70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golovkin A. S., Ponasenko A. V., Khutornaya M. V., et al. Association of TLR and TREM-1 gene polymorphisms with risk of coronary artery disease in a Russian population. Gene. 2014;550(1):101–109. doi: 10.1016/j.gene.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Kutikhin A. G., Ponasenko A. V., Khutornaya M. V., et al. Association of TLR and TREM-1 gene polymorphisms with atherosclerosis severity in a Russian population. Meta Gene. 2016;9:76–89. doi: 10.1016/j.mgene.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponasenko A. V., Khutornaya M. V., Kutikhin A. G., et al. A genomics-based model for prediction of severe bioprosthetic mitral valve calcification. International Journal of Molecular Sciences. 2016;17(9) doi: 10.3390/ijms17091385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 29.Vollmer T., Kleesiek K., Dreier J. Lipopolysaccharide-binding protein (LBP) gene polymorphisms: rapid genotyping by real-time PCR and association with infective endocarditis. Clinical Biochemistry. 2009;42(13-14):1413–1419. doi: 10.1016/j.clinbiochem.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Daga S., Shepherd J. G., Callaghan J. G., et al. Platelet receptor polymorphisms do not influence Staphylococcus aureus-platelet interactions or infective endocarditis. Microbes and Infection. 2011;13(3):216–225. doi: 10.1016/j.micinf.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daga S., Shepherd J. G., Hung R. K., et al. GPIb VNTR C/C genotype may predict embolic events in infective endocarditis. The Journal of Heart Valve Disease. 2013;22(1):133–141. [PubMed] [Google Scholar]
- 32.Durante-Mangoni E., Iossa D., Molaro R., et al. Prevalence and significance of two major inherited thrombophilias in infective endocarditis. Internal and Emergency Medicine. 2015;10(5):587–594. doi: 10.1007/s11739-015-1214-8. [DOI] [PubMed] [Google Scholar]
- 33.Bustamante J., Tamayo E., Flórez S., et al. Toll-like receptor 2 R753Q polymorphisms are associated with an increased risk of infective endocarditis. Revista Española de Cardiología. 2011;64(11):1056–1059. doi: 10.1016/j.recesp.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Araújo I. R., Ferrari T. C., Teixeira-Carvalho A., et al. Cytokine signature in infective endocarditis. PloS One. 2015;10(7, article e0133631) doi: 10.1371/journal.pone.0133631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nucifora G., Badano L. P., Viale P., et al. Infective endocarditis in chronic haemodialysis patients: an increasing clinical challenge. European Heart Journal. 2007;28(19):2307–2312. doi: 10.1093/eurheartj/ehm278. [DOI] [PubMed] [Google Scholar]
- 36.Doulton T., Sabharwal N., Cairns H. S., et al. Infective endocarditis in dialysis patients: new challenges and old. Kidney International. 2003;64(2):720–727. doi: 10.1046/j.1523-1755.2003.00136.x. [DOI] [PubMed] [Google Scholar]
- 37.Weinstock M., Grimm I., Dreier J., Knabbe C., Vollmer T. Genetic variants in genes of the inflammatory response in association with infective endocarditis. PLoS One. 2014;9(10, article e110151) doi: 10.1371/journal.pone.0110151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannitsioti E., Damoraki G., Rokkas C., et al. Impact of haplotypes of TNF in the natural course of infective endocarditis. Clinical Microbiology and Infection. 2014;20(5):459–464. doi: 10.1111/1469-0691.12370. [DOI] [PubMed] [Google Scholar]
- 39.Elbey M. A., Akdağ S., Kalkan M. E., et al. A multicenter study on experience of 13 tertiary hospitals in Turkey in patients with infective endocarditis. Anadolu Kardiyoloji Dergisi. 2013;13(6):523–527. doi: 10.5152/akd.2013.172. [DOI] [PubMed] [Google Scholar]
- 40.Gupta K., Jagadeesan N., Agrawal N., Bhat P., Nanjappa M. C. Clinical, echocardiographic and microbiological study, and analysis of outcomes of infective endocarditis in tropical countries: a prospective analysis from India. The Journal of Heart Valve Disease. 2014;23(5):624–632. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Measurement of plasma tumor necrosis factor-α and interleukin-6 in patients with infective endocarditis at the hospital admission and 7 days postoperation. Two-tailed Student's t-test with the further Tukey's post hoc test to adjust for multiple comparisons, each dot is a measure from one patient, n.s. is for not significant. Supplementary Figure 2. Measurement of plasma interleukin-8 and interleukin-10 in patients with infective endocarditis at the hospital admission and 7 days postoperation. Two-tailed Student's t-test with the further Tukey's post hoc test to adjust for multiple comparisons, each dot is a measure from one patient, n.s. is for not significant.