Abstract
Eukaryotic cell division uses morphologically different forms of mitosis, referred to as open, partially open and closed mitosis, for accurate chromosome segregation and proper partitioning of other cellular components such as endomembranes and cell fate determinants. Recent studies suggest that the spindle matrix provides a conserved strategy to coordinate the segregation of genetic material and the partitioning of the rest of the cellular contents in all three forms of mitosis.
Eukaryotes assemble a microtubule-based spindle apparatus that separates chromosomes into daughter cells during cell division. Although studies of how spindle microtubules capture kinetochores of chromosomes in mitosis have reached the molecular level, there is limited understanding of how chromosome segregation is coordinated with other cellular changes during cell division. For example, it is unclear how, in animal cells, disassembled nuclear membranes are initially absorbed by the endoplasmic reticulum (ER) and then reassembled into a new nuclear envelope. Golgi membranes are known to distribute throughout the mitotic spindle, but the relationship between Golgi and ER and how Golgi membranes are inherited remain subject to debate1. Consequently, it is unclear how signalling pathways use membrane trafficking systems to coordinate their functions in mitosis. Therefore, it is important to broaden the study of mitosis beyond microtubules and kinetochores. Recent efforts towards probing how non-microtubule components of the spindle, collectively called the spindle matrix, behave and function during mitosis indicate that different cellular compartments work together to ensure proper cell division.
I review these studies in the context of the different forms of mitosis that have evolved in eukaryotes — open, partially open and closed mitosis — and propose a unified model for the spindle matrix coordinating the segregation of genetic material, which provides a conceptual framework for studying eukaryotic cell division in a developmental context.
Defining the spindle matrix
Lower eukaryotes such as fungi use closed mitosis for cell division, whereas higher eukaryotes such as metazoans undergo either partially open or completely open mitosis2 (FIG. 1; BOX 1). The idea of an existing spindle matrix was initially suggested by the study of open mitosis. Studies of isolated mitotic spindles from synchronized sea urchin eggs in the 1960s led to the observation that non-microtubule material surrounds mitotic spindles, which are defined by microtubules, over a large area3,4. This non-microtubule material retains a shape that is reminiscent of spindles even after microtubule disassembly5,6. These studies led to the idea that whereas some of the non-microtubule material could be passive passengers that use spindle microtubules for their partitioning into daughter cells, some could function as a spindle matrix to tether mitotic regulators and/or as a mechanical element to hold the spindle microtubules together.
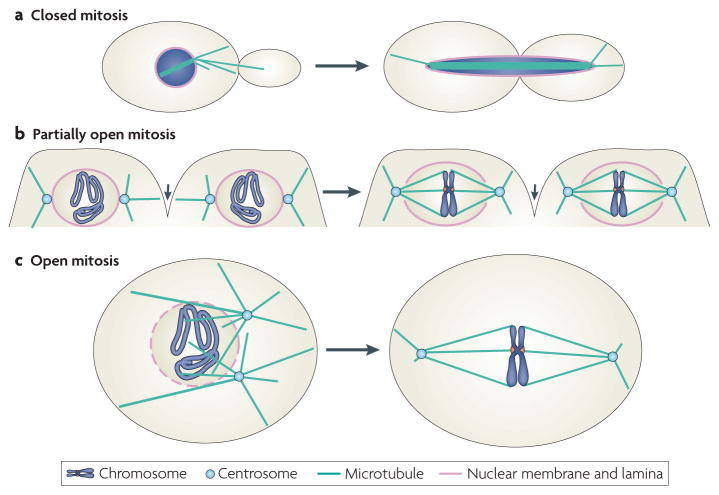
Figure 1. Metaphase spindles in three forms of mitosis.
Eukaryotes have evolved various forms of mitosis to coordinate nuclear morphogenesis with mitotic spindle assembly. a | Closed mitosis, which is found in lower eukaryotes such as the fungus Saccharomyces cerevisiae, refers to mitosis in which nuclear envelope does not breakdown during spindle assembly and chromosome segregation. Spindle pole bodies (not shown) embedded in the nuclear envelope nucleate cytoplasmic microtubules, which interact with the cell cortex, and nuclear microtubules, which interact with one another or kinetochores of chromosomes. After chromosome segregation, the nucleus undergoes karyokinesis to form two daughter nuclei (not shown). b | An example of partially open mitosis can be found in Drosophila melanogaster early syncytial embryos, in which all nuclei occupy the same cytoplasm. During mitosis, plasma membrane invaginations (small arrows) separate spindles formed by different nuclei. Two adjacent nuclei undergoing mitosis are depicted. The microtubules nucleated from the centrosomes invade the nuclear space through two openings on the nuclear envelope to form spindles that are surrounded by the nuclear envelope. During anaphase, the nuclear envelope undergoes further disassembly and is then reassembled during telophase (not shown). c | The assembly of the mitotic spindle during open mitosis, found in vertebrates, is accompanied by a complete disassembly of the nuclear envelope during prometaphase (for more details, see BOX 1).
Box 1. Spindle assembly in open mitosis.
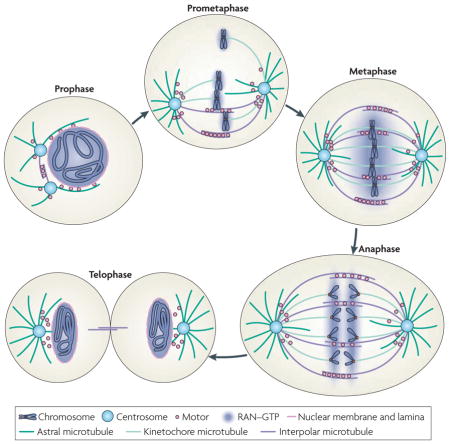
Mitosis can be divided into five stages: prophase, prometaphase, metaphase, anaphase and telophase. As cells prepare for division during prophase, two centrosomes nucleate astral microtubule arrays that interact with the cell cortex and nuclear envelope, while chromatin undergoes condensation to form mitotic chromosomes. During this period, the nuclear envelope remains intact, with the GTP-bound RAN GTPase concentrated inside the nucleus (see the figure). Nuclear envelope breakdown marks the onset of prometaphase. The disassembly of the nuclear envelope allows RAN–GTP to diffuse away from chromosomes, forming a gradient around chromatin that functions to promote microtubule assembly towards condensed chromosomes during spindle assembly. With the help of motor proteins, such as dynein and kinesins, and non-motor microtubule-associated proteins, such as nuclear mitotic apparatus protein (NUMA) and targeting protein for XKLP2 (TPX2), microtubules either interact with each other to form interpolar microtubules or capture kinetochores of condensed chromosomes to form kinetochore microtubules53–59. The two centrosomes (or spindle pole bodies in closed mitosis in fungi) that help to organize minus ends of microtubules to form spindle poles nucleate the astral array of microtubules, which interacts with cytoplasmic structures and the cell cortex to help position the spindle60. Metaphase is characterized by all chromosomes becoming captured by kinetochore microtubules from opposite poles and aligning in the centre of the spindle. The aligned sister chromosomes separate from one another during anaphase. During telophase, two sets of sister chromosomes undergo de-condensation, with RAN GTPase regulating nuclear envelope reassembly, while the two daughter cells remain connected by the midbody, which contains microtubules. The plus and minus ends of microtubules are located distally and proximally to centrosomes, respectively.
Evidence for a continuous elastic spindle matrix
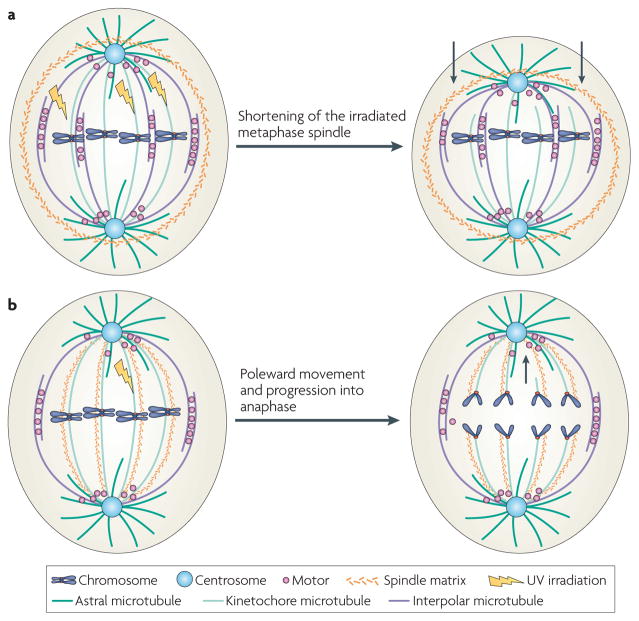
One key feature of a metaphase spindle is its ability to balance all forces within itself to maintain a steady-state length and shape7. Mechanical perturbation experiments have revealed that spindle microtubules seem to be under external compression. When several kinetochore microtubules in one half of a spindle were severed with ultraviolet microbeams, the irradiated half of the spindle shortened, the spindle pole moved towards the kinetochores and the remaining intact kinetochore microtubules bent8 (FIG. 2a). Similar compression of microtubules was also seen using the caged microtubule depolymerizing drug 105D9. The gradual depolymerization of microtubules resulted in spindle shortening and the bending or buckling of remaining microtubules. This suggests that a continuous and elastic spindle matrix exists, which is under mechanical stress created by spindle microtubules. When forces exerted by microtubules are reduced, the spindle matrix contracts, which causes the remaining microtubules to bend and buckle (FIG. 2a).
Figure 2. Two types of spindle matrices in open mitosis, as revealed by physical perturbations.
a | An elastic spindle matrix surrounds spindle microtubules, as revealed by cutting a few kinetochore microtubules in a metaphase spindle by ultraviolet (Uv) irradiation. After irradiating one half of the spindle (top half), the uncut microtubules in this half curve back, causing shortening of the half spindle. This suggests that the elastic spindle matrix is normally stretched by spindle microtubules. When this stretching force is reduced, the spindle matrix would collapse onto spindle microtubules, causing them to bend and buckle. An elastic and continuous spindle matrix that permeates the spindle microtubules could also carry out the same function but has been omitted from this figure for simplicity. The arrows in the right panel show the direction of the compression force from the contracting matrix that causes the shrinking of the half spindle. b | A speculative spindle matrix aligns along kinetochore microtubules. UV irradiation of the indicated kinetochore microtubule did not block this kinetochore from moving to spindle poles during anaphase of insect cell mitosis. The arrow in the right panel shows the direction of movement of the kinetochore with a microtubule stub.
A spindle matrix aligning spindle microtubules
Additional experiments have led to speculation about the nature and function of a spindle matrix that aligns along microtubules in insect cells. When a kinetochore microtubule fibre was severed by irradiation, the kinetochore still moved polewards despite lacking connections to the pole8. Although this movement could be mediated by an interaction of the kinetochore stub with interpolar microtubules, a spindle matrix that aligns along spindle microtubules was also proposed as a mechanical element that mediates the interaction and movement of the broken kinetochore towards the pole8 (FIG. 2b).
Recent studies of the spindle matrix
The above experiments suggest the possibility that certain non-microtubule structures could both surround the spindle and align along microtubules inside the spindle to facilitate cell division. However, for many years the lack of molecular and structural insights about the spindle matrix has made its existence a subject of debate. Below, I discuss the recent progress in studying the spindle matrix in partially open and open mitosis that has provided strong support for the function of this structure.
The spindle matrix in spindle microtubules
Four nuclear proteins — Skeletor, Megator, East, and Chromator — have been found to interact with each other and to localize throughout spindle microtubules in Drosophila melanogaster, which uses partially open mitosis. These proteins remain in spindle-like structures after microtubule depolymerization using nocodazole in D. melanogaster embryos10–13 (FIG. 3a). RnA interference and genetic analyses of Megator, Chromator and East have revealed that reduced levels of each of these proteins lead to mitotic defects, including problems in spindle elongation, chromosome alignment and spindle pole focusing14,15. Interestingly, Megator and its mammalian orthologue, the nucleoporin TPR (translocated promoter region), interact with the spindle assembly checkpoint proteins MAD1 (also known as TXBP181) and MAD2 (REFS 14, 16). Megator, Chromator and TPR facilitate the efficient loading of MAD1 and MAD2 onto unattached kinetochores to strengthen the spindle assembly checkpoint.
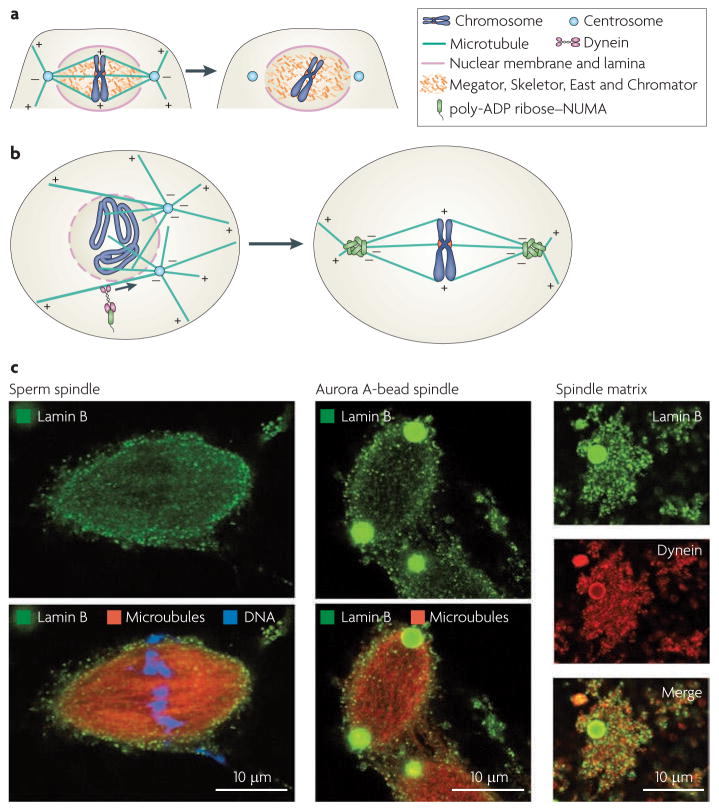
Figure 3. Localization of spindle matrix components.
a | The Drosophila melanogaster spindle matrix. The nuclear proteins skeletor, Megator, East and chromator interact with each other to form an internal spindle matrix that colocalizes with microtubules of the spindle throughout mitosis in D. melanogaster syncytial embryos. The organization of these proteins in the matrix is currently unknown, but after microtubule disassembly by microtubule depolymerization drugs such as nocodazole, this network remains intact. One function of the D. melanogaster spindle matrix is to tether the spindle assembly checkpoint proteins MAD1 (also known as TXBP181) and MAD2 to facilitate their binding to kinetochores. This internal spindle matrix is surrounded by a spindle envelope containing lamin B and the nuclear membrane. The spindle envelope has recently been shown to function as an elastic spindle matrix. b | The spindle pole matrix formed by poly−ADP ribose−modified nuclear mitotic apparatus protein (NUMA) in vertebrates that carry out open mitosis. NUMA localizes to the nucleus in interphase (not shown). In mitosis, poly−ADP ribose polymerase 5A (PARP5A; also known as tankyrase 1) catalyses the covalent modification of NUMA by poly−ADP ribose. Modified NUMA is transported to the minus ends of microtubules in early prometaphase, when the nuclear envelope starts to breakdown (left), to assemble into a network to facilitate the interactions of microtubule minus ends to form focused spindle poles (right). c | Lamin B spindle matrix localizes to spindles assembled from sperm chromatin (left) or Aurora A beads (centre; green). Aurora A spindles were treated with nocodazole and, after microtubule depolymerization, the spindle matrix that remains associated with Aurora A beads was stained using antibodies to lamin B and dynein (right panels; Aurora A beads in green and red). Right and left panels in part c are reproduced, with permission, from Nature Cell Biology REF. 28 © (2009) Macmillan Publishers. All rights reserved.
The studies in D. melanogaster show that a non-microtubule matrix-like structure formed by nuclear proteins does exist in spindle microtubules to functionally tether spindle regulators. It remains unclear whether this structure could interact with kinetochore microtubules to mechanically regulate chromosome segregation. The identification of molecular components should promote further study of the structure of this internal spindle matrix and its interaction with spindle microtubules.
The spindle matrix localized to spindle poles
Nuclear and mitotic apparatus protein (NUMA) is a nuclear protein in interphase and becomes localized to spindle poles during mitosis. NUMA seems to influence higher-order chromatin organization and epithelial differentiation in mammals17. In vertebrate open mitosis, NUMA functions to bundle microtubule minus ends into focused spindle poles18–20. NUMA can both self-associate and associate with other nuclear proteins in interphase21. In mitosis, it directly binds to microtubules and is transported to microtubule minus ends by the motor protein dynein. Oligomerization of NUMA at the minus ends of microtubules seems to facilitate the bundling of microtubules into focused spindle poles.
Interestingly, NUMA also binds the spindle pole-localized poly-ADP ribose polymerase 5a (PARP5A; also known as tankyrase 1) and becomes post-translationally modified by PARP5A to carry poly-ADP ribose chains22. Poly-ADP ribose is present in spindles assembled in Xenopus laevis egg extracts and is concentrated at spindle poles in tissue culture cells23,24. As poly-ADP ribose is required for spindle pole focusing in X. laevis egg extracts, poly-ADP ribose-modified NUMA may form a network at spindle poles (FIG. 3b). Poly-ADP ribose in spindles exhibits slower turnover than tubulins23, suggesting that this network is separate from microtubules and that it functions as a spindle matrix to maintain spindle poles.
Lamin B: part of a membranous spindle matrix
Lamin B in metazoans forms a filamentous network in interphase nuclei called the nuclear lamina25. In interphase, lamin B has well-established functions in regulating transcription, DnA replication, chromatin organization and nuclear integrity25. In partially open mitosis, most of the nuclear lamin B and nuclear envelope remain intact, enclosing spindles from prometaphase through metaphase. In open mitosis, the disassembled lamin B is transported to spindle poles along microtubules during prometaphase. By metaphase, lamin B surrounds the spindle and is concentrated at spindle poles26,27.
Recently, using in vitro spindle assembly assays in X. laevis egg extracts, lamin B was shown to associate with spindles (FIG. 3c) and to have mitosis-specific functions in facilitating both spindle pole focusing and maintaining spindle length28–30. After microtubule depolymerization by nocodazole, lamin B remains in a matrix-like network (FIG. 3c) that can stimulate microtubule assembly from purified tubulins. Perturbation experiments using dominant-negative lamin B suggest that self-association of lamin B, which requires RAN–GTP, is important for its function in maintaining spindle shape and length.
A spindle assembly assay was developed to determine the lamin B spindle matrix proteome from X. laevis egg extracts. This assay uses magnetic beads coated with the mitotic kinase Aurora A to stimulate formation of spindle-like structures in the presence of RAN–GTP in egg extracts, enabling the use of magnets to recover bead-associated spindle matrix after microtubule depolymerization31,32. The isolated spindle matrix can be labelled by membrane dye and is completely disrupted by detergent treatment, suggesting that the matrix is a membranous network29.
Further analyses revealed that the isolated spindle matrix contains lamin B, poly-ADP ribose, nucleoporins and various spindle assembly factors, including mitotic kinesin EG5 (also known as KIF11), NUMA, the motor protein dynein and its regulator Nudel28,29. Interestingly, some of the nucleoporins have recently been shown to regulate spindle morphology in X. laevis egg extracts33. Moreover, dynein interacts with lamin B in part through Nudel. Dynein, Nudel and microtubules are all required for the assembly of lamin B into the membranous spindle matrix that associates with spindles assembled in egg extracts28 (FIG. 4).
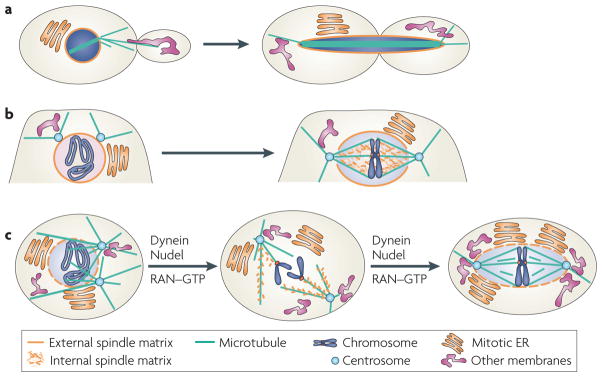
Figure 4. A unified model of the spindle matrix in patterning cell division.
All forms of mitosis can use the spindle matrix to coordinate chromosome segregation with the partitioning of the rest of the cellular components into daughter cells. a | The mitotic nucleus functions as a spindle matrix in closed mitosis. The mitotic nuclear envelope, through its connection with the endoplasmic reticulum (ER), could help partition various cellular components such as the endomembrane system during cell division. b | The spindle matrix in partially open mitosis includes the nuclear envelope, nuclear lamina and the intra−nuclear spindle matrix, which contains Megator, skeletor, chromator and East. This network may organize nuclear content and cytoplasmic membranes for proper division. c | In open mitosis, dynein and its regulator Nudel regulate assembly of membranous lamin B spindle matrix in a microtubule and RAN–GtP−dependent manner. This spindle matrix contains nuclear mitotic apparatus protein (NUMA) and poly−ADP ribose (not shown). Similar to spindle matrix in closed and partially open mitosis, the lamin B−containing matrix associates with the spindle to organize nuclear contents and cytoplasmic membranes, coordinating their partitioning into daughter cells.
Recent studies in D. melanogaster early embryos revealed that the lamin B-containing nuclear envelope surrounds spindles to increase the robustness of motor-mediated balancing forces at spindle microtubules. Either disrupting or over-stabilizing lamin B in this envelope perturbed spindle length maintenance. Mathematical modelling suggests that the lamin B-containing envelope functions as an elastic spring to enhance spindle length control by buffering fluctuations in motor activities34.
Although spindles in open mitosis are not surrounded by a well-defined lamin B envelope, the membranous lamin B network is known to surround spindles in vertebrate tissue culture cells and in X. laevis egg extracts. By analysing spindle morphology and microtubule flux in spindles assembled in egg extracts, we found that lamin B indeed functions in a network surrounding spindles to maintain spindle length and bipolarity by restraining EG5-driven microtubule sliding (B. Goodman and Y.Z., unpublished observations). Therefore, a spindle matrix containing lamin B seems to function similarly in both partially open and open mitosis.
Membranes and the spindle matrix
The membranous nature of the lamin B spindle matrix suggests that proper organization of certain mitotic membranes could be important for spindle morphology. Consistent with this idea, perturbation of several membrane-associated proteins has been shown to cause defects in spindle morphology35–42. Recently, the membrane bending activity of epsin 1, an endocytic adaptor protein, has been shown to regulate both mitotic membrane organization and spindle pole focusing43. As the membrane-bending activity of epsin 1 also facilitates spindle assembly in X. laevis egg extracts, this study provides evidence that proper membrane organization directly influences spindle morphogenesis.
Analyzing the mitotic localization of proteins known to associate with the ER, Golgi or nuclear envelope suggests that epsin 1 facilitates the organization of mitotic membranes that have ER features43. Therefore, proteins such as epsin 1 may be required to organize lamin B-containing ER and/or nuclear membranes around mitotic spindles to facilitate their functions as a spindle matrix. Further study of the mitotic functions of membrane-bending proteins such as epsin 1 should shed light on how mitotic membranes influence microtubule assembly and how microtubules may in turn regulate the segregation of membrane compartments into daughter cells.
The spindle matrix: a unified model
The above studies have revealed that the membranous spindle matrix in partially open and open mitosis contains several nuclear proteins. Whereas some of these proteins, such as Megator, function to tether regulators of mitosis in spindle microtubules, other proteins, such as lamin B and NUMA, could function in a network that surrounds spindles to control spindle length and morphology. In partially open mitosis, the spindle matrix includes both a lamin B-containing envelope that surrounds spindles to balance motor forces and an internal matrix containing nuclear proteins such as Megator and Skeletor that regulates spindle assembly factors in microtubules (FIG. 4b).
In open mitosis, although the nuclear envelope and lamina completely disassemble, RAN–GTP, microtubules and dynein function to drive the assembly and organization of the lamin B-containing membranous network around spindles28. This network has a similar role to the spindle envelope in partially open mitosis (FIG. 4c). This spindle matrix in open mitosis includes NUMA and poly-ADP ribose polymerase (PARP) at spindle poles and the membranous lamin B network surrounding the spindle. Together, they function as a physical barrier to restrain motor-driven microtubule movements in the spindle, which in turn facilitates the maintenance of spindle length and morphology.
Although no internal spindle matrix similar to the one made of the nuclear proteins such as Megator and Skeletor in early embryos (FIG. 4b) has been found in D. melanogaster vertebrate mitosis, I propose that the lamin B spindle matrix could facilitate the spindle microtubule-association of nuclear proteins such as TPR (the orthologue of the D. melanogaster Megator protein) to carry out the same roles. In this regard, it is interesting to note that several nuclear proteins have been shown to directly regulate spindle microtubules in open mitosis44,45. It would be interesting to study whether perturbation of the lamin B spindle matrix could affect the functions of these nuclear proteins in mitosis.
Based on the analogy between the spindle envelope in partially open mitosis and the lamin B spindle matrix in open mitosis, I suggest that the spindle matrix in both mitoses shares a function similar to that of the mitotic nucleus in closed mitosis — to coordinate spindle morphogenesis with cell division (FIG. 4a). This idea could help explain why numerous nuclear and membrane proteins with well-known functions in interphase also influence mitotic spindle assembly. Below, I discuss how the nucleus in closed mitosis can be viewed as a membranous spindle matrix and how the unified idea of this network could help in the study of mitosis in the broader context of cell division.
Similar roles of RAN in different mitoses
One function of the RAN GTPase is to regulate nuclear envelope reassembly in organisms that undergo open mitosis46,47 (BOX 1). Interestingly, studies have shown that RAN–GTP is also required for assembly and maintenance of the lamin B spindle matrix29. As the lamin B spindle matrix contains nucleoporins and other nuclear proteins28, I propose that spindle matrix assembly is the first step towards nuclear re-assembly in open mitosis. Soon after nuclear envelope breakdown, RAN–GTP generated on mitotic chromosomes and dynein facilitate the organization of membranes and proteins involved in nuclear re-assembly around microtubules, leading to the formation of the spindle matrix (FIG. 3c; FIG. 4c). This matrix functions first to maintain spindle morphology and then as a precursor to nuclear envelope reassembly at the end of mitosis.
In closed mitosis, dynein is not essential for spindle assembly but is required for spindle positioning48. This is probably because the nuclear envelope does not breakdown, obviating the need for spindle matrix assembly. Spindle elongation in closed mitosis is accompanied by the expansion of the nuclear envelope, which would require RAN–GTP and the reorganization of the ER and nuclear envelope. Indeed, studies of Schizosaccharomyces pombe, which undergoes closed mitosis, showed that disrupting RAN–GTP production by a mutation in the nuclear exchange factor for RAN results in nuclear envelope rupture as spindles elongate49. Such rupture can be partially rescued by slowing down spindle elongation by tubulin mutations or overexpressing HMG CoA reductase, which stimulates ER membrane expansion49. Interestingly, nuclear envelope rupture can also be remedied by reducing ER curvature through deletion of the ER membrane-bending protein reticulon49, which is also present on the isolated lamin B spindle matrix28. Reduced curvature in the ER may allow more ER to be incorporated into the flat nuclear membrane to accommodate spindle elongation. Therefore, although the nuclear envelope does not breakdown in closed mitosis, changes in the nuclear membrane and its associated ER regulated by RAN–GTPase and membrane-bending proteins do occur. Analogous to the assembly of the membranous spindle matrix, these changes are important for spindle morphogenesis.
A shared tethering function of the spindle matrix
Similar to the membranous spindle matrix in open mitosis, the mitotic nucleus in closed mitosis can tether spindle assembly factors to coordinate spindle assembly and chromosome segregation. This is illustrated by a recent finding in Aspergillus nidulans, a filamentous fungus that undergoes closed mitosis. The A. nidulans nucleoporin Mlp1, like its orthologues D. melanogaster Megator and human TPR, is required to tether a pool of Mad2 close to kinetochores in mitosis50. As the tethering function of Mlp1 is independent of spindle assembly, the authors suggest that Mlp1 functions as part of a spindle matrix in the mitotic nucleus to tether Mad2.
A shared function in patterning cell division
In closed mitosis, the nuclear envelope retains its interphase morphology, which could be used to demarcate cell division. Consistent with this idea, during closed mitosis of the yeast Saccharomyces cerevisiae, a diffusion barrier is formed in the mitotic nuclear envelope by the polymerization of septin proteins to limit the segregation of pre-existing old nuclear pores to the mother cells51. As the old nuclear pores tether the age-causing extrachromosomal ribosomal DNA circles and preferentially segregate them into the older mother cells, the nuclear envelope functions to regulate this unique form of asymmetric cell division, ensuring the production of a younger daughter cell.
By analogy, I propose that assembly of a membranous spindle matrix in open mitosis could allow the organization of membrane compartments, nuclear and cytoplasmic proteins, and RNAs around the spindle for proper partitioning. Indeed, the isolated X. laevis spindle matrix contains signalling molecules and transcription factors known to regulate cell fate specification during development28. By tethering to the spindle matrix, these factors could help to maintain the epigenetic memory of mitotic chromosomes. Such tethering could also allow the factors to be partitioned either symmetrically or asymmetrically into daughter cells. In this context, it is interesting to note that dynein, NUMA and the polarity protein partitioning defective 3 (PAR3), which are all present on the isolated spindle matrix, have been found to regulate spindle positioning and asymmetric cell division in several organisms52. As dynein is required for spindle matrix assembly, it is possible that other proteins involved in asymmetric cell division function in part to regulate assembly of the spindle matrix and spindle morphology during organism development.
Conclusions and perspectives
A mitotic spindle matrix was proposed decades ago as a tensile element and tether that regulates spindle assembly and chromosome segregation in open mitosis. However, the difficulty in defining the molecular nature of this matrix and the prevailing scepticism that spindle assembly requires structures beyond microtubules have resulted in little progress until recently. Studies in the past several years in different systems have provided strong support for the existence of a membranous spindle matrix, which functions alongside microtubules to regulate spindle pole focusing, spindle force balancing and spindle assembly checkpoints.
The emerging theme is that the spindle matrix is a membranous structure that contains several nuclear proteins, including lamin B, NUMA and Megator or its orthologue, TPR. In open mitosis, the assembly of this structure requires RAN–GTP, dynein and microtubules. As a result, the membranous spindle matrix is probably dynamic and could lose many structural features after microtubule depolymerization. In partially open mitosis, the spindle matrix contains nuclear proteins and the partially disassembled nuclear envelope, which contains lamin B. Although there is no nuclear lamina in organisms undergoing closed mitosis, the intact nuclear envelope and nuclear proteins seem to carry out equivalent functions of a membranous spindle matrix. Further study of the assembly, organization and function of the membranous spindle matrix will not only improve our knowledge of chromosome segregation but may also uncover new principles of mitotic cellular organization that allow the transfer or modification of cell fates through successive cell divisions.
Exploring this new frontier will require researchers to broaden the study of mitosis to include proteins with known functions in interphase nuclei, interphase membrane trafficking and signalling. To establish their mitosis-specific roles, use of the existing in vitro spindle assembly system in X. laevis egg extracts and the development of new systems will be vital. New methods to visualize membrane compartments and mitotic chromatin are also needed to explore how different cellular structures affect cell partitioning. Finally, it is important to note that spindle morphology defects caused by a perturbed spindle matrix may not affect equal chromosome segregation, but could affect the ability of mitotic spindles to reorient to ensure asymmetric cell division during development. It is therefore important to begin studying how spindle matrix components could influence mitotic spindle positioning, and symmetric and asymmetric cell divisions in a developmental context. These efforts will fuel new discoveries that should bring the poorly defined mitotic cellular structures into a sharper focus.
Acknowledgments
I thank B. Goodman for spindle drawings and B. Guo, K. Jung and members of the Zheng laboratory for helpful comments. I apologize to colleagues whose work could not be cited owing to space limitations. The work on mitosis in the Y.Z. laboratory is supported by the National Institute of General Medical Sciences (GM056312). Y.Z. is an investigator of the Howard Hughes Medical Institute. The image in the middle panel of figure 3 part c is courtesy of L. Ma.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
UniProtKB: http://www.uniprot.org
Chromator | East | EG5 | epsin 1 | MAD1 | MAD2 | Megator | Mlp1 | Nudel | NUMA | PAR3 | PARP5A | Skeletor | TPR
FURTHER INFORMATION
Y. Zheng’s homepage: http://www.ciwemb.edu/labs/zheng/index.php
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Persico A, Cervigni RI, Barretta ML, Colanzi A. Mitotic inheritance of the Golgi complex. FEBS Lett. 2009;583:3857–3862. doi: 10.1016/j.febslet.2009.10.077. [DOI] [PubMed] [Google Scholar]
- 2.De Souza CP, Osmani SA. Double duty for nuclear proteins — the role of more open forms of mitosis. Trends Genet. 2009;25:545–554. doi: 10.1016/j.tig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman RD, Rebhun LI. The structure and some properties of the isolated mitotic apparatus. J Cell Sci. 1969;4:179–209. doi: 10.1242/jcs.4.1.179. [DOI] [PubMed] [Google Scholar]
- 4.Forer A, Goldman RD. Comparisons of isolated and in vivo mitotic apparatuses. Nature. 1969;222:689–690. doi: 10.1038/222689a0. [DOI] [PubMed] [Google Scholar]
- 5.Leslie RJ, Hird RB, Wilson L, McIntosh JR, Scholey JM. Kinesin is associated with a nonmicrotubule component of sea urchin mitotic spindle. Proc Natl Acad Sci USA. 1987;84:2771–2775. doi: 10.1073/pnas.84.9.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wein H, Bass HW, Cande WZ. DSK1, a kinesin-related protein involved in anaphase spindle elongation, is a component of a mitotic spindle matrix. Cell Motil Cytoskel. 1998;41:214–224. doi: 10.1002/(SICI)1097-0169(1998)41:3<214::AID-CM3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 7.Dumont S, Mitchison TJ. Forces and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickett-Heaps JD, Forer A, Spurck T. Traction fibre: toward a “tensegral” model of the spindle. Cell Motil Cytoskel. 1997;37:1–6. doi: 10.1002/(SICI)1097-0169(1997)37:1<1::AID-CM1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 9.Mitchison TJ, et al. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi H, et al. East interacts with Megator and localizes to the putative spindle matrix during mitosis in Drosophila. J Cell Biochem. 2005;95:1284–1291. doi: 10.1002/jcb.20495. [DOI] [PubMed] [Google Scholar]
- 11.Qi H, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rath U, et al. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein Skeletor. J Cell Biochem. 2004;93:1033–1047. doi: 10.1002/jcb.20243. [DOI] [PubMed] [Google Scholar]
- 13.Walker DL, et al. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J Cell Biol. 2000;151:1401–1411. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lince-Faria M, et al. Spatialtemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Y, et al. Chromator is required for proper microtubule spindle formation and mitosis in Drosophila. Dev Biol. 2009;334:253–263. doi: 10.1016/j.ydbio.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abad PC, et al. NuMA influences higher order chromatin organization in human mammary epithelium. Mol Biol Cell. 2007;18:348–361. doi: 10.1091/mbc.E06-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton D, Cleveland D. NuMA is required for the proper completion of mitosis. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Compton D, Cleveland D. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr Opin Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 20.Merdes A, Ramyar K, Vechio J, Cleveland D. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 21.Fant X, Merdes A, Haren L. Cell and molecular biology of spindle poles and NuMA. Int Rev Cytol. 2004;238:1–57. doi: 10.1016/S0074-7696(04)38001-0. [DOI] [PubMed] [Google Scholar]
- 22.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 24.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 25.Dechat T, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- 27.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 28.Ma L, et al. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nature Cell Biol. 2009;11:247–256. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai MY, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y, Tsai MY. The mitotic spindle matrix: a fibro-membranous lamin connection. Cell Cycle. 2006;5:2345–2347. doi: 10.4161/cc.5.20.3365. [DOI] [PubMed] [Google Scholar]
- 31.Tsai MY, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 32.Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 33.Orjalo AV, et al. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3816. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 36.Cao K, Zheng Y. The Cdc48/p97-Ufd1-Npl4 complex: its potential role in coordinating cellular morphogenesis during M-G1 transition. Cell Cycle. 2004;3:422–424. [PubMed] [Google Scholar]
- 37.Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vong QP, Cao K, li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of Surivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 40.Lehtonen S, et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Mol Biol Cell. 2008;19:2949–2961. doi: 10.1091/mbc.E07-05-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci USA. 2007;104:7939–7944. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. The Sac1 phosphoinositide phosphatase regulates golgi membrane morphology and mitotic spindle organization in mammals. Mol Biol Cell. 2008;19:3080–3096. doi: 10.1091/mbc.E07-12-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Zheng Y. A requirement for epsin in mitotic membrane and spindle organization. J Cell Biol. 2009;186:473–480. doi: 10.1083/jcb.200902071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ducat DC, Kawaguchi S, Liu H, Yates J, 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell. 2008;19:3097–3110. doi: 10.1091/mbc.E07-11-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol. 2009;187:813–829. doi: 10.1083/jcb.200906020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 47.Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- 48.Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle position in a polarized cell. Cell Motil Cytoskel. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez Y, et al. Nuclear shape, growth and integrity in the closed mitosis of fission yeast depends on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J Cell Sci. 2009;122:2464–2472. doi: 10.1242/jcs.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell. 2009;20:2146–2159. doi: 10.1091/mbc.E08-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barrel Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 52.Siller KH, Doe CO. Spindle orientation during assymetric cell division. Nature Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 53.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 54.Kalab P, Pu R, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 55.Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 56.Carazo-Salas RE, et al. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Wirtz D, Zheng Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J Cell Biol. 2003;160:635–644. doi: 10.1083/jcb.200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 2004;18:512–527. doi: 10.1101/gad.1177304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 60.Walczak CE, Heald R. Mechanism of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]