Abstract
Transcriptional enhancers play a key role in cell type-specific gene expression and cell fate transition. Enhancers are marked by histone H3K4 mono- and di-methylation (H3K4me1/2). The tumor suppressor MLL4 (KMT2D) is a major enhancer H3K4 mono- and di-methyltransferase with a partial functional redundancy with MLL3 (KMT2C). However, the functional role of MLL4 enzymatic activity remains elusive. To address this issue, we have generated MLL4 enzyme-dead knock-in (KI) embryonic stem (ES) cells and mice, which carry Y5477A/Y5523A/Y5563A mutations in the enzymatic SET domain of the MLL4 protein. Homozygous MLL4 enzyme-dead KI (Mll4KI/KI) mice are embryonic lethal and die around E10.5, which pheno-copies Mll4 knockout (KO) mice. Interestingly, enzyme-dead MLL4 protein in ES cells is highly unstable. Like Mll4 KO ES cells, Mll4KI/KI ES cells show reduced levels of H3K4me1/2. Further, we show that ectopic expression of histone H3.3 lysine 4 to methionine (K4M) mutant, which reduces endogenous H3K4 methylation levels in ES cells, decreases protein stability of MLL3 and MLL4 but not that of H3K4 methyltransferases SET1A (KMT2F) and SET1B (KMT2G). Taken together, our findings indicate that MLL4 protein stability is tightly regulated by its H3K4 methyltransferase activity.
Keywords: H3K4 methyltransferase, MLL4, MLL3, Enzymatic activity, H3.3K4M
Graphical Abstract
Introduction
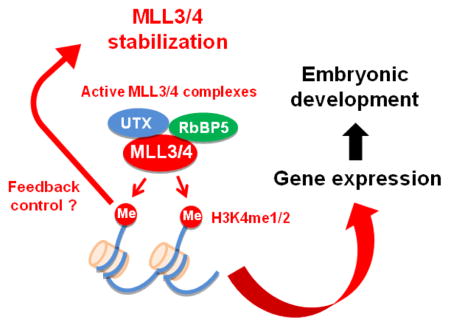
Methylation on lysine 4 of histone H3 (H3K4) correlates with gene activation. H3K4 methylation is catalyzed by the SET domain-containing histone methyltransferase (HMT) Set1 in yeast and Set1-like HMTs in mammals [1]. There are six Set1-like H3K4 HMTs in mammals: MLL1/MLL2 (Mixed-Lineage Leukemia 1/2, also known as KMT2A/B), MLL3/MLL4 (Mixed-Lineage Leukemia 3/4, also known as KMT2C/D) and SET1A/SET1B (also known as KMT2F/G) [1–3]. MLL1 and MLL2 are known as mono- and di-methyltransferases with little tri-methyltransferase activity [4, 5]. MLL3 and MLL4 are major mammalian mono- and di-methyltransferases [2]. SET1A and SET1B are major mammalian H3K4 tri-methyltransferases [6]. The H3K4 methyltransferase activity of mammalian Set1-like HMTs requires the common subunits ASH2L, WDR5, RBBP5 and DPY30, which form the WRAD subcomplex [3, 7–9] (Table 1). Menin is a unique subunit of MLL1/MLL2 complexes [10, 11]. PTIP, PA1, NCOA6 and UTX are unique subunits of MLL3/MLL4 complexes [3]. In yeast, Set1 is responsible for all H3K4 mono-, di- and tri-methylations [1]. Interestingly, yeast Set1 protein levels are linked to H3K4 methylation levels [12]. However, it is unclear whether protein levels of mammalian Set1-like HMTs are also regulated by H3K4 methylation levels.
Table 1.
Yeast Set1 & human Set1-like complexes methylate histone H3K4
| Yeast Set1 | Human Set1-like Complexes | |||
|---|---|---|---|---|
| SET1A/B | MLL1/MLL2 | MLL3/MLL4 | ||
| Enzymatic subunit | Set1 | SET1A/B | MLL1/MLL2 | MLL3/MLL4 |
| Common subunits forming the WRAD subcomplex | Bre2 | ASH2L | ASH2L | ASH2L |
| Swd1 | RbBP5 | RbBP5 | RbBP5 | |
| Swd3 | WDR5 | WDR5 | WDR5 | |
| Sdc1 | DPY30 | DPY30 | DPY30 | |
| Distinct subunits | Swd2 | WDR82 | ||
| Spp1 | CXXC1 | |||
| Menin | PTIP & PA1 | |||
| HCF1 | HCF1/2 | NCOA6 | ||
| UTX | ||||
We previously showed that MLL4 is partially redundant with MLL3 and is a major H3K4 mono- and di-methyltransferase enriched on enhancers in mouse and human cells [2]. MLL3 and MLL4 are required for enhancer activation and cell type-specific gene expression during cell differentiation [2, 13]. MLL3 and MLL4 are dispensable for cell identity maintenance but are required for cell fate transition [13]. Mll3 knockout (KO) mice die around birth with no obvious morphological abnormalities, whereas Mll4 KO mice show early embryonic lethality around E9.5 [2]. By tissue-specific KO of Mll4 in mice, we showed that MLL4 is essential for adipogenesis, myogenesis and heart development in vivo [2, 14]. However, the functional role of MLL4 enzymatic activity needs to be explored.
In the present study, we have identified amino acids critical for the H3K4 methyltransferase activity of MLL4 in cells. By generating MLL4 enzyme-dead KI ES cells and mice, we found that, surprisingly, MLL4 enzymatic activity is required for MLL4 protein stability in cells and for early embryonic development in mice.
Results and Discussion
Identification of amino acids critical for H3K4 methyltransferase activity of MLL4
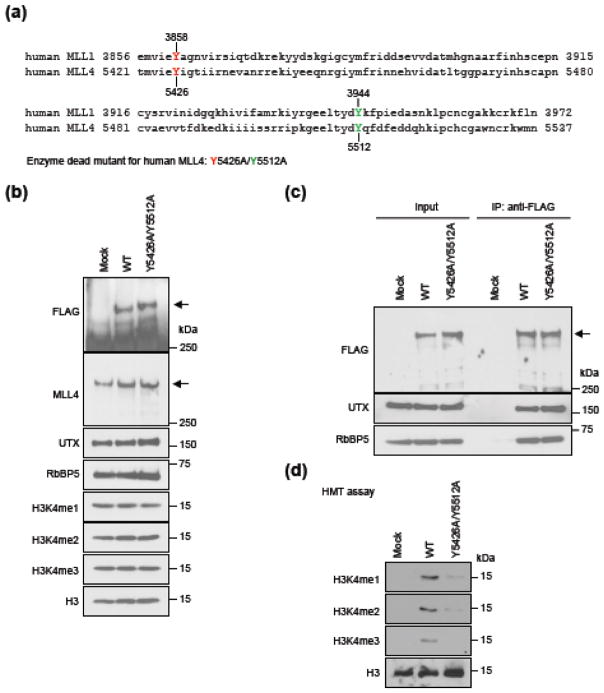
A previous structural study has revealed that Y3858 and Y3942 in the SET domain of human MLL1 are essential for its H3K4 methyltransferase activity [5]. Both Y3944 and Y3942 are involved in H3 recognition and located in the active center [15]. Y3944 is a more conserved residue than Y3942 across MLL family members [5]. We hypothesized that the corresponding residues, Y5426 and Y5512, in the SET domain of human MLL4 could be critical for MLL4 enzymatic activity (Fig. 1a). To test this hypothesis, we generated FLAG-tagged full-length human MLL4 carrying Y5426A/Y5512A mutations by site-directed mutagenesis. 293FT cells were transiently transfected with plasmids expressing FLAG-tagged wild type (WT) MLL4 or the Y5426A/Y5512A mutant. Ectopic expression of the Y5426A/Y5512A mutant MLL4 did not affect global levels of H3K4me1, H3K4me2, H3K4me3, and MLL4-associated proteins UTX and RbBP5 in cells (Fig. 1b). For in vitro histone methyltransferase (HMT) assays, we affinity-purified the MLL4 complex from 293FT cells transiently transfected with plasmids expressing FLAG-tagged WT MLL4 or the Y5426A/Y5512A mutant. Western blot analysis showed that UTX and RbBP5 were present at similar levels between WT and mutant MLL4 complexes (Fig. 1c). In an HMT assay using recombinant histone H3 as the substrate, the mutant MLL4 complex showed a markedly reduced ability to catalyze H3K4me1, H3K4me2 and H3K4me3 (Fig. 1d). These results indicate that the Y5426 and the Y5512 residues are critical for enzymatic activity of human MLL4 in vitro.
Fig. 1. Identification of amino acids critical for H3K4 methyltransferase activity of MLL4.
(a) Alignment of amino acids in the C-terminal SET domain of human MLL1 (KMT2A) and MLL4 (KMT2D). Tyrosine (Y) residues that are potentially critical for enzymatic activities are highlighted in color. (b – d) 293FT cells were transfected with pCMV6-MLL4 plasmid expressing FLAG-tagged wild type (WT) MLL4 or the Y5426A/Y5512A mutant. (b) Nuclear extracts and histone extracts were analyzed by Western blot using antibodies indicated on the left. The arrow indicates the expected position of MLL4 protein. (c) Anti-FLAG M2 agarose was used to immunoprecipitate (IP) from nuclear extracts. The immunoprecipitates were analyzed by Western blot. (d) In vitro histone methyltransferase (HMT) assay. Proteins immunoprecipitated by anti-FLAG M2 agarose from nuclear extracts of 293FT cells expressing FLAG-tagged WT or mutant MLL4 were subjected to HMT assay using recombinant histone H3 as substrate, followed by Western blot.
MLL4 enzymatic activity is essential for early embryonic development
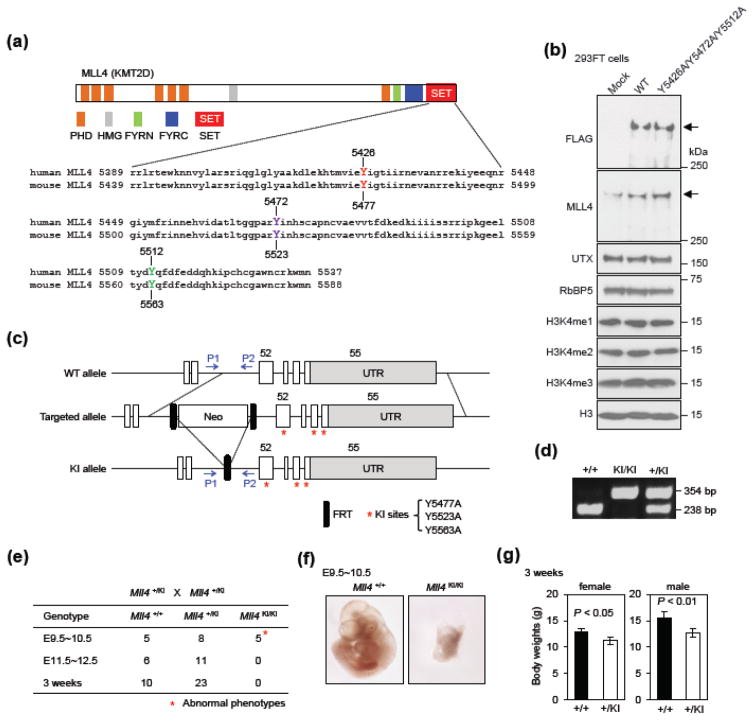
To investigate biological functions of MLL4 enzymatic activity in vivo, we decided to generate MLL4 enzyme-dead knock-in (KI) ES cells and mice. We chose to mutate Y5477 and Y5563 of mouse MLL4, which correspond to Y5426 and Y5512 in human MLL4, to alanine (Fig. 2a). In addition, we mutated mouse Y5523 to ensure MLL4 enzymatic activity was fully inactivated because the corresponding residue in human MLL3 is important for the methyl transfer from the cofactor S-adenosyl-l-methionine to the target histone lysine [15]. Mutating mouse Y5523 to alanine was unlikely to destabilize MLL4 because human MLL4 carrying the corresponding triple mutations (Y5426A/Y5472A/Y5512A) was expressed at similar level as the WT in 293FT cells (Fig. 2b). Also, ectopic expression of the triple-mutated MLL4 did not affect global levels of H3K4me1, H3K4me2 and H3K4me3 in cells (Fig. 2b). Heterozygous and homozygous MLL4 enzyme-dead KI mouse ES cells were generated by conventional gene targeting methods. Y5477, Y5523 and Y5563 were localized in exon 52, 54 and 55 of the mouse Mll4 gene, respectively (Fig. 2c). The genotypes of ES cells and the three mutations in the KI allele were confirmed by genotyping PCR and sequencing (Fig. 2d and data not shown).
Fig. 2. MLL4 enzymatic activity is essential for early embryonic development.
(a) Upper panel shows the domains in human and mouse MLL4 proteins. Lower panel shows the alignment of amino acids in the C-terminal SET domain of human and mouse MLL4. Tyrosine (Y) residues that are potentially critical for enzymatic activity are highlighted in color. (b) 293FT cells were transfected with pCMV6-MLL4 plasmid expressing FLAG-tagged WT human MLL4 or the Y5426A/Y5472A/Y5512A mutant. Nuclear extracts and histone extracts were analyzed by Western blot using antibodies indicated on the left. The arrows indicate the expected position of MLL4 protein. (c) Schematic representation of mouse Mll4 WT, targeted and enzyme-dead knock-in (KI) alleles. Deletion of neo selection cassette from the targeted allele by FLP recombinase generates the KI allele. The KI allele carries Y5477A/Y5523A/Y5563A, represented by the red asterisks. The locations of PCR genotyping primers P1 and P2 are indicated by arrows. (d) PCR genotyping of ES cell lines using P1 and P2 primers. (e) Genotyping results from crossing Mll4+/KI with Mll4+/KI mice. The expected ratio of the three genotypes is 1:2:1. (f) Representative images of E9.5~10.5 embryos. (g) Body weights of 3-week-old Mll4+/KI and Mll4+/+ mice (n = 3 for female and n = 6 for male). Data are presented as mean ± SD and P value by Student’s t-test.
To investigate the role of MLL4 enzymatic activity in mammalian development, we generated MLL4 enzyme-dead KI mice using the Mll4+/KI ES cells. Breeding of heterozygous (Mll4+/KI) males and females failed to produce homozygous (Mll4KI/KI) knock-in mice at weaning age (3 weeks). Further genotyping of younger mice and embryos revealed that Mll4KI/KI embryos died around E9.5 to E10.5 (Fig. 2e). As shown in Fig. 2f, Mll4KI/KI embryos are much smaller and grossly abnormal in morphology, implying that the methyltransferase activity of MLL4 is probably required for the development of multiple organs. This phenotype is similar to what we have reported for the homozygous Mll4 KO mice [2]. Mll4+/KI mice had mildly reduced body weights (Fig. 2g), which were also similar to that of the previously reported Mll4+/− mice [2]. There were no obvious morphological abnormalities at weaning age. Together, these observations suggest that MLL4 enzymatic activity is critical for the role of MLL4 in embryonic development.
H3K4 methyltransferase activity is required for MLL4 protein stability
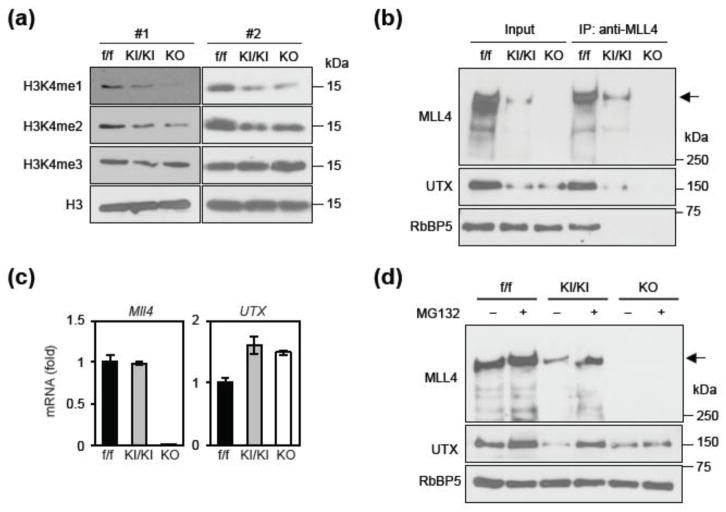
The early embryonic lethality of Mll4KI/KI mice made it difficult to further understand the functions of MLL4 enzymatic activity. Therefore, we turned to Mll4KI/KI ES cells in culture. For control cells, we derived Mll4 conditional KO ES cells (Mll4f/f, hereafter referred to as f/f) from blastocysts of Mll4f/f mice described previously [2]. We also generated Mll4 KO ES cells (Mll4−/−, hereafter referred to as KO) by infecting f/f cells with lentiviruses expressing Cre recombinase. MLL4 is an H3K4me1/2 methyltransferase in cells [11, 16]. By Western blot analysis, we observed global decreases of H3K4me1 and H3K4me2 but not H3K4me3 levels in both KI/KI and KO ES cells (Fig. 3a). This result confirmed that MLL4 enzymatic activity is lost in KI/KI cells. Immunoprecipitation using anti-MLL4 antibody followed by Western blot analysis confirmed the loss of MLL4 protein along with the loss of UTX, a specific subunit of the MLL4 complex, in KO cells (Fig. 3b). This was expected because MLL4 is required for UTX protein stability in cells [2]. Unexpectedly, MLL4 and UTX protein levels also decreased markedly in KI/KI cells compared to f/f cells (Fig. 3b). However, Mll4 mRNA levels were similar between f/f and KI/KI cells but were undetectable in KO cells (Fig. 3c), suggesting that the reduced level of MLL4 protein in KI/KI cells could be due to reduced protein stability. UTX mRNA levels were slightly increased in KI/KI and KO cells compared to the f/f cells (Fig. 3c). Treating ES cells with MG132, a proteasome inhibitor, significantly increased protein levels of MLL4 and UTX in KI/KI cells (Fig. 3d), suggesting that enzyme-dead MLL4 protein is unstable in ES cells.
Fig. 3. H3K4 methyltransferase activity is required for MLL4 protein stability.
(a – d) Characterization of f/f (Mll4f/f), KI/KI (Mll4KI/KI) and KO (Mll4−/−) ES cell lines. (a) Histone extracts prepared from two different cell lines (#1 and #2) for each genotype were analyzed by Western blot using antibodies indicated on the left. (b) Nuclear extracts were incubated with MLL4 antibody. The immunoprecipitates were analyzed by Western blot. The arrow indicates the expected position of MLL4 protein. (c) qRT-PCR of Mll4 and UTX mRNA expression in cells. (d) Cells were treated with 10 μM MG132 for 1 h and the nuclear extracts were analyzed by Western blot.
Depleting cellular H3K4 methylation reduces MLL3/MLL4 but not SET1A/B stability
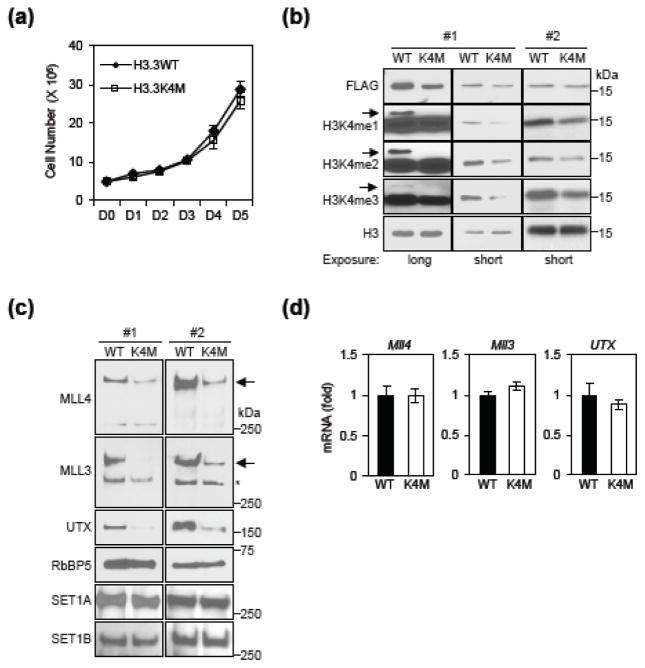
Soares et al. reported that the degradation of yeast SET1, a homolog of mammalian MLL3/MLL4, is triggered by its inactivating mutations [12]. In addition, Soares et al. showed that H3K4 methylation, rather than Set1 enzymatic activity, is required for Set1 protein stability [12]. We thus investigated whether H3K4 methylation levels are also important for MLL4 protein stability in mammalian cells. To test this hypothesis, we used lentiviruses to stably express histone H3.3 carrying lysine 4 to methionine mutation (H3.3K4M) in ES cells. Ectopic expression of H3.3K4M had little effect on ES cell growth (Fig. 4a). Ectopic expression of H3.3K4M has been reported to deplete cellular H3K4me3 [17, 18]. We found that ectopic expression of H3.3K4M reduced global levels of endogenous H3K4me1/2/3 in ES cells (Fig. 4b). Ectopic expression of H3.3K4M also reduced endogenous levels of MLL3, MLL4 and MLL3/MLL4-associated UTX proteins but had no effects on Mll3, Mll4 and UTX mRNA levels in ES cells (Fig. 4c – d). Interestingly, ectopic H3.3K4M did not affect protein levels of SET1A (KMT2F) and SET1B (KMT2G) (Fig. 4c), which are also members of the human Set1-like H3K4 methyltransferase family (Table 1). These data suggest the possibility that cellular H3K4 methylation levels regulate protein stability of MLL3/MLL4 but not SET1A/B.
Fig. 4. Depleting cellular H3K4 methylation reduces MLL3/MLL4 but not SET1A/B stability.
(a – d) FLAG-tagged histone H3.3, either WT or K4M mutant, was ectopically expressed in ES cell lines using a lentiviral vector. (a) To analyze the cell growth, 5 × 105 cells were plated at day 0 and the cumulative cell numbers were determined every day for 5 days. (b) Histone extracts from two different cell lines (#1 and #2) were analyzed by Western blot. The arrows indicate the expected position of FLAG-tagged H3.3. (c) Nuclear extracts were analyzed by Western blot. The arrows indicate expected positions of MLL3 and MLL4. The asterisk indicates a non-specific band. (d) qRT-PCR of Mll4, Mll3 and UTX mRNA expression in cells.
How does H3K4 methyltransferase activity regulate MLL3/MLL4 protein stability? One possibility is that MLL3/MLL4 stability is regulated by cellular H3K4 methylation levels. Three lines of evidence support this possibility. First, H3K4 methylation is required for yeast Set1 protein stability [12]. Second, ectopic expression of H3.3K4M decreases cellular H3K4 methylation levels and leads to destabilization of endogenous MLL3/MLL4 proteins (Fig. 4b and c). Third, unlike the endogenous MLL4 enzyme-dead mutant in KI/KI ES cells, ectopically expressed MLL4 enzyme-dead mutant is stable in 293FT cells where cellular H3K4 methylation levels are not affected (Fig. 2b). Another possibility is that MLL3/MLL4 may auto-methylate themselves or methylate other non-histone proteins to regulate their stability. Finally, we cannot rule out the possibility that the reduced MLL3/MLL4 protein levels in ES cells expressing H3.3K4M are due to decreased expression of genes regulated by MLL3/MLL4. Future work will be needed to find out the mechanism by which H3K4 methyltransferase activity regulates MLL3/MLL4 protein stability.
Materials and Methods
Plasmids
pCMV6-MLL4 plasmid (RC221064) expressing FLAG-tagged wild type (WT) full-length human MLL4 was obtained from OriGene. The Y5426A/Y5512A and the Y5426A/Y5472A/Y5512A mutants of full-length human MLL4 were generated by two or three step site-directed PCR mutagenesis using the Q5 Site-Directed Mutagenesis Kit (E0554S) from NEB. The K4M mutant of FLAG-tagged histone H3.3 (H3.3K4M-FLAG) was generated by PCR mutagenesis using the pQCXIP-H3.3WT-FLAG plasmid (gift from Zhiguo Zhang, Mayo Clinic) as the template. WT or K4M mutant of FLAG-tagged histone H3.3 was subcloned into the lentiviral vector pCDH-EF1-Neo (SBI no. CD533A-1) to generate pCDH-H3.3WT-FLAG or pCDH-H3.3K4M-FLAG. All mutations and plasmids were confirmed by DNA sequencing.
Antibodies and chemicals
The following homemade antibodies have been described: anti-MLL4#3 [19], anti-MLL3#3 [3] and anti-UTX [20]. Anti-RbBP5 antibody (AA300-109A) was obtained from Bethyl Laboratories (Montgomery, TX, USA). Anti-SET1A/B antibodies were described previously [21]. Anti-H3 (ab1791), anti-H3K4me1 (ab8895) and anti-H3K4me2 (ab7766) antibodies were from Abcam (Cambridge, MA, USA). Anti-H3K4me3 antibody (07-473) was from Millipore (Billerica, MA, USA). Anti-FLAG-M2 antibody (F3165), anti-FLAG M2 agarose (A2220), FLAG peptide (F3290) and MG132 (M7449) were from Sigma.
Western blot and immunoprecipitation
Western blot of histone modifications using 0.5 μg acid extracts, Western blot of nuclear proteins using 50 μg nuclear extracts, and immunoprecipitation from 0.5 mg nuclear extracts were done as described [3, 16]. Western blot was performed on Tris-glycine 15 % home-made gels (for detecting histone H3) or NuPAGE Tris-acetate 3–8 % gels from Invitrogen (for detecting MLL3 and MLL4) as described [22].
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA extraction using Trizol (Invitrogen) and qRT-PCR were done as described [19]. qRT-PCR was performed using the following primers: Mll4 (forward 5′-GCTATCACCCGTACTGTGTCAACA-3′ and reverse 5′-CACACACGATACACTCCACACAA-3′), Mll3 (forward 5′-GATTGACGCCACACTCACAG-3′ and reverse 5′-TTTCTGTATCCTCCGGTTGG-3′), UTX (forward 5′-ATACATGAAAGTTCCAGGAGC-3′ and reverse 5′-ACAAACCATTCACAGTCACCTG-3′) and 18S (forward 5′-AGTCCCTGCCCTTTGTACACA-3′ and reverse 5′-CGATCCGAGGGCCTCACTA-3′).
In vitro HMT assay
293FT cells cultured in 10 cm dishes were transfected with 10 μg per dish pCMV6-Entry-MLL4 plasmid expressing FLAG-tagged WT or Y5426A/Y5512A mutant full-length human MLL4 using GenJet (SignaGen SL100489). Two days later, nuclear extracts were prepared. MLL4-associated proteins were immunoprecipitated from nuclear extracts using anti-FLAG M2 agarose, followed by elution with the FLAG peptide as described [3]. For the in vitro HMT assay, 1 μg of MLL4-associated proteins were incubated with 1 μg of recombinant histone H3.1 (NEB M2503S) in the HMT buffer (50 mM Tris pH 8.5, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 4 mM DTT) supplemented with S-adenosyl methionine (NEB, B9003S) at 37 °C for 2 h, followed by Western blot.
Generation of MLL4 enzyme-dead KI, f/f and KO ES cells
To generate MLL4 enzyme-dead KI ES cells, the linearized gene targeting construct (Fig. 2c) was electroporated into wild type ES cells. After selection with G418, surviving clones were expanded for PCR genotyping to identify Mll4KIneo/+ ES cells, which were electroporated with pCAG-Flpe plasmid expressing FLP (Addgene no. 13787) to generate Mll4KI/+ ES cells. Gene targeting was repeated in Mll4KI/+ ES cells to generate homozygous MLL4 enzyme-dead KI (Mll4KI/KI) ES cells. PCR genotyping was performed using the following primers: P1 (5′-GGCCTTGGGGAAGACCTC-3′) and P2 (5′-AAGCTGAGACCCAGATGCCT-3′). MLL4 conditional KO (Mll4f/f) and KO (Mll4−/−) ES cells were generated as described [13]. ES cells were cultured on mouse embryonic fibroblast feeder cells and lentiviral vector-mediated stable gene expression of FLAG-tagged histone H3.3 in ES cells was performed as described [23]. For Western blot and qRT-PCR, ES cells were split off feeder cells and harvested.
Generation of MLL4 enzyme-dead KI mice
Mll4floxneo/+ ES cells were micro-injected into mouse blastocysts following standard procedures. Germline transmitted Mll4KIneo/+ mice were crossed with FLP-expressing mice (Jackson no. 3946) to remove neo cassette and generate Mll4KI/+ mice. Mll4KI/+ mice were crossed with Mll4KI/+ mice to generate Mll4KI/KI mice or embryos. All mouse work was approved by the Animal Care and Use Committee of National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Research Highlights.
MLL4 enzymatic activity is essential for early embryonic development.
H3K4 methyltransferase activity is required for MLL4 protein stability.
MLL3/MLL4 stability may be regulated by cellular H3K4 methylation levels.
Acknowledgments
We thank Eugene Froimchuk for proofreading the manuscript, Zhiguo Zhang for the generous gift of pQCXIP-H3.3WT-FLAG plasmid and David Skalnik for the generous gift of SET1A/B antibodies. This work was supported by the Intramural Research Program of the NIDDK, NIH to KG.
Abbreviations used
- MLL
Mixed-Lineage Leukemia
- HMT
histone methyltransferase
- ES
embryonic stem
- KI
knock-in
- K4M
lysine 4 to methionine mutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho YW, et al. PTIP Associates with MLL3- and MLL4-containing Histone H3 Lysine 4 Methyltransferase Complex. J Biol Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinsky SA, et al. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290(10):6361–75. doi: 10.1074/jbc.M114.627646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Southall SM, et al. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33(2):181–91. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 6.Clouaire T, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26(15):1714–28. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 8.Patel A, et al. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2011;286(5):3359–69. doi: 10.1074/jbc.M110.174524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst P, Vakoc CR. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief Funct Genomics. 2012;11(3):217–26. doi: 10.1093/bfgp/els017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13(4):587–97. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soares LM, et al. Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Rep. 2014;6(6):961–72. doi: 10.1016/j.celrep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1606857113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ang SY, et al. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development. 2016;143(5):810–21. doi: 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016;530(7591):447–52. doi: 10.1038/nature16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–62. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KM, et al. A lesson learned from the H3.3K27M mutation found in pediatric glioma: a new approach to the study of the function of histone modifications in vivo? Cell Cycle. 2013;12(16):2546–52. doi: 10.4161/cc.25625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho YW, et al. Histone Methylation Regulator PTIP Is Required for PPARgamma and C/EBPalpha Expression and Adipogenesis. Cell Metab. 2009;10(1):27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, et al. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282(18):13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–8. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109(38):15324–9. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]