Abstract
Cytokinesis is essential for the survival of all organisms. It requires concerted functions of cell signaling, force production, exocytosis, and extracellular matrix remodeling. Due to the conservation in core components and mechanisms between fungal and animal cells, the budding yeast Saccharomyces cerevisiae has served as an attractive model for studying this fundamental process. In this review, we discuss the mechanics and regulation of distinct events of cytokinesis in budding yeast, including the assembly, constriction, and disassembly of the actomyosin ring, septum formation, abscission, and their spatiotemporal coordination. We also highlight the key concepts and questions that are common to animal and fungal cytokinesis.
1. Introduction to cytokinesis
Cytokinesis is the pivotal process of partitioning cellular constituents from one mother cell into two daughter cells following DNA replication and chromosome segregation. Successful completion of cytokinesis ensures faithful inheritance of genetic codes and is thus essential for maintaining species identity during its propagation [1-3].
Besides its apparent role in increasing cell number via symmetric division, cytokinesis also drives cell diversity via asymmetric division, i.e. a polarized cell dividing to generate two daughters of distinct fate and size. An extreme example is the polar body extrusion during oogenesis in which asymmetric division results in the formation of a large oocyte and a small polar body [4]. Asymmetric division is also exploited by stem cells for self-renewal and differentiation, and loss of the division asymmetry may cause tumorigenesis [5-7]. For example, in Drosophila, neuroblasts undergo asymmetric division to generate each a neuroblast and a smaller daughter cell – the ganglion mother cell (GMC) [5, 8, 9]. The GMC further divides to form two post-mitotic neurons. In the neuroblast, the atypical protein kinase C (aPKC) at the apical cortex promotes self-renewal, while Brain tumor (Brat) and Prospero at the basal cortex inhibit self-renewal and promote differentiation [10-12]. In mutants such as large giant larvae (lgl), the asymmetric localization of these fate determinants is lost, resulting in a tumor-like phenotype - hyperproliferation of neuroblasts at the expense of neurons [10-12].
Cytokinesis is also regulated to control the ploidy of the cells in different tissues and organs. For example, cardiomyocytes undergo nuclear division without cytokinesis, leading to the formation of binucleated cells. This increase in cell size or hypertrophy is believed to drive heart growth after birth [13, 14]. The function or presumed benefit of increasing ploidy is reflected, at least in part, in the observation that mononucleated and binucleated cardiomyocytes display differential electrical activity and calcium dynamics [15]. Hepatocytes also become binucleated postnatally as well as during liver regeneration [16, 17]. The significance of this polyploidization remains unknown, although it has been suggested that its function is to generate genetic diversity and enable adaptation to xenobiotic stress or other environmental insults [16]. Polyploidization due to failed cytokinesis could also have detrimental effects. For example, tetraploid, p53-deficient cells, generated by transient blockage of cytokinesis, caused tumor formation in nude mice, whereas diploid cells did not [18]. The resulting tumors displayed a wide range of genetic abnormalities including aneuploidy, chromosome rearrangements, and gene amplification [18]. This genomic instability is believed to drive tumorigenesis.
Because of its essential role in development and survival of an organism, cytokinesis has been studied in multiple systems for decades. These studies have revealed that cytokinesis requires complex, spatiotemporal coordination of multiple activities including force production, targeted vesicle fusion, and extracellular matrix (ECM) remodeling [1, 3, 19, 20]. In addition, the basic machinery and operating principles of cytokinesis are conserved from yeast to humans. This high level of conservation, coupled with its accessibility to genetic manipulations and live-cell imaging, has made the budding yeast Saccharomyces cerevisiae an attractive model for studying the molecular mechanisms underlying this fundamental process [21].
2. Cytokinesis in S. cerevisiae
Cytokinesis in both animal and fungal cells involves the selection of a division site, cleavage furrow ingression powered by a contractile actomyosin ring (AMR), and localized membrane and ECM remodeling that ultimately leads to abscission. While the core division machinery is conserved through evolution, the mechanisms of division site selection are highly species specific. In animal cells and the fission yeast Schizosaccharomyces pombe, the division site is specified by the mitotic spindle and nuclear position in anaphase and G2, respectively [1]. In S. cerevisiae, the division site, which is the site of budding, is specified in late G1 by Cdc42-controlled cell polarization (Fig. 1) [22]. Cdc42 is also required for polarized organization of actin cables and patches as well as for nascent septin ring assembly at the presumptive bud site. The septin ring then recruits Myo1, the sole myosin-II in budding yeast, via the septin- and myosin-binding protein Bni5 [23-25]. Thus, in all organisms, the specified division site is marked by myosin-II irrespective of the timing of specification during the cell cycle.
Fig. 1.
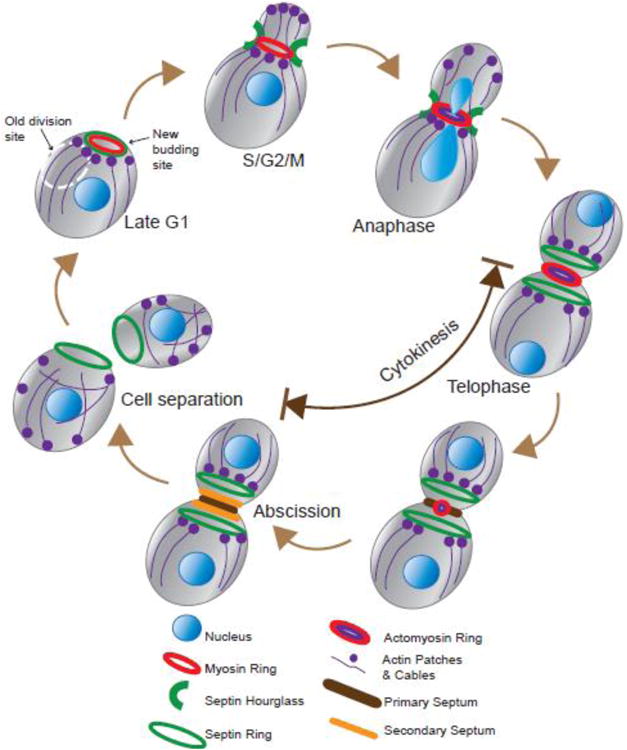
Assembly and execution of key cytokinesis events during the cell cycle.
After bud emergence, the septin ring expands into an hourglass, which, together with the myosin ring, marks the bud neck – the future division site (Fig. 1). Meanwhile, two arrays of actin cables and actin patches are polarized towards the bud cortex and the bud neck to drive bud growth and morphogenesis during S/G2/M phases by mediating exocytosis and endocytosis, respectively (Fig. 1) [22, 26]. Upon the entry into anaphase, actin filaments and myosin-II interact to form an AMR (Fig. 1) [23, 24, 27]. Thus, the timing of AMR assembly in budding yeast and animal cells is strikingly similar, despite their difference in the timing of myosin localization at the division site.
Upon activation of the mitotic exit network (MEN), the septin hourglass is instructed to convert into a double ring, which sandwiches the AMR (Fig. 1) [28, 29]. The AMR begins to constrict centripetally [23, 24]. Meanwhile, actin cables are polarized towards the bud neck to mediate the delivery of post-Golgi vesicles, which fuse with the plasma membrane (PM) to increase surface area as well as release cargoes such as the chitin synthase-II (Chs2) to drive primary septum (PS) formation [30, 31]. AMR constriction is followed closely by PS formation. The AMR guides PS formation in addition to presumed force production [25, 32, 33]. Reciprocally, in the absence of PS formation, the AMR undergoes asymmetric constriction towards one side of the bud neck, which might reflect a partial detachment of the AMR from the PM [31, 34-36]. Thus, the PS is thought to stabilize the AMR during its constriction. Conceptually, PS formation could be considered as the functional counterpart of ECM remodeling during cytokinesis in animal cells [37-39]. Near the end of PS formation, secondary septa (SS), which are synthesized by glucan synthases, begin to form at both sides of the PS [21, 40]. The SS is thought to plug a “tiny hole” at the center of the PS, leading to “abscission” i.e. separation of cytoplasm between mother and daughter cells [41].
Following SS formation, a kinase cascade, named Regulation of Ace2 and Morphogenesis (RAM) network, is activated, leading to the exclusive localization of the transcription factor Ace2 to the daughter cell nucleus [21, 40]. Ace2 controls the expression of cell wall hydrolases that digest the PS and part of the SS, resulting in cell separation.
In this review, we discuss the mechanics and regulation of cytokinesis in budding yeast, highlighting all the key molecules (Fig. 2), concepts, and questions with respect to specific events involved in cytokinesis.
Fig. 2.
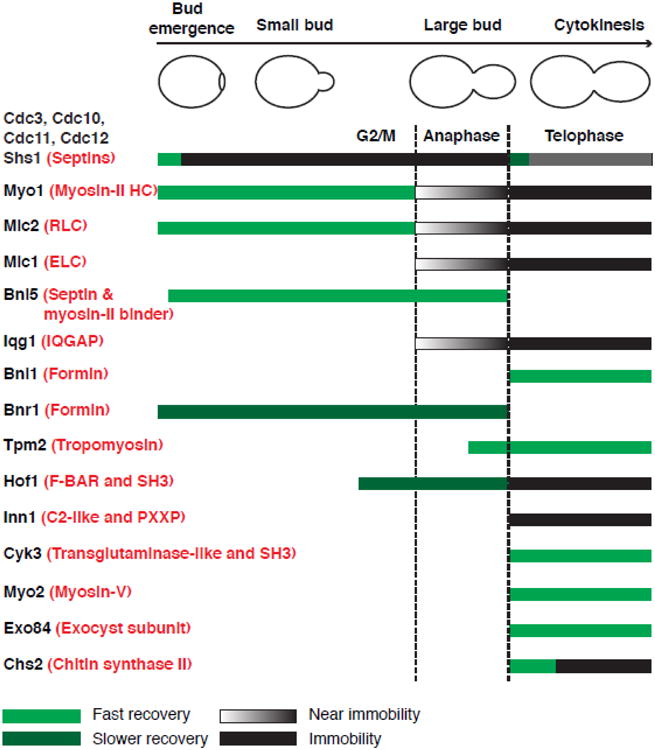
Localization and dynamics of core cytokinesis proteins during the cell cycle. Modified from Wloka et al. 2013, which reports protein dynamics revealed by FRAP analysis.
3. Actomyosin ring assembly
3.1. Septins
3.1.1. Architecture, dynamics, and remodeling of the septin cytoskeleton during the cell cycle
Septins were first identified in budding yeast for their essential role in cytokinesis [42]. Septins are GTP-binding, cytoskeletal proteins that are conserved from yeast to humans and play critical roles in diverse functions including cytokinesis, cell migration, and morphogenesis [43-49]. Defects in septin functions are associated with diverse diseases including neurodegenerative diseases, infertility, and cancer.
Septins form rod-shaped, non-polar heterooligomeric complexes that polymerize end-to-end into linear filaments, which can be further organized into higher-order structures such as rings and gauzes [50, 51]. These structures are thought to impact the above-mentioned processes by acting as a cellular scaffold and/or diffusion barrier. In budding yeast, five mitotic septins form at least two distinct palindromic octameric complexes (Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 and Shs1-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Shs1) (Fig. 2) [52, 53]. These octamers form distinct cellular structures during different phases of the cell cycle [45, 47]. At the beginning of the cell cycle (in late G1), septins are recruited to the presumptive bud site (PBS), where they are assembled into a dynamic cortical ring (Fig. 1). This process depends on Cdc42 [29, 54-56], a small essential GTPase that plays a central role in cell polarization from yeast to humans [57]. Upon bud emergence, the cortical ring expands into a stable hourglass structure at the mother-bud neck, as indicated by fluorescence-recovery-after-photobleaching (FRAP) analysis (Fig. 2) [55, 58]. At the onset of cytokinesis, the hourglass is remodeled into a double ring, which sandwiches the constricting AMR during cytokinesis (Fig. 1). After cell division, the mother and daughter each inherits half of the double ring, which disassembles at the beginning of the next cell cycle while a new ring is formed at the PBS (Fig. 1).
How are the septin filaments packed in different cellular structures, especially in the hourglass and double ring? This key question was addressed by several pioneering studies using different experimental approaches. The original thin-section electron microscopy (EM) and more recent cryo-EM tomography suggest that paired septin filaments are organized into circumferential rings around the bud neck at a spacing of ∼28 nm [59, 60]. In contrast, polarized fluorescence microscopy suggests that septin filaments are aligned along the mother-daughter axis in the hourglass, and circumferentially around the mother-bud neck in the double ring [61, 62]. This approach allows the inference of filament orientation at the population, but not individual-filament, level. Therefore, a detailed architecture of the septin hourglass or double ring remained to be elucidated.
The close proximity of septin filaments to the PM and the high-density ribosomes in the surrounding cytoplasm could complicate the interpretation of filament orientation in the EM studies of thin sections of yeast cells. To overcome this potential problem, rapid-freeze-and-deep-etching EM was used to examine septin filaments associated with cell wall-removed, unroofed yeast cell cortices. This study revealed the presence of rings and gauze-like structures of septin filaments [50]. However, the cells used in this study were unsynchronized. Thus, it is uncertain during which stage of the cell cycle these septin structures are produced. Using the unroofing protocol developed in this study, with some modifications, a platinum-replica EM (PREM), which has been extensively used to analyze the actin and microtubule cytoskeleton in mammalian cells [63, 64], was performed to examine septin structures from cells synchronized at different stages of the cell cycle [51]. It was found that in the early hourglass, paired septin filaments of 300-400nm in length are densely packed along the mother-daughter axis [51]. These filaments likely contain septin octamers with Cdc11 as the terminal subunits [52, 53, 65]. In the late hourglass (before the onset of cytokinesis), a set of single filaments are added perpendicularly to the paired filaments at a regular spacing of ∼29 nm, which is approximately a septin octamer length (Fig. 3A) [51]. It is hypothesized that these filaments contain septin octamers with Shs1 as the terminal subunits [51, 53]. In the double ring, there are both paired and single filaments that are arranged circumferentially (Fig. 3B) [51]. This study provided the first view of the septin hourglass and double ring at the filament level.
Fig. 3.
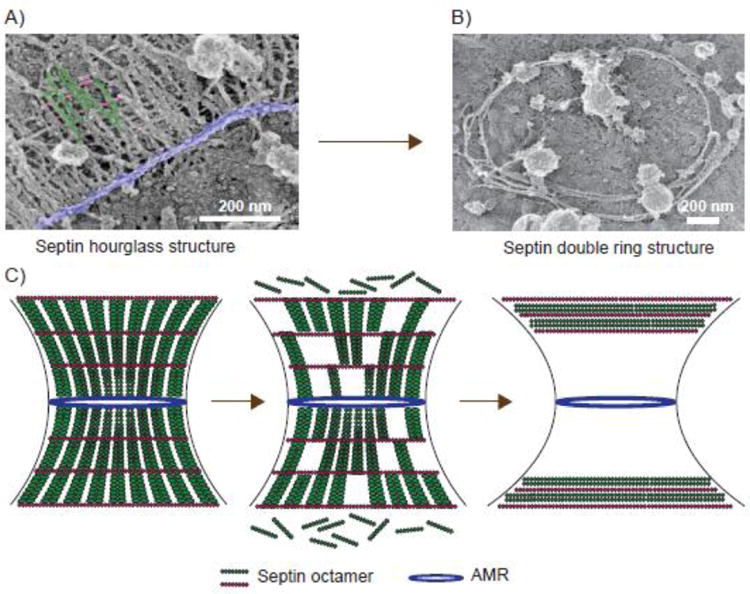
Remodeling of the septin cytoskeleton from an hourglass to a double ring at the onset of cytokinesis. (A and B) Electron micrographs showing orientation of septin filaments in the hourglass and double ring. The paired filaments (green) are perpendicular to the single filaments (magenta), and their orientation with respect to the putative myosin-II filaments (purple) is shown. (C) A model of septin hourglass-to-double ring transition. AMR, actomyosin ring. Modified from Ong et al. 2014.
How is the septin hourglass remodeled into a double ring during the cell cycle? FRAP, photo-activation, and photo-conversion analyses indicate that the septin hourglass-to-double ring transition is accompanied by filament disassembly in the hourglass coupled with a double ring formation at the ends of the hourglass (Fig. 3C) [51]. What triggers filament disassembly and what dictates double ring formation at the ends of the hourglass remains unknown. Bud4, the anillin-like protein in budding yeast, is clearly required for double ring formation [66-68]. It is worth noting that anillin and anillin-like proteins are also involved in septin organization at the division site in other organisms [69-71].
3.1.2. Roles of septins in cytokinesis
Septins play distinct roles in cytokinesis via distinct architectures. They are the first proteins to arrive at the bud neck to scaffold the sequential assembly of the division machinery during the cell cycle. This “scaffolding” role is carried out by the septin hourglass, which also acts as a platform for anchoring, concentrating, and perhaps activating signaling molecules at the bud neck to carry out various cellular functions [45, 72]. From bud emergence to the onset of telophase, the septin hourglass is required for the localization of Myo1 to the bud neck [23]. This targeting event is mediated by the yeast-specific, septin-binding protein Bni5 before anaphase, and by the collective action of Bni5 and the IQGAP Iqg1 during anaphase [25, 73-75]. Actin ring formation begins at the onset of anaphase, and depends on the presence of Iqg1 [24, 27, 76] and Myo1 [23] at the septin hourglass. Thus, a key role of the septins is to scaffold AMR assembly before the onset of cytokinesis via the hourglass architecture.
The septin hourglass is also involved in shaping the AMR. When the hourglass is conditionally disrupted in a septin mutant, Myo1 and its essential light chain (ELC) Mlc1 are still able to localize at the division site in an F-actin-dependent manner, but they no longer form a smooth ring structure [77]. Thus, the septin hourglass is required for the assembly and shaping of the AMR.
At the onset of cytokinesis, the septin hourglass is remodeled into a double ring that sandwiches the AMR. The double ring is thought to act as a diffusion barrier to restrict diffusible factors to the division site during cytokinesis [78]. This barrier role becomes essential only when the AMR is compromised [66].
3.2. Myosin-II
Just like all myosin-IIs in animal cells, the sole myosin-II in budding yeast is a hexamer consisting of a dimer of the myosin-II heavy chain, Myo1, each of which is associated with an ELC (Mlc1), and a regulatory light chain (RLC) (Mlc2) (Fig. 2) [79]. Myo1 forms a two-headed structure with a kink in its tail [25], typical of non-muscle or smooth-muscle myosin-IIs [80]. This kink corresponds to the region containing seven proline residues [25]. Mlc1 and Mlc2 bind to the IQ1 and IQ2 motifs located between the head and tail of Myo1, respectively [79]. Mlc1 is not only the ELC for Myo1, but also a light chain for myosin-Vs (Myo2 and Myo4 in budding yeast) [79, 81, 82] and Iqg1 [76, 83]. While deletion of MYO1 causes pronounced defects in cytokinesis and cell separation, null cells are viable. In contrast, MLC1 is essential for cell viability and cytokinesis [76, 81-83]. This presumably reflects a Myo1-independent role of Mlc1 in targeted membrane trafficking and septum formation during cytokinesis that is mediated by myosin-V and Iqg1, respectively. Mlc2 binds to Myo1, but not Myo2 or Iqg1 [79]. Thus, Mlc2 is a dedicated light chain for Myo1. Deletion of MLC2 causes only a mild defect in Myo1 disassembly during cytokinesis [79, 84]. The number of binding partners of Mlc1 and Mlc2 might explain why ELCs are more conserved than RLCs in sequence and function through evolution for all organisms examined so far [79]. Collectively, these observations indicate that different components of the myosin-II in budding yeast play distinct roles in cytokinesis.
Different components of the myosin-II complex also target to the division site via different mechanisms. Distinct regions of Myo1 tail mediate its localization to the bud neck during different phases of the cell cycle for distinct functions [25]. Myo1 is recruited to the nascent septin ring at the presumptive bud site, and co-localizes with the septin hourglass from bud emergence to the onset of cytokinesis [25, 33, 74]. This Myo1-septin association is mediated by Bni5, which interacts directly with both the septins and the tail of Myo1 [25, 73-75]. In contrast, the localization of Myo1 at the bud neck from the onset of anaphase to the end of cytokinesis is mediated by Iqg1 [25]. Thus, both the Bni5- and Iqg1-based mechanisms contribute to Myo1 localization during anaphase, with the former tapering off while the latter escalating [25]. The Bni5 mechanism may mediate the role of Myo1 in the bud-to-mother retrograde flow of actin cables before anaphase [25, 85] while the Iqg1 mechanism is essential for AMR assembly and function during anaphase and cytokinesis [25], as Iqg1 and Myo1 both are required for actin ring formation [23, 24, 27]. IQGAP is also involved in myosin-II localization at the division site in fission yeast and Dictyostelium [86-89].
The RLC Mlc2 displays an identical localization profile to Myo1, and it localizes to the division site exclusively by binding to the IQ2 motif of Myo1 throughout the cell cycle [79]. In contrast, the ELC Mlc1 begins to accumulate at the division site at the medium-budded stage (∼G2/M phase) and disappears from the bud neck after cytokinesis and cell separation [76, 77, 79, 82, 83]. The neck localization of Mlc1 depends on the septins and actin filaments before and during cytokinesis, which is mediated chiefly by Myo1 and the formin Bni1, respectively [77].
Myosin-II also displays cell cycle-triggered changes in dynamics (Fig. 2). Nearly all the Myo1 molecules are localized to the division site ∼30 min after bud emergence, which corresponds to the small-budded stage [33]. Strikingly, Myo1 is mobile at the division site before the onset of anaphase and is progressively immobilized from anaphase to the onset of telophase and remains immobile during cytokinesis (Fig. 2) [33, 78]. The immobility of Myo1 depends on the “putative assembly domain” at its C-terminus, but not the head domain, the light chain-binding sites, or the actin cytoskeleton [33]. This observation suggests that Myo1 might undergo cell cycle-controlled filament assembly. Not surprisingly, Mlc2 and Myo1 display an identical profile of dynamics during the cell cycle (Fig. 2) [33], as Mlc2 is carried to the division site exclusively by Myo1 [79]. On the other hand, Mlc1 displays an immobility resembling the latter half of Myo1 during the cell cycle (Fig. 2), and this immobility, but not its localization, depends on Myo1 [33]. This and other observations described later suggest that Myo1 might form filaments that scaffold the assembly of the division machinery [33].
3.3. Formins and the actin ring
There are three filamentous actin (F-actin) structures in budding yeast: patches, cables, and the ring [26]. The actin patches consist of branched filaments nucleated by the Arp2/3 complex and are required for endocytosis [26, 90]. The actin cables and ring consist of linear filaments nucleated by the pair of formins, Bni1 and Bnr1 (Fig. 2) [32, 91, 92]. Bni1 and Bnr1 localize to the bud cortex and bud neck before cytokinesis to nucleate the assembly of two polarized arrays of actin cables that mediate vesicle and mRNA transport by myosin-Vs (Myo2 and Myo4 in budding yeast) to drive bud growth [26, 93, 94]. The localization and/or activities of Bni1 and Bnr1 are regulated by the bud cortex-localized small GTPases (Cdc42 and Rho1) and bud neck-localized septins (Shs1) and septin-associated kinases (Gin4), respectively [95-98]. At the onset of cytokinesis, Bni1 changes its location from the bud cortex to the bud neck and remains there until the end of cytokinesis, while Bnr1 disappears from the neck [99, 100]. Cells can survive the loss of either BNI1 or BNR1, but not both [32]. While both formins are collectively required for actin cable and actin ring assembly, Bni1 plays a much more predominant role in this process [32, 91, 92, 101]. Rho1 plays an essential role in actin ring assembly, presumably, by regulating Bni1 activity [101]. The activation of Rho1 at the division site is, in turn, regulated by the Polo kinase Cdc5 via the guanine-nucleotide-exchange factors (GEFs) for Rho1 [102, 103].
The linear actin filaments nucleated by the formins require stabilization by tropomyosins (the major isoform Tpm1 and the minor isoform Tpm2) (Fig. 2) to form actin cables and the actin ring [101, 104, 105]. It remains unknown how the same kind of filaments are shaped into two distinct cellular structures to perform distinct functions.
3.4. IQGAP
IQGAPs are a family of scaffold proteins involved in diverse functions by binding to distinct partners via distinct domains [106, 107]. Mammalian cells have three isoforms, IQGAP1-3, that are expressed either ubiquitously (IQGAP1) or in a tissue-restricted manner (IQGAP2 and 3). While their roles, especially those of IQGAP1, in cell signaling, cytoskeletal organization, and cell-cell adhesion have been analyzed extensively, it remains unclear whether and how they are involved in mammalian cytokinesis. In contrast, the only or major known function of IQGAPs in budding yeast [24, 27], fission yeast [108], Dictyostelium [109], sea urchin [110], and C. elegans [111] is in the process of cytokinesis.
The sole IQGAP, Iqg1, in budding yeast plays at least two distinct roles in AMR assembly. It promotes actin ring formation by binding to actin filaments via its CHD (Calponin Homology Domain) [24, 25, 27, 76, 112], and it is required for the localization or maintenance of Myo1 at the division site during cytokinesis [25]. Both roles appear conserved in fission yeast [86, 87, 89, 113] and Dictyostelium [88, 109, 114].
3.5. A molecular model for AMR assembly
IQGAP (Iqg1) scaffolds AMR assembly by binding to formin (Bni1 and Bnr1)-nucleated actin filaments via its CHD and to myosin-II (Myo1, Mlc1, and Mlc2) in a manner that remains to be elucidated. These molecular complexes and/or assemblies are anchored to and shaped by the septin hourglass into a ring structure.
4. Actomyosin ring constriction
AMR constriction drives ingression of the cleavage furrow and guides vesicle fusion and septum formation. The constriction is powered by motor-dependent and –independent forces. The head domain of Myo1, which includes the motor domain and light chain-binding sites, is responsible for ∼20-30% of the constriction rate [25, 115]. Cells expressing only the tail of Myo1 form a “headless” AMR that can largely direct PS formation [25]. The nature of the “motor-independent” forces remains unclear, but it likely involves PS formation, which presumably drives PM ingression at the division site in the absence of the motor domain [25, 115].
Actin filaments play an essential role in AMR constriction, as their disruption by latrunculin A (LatA) does not block Myo1 localization to the bud neck but completely abolish its constriction during cytokinesis [23, 84]. Actin filaments could directly mediate Myo1 constriction as a part of the AMR, as indicated by the observation that Myo1 displays partial and abnormal constriction in an iqg1 mutant specifically defective in actin ring assembly [25, 112]. The architecture of the AMR remains unknown in any system. Thus, it is unclear how actin filaments and Myo1 interact with each other to produce a contractile force. Actin filaments could also regulate AMR constriction as components of the actin cables, via PS formation, by controlling myosin-V (Myo2)-dependent delivery of Chs2 to the bud neck during cytokinesis. Thus, actin filaments could regulate AMR constriction via two distinct actomyosin systems.
5. Actomyosin ring disassembly
In contrast to muscle contraction where the number of “contractile units” remains the same [116, 117], AMR constriction must be coupled with disassembly [118, 119]. As expected, Myo1 is progressively lost during AMR constriction [66, 84]. Three distinct mechanisms appear to control Myo1 disassembly, which involves its motor domain, its RLC Mlc2, and the IQGAP Iqg1, respectively.
While a “headless AMR” can largely drive cytokinesis in budding yeast, the constriction rate is decreased 20-30% [25, 115]. In addition, Myo1-tail displays a clear disassembly defect as well as a defect in the symmetry of ring constriction during cytokinesis [25]. As Myo1 lacking the light-chain binding sites does not show any similar phenotype [79], these data suggest that the Myo1 motor domain is involved in the control of AMR disassembly. Consistently, inhibition of motor activity of myosin-II in mammalian cells also causes defects in AMR disassembly [120]. However, it remains unknown how the motor domain regulates myosin disassembly during cytokinesis.
Deletion of MLC2 causes a delay in Myo1 disassembly [79]. Similarly, myosin-II purified from RLC-null cells of Dictyostelium discoideum displays a defect in filament disassembly [121]. Strikingly, the cytokinesis defects of RLC-null cells in budding yeast [79], fission yeast [122], and Dictyostelium [123] are suppressed by deletion of the RLC-binding site on the respective myosin heavy chain. These observations suggest that the RLC in these organisms acts to relieve an auto-inhibitory effect of the RLC-binding site on myosin disassembly during cytokinesis.
Iqg1 undergoes anaphase promoting complex/cyclosome (APC/C)-mediated degradation during AMR constriction [84, 124]. It appears to intimately associate with Myo1, Mlc1, and Mlc2 at the bud neck in wild-type cells as well as at the ectopic sites in APC/C mutants where Iqg1 becomes stabilized [84, 124]. Thus, Iqg1 likely controls AMR disassembly via its presumed interaction with Myo1 [25] as well as its demonstrated interaction with Mlc1 [76, 83]. It remains to be seen whether a similar mechanism operates in other systems.
6. Membrane insertion at the division site
Cytokinesis in animal, fungal, and plant cells requires targeted vesicle fusion at the division site [19, 20, 125]. This serves to supply membranes to cover the new cell ends created during cell division and deliver enzymatic cargoes to remodel the ECM at the division site [21]. In budding yeast, the entire growth machinery is switched from the bud cortex to the bud neck during mitotic exit to promote cytokinesis (Fig. 1). This includes polarity regulators, actin cables, myosin-V, and secretory components such as the exocyst, a conserved vesicle-tethering complex [22, 26, 93, 126]. The formin Bni1 plays a dual role in cytokinesis, promoting vesicle transport and AMR constriction by nucleating actin cable and actin ring assembly, respectively [32, 91, 92, 101, 127, 128]. It remains unknown how the growth machinery is targeted to the division site and how vesicle fusion is spatiotemporally coupled to the AMR during cytokinesis.
7. Primary septum formation
Increasing evidence suggests that ECM remodeling is a common feature of fungal and animal cytokinesis. In C. elegans and mice, chondroitin synthesis is essential for embryonic cytokinesis [37, 38]. In sea urchin eggs [129] and human cells [130-132], hyaluronan, a prevalent ECM component, is enriched at the division site. In budding yeast, AMR constriction is followed closely by the centripetal growth of a primary septum, an electron-lucent, chitinous structure that formed only during cytokinesis (Fig. 1). The PS is synthesized by the chitin synthase II (Chs2) [133, 134]. Chitin (polymers of N-acetyl-glucosamine; also exists in Drosophila and zebrafish) is structurally similar to hyaluronan (polymers of glucuronic acid and N-acetyl-glucosamine disaccharide). In addition, both polymers are synthesized by members of the glycosyltransferase-2 family. Thus, the PS can be considered as a yeast counterpart of an animal ECM. In budding yeast, deletion of CHS2 causes rapid and asymmetric AMR constriction, suggesting that the PS might stabilize the AMR during its constriction [31, 34, 35]. This PS-AMR interplay also applies to the fission yeast S. pombe [135]. Similarly, hemicentin, a secreted ECM protein, is required for preventing retraction of the nascent cleavage furrow during cytokinesis [39]. Thus, a common role of ECM remodeling at the division site appears to stabilize the AMR during furrow ingression. Chs2 is delivered to the bud neck by exocytosis at the onset of cytokinesis [30]. The AMR and the septin double ring play distinct and collectively essential roles in restricting Chs2 to the division site [66, 78]. A protein complex consisting of Inn1, Hof1, and Cyk3 is involved in Chs2-mediated PS formation (Fig. 2) [36, 136-139]. Inn1 contains a C2 domain at its N-terminus that is essential for PS formation, and several PXXP motifs at its C-terminus that bind to the SH3 domains of Hof1 and Cyk3 [36]. Hof1 is a F-BAR protein [32, 140-142] and Cyk3 contains a transglutaminase domain [143, 144]. These proteins interact with each other and share an essential role in AMR-independent cytokinesis [41, 137, 138, 143]. Deletion of INN1 completely blocks PS formation but not Chs2 localization, suggesting that Inn1 is involved in Chs2 activation [36]. Overexpression of Cyk3 stimulates PS formation in wild-type cells [145] and suppresses the growth and septal defects of innΔ cells, suggesting that Cyk3 is also involved in Chs2 activation [36]. Thus, a critical question is how Inn1 and Cyk3 promote PS formation by activating Chs2.
8. Secondary septum formation
Secondary septa (SS) are electron-dense structures formed at both sides of the PS (Fig. 1). Their formation is triggered near the end of PS formation [41]. SS not only protect the newly generated cell ends but also “plug” the tiny hole at the center of the PS, leading to complete membrane closure or abscission between the daughter cells [41]. SS are structurally similar to lateral walls, which are composed of mannoproteins and 1,3-β-D-glucan. The glucan is synthesized by the glucan synthases, with Fks1 and Fks2 being their catalytic subunits [146-149]. While Fks1 functions mainly during budding and Fks2 mainly during nutritional starvation, mating, and sporulation, they are collectively essential for cell wall synthesis, SS formation, and cell survival during vegetative growth [147, 148]. The chitin synthase III also plays a role in SS formation, as cells lacking the catalytic subunit Chs3 form abnormal SS [134]. Consistently, Chs3 becomes essential for cytokinesis in the absence of Chs2 (lacking the PS) [134] or Myo1 (lacking the AMR) [41, 103].
SS formation requires Cdc42 inactivation and Rho1 activation during cytokinesis [41, 150, 151]. These GTPases may mutually antagonize each other in part via the paxillin homologue Pxl1 [41, 152, 153] and Gps1/Aim44 [151, 154]. Rho1 controls SS formation by acting as the regulatory subunit of the glucan synthases Fks1 and Fks2 [148, 155]. Thus, Rho1 plays at least two sequential and distinct roles in cytokinesis: promoting actin ring assembly via the formin Bni1 and stimulating SS formation via the glucan synthases. Cyk3 binds to Rho1 and inhibits its activation during PS formation [41]. This ensures that SS are formed only after the PS [41]. Cyk3 also stimulates PS formation [145]. Thus, Cyk3 defines a mechanism for spatiotemporal coordination of PS and SS formation during cytokinesis.
9. Cell separation
Cell separation refers to the physical cut between the mother and daughter cells. This is achieved through degradation of the PS and part of the SS by cell wall-digestive enzymes including the endochitinase Cts1 [156] and the glucanases Dse4/Eng1 and Egt2 [157, 158]. Expression of Cts1 and Eng1 is controlled by the transcriptional factor Ace2 [159-161], whereas the expression of Egt2 is controlled by both Ace2 and its paralog Swi5 [158]. The activation and localization of Ace2 is, in turn, controlled by the RAM pathway, which comprises of Cbk1 kinase, its activating subunit Mob2, and upstream regulatory components [161-163]. Upon activation, Ace2 enters the daughter nucleus to drive the expression of the hydrolytic enzymes. After cell separation, Ace2 exits the nucleus and is kept in the cytoplasm by cyclin-dependent kinases [164]. Cbk1 kinase is evolutionarily conserved [40]. However, it is unknown whether there is a post-abscission or -cytoplasm cleavage process involving ECM remodeling at the division site that is catalyzed by a Cbk1 homologue in mammalian cells.
10. Spatiotemporal coordination of cellular events in cytokinesis
As described above, cytokinesis involves multiple cellular events including AMR constriction, targeted membrane deposition, and septum formation. These events must coordinate in time and space to ensure the execution of cytokinesis with high efficiency and fidelity. Here, we discuss some potential coordination mechanisms.
10.1. Igniting the cytokinesis machine by MEN
The mitotic exit network (MEN) is a conserved GTPase-dependent signaling pathway that coordinates spindle position, mitotic exit, and cytokinesis at the end of the cell cycle (Fig. 4A) [1, 40, 165-167]. The small GTPase Tem1 and its bipartite GTPase-activating protein (GAP) Bub2-Bfa1 preferentially associate with the bud-bound spindle pole body (SPB; counterpart of centrosome in animal cells). Upon the entry of this SPB into the daughter compartment, the bud cortex-localized Lte1 activates Tem1. This is not done by acting as a GEF via its GEF-like domain but by inhibiting the association of the GAP-stimulating kinase Kin4 with the daughter SPB [167-169]. Activated Tem1 binds to its effector, a Hippo-like kinase Cdc15, which, in turn, activates the LATS-like kinase Dbf2. The activation of this kinase cascade ultimately results in the release of the phosphatase Cdc14 from the nucleolus into the nucleus and cytoplasm [170]. Cdc14 then removes the phosphorylation from the substrates of the cyclin-dependent kinase (CDK), culminating in mitotic exit and cytokinesis [1, 40, 167].
Fig. 4.
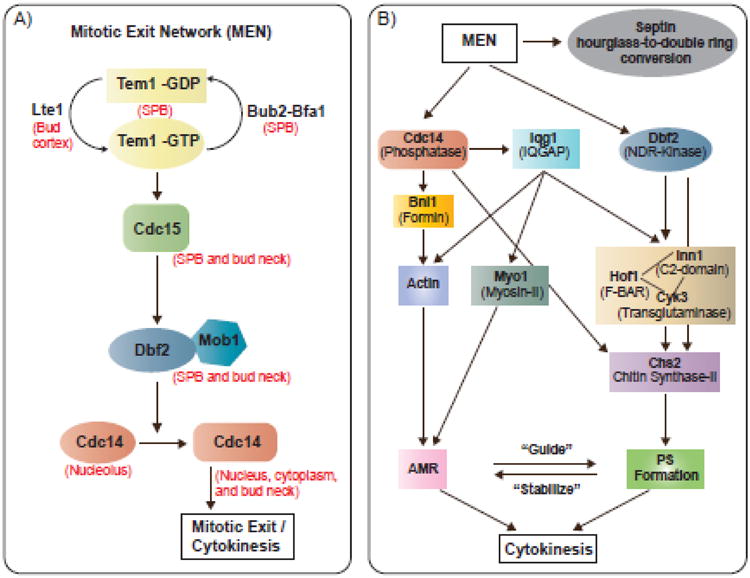
The mitotic exit network and its regulation of cytokinesis. (A) The mitotic exit network. SPB, spindle pole body. (B) Spatiotemporal control of AMR constriction and PS formation by MEN and IQGAP. AMR, actomyosin ring; PS, primary septum.
Several MEN components, including the mitotic kinases Cdc15 and Dbf2 as well as the phosphatase Cdc14, localize to the division site to control cytokinesis [166, 171]. MEN regulates AMR constriction, as the ring can form but fails to constrict in MEN mutants [28, 32]. Several mechanisms could account for this observation. First, MEN controls the remodeling of the septin hourglass into a double ring, which sandwiches the AMR and renders its constriction “permissible”. The AMR is tethered to the “rigid” septin hourglass before the onset of cytokinesis, which makes constriction difficult, if not impossible (Fig. 3C). After the remodeling event, the AMR is tethered to the PM between the septin double ring (Fig. 3C), which permits, but is not necessarily sufficient for, constriction and furrow ingression. Second, MEN controls timely AMR assembly and constriction during the cell cycle (Fig. 4B). CDK phosphorylation of Iqg1 delays actin ring formation until anaphase [172] and Cdc14 dephosphorylation of Iqg1 promotes actin ring assembly from anaphase to, presumably, the end of cytokinesis [173]. As Iqg1 is also essential for Myo1 localization during cytokinesis [25], MEN is likely to control AMR assembly and constriction via Iqg1. MEN also controls AMR assembly via Cdc14 regulation of Bni1 localization during cytokinesis [174]. Currently, it is unclear whether MEN controls AMR constriction independently of its assembly.
MEN also controls PS formation, as the PS fails to form in the MEN mutants [138, 175]. Chs2 is synthesized and retained at the ER via CDK phosphorylation [176]. Cdc14 reverses this phosphorylation, which allows Chs2 to translocate from the ER to the bud neck to drive PS formation (Fig. 4B) [177, 178]. Dbf2 also phosphorylates Chs2 to control its association with the AMR during cytokinesis (Fig. 4B) [145]. Finally, MEN controls the localization of Inn1, Hof1, and Cyk3 to the bud neck during cytokinesis (Fig. 4B) [36, 137, 138]. These proteins are important for PS formation. Thus, MEN controls PS formation via multiple and distinct mechanisms.
10.2. Spatial coordination of actomyosin ring constriction and primary septum formation during cytokinesis
AMR constriction is always followed closely by the centripetal growth of the PS, suggesting that the two processes must be aligned with precision to achieve efficient cytokinesis. These processes are mutually dependent (Fig. 4B) [34, 35]. The AMR guides PS formation in addition to force production [25, 32, 33] whereas the PS stabilizes the AMR during its constriction [31, 36]. A similar scenario involving the interplay between the AMR and localized ECM remodeling at the division site might take place in animal cells [39].
How do the AMR constriction and PS formation spatially coordinate at the molecular level? Perhaps the best understood mechanism involves Iqg1 that links AMR to PS formation (Fig. 4B). Iqg1 is required for actin ring assembly [24, 27] and also for Myo1 localization during cytokinesis [25]. Thus, Iqg1 has a clearly defined role in AMR assembly. Iqg1 is essential for cell viability and cytokinesis whereas Myo1 is not, despite the fact that myoΔ cells display obvious defects in cytokinesis and cell separation [23, 24, 179, 180]. This observation suggests that Iqg1 must play additional role(s) in AMR-independent cytokinesis (Fig. 4B). Indeed, overexpression of Cyk3 rescued the growth and cytokinesis defects of iqgΔ cells without restoring the AMR, supporting a role of Iqg1 and Cyk3 in AMR-independent cytokinesis [143]. Subsequently, it was shown that Cyk3, Hof1, and Inn1 interact with each other to promote PS formation (Fig. 4B) [36, 137, 138]. Both Inn1 and Hof1 also interact with Iqg1 [136, 139, 172, 181]. Taken together, these observations suggest that Iqg1 plays a pivotal role in both AMR constriction and PS formation during cytokinesis.
11. Closing remarks
The collective power of diverse model systems, gene-editing tools, and imaging technologies has led to rapid progress in the field of cytokinesis over the last two decades. However, analysis has been largely focused, and rightfully so, on individual mutants, proteins, and protein complexes. The major challenge in the future is to determine how different proteins are wired together at a systems level to execute cytokinesis with robustness and high fidelity.
Comparative analysis of cytokinesis in different models using diverse experimental approaches has proven to be an effective strategy for illustrating the general principles of cytokinesis, as each model offers distinct experimental advantages and may provide insights from a unique angle. Such an analysis has led to the notion that cytokinesis in animal and fungal cells, including the budding yeast discussed here, entails the same processes (AMR constriction, targeted membrane trafficking, and ECM remodeling) and core components, despite inevitable differences in details. Such an approach should be continuously employed to tackle the fundamental questions regarding cytokinesis: what is the architecture of the AMR in different organisms? Is the core design of the AMR evolutionarily conserved or different despite the common components (actin and myosin filaments)? Is the AMR a pure force-generating machine in animal cells, as currently believed, or is it also involved in guiding membrane traffic and ECM remodeling at the division site, as suggested in budding yeast? How are exocytic vesicles targeted to and fused at the division site? Is abscission mechanistically similar in animal and fungal cells? These compelling questions not only spark the scientific curiosity but also press the need for developing more advanced tools to study the nature and coordination of micro systems pertaining to cytokinesis.
Acknowledgments
We thank Katy Ong, Hiroki Okada, and Kangji for critically reading the manuscript, and the members of Bi laboratory for discussions. We apologize to colleagues whose work is not cited here due to space limitation. Research in the Bi laboratory is supported by the National Institutes of Health Grant GM115420.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–18. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Li R. Cytokinesis in development and disease: variations on a common theme. Cell Mol Life Sci. 2007;64:3044–58. doi: 10.1007/s00018-007-7285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–6. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu XJ. Polar body emission. Cytoskeleton (Hoboken) 2012;69:670–85. doi: 10.1002/cm.21041. [DOI] [PubMed] [Google Scholar]
- 5.Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Wang H, Groth C. Drosophila neuroblasts as a new model for the study of stem cell self-renewal and tumour formation. Biosci Rep. 2014;34 doi: 10.1042/BSR20140008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berika M, Elgayyar ME, El-Hashash AH. Asymmetric cell division of stem cells in the lung and other systems. Front Cell Dev Biol. 2014;2:33. doi: 10.3389/fcell.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jan YN, Jan LY. Asymmetric cell division. Nature (London) 1998;392:775–8. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- 9.Doe CQ, Bowerman B. Asymmetric cell division: fly neuroblast meets worm zygote. Curr Opin Cell Biol. 2001;13:68–75. doi: 10.1016/s0955-0674(00)00176-9. [DOI] [PubMed] [Google Scholar]
- 10.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–9. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature (London) 2006;439:594–8. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 12.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–53. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 13.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–44. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci USA. 2013;110:1446–51. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CF, Chen YC, Yeh HI, Chen SA. Mononucleated and binucleated cardiomyocytes in left atrium and pulmonary vein have different electrical activity and calcium dynamics. Prog Biophys Mol Biol. 2012;108:64–73. doi: 10.1016/j.pbiomolbio.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature (London) 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentric G, Celton-Morizur S, Desdouets C. Polyploidy and liver proliferation. Clin Res Hepatol Gastroenterol. 2012;36:29–34. doi: 10.1016/j.clinre.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature (London) 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 19.Strickland LI, Burgess DR. Pathways for membrane trafficking during cytokinesis. Trends Cell Biol. 2004;14:115–8. doi: 10.1016/j.tcb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–60. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–26. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- 22.Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–87. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, et al. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–12. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998a;140:355–66. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, et al. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin-II. J Cell Biol. 2010;191:1333–50. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–45. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–9. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 28.Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–86. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- 29.Cid VJ, Adamikova L, Sanchez M, Molina M, Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147:1437–50. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- 30.Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VerPlank L, Li R. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol Biol Cell. 2005;16:2529–43. doi: 10.1091/mbc.E04-12-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wloka C, Vallen EA, Thé L, Fang X, Oh Y, Bi E. Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J Cell Biol. 2013;200:271–86. doi: 10.1083/jcb.201208030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi E. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct Funct. 2001;26:529–37. doi: 10.1247/csf.26.529. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- 36.Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, et al. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae J Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuguchi S, Uyama T, Kitagawa H, Nomura KH, Dejima K, Gengyo-Ando K, et al. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature (London) 2003;423:443–8. doi: 10.1038/nature01635. [DOI] [PubMed] [Google Scholar]
- 38.Izumikawa T, Kanagawa N, Watamoto Y, Okada M, Saeki M, Sakano M, et al. Impairment of embryonic cell division and glycosaminoglycan biosynthesis in glucuronyltransferase-I-deficient mice. J Biol Chem. 2010;285:12190–6. doi: 10.1074/jbc.M110.100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Vogel BE. A secreted protein promotes cleavage furrow maturation during cytokinesis. Curr Biol. 2011;21:114–9. doi: 10.1016/j.cub.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss EL. Mitotic exit and separation of mother and daughter cells. Genetics. 2012;192:1165–202. doi: 10.1534/genetics.112.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202:311–29. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–76. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 43.Joo E, Tsang CW, Trimble WS. Septins: traffic control at the cytokinesis intersection. Traffic. 2005;6:626–34. doi: 10.1111/j.1600-0854.2005.00305.x. [DOI] [PubMed] [Google Scholar]
- 44.Hall PA, Russell SEH, Pringle JR. The Septins. John Wiley & Sons, Ltd.; 2008. [Google Scholar]
- 45.McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2011;21:141–8. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bridges AA, Gladfelter AS. Fungal pathogens are platforms for discovering novel and conserved septin properties. Curr Opin Microbiol. 2014;20:42–8. doi: 10.1016/j.mib.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolat L, Hu Q, Spiliotis ET. Septin functions in organ system physiology and pathology. Biol Chem. 2014;395:123–41. doi: 10.1515/hsz-2013-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell. 2005;16:372–84. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong K, Wloka C, Okada S, Svitkina T, Bi E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun. 2014;5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertin A, McMurray MA, Grob P, Park SS, Garcia G, 3rd, Patanwala I, et al. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–9. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia G, 3rd, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, et al. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195:993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gladfelter AS, Bose I, Zyla TR, Bardes ESG, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–26. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–66. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, et al. Role of a cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–25. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–57. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 59.Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976a;69:717–21. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertin A, McMurray MA, Pierson J, Thai L, McDonald KL, Zehr EA, et al. Three-dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23:423–32. doi: 10.1091/mbc.E11-10-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature (London) 2006;443:466–9. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 62.DeMay BS, Bai X, Howard L, Occhipinti P, Meseroll RA, Spiliotis ET, et al. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol. 2011;193:1065–81. doi: 10.1083/jcb.201012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svitkina TM, Borisy G. Correlative light and electron microscopy studies of cytoskeletal dynamics. 3rd. Elsevier; 2006. [Google Scholar]
- 64.Svitkina T. Imaging Cytoskeleton Components by Electron Microscopy. Methods Mol Biol. 2016;1365:99–118. doi: 10.1007/978-1-4939-3124-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, et al. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–49. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, et al. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392:813–29. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- 67.Eluere R, Varlet I, Bernadac A, Simon MN. Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell Cycle. 2012;11:151–8. doi: 10.4161/cc.11.1.18542. [DOI] [PubMed] [Google Scholar]
- 68.Kang PJ, Hood-DeGrenier JK, Park HO. Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci. 2013;126:1218–26. doi: 10.1242/jcs.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–92. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maddox AS, Lewellyn L, Desai A, Oegema K. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev Cell. 2007;12:827–35. doi: 10.1016/j.devcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 72.Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–9. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- 73.Lee PR, Song S, Ro HS, Park CJ, Lippincott J, Li R, et al. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6906–20. doi: 10.1128/MCB.22.19.6906-6920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider C, Grois J, Renz C, Gronemeyer T, Johnsson N. Septin rings act as template for myosin higher-order structures and inhibit redundant polarity establishment. J Cell Sci. 2013 doi: 10.1242/jcs.125302. [DOI] [PubMed] [Google Scholar]
- 75.Finnigan GC, Booth EA, Duvalyan A, Liao EN, Thorner J. The carboxy-terminal tails of septins Cdc11 and Shs1 recruit myosin-II binding factor Bni5 to the bud neck in Saccharomyces cerevisiae. Genetics. 2015;200:843–62. doi: 10.1534/genetics.115.176503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113:4533–43. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- 77.Feng Z, Okada S, Cai G, Zhou B, Bi E. MyosinII heavy chain and formin mediate the targeting of myosin essential light chain to the division site before and during cytokinesis. Mol Biol Cell. 2015;26:1211–24. doi: 10.1091/mbc.E14-09-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–6. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 79.Luo J, Vallen EA, Dravis C, Tcheperegine SE, Drees BL, Bi E. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J Cell Biol. 2004;165:843–55. doi: 10.1083/jcb.200401040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trybus KM. Assembly of cytoplasmic and smooth muscle myosins. Curr Opin Cell Biol. 1991;3:105–11. doi: 10.1016/0955-0674(91)90172-u. [DOI] [PubMed] [Google Scholar]
- 81.Stevens RC, Davis TN. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J Cell Biol. 1998;142:711–22. doi: 10.1083/jcb.142.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wagner W, Bielli P, Wacha S, Ragnini-Wilson A. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 2002;21:6397–408. doi: 10.1093/emboj/cdf650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol. 2000;10:727–30. doi: 10.1016/s0960-9822(00)00539-x. [DOI] [PubMed] [Google Scholar]
- 84.Tully GH, Nishihama R, Pringle JR, Morgan DO. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol Biol Cell. 2009;20:1201–12. doi: 10.1091/mbc.E08-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huckaba TM, Lipkin T, Pon LA. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J Cell Biol. 2006;175:957–69. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–21. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D'Souza V, Tang X, et al. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–72. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 88.Kee YS, Ren Y, Dorfman D, Iijima M, Firtel R, Iglesias PA, et al. A mechanosensory system governs myosin II accumulation in dividing cells. Mol Biol Cell. 2012;23:1510–23. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takaine M, Numata O, Nakano K. Fission yeast IQGAP maintains F-actin-independent localization of myosin-II in the contractile ring. Genes Cells. 2014;19:161–76. doi: 10.1111/gtc.12120. [DOI] [PubMed] [Google Scholar]
- 90.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 91.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- 92.Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- 93.Bretscher A. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J Cell Biol. 2003;160:811–6. doi: 10.1083/jcb.200301035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, et al. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–22. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 96.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, et al. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–55. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dong Y, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J Cell Biol. 2003;161:1081–92. doi: 10.1083/jcb.200212040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buttery SM, Kono K, Stokasimov E, Pellman D. Regulation of the formin Bnr1 by septins anda MARK/Par1-family septin-associated kinase. Mol Biol Cell. 2012;23:4041–53. doi: 10.1091/mbc.E12-05-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15:4971–89. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–38. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12:1864–70. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- 102.Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, et al. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–11. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- 103.Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–23. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–42. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- 105.Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995;128:383–92. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–4. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shannon KB. IQGAP family members in yeast, Dictyostelium, and mammalian cells. Int J Cell Biol. 2012;2012:894817. doi: 10.1155/2012/894817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eng K, Naqvi NI, Wong KC, Balasubramanian MK. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–21. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- 109.Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol. 1997;137:891–8. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nishimura Y, Mabuchi I. An IQGAP-like protein is involved in actin assembly together with Cdc42 in the sea urchin egg. Cell Motil Cytoskeleton. 2003;56:207–18. doi: 10.1002/cm.10146. [DOI] [PubMed] [Google Scholar]
- 111.Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–6. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shannon KB, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10:283–96. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takaine M, Numata O, Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–31. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mondal S, Burgute B, Rieger D, Muller R, Rivero F, Faix J, et al. Regulation of the actin cytoskeleton by an interaction of IQGAP related protein GAPA with filamin and cortexillin I. PLoS One. 2010;5:e15440. doi: 10.1371/journal.pone.0015440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lord M, Laves E, Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell. 2005;16:5346–55. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature (London) 1954;173:973–6. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 117.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–65. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 118.Schroeder TE. The contractile ring II: determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J Cell Biol. 1972;53:419–34. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bi E. Lord of the ring. Nat Rev Mol Cell Biol. 2010;11:606. doi: 10.1038/nrm2953. [DOI] [PubMed] [Google Scholar]
- 120.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 121.Chen P, Ostrow BD, Tafuri SR, Chisholm RL. Targeted disruption of the Dictyostelium RMLC gene produces cells defective in cytokinesis and development. J Cell Biol. 1994;127:1933–44. doi: 10.1083/jcb.127.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Naqvi N, Wong KCY, Tang X, Balasubramanian MK. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat Cell Biol. 2000;2:855–8. doi: 10.1038/35041107. [DOI] [PubMed] [Google Scholar]
- 123.Uyeda TQ, Spudich JA. A functional recombinant myosin II lacking a regulatory light chain-binding site. Science. 1993;262:1867–70. doi: 10.1126/science.8266074. [DOI] [PubMed] [Google Scholar]
- 124.Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, et al. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol Biol Cell. 2007;18:5139–53. doi: 10.1091/mbc.E07-05-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hales KG, Bi E, Wu JQ, Adam JC, Yu IC, Pringle JR. Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol. 1999;11:717–25. doi: 10.1016/s0955-0674(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 126.Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000;10:251–5. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- 127.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, et al. Role of formins in actin assembly: nucleation and barbed end association. Science. 2002;297:612–5. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 128.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–31. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 129.Shuster CB, Burgess DR. Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc Natl Acad Sci USA. 2002;99:3633–8. doi: 10.1073/pnas.052342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tammi R, Tammi M. Correlations between hyaluronan and epidermal proliferation as studied by [3H]glucosamine and [3H]thymidine incorporations and staining of hyaluronan on mitotic keratinocytes. Exp Cell Res. 1991;195:524–7. doi: 10.1016/0014-4827(91)90405-j. [DOI] [PubMed] [Google Scholar]
- 131.Evanko SP, Parks WT, Wight TN. Intracellular hyaluronan in arterial smooth muscle cells: association with microtubules, RHAMM, and the mitotic spindle. J Histochem Cytochem. 2004;52:1525–35. doi: 10.1369/jhc.4A6356.2004. [DOI] [PubMed] [Google Scholar]
- 132.Silverman-Gavrila R, Silverman-Gavrila L, Bendeck MP. Cell division fidelity is altered during the vascular response to injury: its novel role in atherosclerosis progression. Am J Pathol. 2013;182:628–39. doi: 10.1016/j.ajpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 133.Sburlati A, Cabib E. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J Biol Chem. 1986;261:15147–52. [PubMed] [Google Scholar]
- 134.Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, et al. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–23. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cortes JC, Pujol N, Sato M, Pinar M, Ramos M, Moreno B, et al. Cooperation between paxillin-like protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet. 2015;11:e1005358. doi: 10.1371/journal.pgen.1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, et al. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol. 2008;10:395–406. doi: 10.1038/ncb1701. [DOI] [PubMed] [Google Scholar]
- 137.Jendretzki A, Ciklic I, Rodicio R, Schmitz HP, Heinisch JJ. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol Genet Genomics. 2009;282:437–51. doi: 10.1007/s00438-009-0476-0. [DOI] [PubMed] [Google Scholar]
- 138.Meitinger F, Petrova B, Mancini Lombardi I, Bertazzi DT, Hub B, Zentgraf H, et al. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–61. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- 139.Foltman M, Molist I, Arcones I, Sacristan C, Filali-Mouncef Y, Roncero C, et al. Ingression progression complexes control extracellular matrix remodelling during cytokinesis in budding yeast. PLoS Genet. 2016;12:e1005864. doi: 10.1371/journal.pgen.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998b;143:1947–60. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, et al. Interaction of Bnr1p with a novel Src Homology 3 domain-containing Hof1p IMPLICATION IN CYTOKINESIS IN SACCHAROMYCES CEREVISIAE. J Biol Chem. 1998;273:28341–5. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- 142.Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM, Ferguson KM, et al. Comparison of Saccharomyces cerevisiae F-BAR domain structures reveals a conserved inositol phosphate binding site. Structure. 2015;23:352–63. doi: 10.1016/j.str.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10:947–50. doi: 10.1016/s0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- 144.Makarova KS, Aravind L, Koonin EV. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999;8:1714–9. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oh Y, Chang KJ, Orlean P, Wloka C, Deshaies R, Bi E. Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol Biol Cell. 2012;23:2445–56. doi: 10.1091/mbc.E12-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-D-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–11. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Inoue SB, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, et al. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–54. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 148.Mazur P, Baginsky W. In vitro activity of 1,3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J Biol Chem. 1996;271:14604–9. doi: 10.1074/jbc.271.24.14604. [DOI] [PubMed] [Google Scholar]
- 149.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–43. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol. 2013;202:231–40. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G. A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 2013;11:e1001495. doi: 10.1371/journal.pbio.1001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gao XD, Caviston JP, Tcheperegine SE, Bi E. Pxl1p, a paxillin-like protein in Saccharomyces cerevisiae, may coordinate Cdc42p and Rho1p functions during polarized growth. Mol Biol Cell. 2004;15:3977–85. doi: 10.1091/mbc.E04-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mackin NA, Sousou TJ, Erdman SE. The PXL1 gene of Saccharomyces cerevisiae encodes a paxillin-like protein functioning in polarized cell growth. Mol Biol Cell. 2004;15:1904–17. doi: 10.1091/mbc.E04-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wolken DM, McInnes J, Pon LA. Aim44p regulates phosphorylation of Hof1p to promote contractile ring closure during cytokinesis in budding yeast. Mol Biol Cell. 2014;25:753–62. doi: 10.1091/mbc.E13-06-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, et al. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–81. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 156.Kuranda MJ, Robbins PW. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–67. [PubMed] [Google Scholar]
- 157.Baladron V, Ufano S, Duenas E, Martin-Cuadrado AB, del Rey F, Vazquez de Aldana CR. Eng1 p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:774–86. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kovacech B, Nasmyth K, Schuster T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol Cell Biol. 1996;16:3264–74. doi: 10.1128/mcb.16.7.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.King L, Butler G. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr Genet. 1998;34:183–91. doi: 10.1007/s002940050384. [DOI] [PubMed] [Google Scholar]
- 160.O'Conallain C, Doolin MT, Taggart C, Thornton F, Butler G. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol Gen Genet. 1999;262:275–82. doi: 10.1007/s004380051084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–50. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 162.Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, et al. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mazanka E, Weiss EL. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol Biol Cell. 2010;21:2809–20. doi: 10.1091/mbc.E10-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–32. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 166.Meitinger F, Palani S, Pereira G. The power of MEN in cytokinesis. Cell Cycle. 2012;11:219–28. doi: 10.4161/cc.11.2.18857. [DOI] [PubMed] [Google Scholar]
- 167.Scarfone I, Piatti S. Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1. Small GTPases. 2015;6:196–201. doi: 10.1080/21541248.2015.1109023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yoshida S, Ichihashi R, Toh-e A. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J Cell Biol. 2003;161:889–97. doi: 10.1083/jcb.200301128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bertazzi DT, Kurtulmus B, Pereira G. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J Cell Biol. 2011;193:1033–48. doi: 10.1083/jcb.201101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol. 2009;184:527–39. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Juanes MA, Piatti S. The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae Cell Mol Life Sci. 2016;73:3115–36. doi: 10.1007/s00018-016-2220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Naylor SG, Morgan DO. Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci. 2014;127:1128–37. doi: 10.1242/jcs.144097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Miller DP, Hall H, Chaparian R, Mara M, Mueller A, Hall MC, et al. Dephosphorylation of Iqg1 by Cdc14 regulates cytokinesis in budding yeast. Mol Biol Cell. 2015;26:2913–26. doi: 10.1091/mbc.E14-12-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, et al. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–45. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hwa Lim H, Yeong FM, Surana U. Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol Biol Cell. 2003;14:4734–43. doi: 10.1091/mbc.E03-04-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Teh EM, Chai CC, Yeong FM. Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle. 2009;8:2964–74. [PubMed] [Google Scholar]
- 177.Zhang G, Kashimshetty R, Ng KE, Tan HB, Yeong FM. Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother-daughter neck via the secretory pathway in budding yeast. J Cell Biol. 2006;174:207–20. doi: 10.1083/jcb.200604094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chin CF, Bennett AM, Ma WK, Hall MC, Yeong FM. Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol Biol Cell. 2011;23:45–58. doi: 10.1091/mbc.E11-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Watts FZ, Shiels G, Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 1987;6:3499–505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–8. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- 181.Tian C, Wu Y, Johnsson N. Stepwise and cooperative assembly of a cytokinetic core complex in Saccharomyces cerevisiae. J Cell Sci. 2014;127:3614–24. doi: 10.1242/jcs.153429. [DOI] [PubMed] [Google Scholar]


