Abstract
Brown adipose tissue (BAT) represents a promising agent to ameliorate obesity and other metabolic disorders. However, the abundance of BAT decreases with age and BAT paucity is a common feature of obese subjects. As brown adipocytes and myoblasts share a common Myf5 lineage origin, elucidating the molecular mechanisms underlying the fate choices of brown adipocytes versus myoblasts may lead to novel approaches to expand BAT mass. Here we identify MyoD as a key negative regulator of brown adipocyte development. CRISPR/CAS9-mediated deletion of MyoD in C2C12 myoblasts facilitates their adipogenic transdifferentiation. MyoD knockout downregulates miR-133 and upregulates the miR-133 target Igf1r, leading to amplification of PI3K–Akt signaling. Accordingly, inhibition of PI3K or Akt abolishes the adipogenic gene expression of MyoD null myoblasts. Strikingly, loss of MyoD converts satellite cell-derived primary myoblasts to brown adipocytes through upregulation of Prdm16, a target of miR-133 and key determinant of brown adipocyte fate. Conversely, forced expression of MyoD in brown preadipocytes blocks brown adipogenesis and upregulates the expression of myogenic genes. Importantly, miR-133a knockout significantly blunts the inhibitory effect of MyoD on brown adipogenesis. Our results establish MyoD as a negative regulator of brown adipocyte development by upregulating miR-133 to suppress Akt signaling and Prdm16.
Keywords: MyoD, Myogenesis, Repressor, Adipogenesis, Brown adipocytes, CRISPR-Cas9
Highlights
-
•
Loss of MyoD facilitates adipogenic transdifferentiation of myoblasts.
-
•
Overexpression of MyoD transdifferentiate brown preadipocytes to myoblasts.
-
•
MyoD acts partially through miR-133 to suppress brown adipocyte cell fate.
Brown fat burns fat to produce heat, and represents a promising agent to treat obesity and its related disorders. Brown fat cells and muscle cells share a common origin, but what controls the developmental separation of the two cell types is not well understood. This study reports that inhibition of “MyoD” gene in muscle progenitors promotes their differentiation into brown fat cells in mice. Conversely, forced expression of MyoD in brown fat progenitors converts them into muscle cells. This work suggests that inhibition of MyoD may represent a future direction to expand brown fat and alleviate obesity in humans.
1. Introduction
Obesity, caused by an imbalance between energy intake and energy expenditure, has become a global health crisis (Hossain et al., 2007). Brown adipose tissue (BAT) found prominently in rodents and hibernating mammals functions to dissipate energy in the form of non-shivering thermogenesis mediated by the uncoupling protein Ucp1 (Cannon and Nedergaard, 2004, Kopecky et al., 1995). Adult humans also have functional BAT with the capacity to take up and utilize glucose and fatty acids for thermogenesis (Cypess et al., 2009, Nedergaard et al., 2007, Orava et al., 2011, Ouellet et al., 2012, Pfannenberg et al., 2010, van Marken Lichtenbelt et al., 2009, Virtanen et al., 2009). It has been demonstrated that repeated cold treatments activate human BAT in parallel to an increase in energy expenditure (van der Lans et al., 2013, Yoneshiro et al., 2013). As such, BAT represents a promising target in the treatment of obesity and other metabolic disorders. However, BAT abundance decreases with age (Cypess et al., 2009, Nedergaard et al., 2007, Pfannenberg et al., 2010, van Marken Lichtenbelt et al., 2009, Virtanen et al., 2009). Understanding the molecular regulation of BAT biogenesis is key to the development of novel approaches to expand BAT mass.
Brown adipocytes and myoblasts are derived from a common pool of progenitors expressing Myf5 (Seale et al., 2008), the earliest myogenic regulatory factor (MRF) detected in developing muscles (Ott et al., 1991). The fate of brown adipocytes is determined by the expression of PR domain containing protein 16 (Prdm16) (Seale et al., 2008). Ectopic expression of Prdm16 in myoblasts inhibits myogenesis and the expressions of muscle related genes, partly through euchromatic histone-lysine N-methyltransferase 1 and miR-193b-365 (Ohno et al., 2013, Seale et al., 2008, Sun et al., 2011). MRFs, including Myf5, MyoD, Myog and Myf6 (Mrf4) orchestrate the process of myogenesis (Olson and Klein, 1994, Pownall et al., 2002). MyoD acts downstream of Myf5 to commit somite-derived cells to myoblasts (Rudnicki et al., 1993), and to initiate the onset of myogenic differentiation (Rawls et al., 1998). A lineage-tracing study reveals that the MyoD lineage does not give rise to brown adipocytes (Sanchez-Gurmaches and Guertin, 2014), indicating a role of MyoD in myogenic cell fate switch in the common Myf5+ progenitors that give rise to both myoblasts and brown preadipocytes. However, whether loss of MyoD promotes brown adipocyte cell fate conversion of myoblasts, and whether ectopic expression of MyoD in brown preadipocytes inhibits adipogenesis has not been examined. In this paper, we provide gain- and loss-of-function evidence to show that MyoD acts as a key molecular repressor of the brown adipocyte cell fate.
2. Materials and Methods
2.1. Animals
MyoD−/− mice (Rudnicki et al., 1992) were generously provided by Dr. Michael Rudnicki (Ottawa Health Research Institute, Ottawa, Ontario, Canada). The miR-133a1/a2 double knockout mice were generously provided by Dr. Eric Olson (Liu et al., 2008). The mdx mice (stock#001801, RRID: IMSR_JAX:001801) and MyoDiCre/+ mice (stock#014140) were from Jackson lab. Mice were housed in the animal facility with free access to water and standard rodent chow. All procedures involving the use of animals were performed in accordance with the guidelines presented by Purdue University's Animal Care and Use Committee. Unless otherwise indicated, 2-month-old adult mice were used in this study.
2.2. Generation of MyoD-Knockout C2C12 Cell Lines
2.2.1. Generation of Cas9-Overexpressing C2C12 Cells
We utilized the CRISPR-Cas9 technique to mutate the MyoD gene in C2C12 cell line. To examine the efficiency of gRNAs, we generated a C2C12 cell line with Cas9 overexpressing by transfecting cells with the plasmid PX459 (Ran et al., 2013). PX459 contains the Cas9 protein with a FLAG tag and a puromycin resistant gene. Puromycin (1 μg/ml) selection was used to purify cells transfected with PX459, and FLAG antibody was used to verify the expression of Cas9 in C2C12 cells (C2C12Cas9).
2.2.2. Selection of gRNA Targets
We used CRISPR Design (http://crispr.mit.edu/) to choose gRNA targets of MyoD (Ran et al., 2013). The gRNAs were generated by in vitro transcription according to the online protocol (http://www.crisprflydesign.org/grnatranscription/). Then the gRNAs (1 μg for each gRNA/106 cells) were electroporated into C2C12Cas9 cells. For the control sample, we only gave C2C12Cas9 cells electric shocks. One day after the electroporation, we collected the cells, extracted DNA and amplified the region around the target sites by PCR. Clones containing cleaved bands in the gRNA treated sample but not in the control sample were sequenced to confirm correct targeting.
2.2.3. MyoD KO Cell Line Selection
We chose the gRNA target close to the 5′ of the MyoD gene and cloned it into the PX459 plasmid (PX459MyoD). Wildtype C2C12 cells were electroporated with PX459MyoD or PX459 (1 μg/106 cell). Two days after electroporation, puromycin (1 μg/ml) was added to the culture medium. Five days after puromycin selection, the remaining cells electroporated with PX459 were collected as control for the following studies. The remaining cells electroporated with PX459MyoD were detached by 0.25% trypsin and transferred to 48-well plates by serial dilution. When the cell confluency was over 70%, cells were passaged and parts of them were used to do DNA extraction, PCR amplification and sequencing. Cells containing predicted genomic alteration were cloned a second time to achieve homogeneity.
2.2.4. Off-target Detection
The gRNA off-targets were searched by CRISPR Design. None of the 10 most likely off-target sites by the gRNA selected in this study were located in the gene body. The top 5 potential off-target sites (Table S1) were PCR amplified using genomic DNA as templates. The PCR products were subjected to T7EN1 cleavage assay. The potential off-target sites yielding typical cleavage bands were considered as candidates. Primers for PCR amplification of off-target sites were listed in Table S2.
2.3. Primary Adipocyte Isolation, Culture and Differentiation
Interscapular BAT depots from wildtype or miR-133a mutant mice were collected, minced and digested with isolation buffer (DMEM supplemented with 1.5 mg/ml Collagenase I) for proper time at 37 °C on a shaker. The digestion was stopped with DMEM containing 10% FBS, filtered through 100 μm filters, and cells were pelleted at 450 × g for 5 min. The cells were cultured in growth media containing DMEM, 20% FBS and 1% P/S at 37 °C with 5% CO2 for 3 days, and then fresh media was changed every 2 days. Upon confluence, cells were exposed to induction media and then differentiation media. C2C12 cells were exposed to induction media for 2 days and differentiation media for 3 days. Satellite cell-derived myoblasts were exposed to induction media for 6 days and differentiation media for 6 days. Brown preadipocytes were exposed to induction media for 4 days and differentiation media for 4 days. The induction media contains DMEM, 10% FBS, 2.85 μM insulin, 0.3 μM dexamethasone and 0.63 mM 3-isobutyl-1-methylxanthine (Sigma), and the differentiation media contains DMEM, 10% FBS, 200 nM insulin and 10 nM T3. For C2C12 cells and Satellite cell-derived myoblasts, we also add 1 μM Rosiglitazone to the induction media and differentiation media. The miR-133a mirVana miRNA mimic was purchased from Ambion. The Akt dominant negative plasmid, GFP-Akt-K179M was a kind gift from Dr. Xiaoqi Liu (Chen et al., 2015). The PI3K inhibitor (LY294002) was purchased from Sigma and used to treat C2C12 myoblasts during adipogenic induction at 10 μM concentration.
2.4. Oil Red O Staining
Cells or muscle sections were rinsed with PBS and fixed with 10% formaldehyde for 15 min at room temperature. Cells or muscle sections were stained in working solution containing 6 ml of Oil Red O (ORO) stock solution (5 mg/ml in isopropanol) and 4 ml ddH2O for 60 min. After staining, slides were washed with distilled water for 10 min and mounted in glycerol mounting medium.
2.5. Muscle Injury and Myoblasts Transplantation
Muscle regeneration was induced by cardiotoxin (CTX; Sigma-Aldrich) or glycerol (Macron Fine Chemicals) injection. Mice were first anesthetized using a ketamine-xylazine cocktail and then the TA muscles were bilaterally injured with 50 μl of 10 μM CTX or 50 μl 50% glycerol. Two days after the CTX or glycerol injury, we injected 50 μl concentrated adenovirus (1 × 106 cells) into injured TA muscles. For comparable results, the left and right TA muscles were injected with control and MyoDKO C2C12 myoblasts, respectively. The concentrations of triglyceride in TA muscles were measured using the Triglyceride Colorimetric Assay Kit (Cayman). Glycerol is able to degenerate and induce adipogenesis of skeletal muscle (Pisani et al., 2010). TA muscle injection with glycerol was performed to induce adipogenic induction of 2–3 month old wild-type and MyoD−/− mice. Ten days after injection, muscle tissues were harvested for ORO staining and RNA isolation.
2.6. Primary Myoblast Isolation, Culture and Differentiation
TA muscle was harvested, and satellite cell-derived primary myoblasts were isolated as described previously (Motohashi et al., 2014). Briefly, TA muscles of MyoD−/− mice or their wild-type littermates were dissected and minced and then digested in type I collagenase/dispase B mixture (Roche Applied Science). The digestions were filtered from debris for magnetic-activated cell sorting (MACS). Dissociated muscle cells were incubated with Anti-CD45-PE, anti-Sca-1-PE and anti-CD31-PE antibodies (all from eBiosciences) followed by anti-PE beads (Miltenyi Biotec). LD column (Miltenyi Biotec) was used for negative selection to eliminate hematopoietic cells, endothelial cells and other non-muscle cells from the muscle preparation. Flow through fraction was incubated with integrin α-biotin antibody (Miltenyi Biotec) followed by anti-biotin beads (Miltenyi Biotec). MS column (Miltenyi Biotec) was used for primary myoblast isolation as a positive fraction. Isolated primary myoblasts were cultured in growth media (F-10 Ham's media supplemented with 17% FBS, 4 ng/ml basic fibroblast growth factor and 1% penicillin-streptomycin) on collagen-coated dishes, and the growth medium was changed every two days. Primary myoblasts were cultured in normal humidified tissue culture incubators with 5% CO2.
2.7. Adenovirus Generation
The adenovirus with MyoD insertion was generated using the AdEasy system. Briefly, the MyoD ORF was cloned using pairs of primers (MyoD-f and MyoD-r, Table S2), and inserted into pAdTrack-CMV plasmid. The formed pAdTrack-CMV-MyoD (pAdTrack-CMV as the control) plasmids were digested by PmeI and then transfected the DH5a competent cell with pAdEasy-1. The positive recombinant plasmid was detected by PacI digestion. Then, 293A cells (60%–70% confluent) in 10 cm culture dishes was transfected with the 4 μg PacI digested recombinant plasmid using Lipofectamine 2000 (Life Technologies) according to manufacturer's protocol. After 14 days incubation, the recombinant adenovirus was released by four freeze–thaw–vortex cycles. Two more round infections were adapted to amplify the recombinant virus and the titers were determined by the expression of GFP.
2.8. Immunostaining and Image Acquisition
Myoblasts were fixed with 4% paraformaldehyde, and blocked with blocking buffer (5% goat serum, 2% BSA, 0.2% triton X-100, and 0.1% sodium azide in PBS) for at least 1 h. The samples were then incubated with primary antibody [MyoD (M318, RRID: AB_2148870) was from Santa Cruz and Perilipin (D1D8, RRID: AB_10621999) was from Cell Signaling] in blocking buffer overnight. After washing with PBS, the samples were incubated with secondary antibodies and DAPI or BODIPY 493/503 for 45 min at room temperature. Fluorescent images were captured using a Leica DM 6000B fluorescent microscope. As the adenoviral vectors contain GFP and thus express green signals when they are transduced into cells, we calculated the number of different signals only in GFP+ cells.
2.9. Total RNA Extraction, cDNA Synthesis, and Real-Time PCR
Total RNA was extracted from cells using Trizol Reagent according to the manufacturer's protocols. RNA was treated with RNase-free DNase I to remove contaminating genomic DNA. The purity and concentration of total RNA were measured by Nanodrop 3000 (Thermo Fisher). Random primers and Moloney murine leukemia virus reverse transcriptase were used to convert RNA into cDNA. For microRNA, multiple adenosine nucleotides were added to 3′ end of RNAs by E. coli DNA polymerase and cDNAs were synthesized with a specific RT primer (Liu et al., 2013). Real-time PCR was performed using Roche Lightcycler 480 PCR System with SYBR Green Master Mix and gene-specific primers described previously (Sun et al., 2011, Wang et al., 2015). Ct value of 18S rRNA and U6 snRNA was used to normalize mRNA and miRNA expression, respectively. The 2− ΔΔCT method was used to analyze the relative mRNA and miRNA expressions.
2.10. Protein Extraction and Western Blot Analysis
Cultured myoblasts were washed with PBS and homogenized with radioimmune precipitation assay buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein concentrations were determined using Pierce BCA Protein Assay Reagent (Pierce Biotechnology, Rockford, IL). Proteins were separated by 10% SDS-PAGE, electrotransferred onto PVDF membrane (Millipore Corporation, Billerica, MA), and incubated with specific primary antibodies. Igf-1rβ (C20) and Gapdh (6C5, RRID: AB_627679) antibodies were from Santa Cruz; Akt (#9272, RRID: AB_329827) and pAkt (Ser473) (#9271, RRID: AB_329825) antibodies were from Cell Signaling. Immunodetection was performed using enhanced chemiluminescence (ECL) Western blotting substrate (Pierce Biotechnology, Rockford, IL) and detected with FluoChem R imaging system (ProteinSimple, CA).
2.11. Statistical Analysis
The data are presented with mean and standard deviation (SD). P-values were calculated using unpaired two-tailed Student's t-test. P-values < 0.05 were considered to be statistically significant.
3. Results
3.1. Generating MyoD-Knockout C2C12 Clones
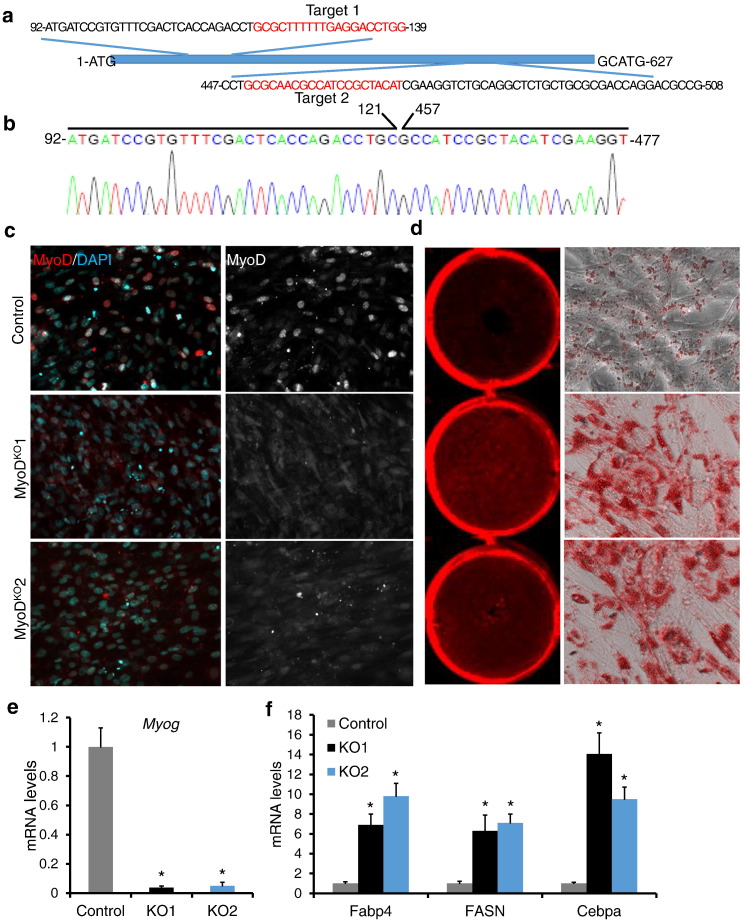
C2C12 cells are well-established myogenic progenitors that also have some limited capacity to differentiate into adipocytes (Teboul et al., 1995). In order to investigate the role of MyoD in myogenic versus adipogenic fate determination, we sought to knockout (KO) MyoD in C2C12 cells using the CRISPR-Cas9 technique. To improve the targeting efficiency, we first established C2C12 cells stably overexpressing Cas9 (C2C12Cas9), and then transfected the cells with two in vitro transcribed guide RNAs (gRNAs) targeting the exon 1 of MyoD (Fig. 1a). Two cleaved bands were only visible in C2C12Cas9 myoblasts transfected with both gRNAs (Fig. S1a). We sequenced the 300 bp band, and found a 336 bp deletion in MyoD (Fig. 1b), indicating that both gRNAs work efficiently with Cas9 to edit MyoD.
Fig. 1.
Loss of MyoD facilitates the adipogenic transdifferentiation of C2C12 myoblasts. (a) Schematic diagram of MyoD partial protein coding region and the 2 targeting loci of MyoD sgRNA. The targeting loci are in red. (b) DNA sequence map around the targeting locus of the cleaved band amplified from myoblasts transfected with both gRNAs. (c) Immunostaining of MyoD (Red) and DAPI (Cyan) in control and MyoDKO C2C12 myoblasts. (d) Representative images of ORO staining of C2C12 myoblasts after adipogenic differentiation. (e and f) qPCR analysis of Myog (e) and adipogenic markers genes (f). Error bars represent mean with SD of five independent biological replicates.⁎P < 0.05 (Student's t-test).
To minimize genome-wide off-target mutations, we only cloned the sequence of Target 1 (Fig. 1a) to the plasmid PX459 to establish MyoD-knockout clones. Of the 21 cell clones established, we found through sequencing that most clones had mutations in one of the two MyoD alleles, with two clones containing 1-bp or 11-bp frame-shifting deletions in both MyoD alleles (Fig. S1b). These mutations resulted in truncated MyoD proteins of 55 and 72 amino acid residues lacking the basic Helix-loop-Helix domain, respectively (Fig. S1b). We further performed immunohistochemistry staining on the MyoDKO cell clones using a well-established MyoD antibody. Control C2C12 myoblasts exhibited strong MyoD signals, whereas both MyoD knockout cell clones lost the MyoD signal (Fig. 1c). The T7EN1 cleavage assays showed that no genetic modification was detected at the selected off-target sites (Fig. S1c). Therefore, we have successfully established two clones of MyoD knockout C2C12 cells. We named them MyoDKO C2C12 cells to distinguish them from MyoD−/− primary myoblasts isolated from skeletal muscles of MyoD mutant mice.
3.2. Loss of MyoD Facilitates Adipogenic Transdifferentiation of C2C12 Myoblasts
Consistent with previous report that MyoD–/– primary myoblasts have reduced differentiation potential (Cornelison et al., 2000, Dedieu et al., 2002, Sabourin et al., 1999), the MyoDKO C2C12 cells showed differentiation defect when induced in myogenic differentiation medium (Fig. S1d). To examine the adipogenic potential of the MyoDKO C2C12 myoblasts, we exposed confluent cultures to an adipogenic differentiation medium. The control cells showed elongated morphology characteristic of myotubes, whereas many MyoDKO cells differentiated into round, lipid droplet-containing adipocyte-like cells (Fig. 1d). Oil Red O (ORO) staining for neutral triglycerides revealed that control cells only had a few oil droplets. In contrast, MyoDKO cells exhibited large regions of cells containing widespread oil droplets (Fig. 1d). The mutation of MyoD also dramatically downregulated the myogenic marker gene Myog (Fig. 1e), but significantly upregulated the adipogenic marker genes Fabp4, Fasn and Cebpα by 6-14 fold (Fig. 1f), indicating loss of MyoD facilitates adipogenic transdifferentiation of C2C12 cells.
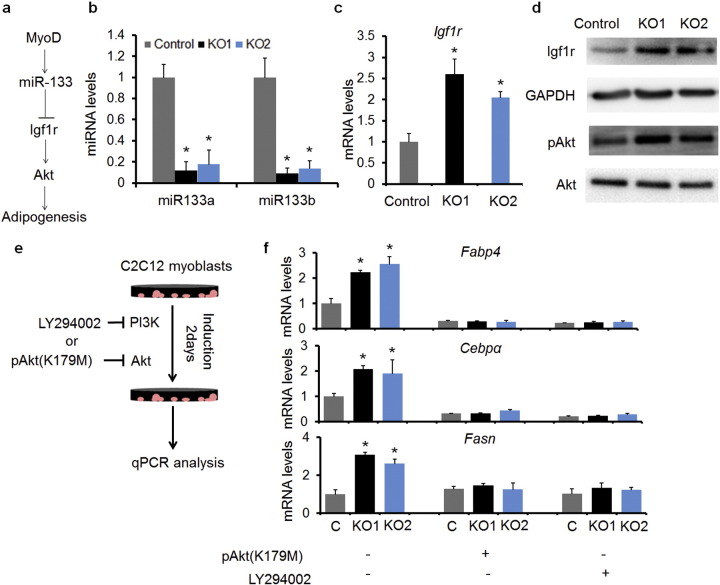
As MyoD regulates miR-133 (Rao et al., 2006), which targets Igf1r (Huang et al., 2011), we hypothesize that loss of MyoD upregulates Igf1r and consequently amplifies the downstream PI3K–Akt signaling to promote adipogenesis (Fig. 2a). Indeed, MyoDKO cells expressed significantly lower levels (~ 90% reduction) of miR-133a and miR-133b (Fig. 2b), accompanied by 2-fold increase in the expression of Igf1r both at mRNA and protein levels (Fig. 2c and d). Within two days after incubation with adipogenic induction media, MyoDKO cells had increased levels of the phosphorylated Akt (pAkt), but normal levels of total Akt (Fig. 2d). In parallel to the activation of Akt, the levels of adipogenic marker genes (Fabp4, Cebpα and Fasn) were 2-3 fold higher in MyoDKO cells than in control cells (Fig. 2e and f).
Fig. 2.
The miR-133/Igf1r/Akt signaling pathway mediates adipogenesis of MyoDKO cells. (a) Hypothetical model showing how MyoDKO induces adipogenic transdifferentiation of C2C12 myoblasts. (b and c) qPCR analysis of miR-133 (b) and Igf1r (c). (d) Western blot analysis of the expressions of Igf1r and pAkt in control and MyoDKO myoblasts. (e) Strategies to examine the role of PI3K and Akt in transdifferentiation of myoblasts to adipocytes. (f) qPCR analysis of adipogenic marker genes under different treatments. Error bars represent mean with SD of five independent biological replicates.⁎P < 0.05 (Student's t-test).
To examine if miR-133 reduction mediates adipogenesis of MyoDKO cells, we overexpressed miR-133a in cultured MyoDKO cells (Fig. S2a). Electroporation-mediated gene transfer resulted in up to 840-fold elevation in the expression of miR-133a (Fig. S2a). As a consequence, the mRNA levels of Igf1r, Fabp4, Fasn and Cebpα in MyoDKO cells were normalized to those in control cells (Fig. S2b). To determine if activation of PI3K–Akt is required for the adipogenic conversion of C2C12 myoblasts, the MyoDKO cells were transfected with a plasmid expressing the dominant negative Akt mutant (K179M), or incubated with a PI3K inhibitor LY294002 (Fig. 2e). Both treatments normalized the mRNA levels of Fabp4, Fasn and Cebpα in MyoDKO to similar levels as the control cells (Fig. 2f), indicating that PI3K–Akt activation is required for the upregulation of adipogenic gene expression in MyoDKO C2C12 cells. Together, loss of MyoD facilitates adipogenic transdifferentiation of C2C12 cells through miR-133/Igf1r/Akt signaling pathway.
3.3. Loss of MyoD Facilitates Brown Adipogenic Transdifferentiation of Satellite Cells
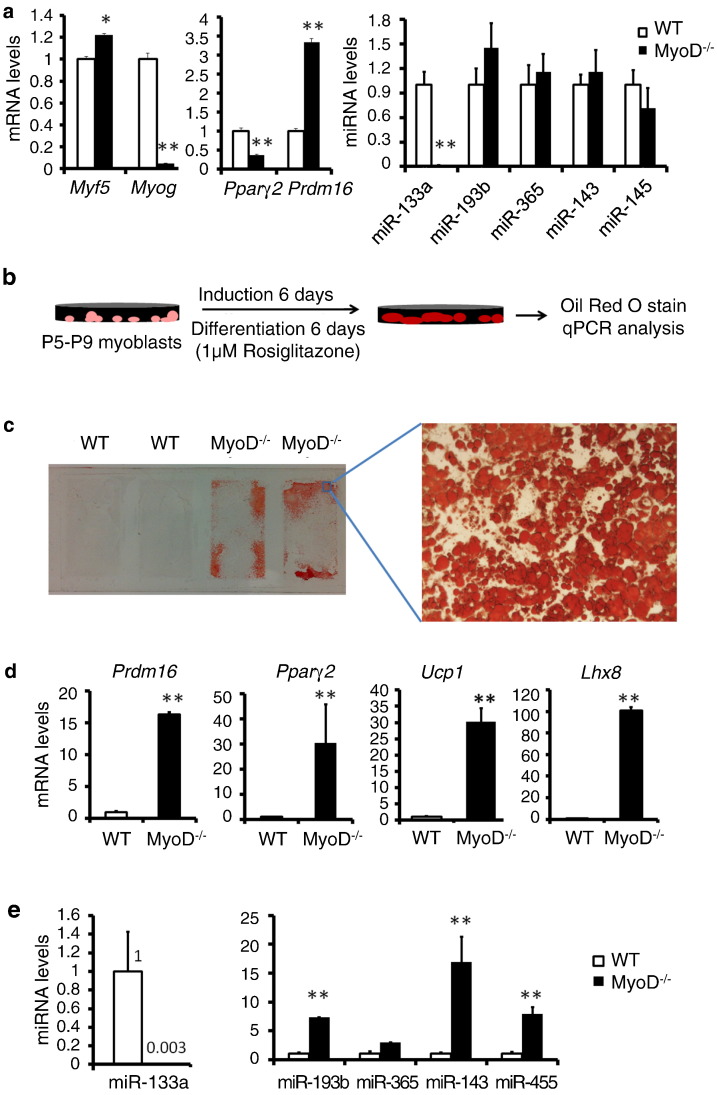
Brown adipocytes and muscle satellite cells share a common developmental origin (Seale et al., 2008). As the loss of MyoD in C2C12 myoblasts facilitates adipogenic transdifferentiation, we hypothesize that loss of MyoD may convert satellite cells to brown adipocytes. To test this hypothesis, we examined cultured primary myoblasts derived from skeletal muscle satellite cells of MyoD−/− mice. Under growth conditions, MyoD−/− myoblasts expressed slightly increased levels of Myf5 and strikingly reduced levels of Myog and miR-133 (Fig. 3a). Consistence with the reports that miR-133 targets Prdm16 (Liu et al., 2013, Trajkovski et al., 2012, Yin et al., 2013), this reduced levels of miR-133 is associated with increased expression of Prdm16 in MyoD−/− myoblasts (Fig. 3a). Interestingly, the expression of Pparγ2, a key transcriptional factor for adipogenesis (Tontonoz et al., 1994), was decreased in MyoD−/− myoblasts (Fig. 3a). However, the expression of several adipogenic marker miRNAs did not change their expression in MyoD−/− myoblasts (Fig. 3a). These results support that MyoD KO does not spontaneously promote adipogenesis in the absence of adipogenic induction (Asakura et al., 2001).
Fig. 3.
Loss of MyoD dramatically downregulates miR-133a, promotes brown adipocyte biogenesis and stimulates transdifferentiation of myoblasts into brown adipocytes. MyoDiCre/+ mice were intercrossed to generate MyoDiCre/iCre (MyoD−/−) knockout mice and WT littermates. The miRNA miR-133a; Prdm16 targets miR-193b and miR-365; adipogenic marker miR-143 and brown adipogenic marker miR-455 were analyzed by qPCR. (a) qPCR analysis of WT and MyoD−/− myoblasts for myogenic markers Myf5 and Myogenin, adipogenic markers Pparγ2 and Prdm16, and the miRNAs. (b) WT and MyoD−/− myoblasts were subjected to adipogenic induction and differentiation as shown in the schematics. (c) Representative images of ORO staining of adipocytes. (d and e) qPCR analysis of adipogenic markers (d) and miRNAs (e). Passage 3 primary myoblasts (purity > 95%) from MyoD−/− and WT mice were used. Error bars represent mean with SD of 3–5 independent biological replicates.⁎P < 0.05, ⁎⁎P < 0.01 (Student's t-test).
We next examined if MyoD−/− myoblasts can transdifferentiate into brown adipocytes under adipogenic differentiation medium containing 1 μM Rosiglitazone, a Pparγ activator (Fig. 3b). Strikingly, MyoD−/− myoblasts robustly differentiated into lipid-filling adipocytes as indicated by ORO staining (Fig. 3c). In contrast, wild-type (WT) myoblasts did not transdifferentiate into adipocytes under identical culture conditions (Fig. 3c). After differentiation, brown adipogenic markers genes Prdm16, Ucp1 and Lhx8 were expressed 16-fold, 30-fold and 100-fold higher in MyoD−/− cells than in WT cells (Fig. 3d). The adipogenic master regulator Pparγ2 was expressed 30-fold higher in MyoD−/− cells as well (Fig. 3d). The level of miR-133a remained low in MyoD−/− cells after adipogenic differentiation (Fig. 3e). However, the Prdm16 target miR-193b was upregulated by 7-fold, and adipogenic miR-143 and miR-455 were 17-fold and 8-fold higher, respectively, in the MyoD−/− cells compared with the WT cells (Fig. 3e). We conclude from these results that the loss of MyoD downregulates miR-133a, upregulates Prdm16, and promotes the transdifferentiation of myoblasts into brown adipocytes under adipogenic conditions.
3.4. Loss of MyoD Facilitates Adipogenic Transdifferentiation of Myoblasts in vivo
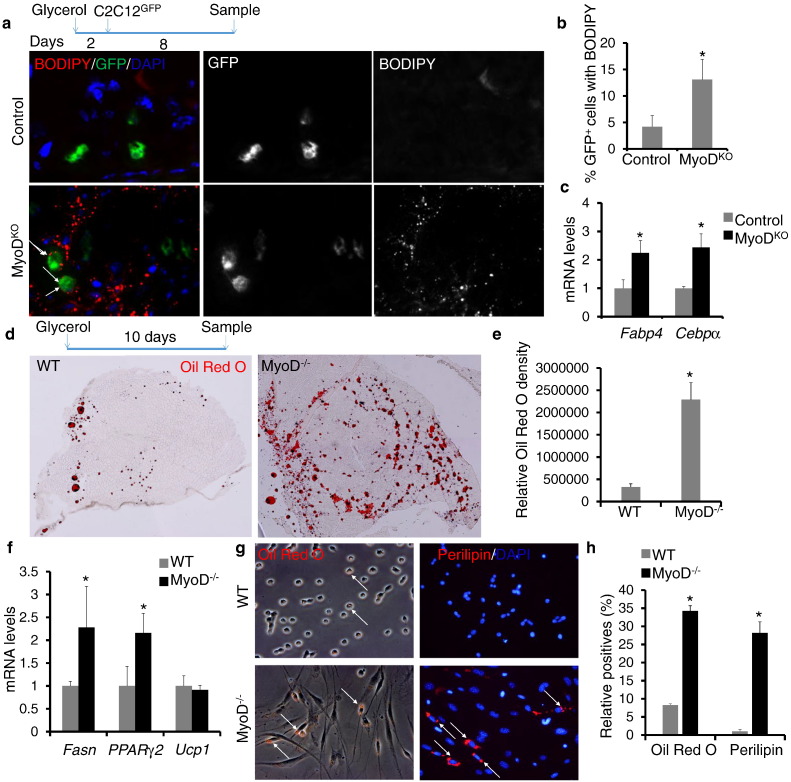
To investigate the adipogenic potential of MyoDKO C2C12 myoblasts in vivo, control and MyoDKO C2C12 cells were injected into injured TA muscles of mdx mice, which lack the expression of dystrophin, a muscle membrane protein. Eight days after cell transplantation, TA muscles injected with MyoDKO C2C12 cells formed fewer dystrophin+ myofibers compared with muscles injected with control C2C12 cells (Fig. S3a and b). Interestingly, the reduced myogenic capacity of MyoDKO cells is accompanied by a concomitant increase in formation of BODIPY-label adipocytes (Fig. S3a). Consistently, the concentration of triglycerides (TG) was 40% higher (1.5 for WT C2C12 cells and 2.1 for MyoDKO C2C12 cells) in muscles grafted with MyoDKO C2C12 cells compared with those injected with WT C2C12 cells (Fig. S3c). To directly trace the fate conversion of injected C2C12 cells, we used adenovirus to overexpress GFP in control and MyoDKO C2C12 cells prior to grafting into glycerol-injected TA muscles (Fig. 4a). Eight days after cell graft, the muscles injected with MyoDKO C2C12 cells had about 3-fold more GFP+ BODIPY+ cells compared with muscles injected with control C2C12 cells (Fig. 4a and b). Consistently, the expression levels of adipogenic marker genes Fabp4 and Cebpα were higher in muscles injected with MyoDKO C2C12 cells than in those injected with control C2C12 cells (Fig. 4c). These results indicate that loss of MyoD facilitates adipogenic transdifferentiation of C2C12 myoblasts in vivo.
Fig. 4.
Loss of MyoD facilitates adipogenic transdifferentiation of myoblasts in vivo. (a) Immunostaining of BODIPY (red) in TA muscles injected with control or MyoDKO C2C12 cells. Arrows indicate GFP+ BODIPY+ cells. (b) The ratio of BODIPY signal in GFP+ cells in TA muscles injected with control or MyoDKO C2C12 cells. (c) qPCR analysis of adipogenic marker genes. (d) Representative images of ORO staining of regenerated TA muscles of WT and MyoD−/− mice. The MyoD−/− mice (Rudnicki et al., 1992) were generously provided by Dr. Michael Rudnicki. (e) Relative ORO intensity on WT and MyoD−/− TA muscle sections. (f) qPCR analysis of adipogenic marker genes. (g) ORO staining and anti-Perilipin staining in myoblasts isolated from glycerol-injured TA muscles of WT and MyoD−/− mice. Arrows indicate ORO+ or Perilipin+ cells. (h) The ratio of ORO+ and Perilipin+ cells in myoblasts isolated from injured WT and MyoD−/− TA muscles. Four sections were analyzed for each sample. Error bars represent mean with SD of three independent biological replicates.⁎P < 0.05 (Student's t-test).
To further investigate the adipogenic transdifferentiation of MyoD−/− satellite cells in vivo, TA muscles of MyoD−/− mice and WT littermates were treated with glycerol to induce muscle degeneration and adipogenesis. On day 10 after glycerol injection, TA muscle sections were stained with ORO to label lipids (Fig. 4d). Strikingly, TA muscles of MyoD−/− mice had much more pronounced ORO labeling (Fig. 4d), with a 7-fold increase in the density of ORO signal compared to WT mice (Fig. 4e). Consistently, mRNA levels of adipogenic marker genes Fasn and Pparγ2 were higher in TA muscles of MyoD−/− mice compared with those in WT mice (Fig. 4f). However, mRNA levels of brown adipogenic marker gene Ucp1 were comparable in TA muscles of WT and MyoD−/− mice (Fig. 4f). To confirm the satellite cell origin of the ORO+ cells in glycerol treated MyoD−/− muscles, primary myoblasts were isolated from TA muscles using magnetic-activated cell sorting (MACS) at 4 days after glycerol injection, prior to adipogenic differentiation (Fig. 4g). Strikingly, the percentage of ORO+ and Perilipin+ myoblasts was 4-fold and 28-fold higher, respectively, in myoblasts isolated from MyoD−/− mice than in the myoblasts isolated from WT mice (Fig. 4h). We conclude that loss of MyoD facilitates adipogenic transdifferentiation of satellite cell-derived myoblasts in vivo.
3.5. Overexpression of MyoD in BAT Preadipocytes Inhibits Adipogenesis
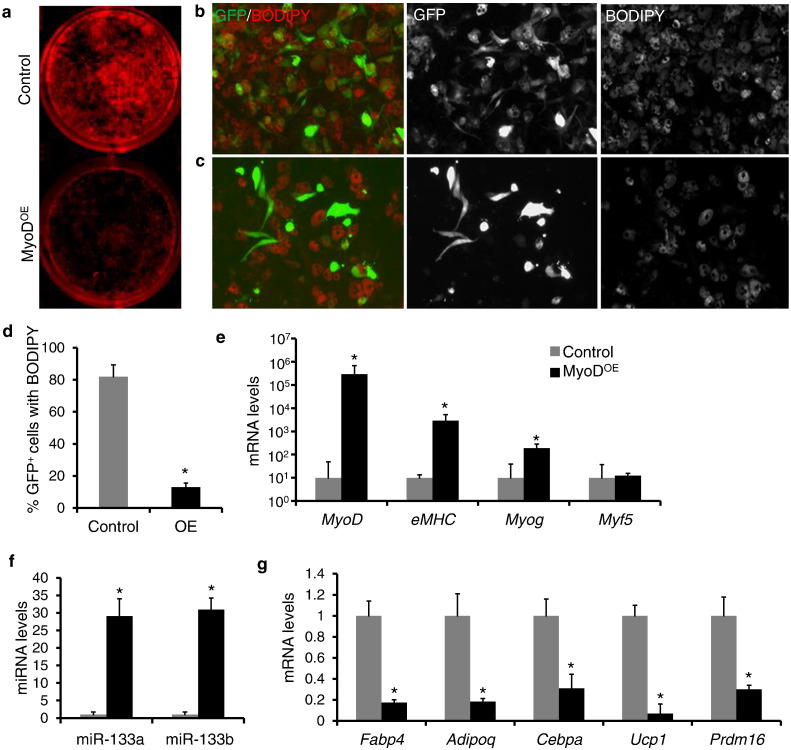
To further examine the role of MyoD in brown adipocyte versus myoblast fate switch, we used adenoviruses to overexpress MyoD-GFP (MyoDOE) or GFP only (control) in BAT SVF preadipocytes isolated from WT mice. Two days after transduction of adenoviral vectors, brown preadipocytes were induced to undergo adipogenesis. ORO staining showed MyoD but not GFP markedly decreased lipid accumulation in brown adipocytes differentiated from BAT SVF cells (Fig. 5a). We also utilized BODIPY to label brown adipocytes (Fig. 5b and c). Whereas 82% (387 out of 473) of GFP+ cells were BODIPY+ in the control group, only 13% (55 out of 426) of the GFP+ (indicative of MyoDOE) cells were BODIPY+ in the MyoDOE group (Fig. 5d), representing a 6-fold decrease. We noticed in the MyoDOE group a negative correlation between GFP and BODIPY signals: there was essentially no BODIPY signal in regions with high densities of GFP signals (Fig. S4a). In addition, a group of MyoDOE brown preadipocytes showed elongated myotube-like morphologies (Fig. S4a), and about 70% (115 out of 165) of the elongated MyoDOE cells expressed myosin heavy chain (MF20+) (Fig. S4b and c), indicating that MyoD transdifferentiates brown preadipocytes into myocyte. At the molecular level, MyoD overexpression was correlated with 18-fold and 288-fold elevation in the expression of Myog and eMHC, respectively (Fig. 5e). In parallel, the expression levels of miR-133a and miR-133b were upregulated by 28-fold and 30-fold, respectively (Fig. 5f). By contrast, brown adipogenic marker genes Prdm16 and Ucp1 were decreased by 92% and 70%, respectively, and other adipogenic marker genes Fabp4, Adipoq and Cebpα were also downregulated by 82%, 81% and 69%, respectively (Fig. 5g). Together, our data indicate that BAT adipogenesis is inhibited by ectopic expression of MyoD.
Fig. 5.
Overexpression of MyoD impairs brown adipogenesis of wildtype BAT SVF cells. (a) ORO staining of brown adipocytes differentiated from wildtype BAT SVF cells transduced with control or MyoDOE adenovirus. (b and c) Immunostaining of BODIPY (red) in control (b) and MyoDOE (c) brown adipocytes (d) The ratio of BODIPY signal in GFP+ cells in control and MyoDOE brown adipocytes. Five different areas analyzed in each experiment. (e–g) qPCR analysis of myogenic markers (e) miRNAs (f) and adipogenic markers (g). Error bars represent mean with SD of three independent biological replicates.⁎P < 0.05 (Student's t-test).
3.6. Deletion of miR-133a Blunts the Inhibitory Effect of MyoD on Brown Adipogenesis
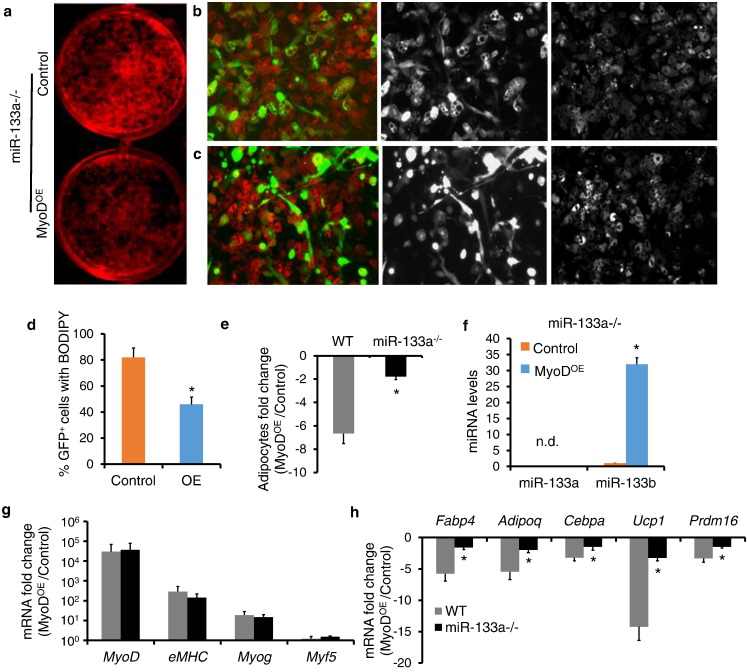
To investigate whether miR-133 mediates the inhibitory effect of MyoD on adipogenesis, we used adenoviral vectors to overexpress MyoD in miR-133a1/a2 double knockout (miR-133a−/−) BAT SVF cells. MyoD overexpression decreased lipid accumulation in miR-133a−/− brown adipocytes differentiated from BAT SVF cells (Fig. 6a), but to a much lesser extent compared with the level of decrease in WT brown adipocytes (Fig. 5a). By labeling brown adipocytes with BODIPY (Fig. 6b and c), we found that 82% (420 out of 512) of GFP+ cells were BODIPY+ in the control group, and 48% (229 out of 476) of the GFP+ cells were BODIPY+ in the MyoDOE group (Fig. 6d), representing < 2-fold decrease (Fig. 6e). By contrast, MyoD decreased the ratio of GFP+/BODIPY+ cells by over 6-fold in WT brown adipocytes (Fig. 6e). MyoD overexpression similarly increased the levels of miR-133b by 30-fold in both WT and miR-133a−/− brown adipocytes (Figs. 6f and 5f). In addition, MyoD overexpression increased the levels of Myog and eMHC to similar extent in WT and miR-133a−/− brown adipocytes (Fig. 6g), suggesting that MyoD induces myogenesis independent of miR-133a. However, the extent of MyoD-induced suppression of adipogenic markers was much lesser in the miR-133a−/− compared to WT brown adipocytes (Fig. 6h). Specifically, the MyoD overexpression induced fold decreases of Fabp4, Adipoq, Cebpα, Ucp1 and Prdm16 were 5-fold, 5-fold, 3-fold, 14-fold and 3-fold, respectively, in WT brown adipocytes (Fig.6h). In contrast, in the absence of miR-133a, MyoD only decreased the expression of these genes by 1.6-fold, 2-fold, 1.5-fold, 3-fold and 1.5-fold, respectively (Fig. 6h). These results demonstrate that MyoD inhibits brown adipogenesis partly through miR-133a.
Fig. 6.
Loss of miR-133a alleviates the inhibitory effect of MyoD on brown adipogenesis. (a) ORO staining of brown adipocytes differentiated from miR-133a−/− BAT SVF cells transduced with control or MyoDOE adenovirus. (b and c) Immunostaining of BODIPY (red) in control (b) and MyoDOE (c) miR-133a−/− brown adipocytes. (d) Ratios of BODIPY signal in GFP+ cells in control and MyoDOE miR-133a−/− brown adipocytes. Five different areas analyzed in each experiment. (e) Fold changes of GFP+ BODIPY+ cells in WT and miR-133a−/− brown adipocytes after MyoD overexpression (f) qPCR analysis of miR-133a and miR-133b. (g and h) Fold changes of myogenic markers (g) and adipogenic markers (h) in WT and miR-133a−/− brown adipocytes after MyoD overexpression. Error bars represent mean with SD of three independent biological replicates.⁎P < 0.05 (Student's t-test).
4. Discussion
We identified MyoD as a negative regulator of brown adipocyte development. This result extends the function of MyoD in addition to its well-established role as a positive regulator of myogenesis. With the CRISPR-Cas9 technique, we successfully established 2 clones of MyoDKO myoblasts and discovered that loss of MyoD facilitates the adipogenic transdifferentiation of C2C12 myoblasts. Consistent with our study is that downregulation of MyoD mRNA by RNaseL in C2C12 myogenic cells led to adipogenic differentiation (Salehzada et al., 2009). We show that the MyoD/miR-133/Igf1r/PI3K/Akt signaling pathway is involved in the promotion of adipogenesis in MyoDKO cells. Insulin, a component of the adipogenic differentiation medium, binds to Igf1r to activate the p85 regulatory subunit of PI3K, which subsequently phosphorylates and activates Akt (Belfiore et al., 2009). Akt then inhibits tuberous sclerosis complex 2 and activate the mammalian target of rapamycin complex 1 to promote insulin-induced adipogenesis (Zhang et al., 2009).
Loss of MyoD is necessary but not sufficient to promote brown adipogenesis of myoblasts. MyoD−/− myoblasts expressed dramatically lower levels of miR-133a, which led to the upregulation of Prdm16, without increasing expression of Pparγ2, a master regulator of adipogenesis. Although Prdm16 primes brown adipogenesis (Seale et al., 2008), the adipogenic conversion of myoblasts requires activation of Pparγ (Yeow et al., 2001). For example, the addition of rosiglitazone in adipogenic medium was required to activate Pparγ to execute the conversion. This also explains why MyoD−/− myoblasts did not result in conversion of myoblasts to brown adipocytes spontaneously in the absence of adipogenic induction. Interestingly, although MyoD−/− satellite cells can transdifferentiate into adipocytes under adipogenic conditions in vivo, these adipocytes are UCP1−. This observation suggests that additional factors, such as Notch signaling and sympathetic innervation (Bartness et al., 2010, Pasut et al., 2016), may be required to induce UCP1 expression in the transdifferentiated MyoD−/− adipocytes. A previous study reported that MyoD mutant mice have normal interscapular BAT mass (Borensztein et al., 2012). This may be due to the relative late activation of MyoD after Pax7/Myf5 during myogenesis and brown adipogenesis (Rudnicki et al., 1993, Sanchez-Gurmaches and Guertin, 2014). Therefore, deletion of MyoD should not affect brown adipogenesis, which takes place before MyoD activation in the Pax7/Myf5 progenitors.
Several myogenic factors including MyoD are expressed in brown preadipocytes, with expression diminishing during differentiation (Timmons et al., 2007), indicating that inhibition of MyoD is required for brown adipogenesis. We demonstrate that overexpression of MyoD dramatically upregulates miR-133 expression in brown adipocytes. This is consistent with the report that MyoD binds to regions upstream of miR-133(Rao et al., 2006). As miR-133 represses Prdm16 (Liu et al., 2013, Trajkovski et al., 2012, Yin et al., 2013), the MyoD/miR-133/Prdm16 signaling cascade explains the inhibitory effect of MyoD on brown adipogenesis. Forced expression of MyoD in adipocyte precursors or white adipocytes similarly inhibits adipogenic differentiation, indicating MyoD inhibits adipogenesis in general regardless of Prdm16 (Kocaefe et al., 2005, Weintraub et al., 1989). This is probably achieved through the transdifferentiation of brown preadipocytes into MF20+ myocytes and miR-133 mediated inhibition of Igf1r which is required for adipogenesis (Boucher et al., 2010).
We show that miR-133 is an important mediator of MyoD-induced inhibition of adipogenesis. There are two mature miR-133 isomers: miR-133a and miR-133b that only differ in one nucleotide. The miR-133a is encoded by two genes (miR-133a1 and miR-133a2), and double mutant mice exhibit cardiac and skeletal muscle myopathy, along with browning of subcutaneous white adipose tissue (Liu et al., 2011, Liu et al., 2008, Liu et al., 2013). By contrast, the miR-133b mutant mice are normal (Boettger et al., 2014), indicating that miR-133a has more important physiological functions than miR-133b. The observation that deletion of miR-133a only partially blocked the inhibitory function of MyoD on brown adipogenesis suggests that additional factors are involved in mediating MyoD function in brown adipocytes. It is possible that the residual miR-133b partially compensated for the miR-133a loss-of-function. Our finding that ectopic expression of MyoD inhibits brown adipogenesis and promotes myogenic transdifferentiation suggests that inhibition of myogenesis is necessary for brown adipogenic commitment of Pax7/Myf5 progenitors. As MyoD KO alone is insufficient to block embryonic myogenesis, the brown adipocyte development is not affected. Supporting this notion, complete blockage of myogenesis in MyoD/Myf5 and MyoD/Igf2 double knockout mice both exhibit more pronounced adipogenesis (Borensztein et al., 2012, Kablar et al., 2003). MyoD inhibits adipogenesis and the expression of Prdm16, and Prdm16 reciprocally inhibits myogenesis and the expression of MyoD (Seale et al., 2008). The mutual exclusion of the two master regulators balances and determines the developmental separation of BAT and muscle progenitors during embryogenesis.
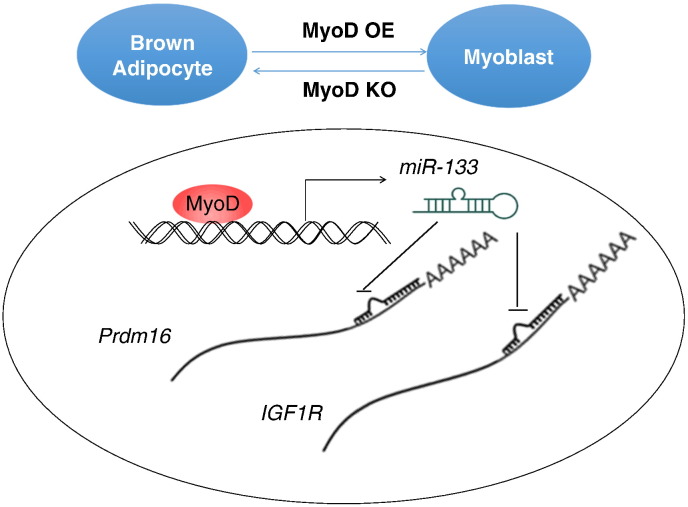
In summary, our results establish MyoD as a negative regulator of brown adipocyte development through upregulating miR-133 to suppress Igf1r and Prdm16 (Fig. 7). Our work suggests that inhibition of MyoD in patient-derived myoblasts may promote their transdifferentiation into brown adipocytes.
Fig. 7.
Diagram summarizing the role of MyoD in brown adipocyte and myoblast fate determination. Loss of MyoD stimulates transdifferentiation of myoblasts into brown adipocytes. Conversely, overexpression of MyoD inhibits brown adipogenesis through myogenic transdifferentiation of brown adipocytes and miR-133-mediated mRNA decay of Prdm16 and Igf1r.
Acknowledgements and Funding Sources
This work was supported by a grant from the US National Institutes of Health (R01AR060652 to S.K.) and Purdue incentive grant from Purdue University Office of Vice President for Research (OVPR, Cat1, 206732) to S.K. We thank Dr. Xiaoqi Liu (Biochemistry, Purdue University) for generously providing pAkt (K179M). Dr. Eric Olson (UT Southwestern Medical Center) for generously providing miR-133a1/a2 double knockout mice. Dr. Michael Rudnicki (Ottawa Health Research Institute) for generously providing MyoD−/− mice. Jun Wu for mouse colony maintenance and members of the Kuang laboratory for valuable comments.
Conflict of Interest Statement
The authors declare no actual or potential competing financial interests.
Author Contributions
C.W. and S.K. conceived the project, designed the experiments and prepared the manuscript. C.W., W.L., Y.N., M.Q., A.A., H.H. and F.Y. performed the experiments.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.01.015.
Appendix A. Supplementary data
Supplementary material
References
- Asakura A., Komaki M., Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- Bartness T.J., Vaughan C.H., Song C.K. Sympathetic and sensory innervation of brown adipose tissue. Int. J. Obes. (Lond) 2010;(34 Suppl. 1):S36–S42. doi: 10.1038/ijo.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009;30:586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- Boettger T., Wust S., Nolte H., Braun T. The miR-206/133b cluster is dispensable for development, survival and regeneration of skeletal muscle. Skelet. Muscle. 2014;4:23. doi: 10.1186/s13395-014-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M., Viengchareun S., Montarras D., Journot L., Binart N., Lombes M., Dandolo L. Double Myod and Igf2 inactivation promotes brown adipose tissue development by increasing Prdm16 expression. FASEB J. 2012;26:4584–4591. doi: 10.1096/fj.12-208496. [DOI] [PubMed] [Google Scholar]
- Boucher J., Tseng Y.H., Kahn C.R. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J. Biol. Chem. 2010;285:17235–17245. doi: 10.1074/jbc.M110.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chen L., Li Z., Ahmad N., Liu X. Plk1 phosphorylation of IRS2 prevents premature mitotic exit via AKT inactivation. Biochemistry. 2015;54:2473–2480. doi: 10.1021/acs.biochem.5b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison D.D., Olwin B.B., Rudnicki M.A., Wold B.J. MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B., Kuo F.C., Palmer E.L., Tseng Y.H., Doria A. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedieu S., Mazeres G., Cottin P., Brustis J.J. Involvement of myogenic regulator factors during fusion in the cell line c2c12. Int. J. Dev. Biol. 2002;46:235–241. [PubMed] [Google Scholar]
- Hossain P., Kawar B., El Nahas M. Obesity and diabetes in the developing world—a growing challenge. N. Engl. J. Med. 2007;356:213–215. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- Huang M.B., Xu H., Xie S.J., Zhou H., Qu L.H. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B., Krastel K., Tajbakhsh S., Rudnicki M.A. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev. Biol. 2003;258:307–318. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Kocaefe Y.C., Israeli D., Ozguc M., Danos O., Garcia L. Myogenic program induction in mature fat tissue (with MyoD expression) Exp. Cell Res. 2005;308:300–308. doi: 10.1016/j.yexcr.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Kopecky J., Clarke G., Enerback S., Spiegelman B., Kozak L.P. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Bezprozvannaya S., Shelton J.M., Frisard M.I., Hulver M.W., McMillan R.P., Wu Y., Voelker K.A., Grange R.W., Richardson J.A. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J. Clin. Invest. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Bi P., Shan T., Yang X., Yin H., Wang Y.X., Liu N., Rudnicki M.A., Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Asakura Y., Asakura A. Isolation, culture, and transplantation of muscle satellite cells. J. Vis. Exp. 2014 doi: 10.3791/50846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J., Bengtsson T., Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Ohno H., Shinoda K., Ohyama K., Sharp L.Z., Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Klein W.H. Bhlh factors in muscle development — dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- Orava J., Nuutila P., Lidell M.E., Oikonen V., Noponen T., Viljanen T., Scheinin M., Taittonen M., Niemi T., Enerback S. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 2011;14:272–279. doi: 10.1016/j.cmet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Ott M.O., Bober E., Lyons G., Arnold H., Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- Ouellet V., Labbe S.M., Blondin D.P., Phoenix S., Guerin B., Haman F., Turcotte E.E., Richard D., Carpentier A.C. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A., Chang N.C., Rodriguez U.G., Faulkes S., Yin H., Lacaria M., Ming H., Rudnicki M.A. Notch signaling rescues loss of satellite cells lacking Pax7 and promotes brown adipogenic differentiation. Cell Rep. 2016;16:333–343. doi: 10.1016/j.celrep.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannenberg C., Werner M.K., Ripkens S., Stef I., Deckert A., Schmadl M., Reimold M., Haring H.U., Claussen C.D., Stefan N. Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes. 2010;59:1789–1793. doi: 10.2337/db10-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D.F., Bottema C.D., Butori C., Dani C., Dechesne C.A. Mouse model of skeletal muscle adiposity: a glycerol treatment approach. Biochem. Biophys. Res. Commun. 2010;396:767–773. doi: 10.1016/j.bbrc.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Pownall M.E., Gustafsson M.K., Emerson C.P. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls A., Valdez M.R., Zhang W., Richardson J., Klein W.H., Olson E.N. Overlapping functions of the myogenic bhlh genes mrf4 and myod revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Schnegelsberg P.N.J., Stead R.H., Braun T., Arnold H.H., Jaenisch R. Myod or myf-5 is required for the formation of skeletal-muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sabourin L.A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M.A. Reduced differentiation potential of primary MyoD −/− myogenic cells derived from adult skeletal muscle. J. Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehzada T., Cambier L., Vu Thi N., Manchon L., Regnier L., Bisbal C. Endoribonuclease L (RNase L) regulates the myogenic and adipogenic potential of myogenic cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Guertin D.A. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun. 2014;5:4099. doi: 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scime A., Devarakonda S., Conroe H.M., Erdjument-Bromage H. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xie H., Mori M.A., Alexander R., Yuan B., Hattangadi S.M., Liu Q., Kahn C.R., Lodish H.F. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul L., Gaillard D., Staccini L., Inadera H., Amri E.Z., Grimaldi P.A. Thiazolidinediones and fatty acids convert myogenic cells into adipose-like cells. J. Biol. Chem. 1995;270:28,183–28,187. doi: 10.1074/jbc.270.47.28183. [DOI] [PubMed] [Google Scholar]
- Timmons J.A., Wennmalm K., Larsson O., Walden T.B., Lassmann T., Petrovic N., Hamilton D.L., Gimeno R.E., Wahlestedt C., Baar K. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Trajkovski M., Ahmed K., Esau C.C., Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- van der Lans A.A., Hoeks J., Brans B., Vijgen G.H., Visser M.G., Vosselman M.J., Hansen J., Jorgensen J.A., Wu J., Mottaghy F.M. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D., Schrauwen P., Teule G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T., Taittonen M., Laine J., Savisto N.J., Enerback S. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu W., Liu Z., Chen L., Liu X., Kuang S. Hypoxia inhibits myogenic differentiation through p53 protein-dependent induction of bhlhe40 protein. J. Biol. Chem. 2015;290:29,707–29,716. doi: 10.1074/jbc.M115.688671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S.J., Davis R.L., Thayer M.J., Adam M.A., Lassar A.B., Miller A.D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow K., Phillips B., Dani C., Cabane C., Amri E.Z., Derijard B. Inhibition of myogenesis enables adipogenic trans-differentiation in the C2C12 myogenic cell line. FEBS Lett. 2001;506:157–162. doi: 10.1016/s0014-5793(01)02900-3. [DOI] [PubMed] [Google Scholar]
- Yin H., Pasut A., Soleimani V.D., Bentzinger C.F., Antoun G., Thorn S., Seale P., Fernando P., van Ijcken W., Grosveld F. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y., Iwanaga T., Saito M. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.H., Huang J., Duvel K., Boback B., Wu S., Squillace R.M., Wu C.L., Manning B.D. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material