Abstract
Phospholipase D (PLD) in plants plays vital roles in growth, development, and stress responses. However, the precise role of PLDs in pineapple remains poorly understood. In this study, 10 putative PLD genes, designated as AcPLD1–AcPLD10, were identified based on the pineapple genome database. The 10 AcPLDs could be clustered into five of the six known PLD families according to sequence characterization. Their deduced amino acid sequences displayed similarities to PLDs from other plant species. Expression analyses of PLD mRNAs from pineapple pulp were performed. The 10 PLDs exhibited differential expression patterns during storage periods of fruits treated with hexaldehyde (a specific PLD inhibitor) which could alleviate internal browning (IB) of pineapple after harvest. Functional subcellular localization signaling assays of two PLD proteins (AcPLD2 and AcPLD9) were performed by fluorescence microscopy. To further detect the potential action mechanism underlying PLD involved in the IB defense response, PLD, hydrogen peroxide (H2O2) and H2O2 associated with antioxidative enzymes such as superoxide dismutase, catalase, NADPH, and ascorbate peroxidase were quantified by enzyme-linked immunosorbent assay. This report is the first to provide a genome-wide description of the pineapple PLD gene family, and the results should expand knowledge of this family.
Keywords: pineapple fruit, phospholipase D, internal browning, gene expression, defense response
Introduction
Phospholipase D (PLD, EC 3.1.4.4) which hydrolyzes different membrane phospholipids (PLs) into phosphatidic acid and various water-soluble head groups, such as choline or ethanolamine, has been detected in bacteria, fungi, plants, and animals (Guo and Wang, 2012; Comoglio et al., 2014). Since plant PLD cDNA was first cloned by Wang et al. (1994), many PLD-encoding genes have been obtained from plants, such as rice (Ueki et al., 1995), Arabidopsis thaliana (Pinosa et al., 2013), tobacco (Lein and Saalbach, 2001), tomato (Whitaker et al., 2001), strawberry (Yuan et al., 2005), and soybean (Zhao et al., 2012). Protein structural analysis has resulted in several conserved domains identified in PLDs, including PLD-C1 and PLD-C2 domains, C2 domain, and PX (phox)/PH (pleckstrin homology) domains. PLD-C1 and PLD-C2 domains are also called HKD domains, which are regarded as catalytic sites and are essential for the catalytic activity of the PLD enzyme (Qin and Wang, 2002). The C2 domain, is known as the Ca2+/PL-binding domain, which can mediate Ca2+-dependent activity through binding to Ca2+ (Kopka et al., 1998). PX/PH domains, which are located at the N-terminus of Ca2+-independent PLDs (Kopka et al., 1998; Qin and Wang, 2002), have been implicated in membrane targeting and polyphosphoinositide signaling of PLD (van Leeuwen et al., 2004).
An increasing number of studies have indicated that PLD, which is considered an important signaling enzyme in various organisms and is widespread in plants (Jang et al., 2012), participates in diverse physiological processes, such as development (Lu et al., 2016); hormone signaling mediated by abscisic acid (Peters et al., 2010), salicylic acid (Janda et al., 2015), jasmonic acid (Zhao et al., 2013), and auxin (Li and Xue, 2007); reactive oxygen generation (Wu et al., 2010); and in the response to abiotic and biotic stresses such as drought (Lu et al., 2012), high salinity (Yu et al., 2010), low temperature (Kargiotidou et al., 2010), and fungal infection (Liu et al., 2013). The diverse cellular functions of PLD suggest that the enzyme is subjected to complex regulation in the cell.
Although the functions of PLD in many plants have been studied extensively, the specific role of different plant PLDs remains unknown. Moreover, the involvement of PLDs in relation to quality of fruits after harvest has received minimal attention. Pineapple (Ananas comosus L.), which is the second most important harvest crop after banana and tasty tropical fruit, is cultivated in over 80 countries worldwide and produced annually with a gross production value approaching nine billion dollars (FAO, 2014). China produced approximately 1.89 million tons of pineapple in 2014, accounting for 7.4% of worldwide production (FAO, 2014). Under natural conditions, pineapples undergo internal browning (IB) during postharvest storage (Zhang et al., 2015). Therefore, the quality and storage potential of this fruit must be improved. Effective methods to delay the development of IB include 1-methylcyclopropene treatment (Selvarajah et al., 2001), controlled atmosphere (Nimitkeatkai et al., 2006), waxing (Hu et al., 2012), and exogenous abscisic acid (Zhang et al., 2015), but the mechanism of the IB remains unclear. The recent completion of the pineapple genome has allowed for the genome-wide identification of a number of important gene families (Ming et al., 2015). Three different PLD inhibitors, namely, 1-butanol (Pinosa et al., 2013), lysophosphatidyl ethanolamine (Li et al., 2015), and hexaldehyde (Paliyath et al., 2003), can all delay the development of IB in pineapple after harvest. Thus, PLDs may be the involved in IB. However, the functions of PLDs in pineapple need to be elucidated. This study was conducted to address the detailed structural analyses of PLDs in pineapple, as well as determine their phylogenetic relationship with other species. The potential regulatory mechanisms of PLD involvement during IB in pineapple are also discussed.
Materials and Methods
Plant Materials and Treatments
Pineapple (A. comosus cv. ‘Comte de Paris’) at stage 3 of maturation as proposed by Selvarajah et al. (2001) was obtained from a commercial orchard in Xuwen County, Guangdong Province in winter. Fruits of uniform size, color, and maturity, as well as being free of physical injury or disease symptoms, were selected, with the crown intact. Subsequently, fruits were immersed for 2 min in 0.05% (w/v) benomyl aqueous solution to control decay, air-dried at 25°C for 2 h and treated as follows. The selected pineapple fruits were immersed into 0.1% (v/v) hexaldehyde containing 0.01% Tween 80 for 5 min. The control fruits were immersed with distilled water, which also contained 0.01% Tween 80. Each treatment was applied to three replications, and each replication contained 100 fruits. Following treatment, all fruits were placed into unsealed plastic bags (0.04 mm thick), and stored at 25°C in the dark with relative humidity at 95% for 12 days. Samples of pulp tissues were collected at days 0, 2, 4, 6, 8, 10, and 12 frozen in liquid nitrogen; and stored at -80°C for future use.
Evaluation of IB Intensity
For IB assessment, at least six fruits for each treatment were halved longitudinally to observe the incidence and severity of IB injury. IB intensity was visually scored from 0 (no IB) to 5 (maximum IB) according to Teisson (1979).
Identification of PLD and Sequence Analysis
A putative pineapple PLD cDNA was identified by searching the pineapple genome database. The homology of predicted PLD protein sequences was checked using Clustal W, with the default parameters set by the DNA Data Bank of Japan1. Domain and motif searches were conducted on the protein sequences in SMART2. Blast searches were also performed against nucleic acid sequence data repositories at the National Center for Biotechnology Information (NCBI3). A phylogenetic tree was generated using Clustal Omega online tools4. The logos of the conserved domains were constructed using MEME program5 with the default parameters. The theoretical isoelectric point (pI) and molecular weight of the pineapple PLD proteins were calculated using PeptideMass program6. Transmembrane helix number was predicted by HMMTOP online tools7.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from pineapple pulp using the hot borate method of Wan and Wilkin (1994). RNA purification and first-strand cDNA synthesis were performed as described in our previous paper (Hong et al., 2016). qRT-PCR analyses were conducted to detect gene expression patterns of the PLD family during the development of IB in fruits treated with PLD inhibitor (hexaldehyde). The oligonucleotide primers for qRT-PCR analysis were designed using Primer 5.0 software and Acactin (accession number HQ148720) was used as a reference gene. Reactions were carried out using SYBR® premix Ex TaqTMII (Takara, Shiga, Japan) on a LightCycler®480 II (Roche, Switzerland). The relative expression levels of AcPLDs were calculated using the method of Livak and Schmittgen (2001). Values represented the average of three biological replicate. The sequences of all primers are listed in Supplemental File 1.
Subcellular Localization Analysis
The coding sequences of AcPLD2 and AcPLD9 without the stop codon were amplified by PCR (primers are listed in Supplemental File 2) and subcloned into the pEAQ-GFP-HT vector to form fusion constructs AcPLD2::GFP and AcPLD9::GFP, under the control of the CaMV 35S promoter. The fusion constructs and a control vector were maintained in Agrobacterium tumefaciens strain EHA105 by electroporation. Transient transformation of tobacco leaves was carried out according to the method described by Sainsbury et al. (2009). Localizations of AcPLD2::GFP and AcPLD9::GFP were investigated with a fluorescence microscope (Zeiss Axioskop 2 Plus). All transient expression assays were repeated three times.
Measurements of Superoxide Dismutase (SOD), Catalase (CAT), NADPH, Ascorbate Peroxidase (APX), PLD, and Hydrogen Peroxide (H2O2) Contents
The pineapple pulp samples (1.0 g) were homogenized in 9.0 ml of phosphate buffered saline (PBS) (pH 7.4) and then centrifuged at 4000 ×g for 15 min. The supernatant solution was collected for analyses of SOD, CAT, NADPH, APX, PLD, and H2O2 contents, which were quantified by enzyme-linked immunosorbent assay (ELISA). ELISA was carried out as follows: (1) The 96-well microplates were coated with 100 μL of the captured specific antibody at 1:2000 dilution in coating buffer and incubated overnight at 4°C. (2) Plates were washed three times with PBS to remove unbound antibody, and each well was filled with 300 μL of blocking buffer and incubated for 30 min at 37°C. (3) Followed by washing, 100 μL sample solution mentioned above was added to each well and incubated for 30 min at 37°C. (4) After washing three times, 100 μL horseradish peroxidase-conjugated the specific antibody was added to each well and incubated at 37°C for 30 min. (5) The plate was washed again, and 100 μL of solution 3,3/5,5/-tetramethylbenzidine was added to each well. (6) After developing the color for 30 min at 37°C, the reaction was terminated after the addition of 50 μL of 2 M H2SO4 per well. (7) The absorbance values at 450 nm were determined, and the values of ELISA were given in relative units (ng.mg-1protein). Protein content was measured by the method of Bradford (1976) using bovine serum albumin as the standard protein.
Statistical Analysis
The experiment was performed in triplicate (n = 3). Data were presented as the mean ± standard error. Least significant difference at the 5% level was analyzed by SPSS software (version 13.0, SPSS Inc., Chicago, IL, United States).
Results
IB Intensity
Control fruits showed signs of IB after 6 days of storage, whereas no sign of IB was detected in fruits treated with hexaldehyde. When the treated fruits began to exhibit the IB at day 8 of storage, the IB intensity reached 1.7 in control samples. IB progressively increased as storage duration increased. The IB intensity was higher for control samples than for treated fruits, and a noticeable difference was observed between them after 6 days of storage (Figure 1).
FIGURE 1.
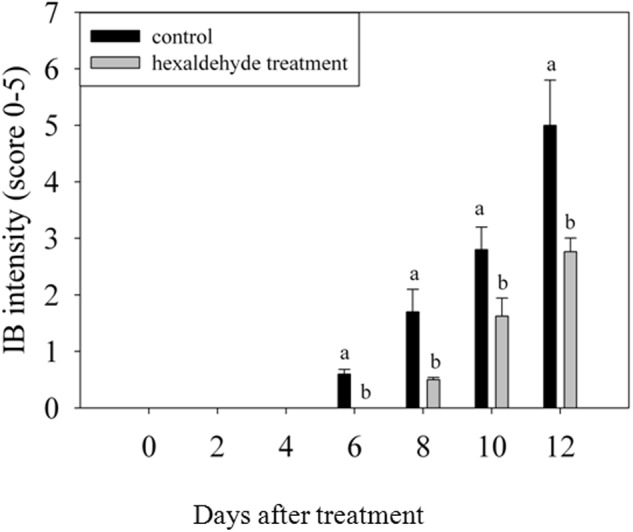
Changes in IB intensity of fruits treated with hexaldehyde. Each value represents the mean ± standard error of three replicates.
Identification of PLD Gene Family Members in Pineapple
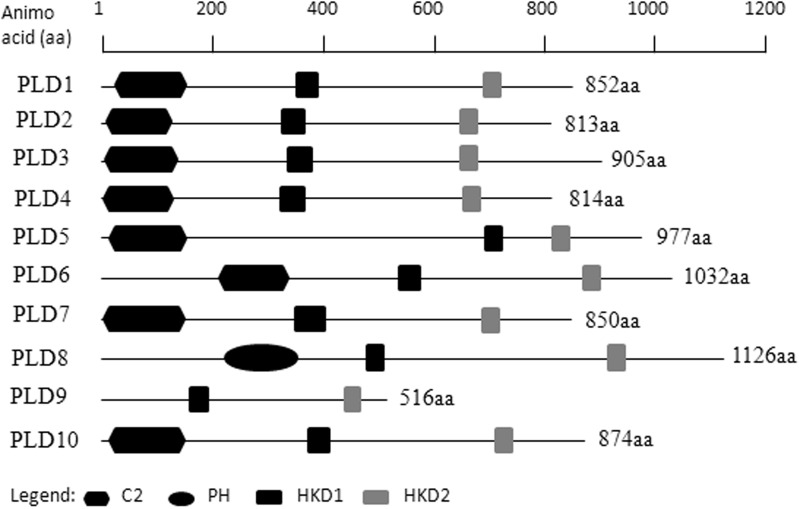
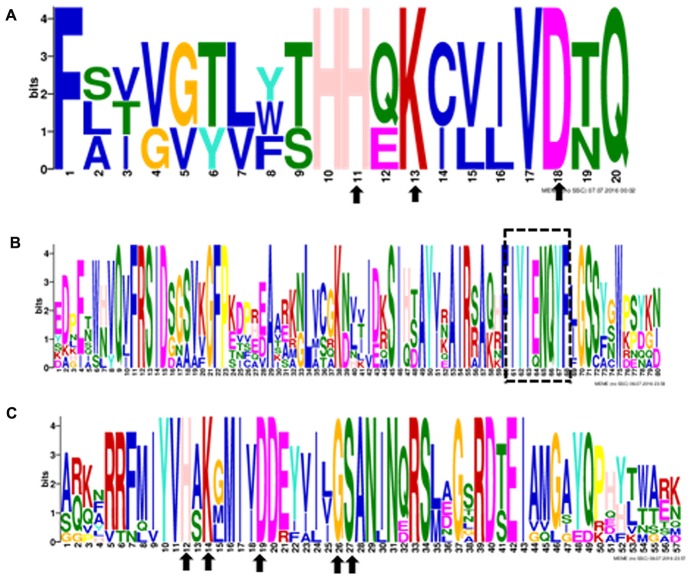
Ten putative PLD genes were identified in pineapples by searching against the pineapple genome database. These genes were located on chromosomes 1, 2, 3, 5, 7, 8, 11, 18, 19, and 25, and their temporary names were designated as AcPLD1–AcPLD10 based on their order on the pineapple chromosome from 1 to 25 (Table 1). AcPLD1–AcPLD10 were predicted to encode proteins of 852, 813, 905, 814, 977, 1032, 850, 1126, 516, and 874 amino acids, respectively, with calculated molecular weights of 96.6, 91.2, 101.7, 92.4, 111.0, 114.5, 96.3, 127.8, 57.3, and 98.7 kDa, respectively, and pI values of 8.00, 5.88, 6.57, 5.80, 6.73, 6.91, 6.78, 6.50, 7.15, and 6.79, respectively. Alignment of the full-length deduced proteins of the AcPLDs clearly showed that the proteins contained two conserved HKD domains (HKD1 and HKD2) in their sequences. HKD1 regions displayed more diversity than HKD2 regions (Figures 2, 3). The two HKD domains were separated by 320 amino acids, except for PLD3, PLD5, PLD8, and PLD9, which demonstrated longer or shorter spacing sequences. The “IYIENQYF” motif, which was found among the 10 pineapple PLDs, with the exception of PLD9, was the most conserved domain with the least diversity. The C2 domain was present in pineapple PLDs, except PLD8 and PLD9. The PH domain was present in pineapple PLD8 but not in other pineapple PLDs (Figures 2, 3 and Tables 1, 2). Alignment analysis indicated that the similarities of the deduced amino acids of AcPLDs varied from 10% (AcPLD4 and AcPLD9; AcPLD8, and AcPLD9) to 73% (AcPLD5 and AcPLD10; Table 3).
Table 1.
The information of AcPLD gene family.
| Name | Gene ID | Chr. | Gene locus | ORF (AA) | HKD1//HKD2 | Transmembrane helix number |
|---|---|---|---|---|---|---|
| PLD1 | Aco018042 | 1 | 20111511-20124381 | 852 | +/320aa/+ | 0 |
| PLD2 | Aco016943 | 2 | 1792303-1797314 | 813 | +/305aa/+ | 1 |
| PLD3 | Aco011986 | 3 | 14132196-14147669 | 905 | +/387aa/+ | 0 |
| PLD4 | Aco014266 | 5 | 108322-114777 | 814 | +/310aa/+ | 0 |
| PLD5 | Aco019548 | 7 | 11601803-11612907 | 977 | +/105aa/+ | 1 |
| PLD6 | Aco013911 | 8 | 9807533-9814878 | 1032 | +/315aa/+ | 1 |
| PLD7 | Aco016554 | 11 | 37825-52472 | 850 | +/320aa/+ | 0 |
| PLD8 | Aco001838 | 18 | 7525795-7540516 | 1126 | +/419aa/+ | 2 |
| PLD9 | Aco008209 | 19 | 8526583-8530850 | 516 | +/262aa/+ | 0 |
| PLD10 | Aco013079 | 25 | 818670-824553 | 874 | +/322aa/+ | 1 |
FIGURE 2.
Structural comparisons of pineapple PLDs domains. Protein structures and domains of pineapple PLD family members PLD1-10. Top line represents amino acid (aa) position. C2, protein kinase C-conserved 2 domain; PH, pleckstrin homology domain; HKD1, HKD1 domain; HKD2, HKD2 domain.
FIGURE 3.
The conserved domains in 10 pineapple PLD proteins. Numbers on the x-axis represents the sequence compositions of each domain. The y-axis represents the information content measured in bits. (A) Alignment of the HKD1 domain. (B) Alignment of the “IYIENQYF” domain. (C) Alignment oftheHKD2 domain.
Table 2.
The structural comparisons of AcPLD proteins containing HKD1, ‘IYIENQYF,’ and HKD2 domains.
| Name | HKD1 | IYIENQYF | HKD2 |
|---|---|---|---|
| PLD1 | HQKCVIMDTQAGGNNRKI | IYIENQYF | HSKGMIVDDEYVIIGSAN |
| PLD2 | HQKTVTLDAPASD—GSSS | IYIQNQYF | HAKLMIVDDEYVIVGSAN |
| PLD3 | HQKCVIVDTQLDTQLSQS | IYIENQYF | HAKGMIIDDEYVILGSAN |
| PLD4 | HQKIVVVDHEMPNEGSQQ | IYIENQYF | HAKMMIVDDEYIIVGSAN |
| PLD5 | HQKCLLVDTQASGNDRKI | IYIENQYF | HAKGMIVDDEYVILGSAN |
| PLD6 | HQKTVIMDADAGHGRRK | IYIENQYF | HSKGMIVDDEFVILGSAN |
| PLD7 | HQKTVIVDYEVPAAGNKK | IYIENQYF | HAKMMIVDDEYIIIGSAN |
| PLD8 | HEKIVIVDNQVCYIGGLD | IYIENQFF | HSKLMIIDDRIALVGSAN |
| PLD9 | HAKVWISDRNSLYIGSAN | — — — — | HGKYAVSDVRAHIGTSN |
| PLD10 | HQKCLLVDTQASRNNRKI | IYIENQYF | HAKGMIIDDEYVILGSAN |
Table 3.
Amino acid sequence comparison between the predicted the full-length phospholipase D (PLD) cDNAs.
| Amino acid similarity (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PLD2 | PLD3 | PLD4 | PLD5 | PLD6 | PLD7 | PLD8 | PLD9 | PLD10 | |
| PLD1 | 36 | 53 | 40 | 59 | 52 | 35 | 13 | 12 | 64 |
| PLD2 | 35 | 48 | 37 | 39 | 45 | 14 | 11 | 33 | |
| PLD3 | 37 | 49 | 39 | 32 | 14 | 12 | 57 | ||
| PLD4 | 41 | 42 | 68 | 14 | 10 | 35 | |||
| PLD5 | 43 | 36 | 12 | 11 | 73 | ||||
| PLD6 | 36 | 12 | 11 | 43 | |||||
| PLD7 | 14 | 11 | 37 | ||||||
| PLD8 | 10 | 14 | |||||||
| PLD9 | 11 | ||||||||
To examine the phylogenetic relationship between the AcPLD proteins and other plant species, a phylogenetic tree was constructed on the basis of their translated amino acid sequences. The whole plant PLD proteins could be grouped into six types: α, β, γ, δ, 𝜀, and ζ (Qin and Wang, 2002). Based on the tree, the 10 PLD genes in pineapple were clustered into five types. AcPLD2, AcPLD4, and AcPLD7 were clustered to α type, and AcPLD6 was clustered to β or γ type. AcPLD1, AcPLD3, AcPLD5, and AcPLD10 were δ type, whereas AcPLD8 and AcPLD9 were ζ type (Figure 4).
FIGURE 4.
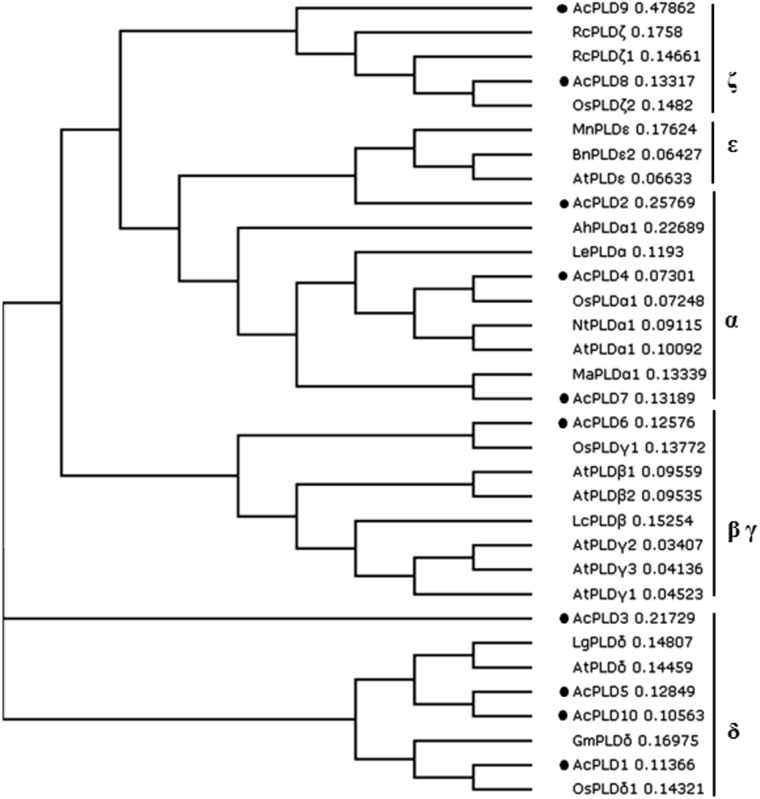
Phylogenetic tree illustrated the genetic relationships between 10 pineapple PLDs (marked with black solid circles) and the other PLDs of 23 species. Sequence name and GenBank accession number in the figure are shown as follows: LgPLDδ (JF791814.1), OsPLDαl (NM_001048688.1), NtPLDα1 (XM_009627397.1), LcPLDβ (HQ833032.1), AhPLDα1 (AB232323.1), MaPLDα1 (XM_009403448.1), AtPLDα1 (NM_112443.2), LePLDα (NM_001302895.2), GmPLDδ (XM_003532746.2), RcPLDζ (XM_002512424.1), AtPLDβ1 (NP_565963.2), AtPLDγ1 (NP_192922.1), AtPLDγ2 (NP_849539.1), BnPLD𝜀2 (XP_013643667.1), AtPLDγ3 (NP_192921.1), AtPLDδ (NP_849501.1), AtPLD𝜀 (NP_175914.1), AtPLDβ2 (NP_567160.1), OsPLDδ1 (XP_015647989.1), OsPLDζ2 (XP_015622124.1), OsPLDγ1 (XP_015614421.1). RcPLDζl (XP_015573381.1), MnPLD𝜀 (XP_010106671.1).
To investigate the potential function of the AcPLD proteins, the transmembrane helix number was analyzed via topology prediction using HMMTOP online tools. The resulting data showed that zero, one and two transmembranes existed in the associated AcPLDs (Table 1).
These data suggested that AcPLD1–AcPLD10 exhibited great structural diversity and possibly exerted different physiological functions.
PLD Gene Expression in Pineapple during IB
To understand the possible role of AcPLD1–AcPLD10 in pineapple during IB, the expression patterns of AcPLD1–AcPLD10 in pulp of fruits treated with hexaldehyde were investigated by qRT-PCR. As shown in Table 4, among the 10 AcPLD genes, AcPLD2 (1.6- to 5.7-fold) was strongly induced after hexaldehyde treatment. By contrast, the transcript level of AcPLD9 (1.9- to 4.0-fold) decreased following hexaldehyde treatment. AcPLD7 and AcPLD8 showed similar expression patterns, and they decreased by 1.3- to 2.5-fold and 1.1- to 2.3-fold in treated fruits, respectively, and did not substantially change between the treated and non-treated fruits. In addition, AcPLD1 and AcPLD3 transcripts remained nearly unchanged, and AcPLD4-6 fluctuated for both control and treated fruits during storage.
Table 4.
qRT-PCR analysis of the 10 PLD genes in hexaldehyde-treated pineapple fruits during storage at 25°C for 12 days.
| Name | Treatment | Storage days |
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | 8 | 10 | 12 | ||
| PLD1 | Control | 1.00 ± 0.05 | 1.19 ± 0.04 | 0.54 ± 0.024 | 0.81 ± 0.065 | 0.48 ± 0.032 | 0.37 ± 0.021 | 0.42 ± 0.021* |
| Hexaldehyde | 1.00 ± 0.04 | 1.20 ± 0.032 | 0.59 ± 0.024 | 0.83 ± 0.025 | 0.46 ± 0.011 | 0.36 ± 0.021 | 0.57 ± 0.018 | |
| PLD2 | Control | 1.00 ± 0.37 | 0.91 ± 0.19* | 0.77 ± 0.15* | 0.79 ± 0.13* | 1.32 ± 0.19* | 2.00 ± 0.088* | 1.98 ± 0.25* |
| Hexaldehyde | 1.00 ± 0.073 | 2.02 ± 0.32 | 2.59 ± 0.18 | 2.49 ± 0.15 | 3.19 ± 0.15 | 3.28 ± 0.15 | 11.29 ± 0.27 | |
| PLD3 | Control | 1.00 ± 0.05 | 1.12 ± 0.15 | 1.18 ± 0.099 | 0.94 ± 0.033 | 1.17 ± 0.018 | 0.58 ± 0.058 | 0.54 ± 0.21 |
| Hexaldehyde | 1.00 ± 0.04 | 1.20 ± 0.06 | 1.09 ± 0.035 | 1.03 ± 0.078 | 1.11 ± 0.095 | 0.62 ± 0.027 | 0.57 ± 0.035 | |
| PLD4 | Control | 1.00 ± 0.039 | 0.63 ± 0.084 | 0.60 ± 0.021* | 0.16 ± 0.038 | 0.17 ± 0.048 | 0.34 ± 0.015 | 0.35 ± 0.036 |
| Hexaldehyde | 1.00 ± 0.06 | 0.22 ± 0.028* | 0.73 ± 0.049 | 0.14 ± 0.021 | 0.18 ± 0.012 | 0.10 ± 0.046* | 0.26 ± 0.013* | |
| PLD5 | Control | 1.00 ± 0.021 | 0.92 ± 0.042 | 1.15 ± 0.01 | 1.13 ± 0.008 | 1.12 ± 0.007 | 0.74 ± 0.051* | 0.72 ± 0.042 |
| Hexaldehyde | 1.00 ± 0.034 | 0.88 ± 0.035 | 1.16 ± 0.008 | 1.03 ± 0.041* | 1.06 ± 0.01 | 1.01 ± 0.006 | 0.78 ± 0.042 | |
| PLD6 | Control | 1.00 ± 0.15 | 1.80 ± 0.19* | 3.89 ± 0.19 | 4.39 ± 0.28* | 3.67 ± 0.15 | 4.55 ± 0.26 | 4.32 ± 0.44 |
| Hexaldehyde | 1.00 ± 0.14 | 2.11 ± 0.16 | 2.63 ± 0.55* | 5.78 ± 0.25 | 3.53 ± 0.16 | 1.64 ± 0.63* | 2.89 ± 0.17* | |
| PLD7 | Control | 1.00 ± 0.088 | 1.79 ± 0.12* | 4.59 ± 0.34 | 4.28 ± 0.28 | 2.37 ± 0.15 | 1.97 ± 0.12 | 3.05 ± 0.27 |
| Hexaldehyde | 1.00 ± 0.079 | 2.23 ± 0.18 | 1.86 ± 0.15* | 3.19 ± 0.17* | 2.49 ± 0.14 | 2.03 ± 0.13 | 2.42 ± 0.24* | |
| PLD8 | Control | 1.00 ± 0.045 | 1.98 ± 0.086 | 3.87 ± 0.15 | 3.90 ± 0.16 | 3.12 ± 0.15 | 1.74 ± 0.15* | 2.48 ± 0.16 |
| Hexaldehyde | 1.00 ± 0.057 | 1.34 ± 0.086* | 1.66 ± 0.15* | 3.42 ± 0.20* | 3.01 ± 0.19 | 3.48 ± 0.19 | 2.52 ± 0.096 | |
| PLD9 | Control | 1.00 ± 0.19 | 1.25 ± 0.049 | 2.05 ± 0.14 | 2.36 ± 0.038 | 1.89 ± 0.18 | 1.04 ± 0.16 | 1.60 ± 0.12 |
| Hexaldehyde | 1.00 ± 0.054 | 0.49 ± 0.024* | 0.51 ± 0.042* | 0.67 ± 0.051* | 0.80 ± 0.067* | 0.55 ± 0.042* | 0.53 ± 0.025* | |
| PLD10 | Control | 1.00 ± 0.031 | 1.49 ± 0.061 | 2.07 ± 0.14 | 4.21 ± 0.15 | 1.33 ± 0.069* | 1.88 ± 0.14 | 2.15 ± 0.12 |
| Hexaldehyde | 1.00 ± 0.18 | 1.19 ± 0.14* | 0.87 ± 0.038* | 2.38 ± 0.087* | 1.83 ± 0.041 | 1.65 ± 0.079 | 1.94 ± 0.057 | |
∗Significant difference between control and hexaldehyde treatment at the P ≤ 0.05 level by t-test.
Subcellular Localization of AcPLD2 and AcPLD9 Proteins
AcPLD2 and AcPLD9 were selected for subcellular localization analysis due to their gene expression patterns with obvious characteristics during IB. To detect the potential role of the two proteins (AcPLD2 and AcPLD9), we assayed their localizations in the cell. The results indicated that the fluorescent signals from the AcPLD2-GFP and AcPLD9-GFP constructs were observed in the plasma membrane of tobacco leaf epidermal cells, whereas the fluorescence from the GFP control was distributed within the cells (Figure 5).
FIGURE 5.
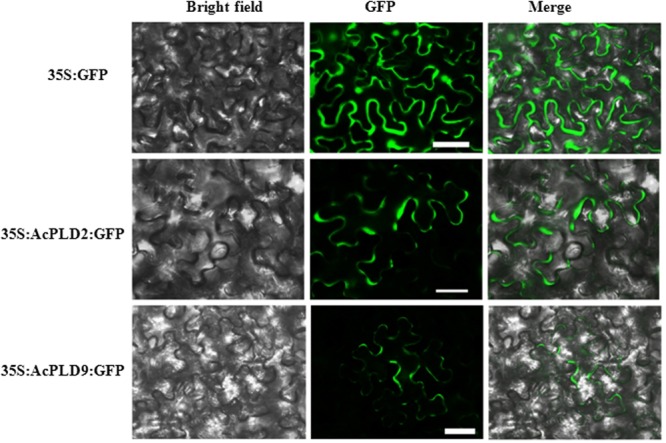
Subcellular localization of AcPLD2 andAcPLD9 in tobacco (N. benthamiana) leaf epidermal cells. Tobacco epidermal cells were transiently transformed with constructs containing either fusion plasmids (AcPLD2-GFP and AcPLD9-GFP) or control (GFP alone) via Agrobacterium inoculation, GFP fluorescence was detected 24 h after inoculation. Bar = 50 μrn.
Contents of H2O2 and Enzymes Related to H2O2
To further investigate the potential regulatory mechanism of hexaldehyde treatment alleviating IB, we analyzed the contents of H2O2 and enzymes related to H2O2. As shown in Table 5, the SOD content was decreased before the eighth day of storage and then increased in the treated fruits compared with the control samples. The contents of CAT, NADPH, and APX abundances showed similar patterns; they increased and peaked after 6 days of storage and then decreased in treated and non-treated fruits. However, the CAT, NADPH, and APX levels were suppressed at day 2, and their contents were induced in treated fruits than in non-treated samples. The H2O2 content clearly fluctuated after hexaldehyde treatment during storage, increased at days 4 and 6, decreased at day 8, and increased again at day 12 in treated fruits. A significant difference was observed between the treated and non-treated fruits.
Table 5.
Changes in SOD, CAT, NADPH, APX, H2O2, and PLD contents (ng.mg-1protein) in hexaldehyde-treated pineapple fruits during storage at 25°C for 12 days.
| Name | Treatment | Storage days |
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 6 | 8 | 10 | 12 | ||
| SOD | Control | 0.557 ± 0.0012 | 0.389 ± 0.0053 | 0.307 ± 0.0069* | 0.774 ± 0.019 | 0.602 ± 0.028 | 0.239 ± 0.0019* | 0.223 ± 0.011* |
| Hexaldehyde | 0.557 ± 0.0012 | 0.346 ± 0.0039* | 0.510 ± 0.021 | 0.497 ± 0.0049* | 0.498 ± 0.0035* | 0.854 ± 0.034 | 0.445 ± 0.015 | |
| CAT | Control | 2.714 ± 0.108 | 3.735 ± 0.174 | 2.591 ± 0.0059 | 5.139 ± 0.076* | 2.998 ± 0.188* | 2.502 ± 0.088* | 2.409 ± 0.309* |
| Hexaldehyde | 2.714 ± 0.108 | 1.633 ± 0.122* | 2.373 ± 0.384 | 6.145 ± 0.073 | 3.891 ± 0.0068 | 4.059 ± 0.250 | 3.188 ± 0.336 | |
| NADPH | Control | 0.338 ± 0.0287 | 0.326 ± 0.0133 | 0.207 ± 0.0032* | 0.483 ± 0.0228 | 0.399 ± 0.0466 | 0.236 ± 0.0206* | 0.159 ± 0.0044* |
| Hexaldehyde | 0.338 ± 0.0287 | 0.166 ± 0.0287* | 0.398 ± 0.0223 | 0.494 ± 0.0509 | 0.417 ± 0.0117 | 0.369 ± 0.0367 | 0.292 ± 0.0376 | |
| APX | Control | 8.404 ± 0.186 | 7.343 ± 0.0698 | 4.843 ± 0.0415* | 11.452 ± 0.497 | 5.487 ± 0.585* | 3.901 ± 0.259* | 4.039 ± 0.258 |
| Hexaldehyde | 8.404 ± 0.186 | 4.897 ± 0.182* | 6.614 ± 0.426 | 10.750 ± 0.378 | 7.628 ± 0.259 | 7.607 ± 0.284 | 3.791 ± 0.186 | |
| H2O2 | Control | 0.0089 ± 0.0004 | 0.010 ± 0.00011 | 0.007 ± 0.0003* | 0.010 ± 0.0063* | 0.011 ± 0.00065 | 0.0099 ± 0.0005 | 0.005 ± 0.0002* |
| Hexaldehyde | 0.0089 ± 0.0004 | 0.0010 ± 0.0002 | 0.0099 ± 0.0001 | 0.013 ± 0.0044 | 0.008 ± 0.0005* | 0.001 ± 0.0006 | 0.0082 ± 0.0004 | |
| PLD | Control | 0.043 ± 0.00026 | 0.025 ± 0.00087 | 0.020 ± 0.0009* | 0.047 ± 0.0023 | 0.041 ± 0.00019 | 0.030 ± 0.0005* | 0.032 ± 0.0001* |
| Hexaldehyde | 0.043 ± 0.00026 | 0.026 ± 0.00021 | 0.044 ± 0.00019 | 0.046 ± 0.0068 | 0.037 ± 0.0007* | 0.037 ± 0.0003 | 0.045 ± 0.00035 | |
∗Significant difference between control and hexaldehyde treatment at the P ≤ 0.05 level by t-test.
PLD Content
Hexaldehyde-treated fruits showed slightly higher PLD contents than control samples on day 2. However, on day 4, the PLD content was significantly higher in the former than in the latter. Hexaldehyde-treated fruits had lower PLD content than control samples, and treated fruits had relatively higher PLD content than non-treated fruits. Significant differences were noted between the groups (Table 5).
Discussion
Phospholipase D is an important signaling enzyme that has been identified in various species. A total of 12, 17, 11, and 18 PLD members have been characterized in Arabidopsis (Qin and Wang, 2002), in rice (Oryza sativa) (Li et al., 2007), grape (Vitis vinifera), and poplar (Populus trichocarpa) (Liu et al., 2010) genomes, respectively. Genome-wide identification of PLD genes in pineapples has not been reported. The recent completion of pineapple genome sequencing allows us to identify all PLD protein coding genes in pineapples via homology analyses. In the present study, 10 putative proteins in the pineapple genome database exhibited significant sequence identity with PLD homolog upon comparison with the NCBI data bank. We performed comprehensive analyses of the 10 AcPLD proteins with 23 other species such as rice and Arabidopsis of PLD protein for reference. Phylogenetic analysis indicated that the 10 identified AcPLD proteins could be classified into five major orthologous types (α, β, γ, δ, and ζ). The results of our study were inconsistent with previous reports on rice (α, β, κ, δ, φ, and ζ) (Li et al., 2007) and Arabidopsis (α, β, γ, δ, 𝜀, and ζ) (Pinosa et al., 2013) based on sequence similarity, thereby indicating the evolution of PLDs in different species. Phylogenetic analysis also revealed the evolutionary relationships among the AcPLD proteins, and the δ type contained the largest number (4) of PLD protein family members.
The involvement of the PLD gene in various biological processes including seed germination (Li et al., 2007), cold acclimation (Kargiotidou et al., 2010), resistance against pathogens (Zhao et al., 2013), and cold stress responses (Margutti et al., 2017), has been reported. In this study, the AcPLDs exhibited differential expression patterns during IB of pineapple. Among the 10 AcPLD genes, AcPLD6, AcPLD7, AcPLD8, AcPLD9, and AcPLD10 were induced prior to IB (6 days). Furthermore, AcPLD9 was downregulated by hexaldehyde treatment, which implied that AcPLD9 protein functioned as a positive regulator during IB. Our results were similar to the findings a previous report on PLD in Arabidopsis that showed a positive effect on pathogen defense (Pinosa et al., 2013). Notably, AcPLD2 was upregulated in the presence of hexaldehyde, thereby indicating that AcPLD2 protein could function as a repressor in response to hexaldehyde treatment in pineapple. Similarly, Yamaguchi et al. (2009) demonstrated that OsPLDβ1 negatively regulates resistance against pathogens. PLD proteins are suggested to act as a membrane protein (Pinosa et al., 2013) and most isolated PLD proteins are localized in the plasma membrane (Wang and Wang, 2001; Pinosa et al., 2013). Further subcellular localization analyses demonstrated that AcPLD2 and AcPLD9 proteins were localized in the membrane. Thus, the two proteins may function within the plasma membrane.
A growing number of studies have suggested that PLD plays vital roles in response to environmental stresses in plants (Li et al., 2004; Wang, 2005; Liu et al., 2013). Activated PLD can hydrolyze phosphatidylcholine to produce PA, which stimulates the NADPH oxidase complex to generate H2O2 (Stahelin et al., 2003). Additionally, positive feedback exists between PLD and H2O2 (Xiao et al., 2005). However, little is known about the relationship between PLD and H2O2 during IB of pineapple. In this study, we showed that the production patterns of PLD and H2O2 were similar. Further hexaldehyde treatment suppressed PLD and H2O2 production in pineapple during late storage. These similarities suggested that PLD may inhibit H2O2 production. Similarly, PLD activity could specifically suppress H2O2-induced apoptosis in rats (Lee et al., 2000) and cell death in Arabidopsis (Zhang et al., 2003). Stahelin et al. (2003) reported a similar pattern of NADPH and H2O2 changes, thereby suggesting that NADPH may be a key enzyme in the induction of production H2O2 production. In view of the present observations, we propose that hexaldehyde treatment resulted in decreased PLD gene expression and diminished PLD enzyme abundance, decreasing H2O2 generation. However, our findings require verification.
In summary, a total of 10 putative PLD gene family members were identified in the pineapple genome. The 10 PLD genes were characterized based on their sequence identity, gene structure, conserved domains, phylogenetic analysis and gene expression profiles during IB by qRT-PCR. Data from this current study will assist in identifying AcPLD genes and provide a foundation for further study on the biological functions of PLD proteins in pineapples.
Author Contributions
KH carried out the experiment. LZ participated in the research design, analyzing the data and drafting the manuscript, and initiated the project, designed the research framework, coordinated the research and reviewed the manuscript. RZ, BH, KS, and ZJ participated in data analysis and supported technically. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by Natural Science Foundation of Hainan Province (No. 20153043) and Central Public-interest Scientific Institution Basal Research Program for Scientific Research Innovation Team (No. 1630062017032; No. 1630062017008).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00913/full#supplementary-material
References
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Comoglio Y., Levitz J., Kienzler M. A., Lesage F., Isacoff E. Y., Sandoz G. (2014). Phospholipase D2 specifically regulates TREK potassium channels via direct interaction and local production of phosphatidic acid. Proc. Natl. Acad. Sci. U.S.A. 111 13547–13552. 10.1073/pnas.1407160111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2014). Food and Agriculture Organization of the United Nations. Available at: http://www.fao.org/faostat/en/#data/QC [Google Scholar]
- Guo L., Wang X. M. (2012). Crosstalk between phospholipase D and sphingosine kinase in plant stress signaling. Front. Plant Sci. 3:51 10.3389/fpls.2012.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K. Q., Gong D. Q., Zhang L. B., Hu H. G., Jia Z. W., Gu H., et al. (2016). Transcriptome characterization and expression profiles of the related defense genes in postharvest mango fruit against Colletotrichum gloeosporioides. Gene 576 275–283. 10.1016/j.gene.2015.10.041 [DOI] [PubMed] [Google Scholar]
- Hu H. G., Li X. P., Dong C., Chen W. X. (2012). Effects of wax treatment on the physiology and cellular structure of harvested pineapple during cold storage. J. Agric. Food Chem. 60 6613–6619. 10.1021/jf204962z [DOI] [PubMed] [Google Scholar]
- Janda M., Šašek V., Chmelařová H., Andrejch J., Nováková M., Hajšlová J., et al. (2015). Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front. Plant Sci 6:59 10.3389/fpls.2015.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. H., Lee C. S., Hwang D., Ryu S. H. (2012). Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lip. Res. 51 71–81. 10.1016/j.plipres.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Kargiotidou A., Kappas I., Tsaftaris A., Galanopoulou D., Farmaki T. (2010). Cold acclimation and low temperature resistance in cotton: gossypium hirsutum phospholipase Da isoforms are differentially regulated by temperature and light. J. Exp.Bot. 61 2991–3002. 10.1093/jxb/erq124 [DOI] [PubMed] [Google Scholar]
- Kopka J., Pical C., Hetherington A. M., Muller-Rober B. (1998). Ca2+/phospholipid-binding (C2) domain in multiple plant proteins: novel components of the calcium-sensing apparatus. Plant Mol. Biol. 129 908–925. [DOI] [PubMed] [Google Scholar]
- Lee S. D., Lee B. D., Han J. M., Kim J. H., Kim Y., Suh P. G., et al. (2000). Phospholipase D2 activity suppresses hydrogen peroxide-induced apoptosis in pc12 cells. J. Neurochem. 75 1503–1509. 10.1046/j.1471-4159.2000.0751053.x [DOI] [PubMed] [Google Scholar]
- Lein W., Saalbach G. (2001). Cloning and direct G-protein regulation of phospholipase D from tobacco. Biochim. Biophys. Acta. 1530 172–183. 10.1016/S1388-1981(00)00182-7 [DOI] [PubMed] [Google Scholar]
- Li G., Lin F., Xue H. W. (2007). Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLDβ1 in seed generation. Cell Res. 17 881–894. 10.1038/cr.2007.77 [DOI] [PubMed] [Google Scholar]
- Li G., Xue H. W. (2007). Arabidopsis PLDζ2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19 281–295. 10.1105/tpc.106.041426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li J. M., Sun J., Li C. B., Sheng J. F., Zheng F. J., et al. (2015). Effects of 2-butanol on quality and physiological characteristics of longan fruit stored at ambient temperature. Postharvest Biol. Technol. 101 96–102. 10.1016/j.postharvbio.2014.12.002 [DOI] [Google Scholar]
- Li W., Li M., Zhang W., Welti R., Wang X. (2004). The plasma memebrane-bound phospholipase D delta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22 427–433. 10.1038/nbt949 [DOI] [PubMed] [Google Scholar]
- Liu J., Ding P., Sun T., Nitta Y., Dong O., Huang X., et al. (2013). Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 161 2146–2158. 10.1104/pp.112.212431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang C., Yang Y., Hu X. (2010). Genome-wide and molecular evolution analysis of the phospholipase D gene family in poplar and grape. BMC Plant Biol. 10:117 10.1186/1471-2229-10-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S. P., Bahn S. C., Qu G., Qin H. Y., Hong Y., Xu Q. P., et al. (2012). Increased expression of phospholipase Dα1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol. J. 11 380–389. 10.1111/pbi.12028 [DOI] [PubMed] [Google Scholar]
- Lu S. P., Yao S. B., Wang G. L., Guo L., Zhou Y. M., Hong Y. Y., et al. (2016). Phospholipase D𝜀 enhances Braasca napus growth and seed production in response to nitrogen availability. Plant Biotechnol. J. 14 926–937. 10.1111/pbi.12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margutti M. P., Reyna M., Meringer M. V., Racagni G. E., Villasuso A. L. (2017). Lipid signalling mediated by PLD/PA modulates proline and H2O2 levels in barley seedlings exposed to short- and long-term chilling stress. Plant Physiol. Biochem. 113 149–160. 10.1016/j.plaphy.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Ming R., VanBuren R., Wai C. M., Tang H. B., Schatz M. C., Bowers J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47 1435–1442. 10.1038/ng.3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimitkeatkai H., Srilaong V., Kanlayanarat S. (2006). “Effect of semi-active modified atmosphere on internal browning of cold stored pineapple,” in Proceedings of the IVth International Conference on Managing Quality in Chains, The Integrated View on Fruits and Vegetables Quality, Vol. 1 and 2 eds Purvis A. C., McGlasson W. B., Kanlayanarat S. (Leuven: International Society for Horticultural Science; ), 649–653. 10.17660/actahortic.2006.712.80 [DOI] [Google Scholar]
- Paliyath G., Yada R., Muir D. P., Pinhero R. G. (2003). Inhibition of phospholipase D: United States Patent 6514914. Available at: http://www.freepatentsonline.com/6514914.html [Google Scholar]
- Peters C., Li M., Narasimhan R., Roth M., Welti R., Wang X. (2010). Nonspecificphospholipase C NPC4 promotes responses to abscisic acid and tolerance tohyperosmotic stress in Arabidopsis. Plant Cell 22 2642–2659. 10.1105/tpc.109.071720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinosa F., Buhot N., Kwaaitaal M., Fahlberg P., Thordal-Christensen H., Ellerström M., et al. (2013). Arabidopsis phospholipase Dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 163 896–906. 10.1104/pp.113.223503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Wang X. (2002). The Arabidopsis phospholipase D family characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol. 128 1057–1068. 10.1104/pp.010928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E. C., Lomonossoff G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7 682–693. 10.1111/j.1467-7652.2009.00434.x [DOI] [PubMed] [Google Scholar]
- Selvarajah S., Bauchot A. D., John P. (2001). Internal browning in cold stored pineapples is suppressed by a postharvest application of 1-methylcyclopropene. Postharvest Biol. Technol. 23 167–170. 10.1016/S0925-5214(01)00099-0 [DOI] [Google Scholar]
- Stahelin R. V., Burian A., Bruzik K. S., Murray D., Cho W. (2003). Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J. Biol. Chem. 278 14469–14479. 10.1074/jbc.M212579200 [DOI] [PubMed] [Google Scholar]
- Teisson C. (1979). Internal browning of pineapples: I – History, II – materials and methods. Fruits 34 245–261. [Google Scholar]
- Ueki J., Morioka S., Komari T., Kumashiro T. (1995). Purification and characterization of phospholipase D (PLD) from (Oryza sativa L.) and cloning of cDNA for PLD from rice and maize (Zea mays L.). Plant Cell Physiol. 36 903–914. 10.1093/oxfordjournals.pcp.a078837 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W., Okresz L., Bogre L., Munnik T. (2004). Learning the lipid language of plant signaling. Trends. Plant Sci. 9 378–384. 10.1016/j.tplants.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Wan C. Y., Wilkin T. A. (1994). A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 223 7–12. 10.1006/abio.1994.1538 [DOI] [PubMed] [Google Scholar]
- Wang C. X., Wang X. M. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127 1102–1112. 10.1104/pp.010444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu L., Zheng L. (1994). Cloning and expression of phosphatidylcholine- hydrolyzing phospholipase D from Ricinus communis L. J. Biol. Chem. 269 20312–20317. [PubMed] [Google Scholar]
- Wang X. M. (2005). Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139 566–573. 10.1104/pp.105.068809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker B. D., Smith D. L., Green K. C. (2001). Cloning, characterization and functional expression of a phospholipase Dα cDNA from tomato fruit. Physiol. Plant. 112 87–94. 10.1034/j.1399-3054.2001.1120112.x [DOI] [PubMed] [Google Scholar]
- Wu M., Wang Q., Luo J. Y., Jiang B., Li X. Y., Chen R. K., et al. (2010). Activation of phospholipase D involved in both injury and survival in A549 alveolar epithelial cells exposed to H2O2. Toxicol. Lett. 196 168–174. 10.1016/j.toxlet.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Xiao N., Du G., Frohman M. A. (2005). Peroxiredoxin II functions as a signal terminator for H2O2-activated phospholipase D1. FEBS J. 272 3929–3937. 10.1111/j.1742-4658.2005.04809.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Kuroda M., Yamakawa H., Ashizawa T., Hirayae K., Kurimoto L., et al. (2009). Suppression of a phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 150 308–319. 10.1104/pp.108.131979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188 762–773. 10.1111/j.1469-8137.2010.03422.x [DOI] [PubMed] [Google Scholar]
- Yuan H. Y., Chen L. G., Paliyath G., Sullivan A., Muir D. P. (2005). Characterization of microsomal and mitochondrial phospholipase D activities and cloning of a phospholipase D alpha cDNA from strawberry fruits. Plant Physiol. Biochem. 31 1–8. 10.1016/j.plaphy.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Liu Y. L., He C. C., Zhu S. J. (2015). Postharvest exogenous application of abscisic acid reduces internal browning in pineapple. J. Agric. Food Chem. 63 5313–5320. 10.1021/jf506279x [DOI] [PubMed] [Google Scholar]
- Zhang W. H., Wang C. X., Qin C. B., Wood T., Olafsdottir G., Welti R., et al. (2003). The oleate-stimulated phospholipase D, PLD, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15 2285–2295. 10.1105/tpc.013961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Devaiah S. P., Wang C., Li M., Welti R., Wang X. (2013). Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol. 199 228–240. 10.1111/nph.12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhou D., Zhang Q., Zhang W. (2012). Genomic analysis of phospholipase D family and characterization of GmPLDαs in soybean (Glycine max). J. Plant. Res. 125 569–578. 10.1007/s10265-011-0468-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.