Abstract
Methyl-CpG binding protein 2 (MeCP2) has recently been characterized as an oncogene frequently amplified in several types of cancer. However, its precise role in gastric cancer (GC) and the molecular mechanism of MeCP2 regulation are still largely unknown. Here we report that MeCP2 is highly expressed in primary GC tissues and the expression level is correlated with the clinicopathologic features of GC. In our experiments, knockdown of MeCP2 inhibited tumor growth. Molecular mechanism of MeCP2 regulation was investigated using an integrated approach with combination of microarray analysis and chromatin immunoprecipitation sequencing (ChIP-Seq). The results suggest that MeCP2 binds to the methylated CpG islands of FOXF1 and MYOD1 promoters and inhibits their expression at the transcription level. Furthermore, we show that MeCP2 promotes GC cell proliferation via FOXF1-mediated Wnt5a/β-Catenin signaling pathway and suppresses apoptosis through MYOD1-mediated Caspase-3 signaling pathway. Due to its high expression level in GC and its critical function in driving GC progression, MeCP2 represents a promising therapeutic target for GC treatment.
Abbreviations: AJCC, American Joint Committee on Cancer; Aza, 5-aza-2′-deoxycytidine; ChIP-Seq, chromatin immunoprecipitation sequencing; DMSO, dimethyl sulphoxide; DTGs, direct target genes; FFPE, Formaldehyde-fixed paraffin-embedded; GC, gastric cancer; MBD, methyl-CpG-binding domain; MeCP2, Methyl-CpG binding protein 2; MT, mutation type; PI, propidium iodide; TCGA, Cancer Genome Atlas; TRD, transcriptional repression domain; TSS, transcription start site; WT, wild type
Keywords: MeCP2, Gastric cancer, FOXF1, MYOD1, Wnt5a, Caspase-3
Highlights
-
•
MeCP2 inhibits FOXF1 and MYOD1 transcription by binding to their promoter regions.
-
•
MeCP2 promotes GC cell proliferation via FOXF1-mediated Wnt/β-Catenin signaling pathway.
-
•
MeCP2 suppresses GC cell apoptosis through MYOD1-mediated Caspase-3 signaling pathway.
Gastric cancer is the fourth most common malignant cancer and the third most frequent cause of cancer-related deaths worldwide. The molecular mechanism underlying gastric carcinogenesis and progression is still unknown. Methyl-CpG binding protein 2 (MeCP2) has recently been characterized as an oncogene frequently amplified in several types of cancer. However, its precise role and the molecular mechanism of MeCP2 regulation in gastric cancer are largely unknown. Our results show that MeCP2 promotes gastric cancer cell proliferation via FOXF1-mediated Wnt5a/β-Catenin signaling pathway and suppresses cell apoptosis through MYOD1-mediated Caspase-3 signaling pathway. MeCP2 represents a promising therapeutic target for gastric cancer treatment.
1. Introduction
Gastric cancer (GC) is the fourth most common malignant cancer and the third most frequent cause of cancer-related deaths worldwide (Fock, 2014). Although the incidence of GC has decreased significantly in the past several decades, there remain approximately 723,000 GC-related deaths every year (Tan and Yeoh, 2015). This cancer is especially common in developing countries, particularly in Asia (Fock, 2014). At present, a combination of surgery, chemotherapy, and radiotherapy is used to treat patients with GC, yet a satisfactory therapeutic effect has not been achieved because it is a highly complex disease. This complexity makes it challenging to investigate the molecular mechanisms underlying gastric carcinogenesis and progression, which are multistep processes involving numerous genetic and environmental factors. Understanding the molecular regulation of GC development is crucial for GC diagnosis and treatment.
Methyl-CpG binding protein 2 (MeCP2), a member of methyl-CpG-binding domain (MBD) family, is a plentiful mammalian protein with two main domains: a MBD and a transcriptional repression domain (TRD) (Wakefield et al., 1999, Free et al., 2001, Adkins and Georgel, 2011, Vieira et al., 2015). As a key epigenetic regulator, MeCP2 regulates chromatin organization and gene transcription by binding to methylated DNA (Yasui et al., 2007, Hite et al., 2009), or gene promoters (Chahrour et al., 2008, Mellén et al., 2012). MeCP2 is a genetic cause of a variety of neurological disorders, such as Rett syndrome, and its role in neuronal systems has been well studied (Gadalla et al., 2011). It is reported to be a master regulator of gene expression. On the one hand, MeCP2 functions as a transcriptional repressor by binding methylated CpG dinucleotides and recruiting co-repressors, such as HDAC and Sin3A, to the promoter region to inhibit the expression of a variety of genes, such as BDNF and Cdkl5 (Ballas et al., 2005, Adams et al., 2007, Carouge et al., 2010). On the other hand, it acts as a transcriptional activator by binding methylated CpG islands and recruiting activators such as CREB1 (Chahrour et al., 2008, McGraw et al., 2011, Zachariah and Rastegar, 2012, Baker et al., 2013, Shin et al., 2013, Gabel et al., 2015).
The role of MeCP2 in malignancy has not been extensively researched. Emerging evidence suggests MeCP2 as a key oncogene in cancer development. It was reported a higher level of MeCP2 expression in neoplastic breast tissues than in non-neoplastic tissues (Müller et al., 2003). We previously reported an up-regulated MeCP2 expression in human hepatocellular carcinoma tissues and demonstrated the ability of MeCP2 to promote cell proliferation (Zhao et al., 2013). More recently, Neupane and colleagues identified MeCP2 as a frequently amplified oncogene in several cancer types, including breast cancer, lung cancer, cervical cancer and uterine cancer, and it is found to be able to drive breast cancer development (Neupane et al., 2016). However, the role of MeCP2 in many other types of cancer, including GC, has not been studied precisely. In particular, the molecular mechanism accounting for the ability of MeCP2 to promote tumor development remains unknown.
In the present study, we examined the expression of MeCP2 in 76 GC patients and investigated the molecular mechanism of MeCP2 in regulating GC progression. We found that MeCP2 expression was significantly up-regulated in GC and the expression level was correlated with the clinicopathologic features, and MeCP2 promoted GC cell growth. Through microarray analysis and chromatin immunoprecipitation sequencing (ChIP-Seq), we found that MeCP2 promoted GC cell proliferation by inhibiting FOXF1-mediated Wnt5a/β-Catenin signaling pathway and inhibited GC cell apoptosis by suppressing MYOD1-mediated Caspase-3 signaling pathway.
2. Materials and Methods
2.1. Specimens From Patients With GC
Formaldehyde-fixed paraffin-embedded (FFPE) GC tumor specimens and adjacent nontumor tissues were randomly collected from 76 GC patients treated at the Department of Oncology Surgery, the First Affiliated Hospital of Medical College in Xi'an Jiaotong University, PR China, between April 2012 and November 2013. None of the patients had been pretreated with radiotherapy or chemotherapy prior to surgery. Clinicopathological data such as age and gender, as well as histological data, tumor size, lymph node metastasis status, lymphatic and venous invasion status, T stage, and pTNM stage were obtained by reviewing their pathology records. Tumor stage was determined according to the American Joint Committee on Cancer (AJCC) staging criteria. The study was approved by the Ethical Committee of Xi'an Jiaotong University, and guidelines of the committee were followed. Informed consent was obtained from all patients before collection of samples.
2.2. Animals
Five-week-old male BALB/c nude mice (Central Laboratory of Animal, Xi'an Jiaotong University Health Science Center) were bred under specific pathogen-free conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University and were performed according to the institution's guidelines for the use of laboratory animals.
2.3. Cell Culture
Human GC cell lines BGC-823, AGS, MKN-45, SGC-7901, and normal human gastric epithelial cell line GES-1 were obtained from the Cell Bank (Shanghai Genechem Co., Ltd., Shanghai, China). All the cell lines have been tested and authenticated by the Cell Bank. For verification, mycoplasma tests were performed in our laboratory, and the cell morphology and behavior were proved consistent with the Cell Bank descriptions. Cells (1 × 105 cells/ml) were cultured in RPMI-1640 medium (Gibco BRL, NY, USA) supplemented with 10% fetal bovine serum (Gibco) at 37 °C in a humidified incubator containing 5% CO2.
2.4. Immunohistochemistry (IHC) and Analysis
The FFPE tissue samples, including GC patient specimens and transplantation tumor tissues, were sectioned at 4-μm thickness. The sections were deparaffinized with xylene and hydrated with graded alcohol for antibody staining. Antigen retrieval and blocking were then performed. The slides were incubated with primary antibodies against MeCP2 (Santa Cruz, CA, USA) at a dilution of 1:200, followed by incubation with secondary antibodies. Examination was performed using 3, 3′-diaminobenzidine (DAB) and hematoxylin. If the proportion of positive cells was > 50% in 5 random fields, the specimen was considered to show high MeCP2 expression.
2.5. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from the cell lines and frozen tissues was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The FFPE tissue samples (10 sections) were deparaffinized by incubating in xylene for 10 min and in 100% ethanol for 5 min, and washed with distilled water for 30 s, followed by RNA extraction using the Qiagen FFPE Rneasy Kit (Valencia, CA, USA) according to its manual. The RNA samples were measured spectrophotometrically using Nanodrop (Thermo Fisher Scientific Inc., DE, USA). cDNA was synthesized according to the manufacturer's protocol (Takara, Dalian, China). qRT-PCR was performed using the SYBR Green PCR kit (Takara Biotechnology, Takara, Dalian, China). The primers are listed in Table S1. All qRT-PCR reactions were performed in triplicate for each sample using an IQ5 Multicolor qRT-PCR Detection System (Bio-Rad, USA). β-Actin and U6 were used as control for mRNA and miRNA, respectively. The 2− ΔΔCt method was employed in the qRT-PCR analysis.
2.6. Plasmid Construction and Transient Transfection
Full-length human MeCP2 complementary DNA was cloned into pCMV2-GV146 vector. We constructed pCMV2-GV146-GFP-MeCP2 plasmid (WT), pCMV2-GV146-GFP-Mutation type 1 plasmid (MT1) and pCMV2-GV146-GFP-Mutation type 2 plasmid (MT2, Table S2). The reporter plasmid pGL3-FOXF1 containing a 217-bp fragment spanning from 86543672 to 86543888 relative to the TSS of FOXF1 promoter, placed upstream of the Firefly Luciferase reporter gene (pGL3-FOXF1-luc), the reporter plasmid pGL3-MYOD1 containing a 236-bp fragment spanning from 17742867 to 17743102 relative to the TSS of MYOD1 promoter (pGL3-MYOD1-luc) (Genechem Co. Ltd., Shanghai, China). BGC-823 and AGS cells were seeded in RPMI-1640 medium without antibiotics for 24 h. Then, pCMV2-GV146 vector, or pCMV2-GV146-MeCP2 vector was transiently transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were transfected and cultured for 48 h before performing assays.
2.7. Immunofluorescence Microscopy
BGC-823 and AGS cells were cultured on a polylysine-coated coverslip in 6-well plates. After culturing for 24 h, cells were transfected with pCMV2-GV146-GFP plasmid and pCMV2-GV146-GFP-MeCP2 plasmid, respectively, for 48 h. For immunofluorescent staining, cells were fixed with chilled methanol and acetone for 20 min and then blocked with 10% normal goat serum in PBS containing 0.3% Triton X-100 for 1 h at room temperature. The cells were incubated with the primary antibodies (GFP and MeCP2, Table S3). Negative controls lacking the primary antibody were used to eliminate nonspecific staining. Immunostained cells were visualized by indirect fluorescence under a fluorescent microscope (Olympus BX51; Olympus, Tokyo, Japan) equipped with a DP70 digital camera and the DPManager (DPController) software (Olympus).
2.8. siRNA Synthesis and Transfection
siRNAs were pre-designed for MeCP2, FOXF1, and MYOD1 gene silencing, and were synthesized by GenePharma Corporation (SGC, Shanghai, China). Scramble siRNA was used as negative control (named NC-siRNA). The sequences are listed in Table S4. After culturing BGC-823 and AGS cells for 24 h in plates, the siRNAs were transiently transfected into the cells using Lipofectamine TM-2000 (Invitrogen) according to the manufacturer's protocol.
2.9. Lentiviral Construction and Cell Transfection
MeCP2 shRNA-1 and shRNA-2 lentiviral vectors (Genechem Company Ltd.) were used to knockdown MeCP2 expression. The sequences are listed in Table S5. The negative control lentiviral vector contained non-silencing short hairpin RNA (named sh-Ctrl). BGC-823 cells were seeded in a 6-well plate and infected with 1 ml viral stock for 10 h at 37 °C, after which the medium was replaced with normal culture medium. The efficiency of MeCP2 mRNA interference was determined by qRT-PCR.
2.10. MTT Assays
BGC-823, AGS and GES-1 cells were seeded at a density of 5000 cells/well in 96-well plates. Cell viability was analyzed using MTT assays (Sigma, St. Louis, MO, USA) at 24 h, 48 h, and 72 h after transfection. At the end of the culturing period, 20 μl of MTT solution was added to each well and cells were incubated for 4 h at 37 °C. After that, supernatants were discarded and formazan crystals were dissolved in 150 μl dimethylsulfoxide (DMSO). Absorbance was measured at a wavelength of 492 nm using a multi-microplate test system (POLARstar OPTIMA, BMG Labtechnologies, Germany).
2.11. Colony Formation Assays
BGC-823, AGS and GES-1 cells were seeded at a density of 500 cells/well in 12-well plates 24 h after transfection and were cultured for 14 days. Cell colonies were fixed, stained with 0.1% crystal violet, counted, and normalized to the control. Five parallel wells were used for each assay. Results were obtained from three independent experiments.
2.12. Cell Cycle Analysis
At 48 h after transfection, cells were harvested for analysis by trypsinization. The cells were washed twice with PBS and fixed with 70% ice-cold ethanol at 4 °C overnight. After washing twice again, the cells were incubated with 0.1 mg/ml Rnase A and 0.05 mg/ml propidium iodide (PI) for 15 min at room temperature. The distribution of cell-cycle stages was examined by flow cytometry (FACSCalibur, BD Biosciences, CA, USA).
2.13. Apoptosis Analysis
BGC-823, AGS and GES-1 cells cells were harvested 48 h after transfection. The cells were washed twice with PBS and stained using the Annexin V-FITC/PI Apoptosis Detection kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Flow cytometry was performed and cell apoptosis levels were quantified by counting the number of stained cells.
2.14. Tumorigenicity Assays in Nude Mice
All in vivo experiments were approved by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University. Five-week-old male BALB/C nude mice were used to examine tumorigenicity. BGC-823 cells were infected with sh-Ctrl, MeCP2 shRNA-1, or MeCP2 shRNA-2, and were then resuspended with PBS. The cells (1 × 106) were injected subcutaneously into both posterior flanks of the mice (n1 = 5, n2 = 5). Tumor growth was analyzed using vernier calipers every 3 days for a total of 28 days. Tumor volume (V) was calculated by measuring the length (L) and width (W) of tumor and using the formula: V = (L × W2)/2. Bioluminescence imaging in vivo was obtained using the Xenogen IVIS Spectrum imaging system (Xenogen, Alameda, CA, USA). After this, tumors were removed and weighed. The tissues were frozen for qRT-PCR and Western blot, and embedded in paraffin for immunohistochemistry.
2.15. Western Blot
GC tissues, GC cells, transfected cells, and grafting tumor cells were lysed for whole-cell extracts with RIPA buffer (Cell Signaling Technology, Boston, MA) supplemented with protease inhibitors (Roche, Indianapolis, IN, USA). Mitochondrial and cytosolic proteins were isolated with the ApoAlert Cell Fractionation kit (Takara Biotechnology, Takara, Dalian, China) following the manufacturer's protocols. Nuclear proteins were extracted with the CelLytic™ NuCLEAR™ Extraction Kit (Sigma, St. Louis, MO, USA). Equal amounts of protein lysates were run on 10% SDS-PAGE gels and transferred to PVDF membranes. The membranes were blocked with 5% nonfat milk in Tris-buffered saline Tween-20 (TBST) for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C; they were then incubated with the corresponding secondary antibodies on the second day for 1 h at room temperature. After that, the membranes were incubated with ECL (Pierce, Rockford, IL, USA) for chemiluminescence detection. Luminescent signals were detected and recorded by Syngene GBox (Syngene, Cambridge, UK). The primary antibodies used are listed in Table S3.
2.16. Microarray Analysis
BGC-823 cells were transfected with sh-Ctrl or MeCP2 shRNA for 48 h. Total RNA was extracted using TRIzol reagent and purified using the PureLink™ RNA mini kit (Invitrogen, Carlsbad, CA). Global gene expression was analyzed using the Human Gene Expression 4x44K v2 Microarray Kit (Agilent, Santa Clara, CA). The extracted total RNA from BGC-823 cells was used for cDNA synthesis. The labeled cDNA was purified and hybridized to the microarray, and the arrays were then washed and stained following the manufacturer's instructions. The slides were then scanned using an Agilent DNA Microarray Scanner (Agilent, Santa Clara, CA), and microarray experiments were performed by KangChen Bio-tech (KangChen, Shanghai, China). Data were obtained using the Agilent Feature Extraction software. Volcano plot filtering (fold change ≥ 2.0; p < 0.05) was used for evaluation of significant differential expressions of mRNAs, and clustering was performed based on the different expressions of mRNAs using Cluster Treeview software from Stanford University (Stanford, CA, USA).
2.17. Chromatin Immunoprecipitation (ChIP), ChIP-Seq and ChIP-qRT-PCR
BGC-823 cells were transfected with empty plasmid, pCMV2-GV146-GFP-MeCP2 plasmid (WT), pCMV2-GV146-GFP-Mutation type 1 plasmid (MT1) and pCMV2-GV146-GFP-Mutation type 2 plasmid (MT2) and performed ChIP. ChIP was conducted as previously described (Kasowski et al., 2010). Briefly, BGC-823 cells were cross-linked with 1% formaldehyde for 15 min at room temperature and quenching was performed using glycine (125 mM). Nuclear lysates were sonicated using a cell cracker. The chromatin was sonicated into 200-bp (approx.) fragments. The lysates were divided into two portions and incubated respectively with 5 μg antibodies against MeCP2 or IgG (Abcam, Cambridge, MA, USA, Table S3) overnight at 4 °C. DNA-protein complexes were captured using Dynabeads Protein A (Invitrogen, Carlsbad, CA, USA) and eluted in TE buffer at 65 °C. Crosslinking was reversed for 8 h at 65 °C. After that, DNA was extracted using the QIA quick PCR purification kit (QIAGEN, Germany) and sequenced on Illumina HiSeq 2000 using the TruSeq Rapid SBS Kit (Illumina, USA, FC-402-4002), according to the manufacturer's instructions. The locations of ChIP enriched DNA present in the library were based on the Human Feb 2009 assembly and visualized using the genome browser of the University of California. Peak calling in the mapped ChIP-Seq data was performed with ChIP-Peak and subjected to further bioinformatics analysis. ChIP-Seq experiments were conducted by KangChen Bio-tech (KangChen, Shanghai, China), and analysis of DNA via qRT-PCR or RT-PCR was performed using gene-specific primers (Table S6).
2.18. Luciferase Reporter Assay
BGC-823 cells were seeded into 96-well culture plates, four wells per group. pGL3-MYOD1-luc and pGL3-FOXF1-luc were amplified in DH5α. The two plasmids were treated with CpG methyltransferase M.Sssl (M0226S, NEB, USA) for 48 h and were used as the pGL3-MYOD1-luc + Methylation and pGL3-FOXF1-luc + Methylation groups. The cells were transfected with pGL3-luc, pGL3-MYOD1-luc, pGL3-MYOD1-luc + Methylation, pGL3-FOXF1-luc, or pGL3-FOXF1-luc + Methylation plasmids for 48 h. After transfection (except with pGL3-luc), the cells were treated with NC-siRNA, MeCP2 siRNA-1/2, dimethyl sulphoxide (DMSO) or methylation inhibitors 5-aza-2′-deoxycytidine (Aza), respectively, for 48 h. Then, the luciferase activity per 1000 cells (trypan blue staining) was measured. A Dual-Luciferase Reporter Assay System (Promega, Madison, WI) was used to measure the experimental results.
2.19. TCF Reporter Assay
TCF luciferase construct (pTOPFLASH; 0.1 μg/well) containing TCF-binding sites, with an internal control (0.01 μg/well pRL-TK Renilla luciferase vector), were cotransfected with NC-siRNA, MeCP2 siRNA-1, MeCP2 siRNA-2, empty vector, MeCP2 overexpression vector, or MeCP2 siRNA-1 + FOXF1 siRNA-1 into BGC-823 and AGS cells in a 96-well plate. After 48 h, the cells were lysed in 1 × passive lysis buffer (Dual Luciferase kit; Promega). The cell lysates were transferred into an OptiPlate 96-well plate and analyzed in a 1420-Multilabel counter luminometer, VICTOR3 using the Dual-Luciferase kit (Promega). Relative TOPFLASH luciferase units were examined and normalized against Renilla luciferase activity. Five independent replicates were performed for each assay and the assay was performed at least three times.
2.20. Statistical Analysis
All experiments were performed at least in triplicate unless otherwise stated. Statistical analyses were carried out using SPSS software (Abbott Laboratories, Chicago, IL). Student's t-test was used to analyze the difference between two independent groups. Chi-square test was employed to examine the relationships between MeCP2 expression and clinicopathologic characteristics, and Pearson's correlation analyses were conducted to determine the association of MeCP2 with FOXF1 and MYOD1. Mean ± SEM are reported. p values < 0.05 were considered to indicate statistical significance.
3. Results
3.1. MeCP2 is Significantly up-regulated in GC Samples and is Correlated With the Clinicopathologic Features of GC
We examined the mRNA and protein levels of MeCP2 expression by qRT-PCR and IHC staining, respectively, in GC tissue samples and adjacent normal (nontumor) gastric tissue samples from 76 GC patients. Consistent with our previous results from 21 samples (Tong et al., 2016), the expression of MeCP2 protein was significantly higher in GC tissues than in normal gastric tissues (Fig. 1A–C). In addition, this study revealed that MeCP2 was expressed in cytoplasm and nucleus of GC cells, and MeCP2 staining was negative in lymphocytes infiltrating gastric mucosa. No significant difference was observed in MeCP2 expression between G2 and G3 cancers. The new data suggested a correlation between MeCP2 expression and clinicopathologic features. The correlations between the MeCP2 protein levels and clinicopathologic characteristics of the involved GC patients are summarized in Table S7. High MeCP2 expression was associated with poor tumor histology [well: 44.4% (16/36); moderate: 83.3% (15/18); poor: 95.5% (21/22)] (Fig. 1A and B) and tumor size [tumor size < 50 mm: 45.5% (15/33); tumor size ≥ 50 mm: 86% (37/43)] (Fig. 1C). However, the expression was not associated with age, gender, lymph node metastasis, lymphatic invasion, venous invasion, T stage, and TNM stage. The mRNA expression of MeCP2 in normal tissues gradually increased from G1 to G3, but that in GC tissues, no matter what grade, was evidently up-regulated compared with the expression in normal tissues (Fig. 1D). The Cancer Genome Atlas (TCGA) data showed MeCP2 expression was associated with poor tumor histology and T stage (Fig. 1E and F). The correlative evidence suggested that up-regulated MeCP2 expression was involved in the progression of human GC. This trend was further verified by the examination of some established GC cell lines, including MKN-45, SGC-7901, BGC-823 and AGS. The results showed that MeCP2 mRNA expression in GC cells was significantly higher than that in normal human gastric epithelial cell line (GES-1), and MeCP2 protein expressions of whole-cell and nuclear was up-regulated (Fig. 1G–I). The MeCP2 protein expressions of whole-cell and nuclear increased in 5 pairs of GC tissues compared with normal gastric tissues (Fig. 1H and I). It was also observed that the levels of Cyclin D1, Bcl-2 and Bcl-xL were up-regulated and those of active Caspase-9 and Caspase-3 were down-regulated in GC tissues (Fig. 1J).
Fig. 1.
MeCP2 overexpression is correlated with clinicopathologic features of GC. (A) MeCP2 protein expressions in various histological types of GC samples and normal tissues. (B) MeCP2 protein expression in various histological grades of GC samples, expressed in percentages. Tumor histological grade was assigned according to the AJCC criteria: grade 1 (G1), well differentiated; grade 2 (G2), moderately differentiated; and grade 3 (G3), poorly differentiated. Data are shown as mean ± SEM (p < 0.05, Chi-square test). (C) MeCP2 protein expression in various-sized tumors of GC samples, expressed in percentages. For B and C, whiskers represent the 5th and 95th percentiles. Data are shown as mean ± SEM (p < 0.01, Chi-square test). (D) MeCP2 mRNA expression in G1, G2 and G3 GC tissues versus normal tissues. Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (E) Correlation between MeCP2 expression and poor tumor histology in GC patients using data from TCGA. Data are shown as mean ± SEM (p < 0.01, Chi-square test). (F) Correlation between MeCP2 expression and T stage in GC patients using data from TCGA. Data are shown as mean ± SEM (p < 0.01, Chi-square test). (G) MeCP2 mRNA expression in GC cell lines (BGC-823, AGS, SGC-7901 and MKN-45) and normal human gastric epithelial cell line (GES-1). Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (H) MeCP2 protein expressions in GC cell BGC-823, AGS, SGC-7901, MKN-45, human gastric epithelial cell line GES-1, 5 pairs of GC tissues, and their corresponding normal counterparts in whole-cell extracts, with β-Actin as an internal control. (I) MeCP2 protein expressions in GC cell BGC-823, AGS, SGC-7901, MKN-45, GES-1, 5 pairs of GC tissues, and their corresponding normal counterparts in nuclear extracts, with Lamin A as an internal control. (J) The expressions of Cyclin D1, Bcl-2, Bcl-xL, active Caspase-9 and active Caspase-3 in GC tissues.
3.2. MeCP2 Promotes GC Cell Proliferation in vitro and in vivo
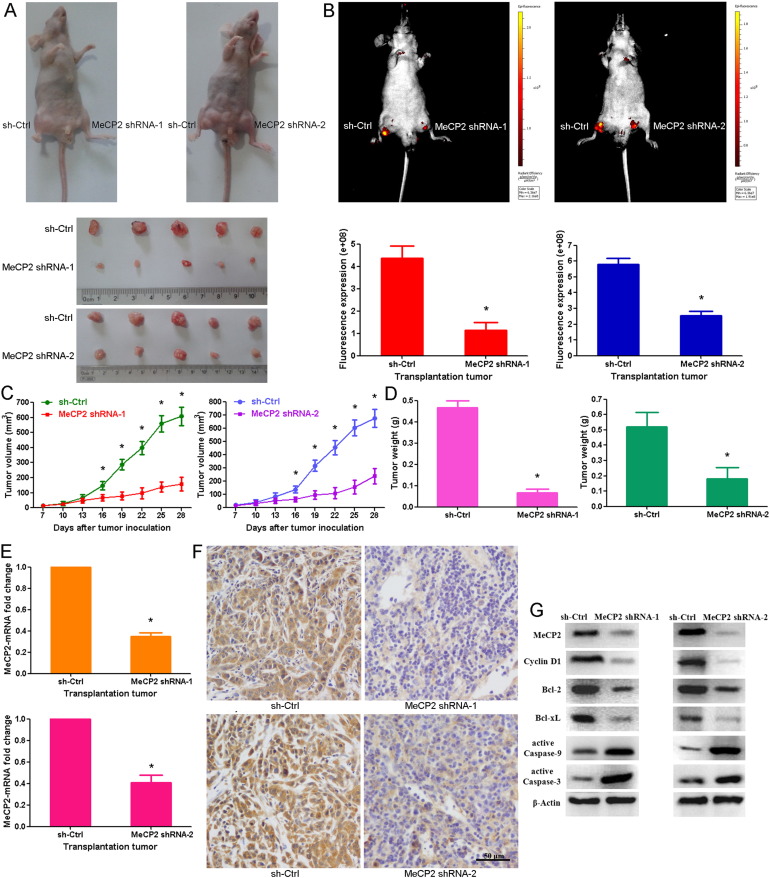
To examine the role of MeCP2 in GC progression in vivo, we constructed artificial shRNAs lentiviral vector containing a selected MeCP2-targeting sequence (LV-MeCP2-shRNA-1 and LV-MeCP2-shRNA-2) and generated a stable BGC-823 clone. qRT-PCR and Western blot showed that the MeCP2 shRNA-1 and shRNA-2 significantly down-regulated the MeCP2 expression at both mRNA and protein levels in BGC-823 cell (Fig. S1A and B). MeCP2 shRNAs-induced decrease of MeCP2 expression significantly inhibited GC cell proliferation, as evidenced by both cell viability and colony formation assays (Fig. S1C and D). MeCP2 shRNAs also led to an increase in cells of the G1 phase and a decrease in cells of the S phase in BGC823 cells (Fig. S1E). MeCP2 shRNA increased early and late apoptotic cells in BGC-823 (Fig. S1F). We inoculated BGC-823 cells transduced with LV-MeCP2-shRNA-1 and LV-MeCP2-shRNA-2 viruses, together with control cells transduced with a LV-sh-Ctrl vector, into nude mice and monitored tumor progression over 4 weeks. Our results showed that the tumor growth, based on both tumor weight and size, were remarkably suppressed by MeCP2 shRNA-1 and MeCP2 shRNA-2 (Fig. 2A–D). The down-regulation of MeCP2 in tumors derived from LV-MeCP2-shRNAs-transduced BGC-823 cells was confirmed at both mRNA and protein levels (Fig. 2E–G). Our data suggest that MeCP2 promotes the proliferation of GC cells, which is consistent with our previous results. Our previous study showed that MeCP2 promotes the growth of gastric cancer cells by suppressing miR-338 (Tong et al., 2016). However, tumorigenesis and progression of GC are complex and are multistep processes involving numerous genetic factors and molecular networks. Therefore, we further investigate the molecular mechanism of MeCP2 regulation in GC progression. The results showed that Cyclin D1, a G1 cell cycle regulator, was down-regulated by MeCP2 shRNA-1 and shRNA-2 in vivo. In addition, MeCP2 shRNA-1 and shRNA-2 also inhibited Bcl-2 and Bcl-xL expressions, and promoted Caspase-9 and Caspase-3 activation in xenograft tumor (Fig. 2G).
Fig. 2.
MeCP2 silencing suppresses tumor growth in vivo. (A) Gross morphology of tumors measured 28 days after being injected with either LV-MeCP2-shRNA-1, LV-MeCP2-shRNA-2 or LV-sh-Ctrl cells (n = 5). (B) Small animal imaging analysis was used to assess tumor volume in situ at day 28 during tumor development. (C) Growth curves of tumor volume generated every 3 days for 21 days. Data are shown as mean ± SEM (⁎p < 0.001, Student's t-test). (D) Tumor weight at day 28 after the initial injection. Data are shown as mean ± SEM (⁎p < 0.001, Student's t-test). (E) MeCP2 expression in tumor xenografts were quantified by qRT-PCR. Data are shown as mean ± SEM (⁎p < 0.001, Student's t-test). (F) IHC staining of MeCP2 in tumor tissues from mice injected with LV-sh-Ctrl, LV-MeCP2-shRNA-1 or LV-MeCP2-shRNA-2. (G) MeCP2 regulated the expressions of Cyclin D1, Bcl-2, Bcl-xL, active Caspase-9 and active Caspase-3 in vivo.
Next, we designed and synthesized MeCP2-targeting siRNAs and constructed a MeCP2-overexpressing plasmid. In cell lines BGC-823, AGS and GES-1, the plasmid efficiently up-regulated the MeCP2 levels (Fig. S2A–C), and the selected siRNAs efficiently down-regulated the MeCP2 expression (Fig. 2SD–E). Immunofluorescent assay showed that the exogenous level of MeCP2 increased in GFP-MeCP2-vector group compared with in GFP-vector group in both BGC-823 and AGS cells (Fig. S3A and B). Based on cell viability and colony formation, up-regulation of MeCP2 significantly promoted GES-1 cell growth (Fig. 3A and B) and down-regulation of MeCP2 resulted in suppression of BGC-823 and AGS cell growth (Fig. S4A and B). The effect of MeCP2 knockdown on cell proliferation was rescued by up-regulating MeCP2. Impact of MeCP2 expression on cell cycle was examined by flow cytometry and it was found that MeCP2 overexpression decreased G1 phase GES-1 cells and increased S phase GES-1 cells (Fig. 3C). Consistently, silencing MeCP2 induced a significant increase of G1 phase and a concomitantly remarkable decrease of S phase in both BGC-823 and AGS cells, and the effect was then rescued by MeCP2 overexpression (Fig. S4C). As for the impact of MeCP2 expression on cell apoptosis, MeCP2 overexpression significantly reduced early and late apoptotic GES-1 cells (Fig. 3D); in contrast, silencing the expression of MeCP2 induced a remarkable increase in both early and late apoptotic cells in BGC-823 and AGS, and the impact was cancelled then by MeCP2 overexpression (Fig. S4D). Further analysis revealed that MeCP2 promoted G1-S cell-cycle transition by increasing the expression of Cyclin D1 (Fig. 3E–G). It promoted key apoptosis regulators Bcl-2 and Bcl-xL, and suppressed activation of Caspase-9 and Caspase-3 (Fig. 3E–G, Fig. S4E and F). Our results suggested that MeCP2 promoted GC cell proliferation and G1-S cell cycle transition through regulating Cyclin D1, and inhibited GC cell apoptosis via modulating Caspase-3 signaling pathway.
Fig. 3.
MeCP2 promotes human gastric epithelial cell line GES-1 cell proliferation and regulates Cyclin D1 expression and Caspase-3 signaling pathway. (A) MTT assay of GES-1 cell proliferation at 24 h, 48 h, and 72 h after transfection with MeCP2 MeCP2 overexpression vector. Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (B) GES-1 cell colonies examined 12 days after transfection. (C) Flow cytometry analysis of cell cycle visualized via PI staining. The histograms showed the percentages of cells in the G1/G0, S, and G2/M phases after transfection. Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (D) Flow cytometry analysis of apoptosis visualized using Annexin-V/PI staining. The percentages of early-apoptotic and late-apoptotic cells were presented. Data are shown as mean ± SEM (⁎p < 0.05, Student's t-test). (E) MeCP2 overexpression promoted the expressions of Cyclin D1, Bcl-2 and Bcl-xL, and inhibited Caspase-9 and Caspase-3 activation in GES-1, BGC-823 and AGS cells. (F) Silencing MeCP2 down-regulated the expressions of Cyclin D1, Bcl-2 and Bcl-xL, and promoted Caspase-9 and Caspase-3 activation in BGC-823 and AGS cells.
3.3. MeCP2 Inhibits FOXF1 and MYOD1 Transcription by Binding Their Promoter
To explore how MeCP2 regulates Cyclin D1 and Caspase-3 signaling pathway, we identify the genes regulated by MeCP2. Firstly, we profiled the global gene expression in shRNA-treated cells using microarray. Our results showed that 1553 genes were significantly up-regulated and 1468 genes were significantly down-regulated after transfection with MeCP2 siRNA (fold change ≥ 2.0) (Fig. S5A). Pathway analysis was performed using the KEGG database (http://www.genome.jp/kegg/) and the top 10 up-regulated and the top 10 down-regulated pathways were showed in Fig. S5B and C. Next, we performed ChIP-Seq assay to identify MeCP2-bound genes in GC cells. A total of 8128 ChIP-Seq peaks with various fold enrichments were obtained, among which 1726 peaks exhibited over 2 folds enrichment. The number of peaks in a region from − 40 kb upstream of the transcription start site (TSS) to + 40 kb downstream of the TSS is shown in Fig. S5D. Interestingly, MeCP2 protein was found to localize around the TSS. Further analysis showed that 5.8% (220/3794) of the peaks were located in the promoter regions of genes (Fig. S5E). In order to characterize the chromosome distribution of ChIP peaks, the number of peaks in the promoter regions of genes with over 2 folds enrichment present on each chromosome was counted. The peak density appeared to have no significant difference between chromosomes. MeCP2 exhibited the highest binding strength to chromosome 19 and the lowest binding strength to chromosome Y (Fig. S5F). The GO functions of ChIP-enriched genes showed that MeCP2 regulated the pathways involved in DNA replication, nucleotide excision repair, Mucin type O-Glycan biosynthesis, and Base excision repair (Fig. S6).
In order to identify direct target genes (DTGs) of MeCP2, genes that were up-regulated or down-regulated by MeCP2 siRNA were compared with those bound by MeCP2. As a result, 23 DTGs were initially identified, in which 15 genes were up-regulated and 8 genes were down-regulated by MeCP2 siRNA. The DTGs included FOXF1, MYOD1, NFE2, ZNF329, SRRM2, PPIF, MYL5, HIST1H4H, HIST1H2AK, BDNF, FAM90A1, DEFB103A, UBC, MAPK6, ACRBP, SLC43A1, R3HDM2, PLS1, OR2A9P, GALNT12, ATHL1, GIT1 and ABAT (Fig. 4A). The sites and fold enrichment of MeCP2 binding these 23 genes are shown in Table S8. Then, we predicted CpG islands at the MeCP2 binding sites using MethPrimer and found that MeCP2 binding sites in the promoter regions of 10 genes contained CpG islands, including FOXF1, MYOD1, MAPK6, UBC, ZNF329, SRRM2, HIST1H4H, HIST1H2AK, FAM90A1 and GIT1 (Table S8). ChIP qRT-PCR showed that MeCP2 directly bound the promoters of these 10 genes (Fig. S7A). We analyzed the mRNA levels of these genes after transfection with MeCP2 siRNA or MeCP2 overexpression vector, and found that MeCP2 repressed the expressions of FOXF1, MYOD1, MAPK6, UBC, SRRM2, HIST1H4H and HIST1H2AK, and activated the expression of GIT1 in BGC-823 and AGS cells (Fig. S7B). As shown in Fig. 4B, MeCP2 bound the CpG islands of promoter regions of FOXF1 and MYOD1. ChIP RT-PCR also confirmed that MeCP2 binds the CpG islands (Fig. 4C).
Fig. 4.
MeCP2 binds the promoter regions of FOXF1 and MYOD1. (A) Changed genes after MeCP2 siRNA transfection compared with the 220 genes corresponding to the ChIP-Seq peaks located in the promoter. (B) MeCP2 binding sites relative to CpG islands location. (C) ChIP RT-PCR of FOXF1 and MYOD1 were performed with anti-MeCP2 antibody. (D) ChIP RT-PCR of FOXF1 and MYOD1 were performed with anti-GFP antibody after transfection with GFP-MeCP2 plasmid. (E) ChIP RT-PCR of FOXF1 and MYOD1 were performed with anti-GFP antibody after transfection with Ctrl (GFP plasmid), WT (GFP-MeCP2 plasmid), MT1(GFP- Mutation type 1) and MT2 (Mutation type 2). (F) BGC-823 cells were transfected with pGL3-FOXF1-luc (target sequences of promoter regions of FOXF1) and pGL3-FOXF1-luc + Methylation; luciferase activity was determined at 48 h post-transfection. Renilla luciferase served as the internal control. Data are shown as mean ± SEM (⁎p < 0.001, as compared with pGL3-luc; #p < 0.001, as compared with pGL3-MYOD1-luc; Student's t-test). (G) BGC-823 cells were transfected with pGL3-MYOD1-luc (target sequences of promoter regions of MYOD1) and pGL3-MYOD1-luc + Methylation; luciferase activity was determined at 48 h post-transfection. Data are shown as mean ± SEM (⁎p < 0.001, as compared with pGL3-luc; #p < 0.001, as compared with pGL3-MYOD1-luc; Student's t-test). (H) BGC-823 cells were treated with pGL3-FOXF1-luc, pGL3-FOXF1-luc + Methylation, MeCP2 siRNAs and methylation inhibitors 5-aza-2′-deoxycytidine (Aza); luciferase activity was determined. Data are shown as mean ± SEM (⁎p < 0.001, as compared with NC-siRNA; #p < 0.001, as compared with DMSO; Student's t-test). (I) BGC-823 cells were treated with pGL3-MYOD1-luc, pGL3-MYOD1-luc + Methylation, MeCP2 siRNAs and methylation inhibitors 5-aza-2′-deoxycytidine (Aza); luciferase activity was determined. Data are shown as mean ± SEM (⁎p < 0.01, as compared with NC-siRNA; #p < 0.001, as compared with DMSO; Student's t-test). (J) MeCP2 inhibited FOXF1 and MYOD1 protein expression in vitro and in vivo.
We constructed GFP-MeCP2 plasmid including wild type (WT) and mutation type (MT). After they were transfected into BGC-823 cells, ChIP RT-PCR was performed with anti-GFP antibody. The results showed that exogenous MeCP2 bound to the CpG islands of promoter regions of FOXF1 and MYOD1 (Fig. 4D). GFP plasmid (Ctrl), GFP-MT1 and GFP-MT2 did not bind to the islands, but GFP-WT did (Fig. 4E). Then, a promoter reporter assay was conducted to determine whether MeCP2 bound to the CpG islands of promoter regions of FOXF1 and MYOD1. The target sequences of promoter regions of FOXF1 and MYOD1 from ChIP-Seq were subcloned into downstream of the luciferase gene in the pGL3 reporter plasmid (Table S8). BGC-823 cells was transfected with each of the constructed plasmids and luciferase activity was measured at 48 h post-transfection. The data showed that luciferase activity significantly decreased in the pGL3-FOXF1-luc and pGL3-FOXF1-luc + Methylation groups as compared with in the pGL3 group, and the activity was lower in pGL3-FOXF1-luc + Methylation than in pGL3-FOXF1-luc (Fig. 4F); similarly, it declined in the pGL3-MYOD1-luc and pGL3-MYOD1-luc + Methylation groups as compared with in the pGL3 group, and was lower in pGL3-MYOD1-luc + Methylation than in pGL3-MYOD1-luc (Fig. 4G). When pGL3-FOXF1-luc plasmid was transfected, luciferase activity increased in the MeCP2 siRNA-1 and siRNA-2 groups as against the NC-siRNA group, and in the methylation inhibitors 5-aza-2′-deoxycytidine (Aza) group as against the DMSO group. Transfection with pGL3-FOXF1-luc + Methylation plasmid induced similar results, but the luciferase activity was lower than those in the pGL3-FOXF1-luc groups (Fig. 4H). When pGL3-MYOD1-luc and pGL3-MYOD1-luc + Methylation were transfected, the treated group results were similar with pGL3-FOXF1-luc (Fig. 4I). We also found that the protein levels of FOXF1 and MYOD1 increased in BGC-823 and AGS cells after transfection with MeCP2 siRNA-1 and siRNA-2, and decreased after transfection with MeCP2 overexpression vector (Fig. 4J). In xenograft tumor, the protein expressions of FOXF1 and MYOD1 were also promoted by MeCP2 shRNA-1 and shRNA-2 (Fig. 4J). These results demonstrated MeCP2 as a regulator of FOXF1 and MYOD1 in GC cells.
3.4. FOXF1 and MYOD1 are Frequently Reduced in Human GC Tissues and are Correlated With MeCP2 Expression
To further validate this finding, we examined the expressions of FOXF1 and MYOD1 in the tissue samples. The mRNA levels of FOXF1 and MYOD1 were significantly lower in G1, G2 and G3 GC tissues than in normal gastric tissues, and gradually decreased in G1, G2 and G3 normal tissues (Fig. 5A and B). There was a significant inverse correlation between MeCP2 mRNA and FOXF1 mRNA (Fig. 5C), and the extent of MeCP2 up-regulation inversely correlated with the extent of FOXF1 down-regulation (Fig. 5D). The protein levels of FOXF1 and MYOD1 were also lower in GC tissues (Fig. 5E). Taken together, our data suggested that MeCP2 repressed the expressions of FOXF1 and MYOD1 in GC by binding CpG islands at their promoters.
Fig. 5.
MeCP2 suppresses FOXF1 and MYOD1 transcription by binding their promoter regions. (A) FOXF1 mRNA expression in G1, G2 and G3 GC tissues versus normal tissues. Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (B) MYOD1 mRNA expression in G1, G2 and G3 GC tissues versus normal tissues. Data are shown as mean ± SEM (⁎p < 0.01, Student's t-test). (C) MeCP2 and FOXF1 expressions were inversely correlated. The 2− ΔΔCt values of MeCP2 and FOXF1 mRNA were subjected to a Pearson correlation analysis (r = − 0.685, n = 76, p < 0.001, Pearson's correlation). (D) The expression levels of MeCP2 and MYOD1 were inversely correlated (r = − 0.677, n = 76, p < 0.0001, Pearson's correlation). (E) Western blot of FOXF1 and MYOD1 protein expressions in GC tissues. β-Actin was used as an internal control. n = 5.
3.5. MeCP2 Promotes GC Cell Proliferation Through Regulating the FOXF1/Wnt5a/β-Catenin Signaling Pathway
To further confirm that MeCP2 promotes tumor growth by suppressing FOXF1 expression, we synthesized FOXF1 siRNA-1 and siRNA-2 for efficient knockdown of FOXF1 (Fig. S8A and B). FOXF1 siRNA-1 was co-transfected with MeCP2 siRNA-1 into BGC-823 or AGS cells. Cell viability and colony formation assays revealed that down-regulation of MeCP2 expression resulted in suppression of cell proliferation in vitro, and silencing of FOXF1 rescued the effect of MeCP2 knockdown on cell proliferation (Fig. 6A and B). Cell cycle assay also showed that silencing MeCP2 induced a significant increase of G1 phase cells and a concomitantly remarkable decrease of S phase cells in both BGC-823 and AGS cells, the cells co-transfected with MeCP2 siRNA-1 and FOXF1 siRNA-1 were able to re-enter the S-phase (Fig. 6C and S9A). We found that silencing the expression of MeCP2 induced a remarkable increase in both early and late apoptotic cells in BGC-823 and AGS cells, there were no significant differences between cells co-transfected with MeCP2 siRNA-1 and FOXF1 siRNA-1 and those transfected with MeCP2 siRNA-1 alone (Fig. 6D and S9B).
Fig. 6.
MeCP2 promotes GC cell proliferation by regulating the FOXF1/Wnt5a/β-Catenin signaling pathway. (A) MTT assay showed GC cell proliferation after co-transfection with MeCP2 siRNA-1 and FOXF1 siRNA-1 in BGC-823 and AGS cells. Data are shown as mean ± SEM (⁎p < 0.01, as compared with NC-siRNA-transfected cells; #p < 0.01, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (B) Cell colonies were examined 12 days after co-transfection with MeCP2 siRNA-1 and FOXF1 siRNA-1. (C) The percentages of cells in the G1/G0, S, and G2/M phases after co-transfection with MeCP2 siRNA-1 and FOXF1 siRNA-1 was analyzed. Data are shown as mean ± SEM (⁎p < 0.01, as compared with NC-siRNA-transfected cells; #p < 0.01, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (D) The data showed the percentages of early-apoptotic and late-apoptotic cells after co-transfection with MeCP2 siRNA-1 and FOXF1 siRNA-1. Data are shown as mean ± SEM (⁎p < 0.01, as compared with NC-siRNA-transfected cells; #p < 0.01, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (E) FOXF1/Wnt5a/β-Catenin signaling pathways were detected after co-transfection with MeCP2 siRNA-1 and FOXF1 siRNA-1. (F) MeCP2 siRNA-1 and FOXF1 siRNA-1 affects TOPFLASH TCF-reporter construct activity in BGC-823 and AGS cells. Data are shown as mean ± SEM (⁎p < 0.01, as compared with NC-siRNA-transfected cells; #p < 0.01, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3).
Pathway analysis of mRNA microarray showed that the Wnt signaling pathway is one of the most important pathways involved in GC progression (Fig. S5B). It was previously reported that FOXF1 is involved in the regulation of Wnt signaling pathway (Ormestad et al., 2006). We further performed Western blot analyses to determine the downstream regulators involved in the promotion of proliferation by MeCP2. We found that down-regulation of MeCP2 also increased BMP4 expression. Compared with that in cells transfected with MeCP2 siRNA-1 alone, the FOXF1 and BMP4 expressions were found significantly down-regulated in the co-transfection cells, whereas Wnt5a, β-Catenin, c-Myc, and cyclin D1 expressions were significantly up-regulated in whole-cells, the nuclear β-Catenin level change was consistent with the whole-cell change (Fig. 6E). In accordance with this result, the luciferase activity of TCF luciferase reporter construct TOPFLASH was significantly decreased in MeCP2 siRNA-1 group compared with NC-siRNA group, but not in MeCP2 siRNA-1 + FOXF1 siRNA-1 group (Fig. 6F). This result further confirmed that MeCP2 regulates Wnt5a/β-Catenin signaling pathway through suppressing FOXF1 expression. The protein levels of Wnt5a, β-Catenin, and c-Myc decreased in BGC-823 and AGS cells transfected with MeCP2 siRNA-1 and siRNA-2; an opposite trend was observed in these cells transfected with the MeCP2 overexpression construct; MeCP2 shRNA-1 and shRNA-2 inhibited the protein levels of Wnt5a, β-Catenin, and c-Myc in xenograft tumor (Fig. S10A). In addition, the luciferase activity significantly decreased in siRNA-1 and siRNA-2 groups, but increased in MeCP2 overexpression group (Fig. S10B). These results further verified that MeCP2 regulates Wnt5a/β-Catenin signaling pathway. Our results confirmed that MeCP2 promotes GC cell growth and G1-S cell-cycle transition by suppressing the expression of FOXF1, thereby activating Wnt5a/β-Catenin signaling pathway.
3.6. MeCP2 Suppresses GC Cell Apoptosis via Inhibiting MYOD1/Caspase-3 Signaling Pathway
To determine the role of MYOD1, we synthesized MYOD1-targeting siRNAs and found that MYOD1 siRNA-1 and siRNA-2 efficiently knocked down the expression of MYOD1 (Fig. S11A and D). Both cell viability and colony formation assays demonstrated that silencing MYOD1 attenuated the effect of MeCP2 knockdown on GC cell proliferation (Fig. 7A and B). No significant differences were observed in the distribution of cell cycle stages between cells co-transfected with MeCP2 siRNA-1 and MYOD1 siRNA-1 and those transfected with MeCP2 siRNA-1 alone (Fig. 7C and S12A). However, apoptosis levels were significantly lower in co-transfected cells with MeCP2 siRNA-1 and MYOD1 siRNA-1 than in cells transfected with MeCP2 siRNA-1 alone (Fig. 7D and S12B).
Fig. 7.
MeCP2 inhibits GC cell apoptosis through suppressing MYOD1/Caspase-3 signaling pathway. (A) MTT assay showed GC cell proliferation after co-transfection with MeCP2 siRNA-1 and MYOD1 siRNA-2 in BGC-823 and AGS cells. Data are shown as mean ± SEM (⁎p < 0.001, as compared with NC-siRNA-transfected cells; #p < 0.001, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (B) Cell colonies were examined 12 days after co-transfection. (C) Percentages of cells in the G1/G0, S, and G2/M phases after co-transfection. Data are shown as mean ± SEM (⁎p < 0.001, as compared with NC-siRNA-transfected cells; #p < 0.001, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (D) Percentages of early-apoptotic and late-apoptotic cells after co-transfection. Data are shown as mean ± SEM (⁎p < 0.001, as compared with NC-siRNA-transfected cells; #p < 0.01, as compared with MeCP2 siRNA-1-transfected cells; Student's t-test, n = 3). (E) MYOD1/Caspase signaling pathways were examined after co-transfection with MeCP2 siRNA-1 and MYOD1 siRNA-1. Mitochondrion (Mito), Cytoplasm (Cyto). (F) Proposed model for the effects of MeCP2 on GC progression. MeCP2 facilitates GC cell proliferation by binding to a methylated CpG island in the FOXF1 promoter region and suppressing FOXF1 expression, thereby further down-regulating the Wnt5a/β-Catenin signaling pathway. Meanwhile, MeCP2 inhibits apoptosis by suppressing MYOD1 expression, thereby inhibiting the Caspase-3 signaling pathway.
It was previously reported that MYOD1 regulates the Caspase-3 apoptosis signaling pathway through inhibiting Pax3 expression (Hirai et al., 2010). We analyzed major factors in the Caspase-3 apoptosis signaling pathway and found that the expressions of miR-1, miR-206, active Caspase-9 and active Caspase-3 were down-regulated after transfection with MYOD1 siRNA-1 and siRNA-2, the expressions of Pax3, Bcl-2 and Bcl-xL were up-regulated (Fig. S11B–D). The results are in accordance with previous study (Hirai et al., 2010). In addition, the expressions of MYOD1, miR-1, miR-206, active Caspase-9 and active Caspase-3 were up-regulated in MeCP2 siRNA-1 cells compared with NC-siRNA cells, the expressions of Pax3, Bcl-2 and Bcl-xL were down-regulated, but co-transfected cells with MeCP2 siRNA-1 and MYOD1 siRNA-1 rescued these factors expression level reduced or increased by MeCP2 siRNA-1 (Fig. 7E; Fig. S13A and B). The Cyt c expression was down-regulated in mitochondrion in MeCP2 siRNA-1 cells compared with NC-siRNA cells, but the Cyt c expression was up-regulated in cytoplasm; and co-transfected cells eliminated the effects of MeCP2 siRNA-1 on GC cells (Fig. 7E). These data suggested that MeCP2 suppresses GC cell apoptosis by inhibiting the expression of MYOD1, thereby inhibiting the Caspase-3 signaling pathway.
4. Discussion
As a master regulator of gene expression, MeCP2 has recently emerged as a critical oncogene that is frequently amplified in many types of cancer (Müller et al., 2003, Neupane et al., 2016). It is reported that MeCP2 regulates carcinogenesis and progression of osteosarcoma and neuroblastoma (Murphy et al., 2011, Meng et al., 2014). Knock-down of MeCP2 decreases cell proliferation in human prostate transformed cells (Babbio et al., 2012). Our previous study has demonstrated that the expression of MeCP2 is remarkably increased in hepatocellular carcinoma, which promotes hepatocellular carcinoma cell proliferation in vitro (Zhao et al., 2013). The present study aims to determine the role and the molecular mechanism of MeCP2 in GC. Our results present a strong up-regulation of MeCP2 in primary GC as well as a close association of high MeCP2 level with poor tumor histology and tumor size. These findings suggest that MeCP2 may play an importance role in gastric carcinogenesis.
Our results show that down-regulation of MeCP2 significantly inhibits GC cell proliferation and slows down tumor progression, and MeCP2 promotes cell growth by driving G1-S cell cycle transition through regulation of cyclin D1, a critical regulator governing cell cycle transition from G1 phase to S node (Wang et al., 2016). In mammalian, cell cycle regulatory gene cyclin D1 is involved in the proliferation of a range of cells. The overexpression of cyclin D1 is observed in many types of tumor tissues, and it is also an oncogene related to tumor occurrence. In addition, cell transformation requires amplification of cyclin D1. Our study also shows that MeCP2 inhibits GC cell apoptosis through up-regulation of antiapoptotic gene Bcl-2 and down-regulation of proapoptotic genes, including Bax, Caspase-9 and Caspase-3. The Bcl-2 family, an apoptosis related gene family, can be divided into antiapoptotic genes and proapoptotic genes. Bcl-2 can suppress apoptosis through forming heterodimers with Bax after inactivating it. There is a balance between Bcl-2 and Bax. An increase in Bax hurries apoptosis, but excessive Bcl-2 inhibits apoptosis. The ratio of Bax/Bcl-2 in apoptosis is considered as to play a key role in cancer progression. Caspases family is the executor in the process of cell apoptosis. Caspase-3, a critical enzyme in the apoptotic pathway, activates poly (ADP-ribose) polymerase which leads to subsequent apoptotic events, and is located downstream of a series of cascades (Zhang et al., 2010). As we know, apoptosis is the result of a series of highly regulated caspase cascades. In these cascades, Caspase-3 plays a critical role. Activated caspase-3 can shear DNA and inactivate the related protease in DNA damage repair, thus resulting in apoptosis. Taken together, our study demonstrates the mechanism that MeCP2 suppresses GC cell apoptosis by repressing Caspase-3 signaling pathway.
To reveal the molecular mechanism of MeCP2 regulation, we performed a combination of microarray analysis and ChIP-Seq, through which we identified FOXF1 and MYOD1 as MeCP2 targeting genes. We confirmed that MeCP2 binds to the methylated CpG islands in the promoter regions of both genes, leading to down-regulation of FOXF1 and MYOD1 expression. FOXF1 is a member of the forkhead transcription factor family that regulates cell proliferation and differentiation, tissue repair, and embryo early development (Ormestad et al., 2006). Recent studies have demonstrated that FOXF1 potentially acts as a tumor suppressor in breast cancer, lung cancer, and esophageal carcinoma (Lo et al., 2010, Saito et al., 2010, Dura et al., 2013). In breast cancer, it was reported that FOXF1 is epigenetically silenced and re-expression of FOXF1 suppresses the growth and tumorigenicity of breast cancer cells (Lo et al., 2010). FOXF1 functions through interaction with BMP4 which suppresses epithelial proliferation by inhibiting the Wnt5a/β-Catenin signaling pathway (Ormestad et al., 2006, Madison et al., 2009), a key pathway regulating cancer progression (Gougelet et al., 2016, Jiang et al., 2015a, Jiang et al., 2015b, Ma et al., 2016, Qi et al., 2016). MYOD1 is a transcription factor that regulates cell proliferation, differentiation, and satellite cell activation (Hirai et al., 2010) and is shown to play an important role in cancer development (Orbach, 2014, Jiang et al., 2015a, Jiang et al., 2015b, Sood et al., 2015). Recently, it is reported that MYOD1 is involved in apoptosis by regulating downstream molecules such as Bcl-2, caspase-3, and caspase-9 (Ikeda et al., 2009, Narasimhan et al., 2014). Caspases are a family of cysteine proteases that regulate apoptosis. Apoptosis signaling cascades are involved in numerous pathways in the endoplasmic reticulum, death receptors, and mitochondria. In particular, endoplasmic reticulum stress triggers a specific cascade involving caspase-9 and caspase-3 (Ma et al., 2014). Our gene knock-down and rescue studies suggested that MeCP2 promotes GC cell proliferation through FOXF1-mediated inhibition of the Wnt5a/β-Catenin signaling pathway, and inhibits GC cell apoptosis through MYOD1-mediated down-regulation of the Caspase-3 signaling pathway (see model Fig. 7F).
During the course of our study, Neupane and colleagues published their work in which they identified MeCP2 as a frequently amplified oncogene (Neupane et al., 2016). Our work is similar to theirs in that both studies demonstrate the oncogenetic function of MeCP2. Oncogenesis is a multiple-factor and -stage network regulatory process, involving in many different types of signaling pathways. Neupane and colleagues found MeCP2 activates the MAPK and PI3K signaling in breast cancer, cervical cancer and lung cancer. However, our work is different from theirs, not only because we characterized the precise role of MeCP2 in GC that has not been studied before, but, more importantly, because we revealed different molecular mechanism of MeCP2 regulation in GC.
In summary, the present study demonstrate that MeCP2 is an oncogene highly expressed in GC and that MeCP2 drives GC progression through regulation of FOXF1-mediated Wnt5a/β-Catenin signaling and MYOD1-mediated Caspase-3 signaling. Due to its high expression in GC and its critical function in promoting GC progression, MeCP2 represents a promising therapeutic target for the treatment of this disease.
Funding sources
This work was supported by the National Nature Science Foundation of China (31400730 and 81401137), China Postdoctoral Science Foundation (2013M542358), Shaanxi Province Natural Science Foundation (2014JM4122), Xi'an Jiaotong University Biomedical Sharing Platform Project (2015FWPT-14), Shaanxi Province Social Development Of Science and Technology Project (2016SF-190), and Yanan City Science and Technology Research Development Planning Project (2016KS-06).
Conflicts of interest
The authors declare no competing financial interests.
Author contributions
L.Z. and C.H. designed the experiments. D.T., N.D., J.H., X.Z., and L.W. collected clinical data and sample. L.Z., Y.L., Y.Q., L.L., M.X., B.G., N.H., Y.C., S.L., and N.L. performed the experiments. L.Z., C.H., Y.L., and J.Y. performed the statistical analysis. L.Z., C.H., and J.Z. wrote and edited the manuscript. C.H. supervised the work.
Acknowledgments
We thank all the members of the key laboratory of ministry of education of china for discussions and support and for critical reading of the manuscript. The authors also want to thank Qi Chen, a teacher of English in Xi'an Jiaotong University, for contributing to language revision of the manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.01.021.
Appendix A. Supplementary data.
Supplementary material
References
- Adams V.H., McBryant S.J., Wade P.A., Woodcock C.L., Hansen J.C. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- Adkins N.L., Georgel P.T. MeCP2: structure and function. Biochem. Cell Biol. 2011;89:1–11. doi: 10.1139/O10-112. [DOI] [PubMed] [Google Scholar]
- Babbio F., Castiglioni I., Cassina C., Gariboldi M.B., Pistore C., Magnani E., Badaracco G., Monti E., Bonapace I.M. Knock-down of methyl CpG-binding protein 2 (MeCP2) causes alterations in cell proliferation and nuclear lamins expression in mammalian cells. BMC Cell Biol. 2012;11:13–19. doi: 10.1186/1471-2121-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.A., Chen L., Wilkins A.D., Yu P., Lichtarge O., Zoghbi H.Y. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Carouge D., Host L., Aunis D., Zwiller J., Anglard P. CDKL5 is a brain MeCP2 target gene regulated by DNA methylation. Neurobiol. Dis. 2010;38:414–424. doi: 10.1016/j.nbd.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura P., van Veen E.M., Salomon J., te Morsche R.H., Roelofs H.M., Kristinsson J.O., Wobbes T., Witteman B.J., Tan A.C., Drenth J.P., Peters W.H. Barrett associated MHC and FOXF1 variants also increase esophageal carcinoma risk. Int. J. Cancer. 2013;133:1751–1755. doi: 10.1002/ijc.28160. [DOI] [PubMed] [Google Scholar]
- Fock K.M. Review article: the epidemiology and prevention of gastric cancer. Aliment. Pharmacol. Ther. 2014;40:250–260. doi: 10.1111/apt.12814. [DOI] [PubMed] [Google Scholar]
- Free A., Wakefield R.I., Smith B.O., Dryden D.T., Barlow P.N., Bird A.P. DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., Greenberg M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla K.K., Bailey M.E., Cobb S.R. MeCP2 and Rett syndrome: reversibility and potential avenues for therapy. Biochem. J. 2011;439:1–14. doi: 10.1042/BJ20110648. [DOI] [PubMed] [Google Scholar]
- Gougelet A., Sartor C., Bachelot L., Godard C., Marchiol C., Renault G., Tores F., Nitschke P., Cavard C., Terris B., Perret C., Colnot S. Antitumour activity of an inhibitor of miR-34a in liver cancer with β-catenin-mutations. Gut. 2016;65:1024–1034. doi: 10.1136/gutjnl-2014-308969. [DOI] [PubMed] [Google Scholar]
- Hirai H., Verma M., Watanabe S., Tastad C., Asakura Y., Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J. Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite K.C., Adams V.H., Hansen J.C. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Kanazawa T., Otsuka S., Ichii O., Hashimoto Y., Kon Y. Expression of caspase family and muscle- and apoptosisi-specific genes during skeletal myogenesis in mouseembryo. J. Vet. Med. Sci. 2009;71:1161–1168. doi: 10.1292/jvms.71.1161. [DOI] [PubMed] [Google Scholar]
- Jiang H., Wang H., Wang S., Pei Z., Fu Z., Fang C., Wang J., Lu Q., Wang E., Li J. Expression of ERCC1, TYMS, RRM1, TUBB3, non-muscle myosin II, myoglobin and MyoD1 in lung adenocarcinoma pleural effusions predicts survival in patients receiving platinum-based chemotherapy. Mol. Med. Rep. 2015;11:3523–3532. doi: 10.3892/mmr.2014.3141. [DOI] [PubMed] [Google Scholar]
- Jiang H.L., Jiang L.M., Han W.D. Wnt/β-catenin signaling pathway in lung cancer stem cells is a potential target for the development of novel anticancer drugs. J. BUON. 2015;20:1094–1100. [PubMed] [Google Scholar]
- Kasowski M., Grubert F., Heffelfinger C., Hariharan M., Asabere A., Waszak S.M., Habegger L., Rozowsky J., Shi M., Urban A.E., Hong M.Y., Karczewski K.J., Huber W., Weissman S.M., Gerstein M.B., Korbel J.O., Snyder M. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P.K., Lee J.S., Liang X., Han L., Mori T., Fackler M.J., Sadik H., Argani P., Pandita T.K., Sukumar S. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70:6047–6058. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Zou C., Guo L., Seneviratne D.S., Tan X., Kwon Y.K., An J., Bowser R., DeFrances M.C., Zarnegar R. Novel death defying domain in met entraps the active site of caspase-3 and blocks apoptosis in hepatocytes. Hepatology. 2014;59:2010–2021. doi: 10.1002/hep.26769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Yang Y., Wang F., Moyer M.P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N., Wang H., Wang H., Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signaling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- Madison B.B., McKenna L.B., Dolson D., Epstein D.J., Kaestner K.H. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J. Biol. Chem. 2009;284:5936–5944. doi: 10.1074/jbc.M808103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw C.M., Samaco R.C., Zoghbi H.Y. Adult neural function requires MeCP2. Science. 2011;333:186. doi: 10.1126/science.1206593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G., Lv Y., Dai H., Zhang X., Guo Q.N. Epigenetic silencing of methyl-CpG-binding protein 2 gene affects proliferation, invasion, migration, and apoptosis of human osteosarcoma cells. Tumour Biol. 2014;35:11819–11827. doi: 10.1007/s13277-014-2336-8. [DOI] [PubMed] [Google Scholar]
- Müller H.M., Fiegl H., Goebel G., Hubalek M.M., Widschwendter A., Müller-Holzner E., Marth C., Widschwendter M. MeCP2 and MBD2 expression in human neoplastic and non-neoplastic breast tissue and its association with oestrogen receptor status. Br. J. Cancer. 2003;89:1934–1939. doi: 10.1038/sj.bjc.6601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.M., Buckley P.G., Das S., Watters K.M., Bryan K., Stallings R.L. Co-localization of the oncogenic transcription factor MYCN and the DNA methyl binding proteinMeCP2 at genomic sites in neuroblastoma. PLoS One. 2011;6:e21436. doi: 10.1371/journal.pone.0021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan M., Hong J., Atieno N., Muthusamy V.R., Davidson C.J., Abu-Rmaileh N., Hoidal J.R., Rajasekaran N.S. Nrf2 deficiency promotes apoptosis and impairs PAX7/MyoD expression in aging skeletal muscle cells. Free Radic. Biol. Med. 2014;71:402–414. doi: 10.1016/j.freeradbiomed.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane M., Clark A.P., Landini S., Birkbak N.J., Eklund A.C., Lim E., Culhane A.C., Barry W.T., Schumacher S.E., Beroukhim R., Szallasi Z., Vidal M., Hill D.E., Silver D.P. MECP2 is a frequently amplified oncogene with a novel epigenetic mechanism that mimics the role of activated RAS in malignancy. Cancer Discov. 2016;6:45–58. doi: 10.1158/2159-8290.CD-15-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach D. Discovery of a new mutation in MYOD1 characterizes a subset of aggressive embryonal rhabdomyosarcoma partnering with mutations of the PI3K-AKT pathway. Bull. Cancer. 2014;101:776–777. doi: 10.1684/bdc.2014.2017. [DOI] [PubMed] [Google Scholar]
- Ormestad M., Astorga J., Landgren H., Wang T., Johansson B.R., Miura N., Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- Qi J., Yu Y., Akilli Öztürk Ö., Holland J.D., Besser D., Fritzmann J., Wulf-Goldenberg A., Eckert K., Fichtner I., Birchmeier W. New Wnt/β-catenin target genes promote experimental metastasis and migration of colorectal cancer cells through different signals. Gut. 2016;65:1690–1701. doi: 10.1136/gutjnl-2014-307900. [DOI] [PubMed] [Google Scholar]
- Saito R.A., Micke P., Paulsson J., Augsten M., Peña C., Jönsson P., Botling J., Edlund K., Johansson L., Carlsson P., Jirström K., Miyazono K., Ostman A. Forkhead box F1 regulates tumor-promoting properties of cancer-associated fibroblasts in lung cancer. Cancer Res. 2010;70:2644–2654. doi: 10.1158/0008-5472.CAN-09-3644. [DOI] [PubMed] [Google Scholar]
- Shin J., Ming G.L., Song H. By hook or by crook: multifaceted DNA-binding properties of MeCP2. Cell. 2013;152:940–942. doi: 10.1016/j.cell.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S., Patel F.D., Ghosh S., Arora A., Dhaliwal L.K., Srinivasan R. Epigenetic alteration by DNA methylation of ESR1, MYOD1 and hTERT gene promoters is useful for prediction of response in patients of locally advanced invasive cervical carcinoma treated by chemoradiation. Clin. Oncol. (R. Coll. Radiol.) 2015;27:720–727. doi: 10.1016/j.clon.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Tan P., Yeoh K.G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149:1153–1162. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- Tong D., Zhao L., He K., Sun H., Cai D., Ni L., Sun R., Chang S., Song T., Huang C. MECP2 promotes the growth of gastric cancer cells by suppressing miR-338-mediated antiproliferative effect. Oncotarget. 2016;7:34845–34859. doi: 10.18632/oncotarget.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J.P., Lopes F., Silva-Fernandes A., Sousa M.V., Moura S., Sousa S., Costa B.M., Barbosa M., Ylstra B., Temudo T., Lourenço T., Maciel P. Variant Rett syndrome in a girl with a pericentric X-chromosome inversion leading to epigenetic changes and overexpression of the MECP2 gene. Int. J. Dev. Neurosci. 2015;46:82–87. doi: 10.1016/j.ijdevneu.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Wakefield R.I., Smith B.O., Nan X., Free A., Soteriou A., Uhrin D., Bird A.P., Barlow P.N. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- Wang K., Liang Q., Li X., Tsoi H., Zhang J., Wang H., Go M.Y., Chiu P.W., Ng E.K., Sung J.J., Yu J. MDGA2 is a novel tumour suppressor cooperating with DMAP1 in gastric cancer and is associated with disease outcome. Gut. 2016;65:1619–1631. doi: 10.1136/gutjnl-2015-309276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., Lasalle J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah R.M., Rastegar M. Linking epigenetics to human disease and Rett syndrome: the emerging novel and challenging concepts in MeCP2 research. Neural Plast. 2012;2012:415825. doi: 10.1155/2012/415825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.X., Zhao P.T., Xia L., Liu L.L., Liang J., Zhai H.H., Zhang H.B., Guo X.G., Wu K.C., Xu Y.M., Jia L.T., Yang A.G., Chen S.Y., Fan D.M. Potent inhibition of human gastric cancer by HER2-directed induction of apoptosis with anti-HER2 antibody and caspase-3 fusion protein. Gut. 2010;59:292–299. doi: 10.1136/gut.2008.155226. [DOI] [PubMed] [Google Scholar]
- Zhao L.Y., Zhang J., Guo B., Yang J., Han J., Zhao X.G., Wang X.F., Liu L.Y., Li Z.F., Song T.S., Huang C. MECP2 promotes cell proliferation by activating ERK1/2 and inhibiting p38 activity in human hepatocellular carcinoma HEPG2 cells. Cell. Mol. Biol. (Noisy-le-Grand) 2013;(Suppl. 59):OL1876–OL1881. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material