Abstract
The complexity of the traumatic brain injury (TBI) pathology, particularly concussive injury, is a serious obstacle for diagnosis, treatment, and long-term prognosis. Here we utilize modern systems biology in a rodent model of concussive injury to gain a thorough view of the impact of TBI on fundamental aspects of gene regulation, which have the potential to drive or alter the course of the TBI pathology. TBI perturbed epigenomic programming, transcriptional activities (expression level and alternative splicing), and the organization of genes in networks centered around genes such as Anax2, Ogn, and Fmod. Transcriptomic signatures in the hippocampus are involved in neuronal signaling, metabolism, inflammation, and blood function, and they overlap with those in leukocytes from peripheral blood. The homology between genomic signatures from blood and brain elicited by TBI provides proof of concept information for development of biomarkers of TBI based on composite genomic patterns. By intersecting with human genome-wide association studies, many TBI signature genes and network regulators identified in our rodent model were causally associated with brain disorders with relevant link to TBI. The overall results show that concussive brain injury reprograms genes which could lead to predisposition to neurological and psychiatric disorders, and that genomic information from peripheral leukocytes has the potential to predict TBI pathogenesis in the brain.
Keywords: Traumatic brain injury, Cognition, Hippocampus, Leukocytes, Gene networks, Transcriptome, DNA methylome
Highlights
-
•
TBI disrupts gene programming involving epigenomic, transcriptomic, and gene network alterations.
-
•
Homology of genomic signals between brain and blood leukocytes was identified.
-
•
Genomic signatures altered by TBI are enriched for genetic risk factors for psychiatric and neurological disorders in humans.
We report that TBI, a common injury in sport, military, and domestic environments, alters the fundamental aspects of gene regulation, which could explain mechanisms by which the TBI pathology can divert into other brain disorders. TBI affected the network organization and the epigenetic and transcriptional program of genes in the brain and blood governing critical aspects of cell communication, metabolism, inflammation, and blood functions. These genes intersect with those previously linked to psychiatric and neurological diseases in humans. These results provide new genomic and epigenomic evidence of mechanisms underlying the TBI pathology, infer the risk posed by TBI on the pathogenesis of related brain disorders, and emphasize the potential of leukocyte genomics as peripheral biomarkers of TBI, which could help guide the design of personalized medicine strategies.
1. Introduction
Traumatic brain injury (TBI) accounts for over 90% of the brain injuries in USA, and is common in sport related injuries, military, and domestic environments. TBI is characterized by a high level of complexity based on the multiple components involved in the onset and progression of the pathology. TBI compromises neuronal function and cognitive abilities that can last for years after the first incident (Levin et al., 1988, Levin and Diaz-Arrastia, 2015, Rabinowitz and Levin, 2014). Mild TBI such as concussive injury is particularly difficult to diagnose since several of its symptoms such as blurred vision, headache, nausea, dizziness could be caused by other reasons (Levin and Diaz-Arrastia, 2015, Rabinowitz and Levin, 2014), though about 20% of patients develop persistent symptoms that can last for years. Neurons that survive the initial insult show a decline in function (Yamaki et al., 1996, Bergsneider et al., 2000), but the underlying regulatory mechanisms have proven difficult to grasp based on information derived from isolated molecular events. The lack of comprehensive mechanistic information has left unanswered one of the most intriguing aspects of TBI, which is why many patients become vulnerable to other neurological disorders such as chronic traumatic encephalopathy (CTE), Alzheimer's disease (AD), and posttraumatic stress disorder (PTSD) (Levin and Diaz-Arrastia, 2015, Rabinowitz and Levin, 2014). The complexity of the TBI pathology becomes even a larger limiting factor for the design of strategies to diagnose and to predict the outcome of concussive injury (Crawford et al., 2012, Kulbe and Geddes, 2016).
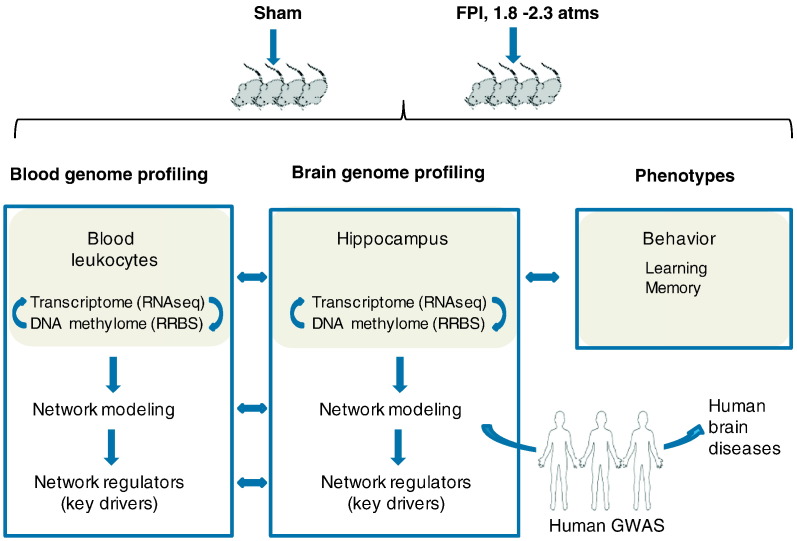
To address these knowledge gaps in the pathogenesis of TBI, we conducted a comprehensive systems biology study (Fig. 1) focusing on the impact of concussive injury on fundamental gene regulatory mechanisms. Unlike previous studies based on microarrays (Von Gertten et al., 2005, Rojo et al., 2011, Samal et al., 2015, Redell et al., 2013) and more recently RNA sequencing (Lipponen et al., 2016) which have reported changes in gene expression in response to TBI, in the current study we leveraged the power of system biology to combine genome-wide transcriptome and DNA methylome analyses in the hippocampus (a main site of cognitive dysfunction in TBI pathology) with modern data-driven gene network modeling approaches, which allowed us to extract information about crucial gene regulatory mechanisms (expression level, alternative splicing, epigenetic regulation) underlying TBI pathogenesis and to model tissue-specific gene-gene interactions which have the power to predict essential regulatory points. Gene networks are graphical models that depict genes as nodes and connections (reflecting regulatory relations or interactions) between genes as edges. Networks can be used to identify key driver genes that connect a large number of genes in a network affected by TBI, and are likely potent therapeutic targets (Makinen et al., 2014, Meng et al., 2016, Schadt et al., 2009, Wang et al., 2012, Zhang et al., 2013). To infer translatability to human pathophysiology, we analyzed the intersection of the molecular signals from our TBI rodent model with human genome-wide association studies (GWAS) of brain disorders.
Fig. 1.
Overall genomic study design. We assessed transcriptomic and epigenomic profiling of hippocampus and blood leukocytes, followed by network modeling and key driver identification. Transcriptomic and epigenomic profiling of blood samples was compared to that of hippocampus to identify genomic features in concordance with the brain signals for biomarker identification. We characterized the functional relevance of the brain genomic signatures and networks to TBI characteristics and behavior outcomes (learning and memory). By intersecting with human genome-wide association studies (GWAS), many TBI signature genes and network regulators identified in the rodent model were found to be causally associated with brain disorders with relevant link to TBI.
We also took advantage of our system biology paradigm to uncover a possible association between genomic changes in brain and periphery driven by TBI, which could shed light on the possible implementation of blood genomic biomarkers of TBI pathology in the brain. The current standard for biomarker research focuses on individual protein products in the cerebrospinal fluid or blood plasma (Zetterberg and Blennow, 2016, Zetterberg et al., 2013) that are by-products of TBI pathology and do not capture central regulatory mechanisms driving pathogenesis. Protein biomarkers also impose technical challenges due to limitations in the sensitivity of bioassays (Zetterberg and Blennow, 2016, Zetterberg et al., 2013). Additionally, blood leukocytes are abundant in the circulation and have been lately found to serve as a source of surrogate markers for AD (Rezai-Zadeh et al., 2009) and Parkinson's disease (PD) (Masliah et al., 2013). Comparison of the genomic signals between hippocampus and blood leukocytes facilitates the extraction of blood genomic markers that can trace the hidden aspects of the brain pathology of TBI.
2. Materials and methods
2.1. Fluid percussion injury (FPI) as a rodent model of concussive injury
Adult male Sprague–Dawley rats (Charles River Laboratories, Inc., MA, USA) weighing 200–220 g were maintained under standard housing conditions (room temperature 22–24 °C) with 12 h light/dark cycle (Agrawal and Gomez-Pinilla, 2012). The rats were kept individually in polyacrylic cages with free access to food and water. After acclimatization for one week on standard rat chow, rats were randomly assigned to sham control (n = 10) and FPI (n = 10) groups. FPI was performed by making a 3-mm diameter craniotomy with a trephine centered on the sagittal suture between bregma and lambda. A Teflon disk (1 mm) was placed over the craniotomy, and the cap was filled with 0.9% saline solution. At the first sign of hind-limb withdrawal to a paw pinch, a moderate fluid percussion pulse (approx. 1.8–2.3 atm) was administered using a FPI device (Virginia Commonwealth University). Apnea and unconsciousness times were determined. Sham animals underwent an identical preparation with the exception of FPI. All experiments were performed in accordance with the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the University of California at Los Angeles (UCLA) Chancellor's Animal Research Committee (ARC). Rats were sacrificed at two post-injury time points and samples from both 24 h and 7 d were used for molecular analysis. The acute period of TBI (24 h) is crucial for biomarker discovery (Kulbe and Geddes, 2016, Zetterberg and Blennow, 2016), and was used to predict the pathology that is further developed by the sub-acute period (7 d) including inflammation, degeneration, and early regeneration. In addition, we used the gene profile from 7 d to assess the overlap between signature genes and candidate genes of brain disorders since at this stage the pathology is underway.
2.2. Barnes maze test
Rats were tested with the Barnes maze test before and after TBI. Animals were trained to locate a dark escape chamber, hidden underneath a hole positioned around the perimeter of a disk, brightly illuminated by four overhead halogen lamps to provide a low level aversive stimulus. Animals were trained before TBI with two trials per day for five consecutive days. A trail started by placing the animal in the center of the maze covered under a cylindrical start chamber; after a 10-s delay, the start chamber was raised. A training session ended after the animal has entered the escape chamber or when a pre-determined time (5 min) has elapsed. In order to assess memory retention, two trials were given on day 7 post-TBI. We recorded and quantified behavior using AnyMaze video tracking software (San Diego Instruments).
2.3. Tissue collection
Right after the last memory test, rats were sacrificed by decapitation and hippocampus were dissected out, frozen on dry ice, and stored at − 70 °C for later use. Blood samples were collected retro-orbitally under isoflurane anesthesia into EDTA-coated Vacutainers (BD Diagnostics, Franklin Lakes, New Jersey) 24-h post FPI, and immediately centrifuged at 1600g for 10 min at 4 °C. The buffy coat layer containing leukocytes and platelets was carefully transferred to a new tube without disturbing the underneath leukocytes, and washed twice with 15 ml of EL Buffer (QIAGEN GmbH, Hilden, Germany) for 15 min at 4 °C. Total cellular DNA and RNA were extracted using the AllPrep DNA/RNA mini kit (QIAGEN GmbH, Hilden, Germany).
2.4. RNA sequencing (RNA-Seq) and data analyses
RNA samples from hippocampus and leukocytes (n = 5 per treatment group per tissue) were used for RNA-Seq to assess alterations in the transcriptome. RNA was first processed via poly-A selection and fragmentation, reverse transcribed into cDNA, and sequencing adapters ligated using the Illumina Paired-End sample prep kit. Fragments of 250-400 bp were isolated, amplified, and sequenced on an Illumina Hiseq2500 System. Paired-end RNA-Seq reads were mapped using TopHat2/Bowtie2 (Langmead et al., 2009). Differentially expressed genes, transcripts, and alternative splicing events were further identified using Cufflinks and Cuffdiff (Trapnell et al., 2012, Trapnell et al., 2010). A statistical cutoff of p < 0.01 was used to define TBI gene signatures. Multiple testing was corrected using false discovery rate (FDR) estimated with the Benjamini-Hochberg (BH) method (Benjamini and Hochberg, 1995). The RNA-Seq data was deposited to Gene Expression Ominbus (GEO) with accession number GSE64978 (hippocampus) and GSE68207 (leukocytes).
2.5. Quantitative real-time PCR (qPCR) to confirm top transcriptome signals
We chose to validate 7 differentially expressed genes (F5, Lgmn, Pls1, Cd63, Cdr2, Gch1, Vwf) that were common between hippocampus and leukocytes using qPCR. First-strand complementary DNA was synthesized using the iScript cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, California) and 100 ng total RNA according to the manufacturer's instructions. qPCR was carried out using the SsoFast EvaGreen Supermix kit (Bio-Rad Laboratories, Hercules, California) to amplify the candidate genes in the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, California), with Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as an endogenous control. Each experiment was repeated twice independently.
2.6. Functional annotation of genes altered by TBI
Signature genes identified from RNA-Seq analyses were classified for their biological functions using Molecular Signatures Database (MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp) (Subramanian et al., 2005), which catalogs canonical pathways or functional categories from various databases including Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html), BioCarta (http://www.biocarta.com/genes/index.asp), Reactome (http://www.reactome.org/), and gene sets derived from Gene Ontology (GO, http://www.geneontology.org/). Fisher's exact test was performed to calculate the enrichment p-values for each pathway or functional category within each gene signature. Results were corrected for multiple comparisons of hundreds of pathways using Bonferroni correction and Bonferroni-corrected p < 0.05 was considered significant.
2.7. Relationship between gene expression and behavior phenotypes
Gene expression levels of the signature genes identified from RNA-Seq analysis were tested for the association with behavior traits (latency time in the Barnes Maze test) by Pearson correlation. p value < 0.05 was considered significant.
2.8. Reduced representation bisulfite sequencing (RRBS)
RRBS libraries were constructed from the DNA samples of hippocampus and leukocytes (n = 5 per treatment group per tissue) (Meissner et al., 2005) by digesting 1 μg of genomic DNA with MspI. Digested DNA was then purified, end-repaired, and adenylated, followed by ligation to Illumina TruSeq barcode adapters. Fragments of size 150–300 bp were selected, bisulfite-treated, and sequenced using an Illumina Hiseq 2500 System. Sequences were called using standard Illumina software and bisulfite-converted reads were aligned to the genome using BS Seeker (Chen et al., 2010). Methylation percentages at each cytosine were computed and compared between treatment groups using methylKit, an R package implanted with logistic regression test, and the sliding linear model (SLIM) (Wang et al., 2011) to adjust p-values to q-values to correct for multiple testing. Loci with methylation level differing by > 25% between groups and q < 0.05 were considered significant, and defined as differentially methylated loci (DMLs). Differential genes from RNA-Seq and local DMLs within 10 kb distance were compared for overlap and correlated using Pearson correlation. RRBS data was deposited to GEO with accession numbers GSE64984 (hippocampus) and GSE79270 and GSE83776 (leukocytes).
2.9. Assessing overlap between TBI signatures and candidate genes of brain disorders from human GWAS
We tested the consistency between the genes affected by TBI from our study and those from human GWAS related to brain functions in two ways. First, we cross-checked genes identified in our study with top GWAS candidate genes reported in the GWAS catalog (Hindorff et al., 2009) (http://www.ebi.ac.uk/gwas/) for direct overlap. Second, we used a SNP Set Enrichment Analysis (SSEA) (Makinen et al., 2014), which goes beyond the top GWAS hits and tests the overall enrichment of the genes affected in our TBI animal model for SNPs that demonstrated disease association in human GWAS using Kolmogorov-Smirnov (KS) test and Fisher's exact test. Bonferroni-corrected p < 0.05 by either KS test or Fisher's exact test was considered significant.
2.10. Network models of TBI genomic signatures
We constructed tissue-specific Bayesian gene networks (BN) maps based on global gene expression patterns to reflect gene-gene relationship and organization. BNs are graphical models that depict genes as nodes and connections between genes as edges defined based on global genomic data. Network connections among genes in BNs have been shown to accurately capture the functional and mechanistic relationship of genes that can inform on disease mechanisms and help define composite sets of molecular markers (Chen et al., 2008, Makinen et al., 2014, Meng et al., 2016, Wang et al., 2012, Yang et al., 2009, Yang et al., 2010, Zhang et al., 2013, Zhu et al., 2008). BNs for brain and blood were constructed based on genetic and transcriptomic data from several large-scale human and mouse studies (Tu et al., 2012, Wang et al., 2012, Millstein et al., 2011, Derry et al., 2010, Emilsson et al., 2008) each involving hundreds of individuals, using a previously established method by Zhu et al. (Zhu et al., 2008, Zhu et al., 2007). These studies are all population-based and reflect broad spectrums of physiological states and, therefore, can capture intrinsic gene-gene interactions across conditions. The large sample sizes involved in these studies also ensure the statistical power to infer accurate networks from each study. The edges in the BNs are defined by conditional probabilities that characterize the distribution of states of each gene given the state of its parents (Pearl, 1988). The network topology defines a partitioned joint probability distribution over all genes in a network. The likelihood of a BN network model given observed transcriptomic data was determined using the Bayes formula. For each dataset, 1000 Bayesian networks, each using different random seeds, were reconstructed using Monte Carlo Markov Chain simulation (Madigan, 1995). Bayesian Information Criteria was used to determine the model with the best fit for each network. From the resulting set of 1000 networks, edges that appeared in > 30% of the networks were used to define a consensus network. To infer causal directions between genes in a network, genetic information was used as priors (Zhu et al., 2004). For each tissue, the union of nodes and edges from BNs of multiple studies was used as the tissue-specific network, and the predictive power of multi-study union networks has been supported previously (Makinen et al., 2014, Meng et al., 2016, Wang et al., 2012). To derive gene subnetworks affected by TBI, we used the signature genes identified from the RNA-Seq analysis as seeds to extract the top most connected subnetworks within the BNs as described previously (Yang et al., 2009). Network visualization was carried out using Cytoscape (Smoot et al., 2011).
2.11. Identification of key drivers (KD) of TBI signatures
A previously established Key Driver Analysis (KDA) was used to identify the potential key regulators, or key drivers (KDs) for the TBI signature genes based on the topology of BN network models (Wang et al., 2012, Yang et al., 2010). Conceptually, KDA aims to identify genes whose network neighbors are more likely to be genes affected by TBI than expected by random chance. Because these genes act as “hubs” that are surrounded by TBI genes, it is more likely that they regulate the activities of genes affected by TBI. Statistically, KDA takes as input a set of genes G (i.e., genes affected by TBI in each tissue) and a gene network N (BNs described above). For every gene Nk in the network N, the neighboring genes of Nk within 3-edge distance were tested for enrichment of genes in G using Fisher's exact test. Network genes that reach Bonferroni-adjusted p < 0.05 were reported as KDs.
3. Results
3.1. TBI promotes large-scale alterations in the transcriptional activities of genes
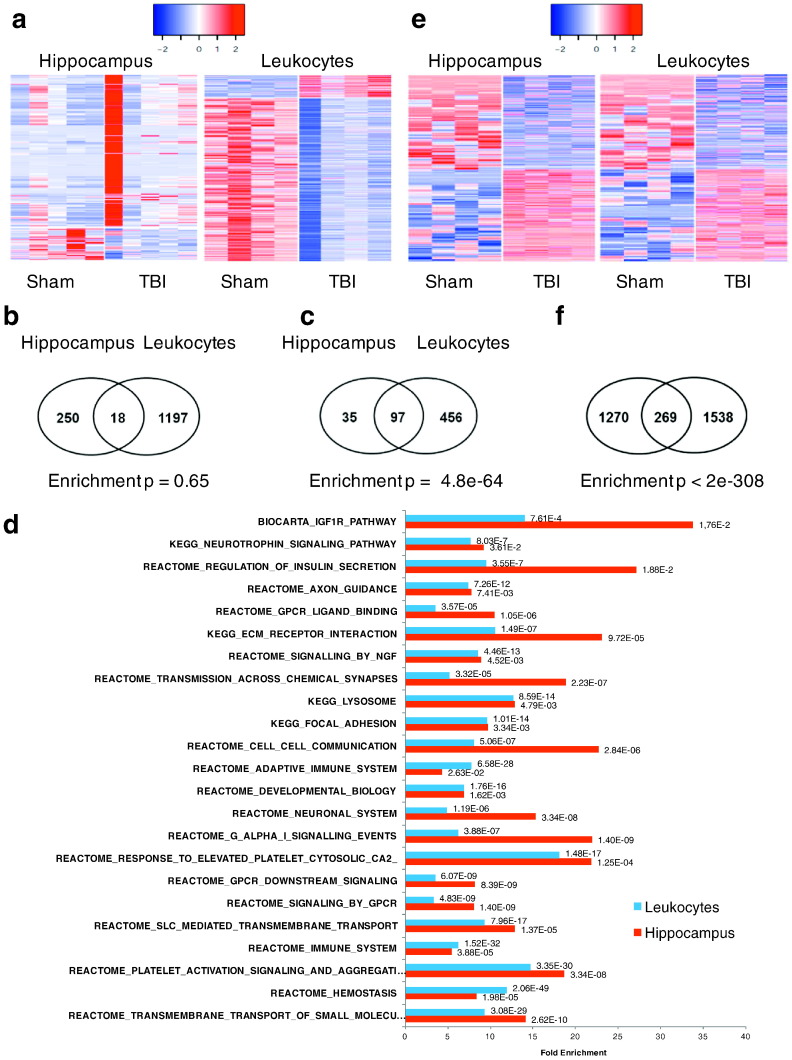
We conducted RNA-Seq analysis of hippocampus and leukocyte samples from rats exposed to TBI (n = 5) or sham surgery (n = 5). At p < 0.01, we identified 240 and 1052 differentially expressed genes, 111 and 739 differentially expressed transcripts, and 23 and 93 genes showing alternative exon usage in hippocampus and leukocytes, respectively (Table S1 in Dataset S1). We pooled all these genes to define 268 (121 pass false discovery rate or FDR < 10%) and 1215 (1092 pass FDR < 10%) unique genes as gene “signatures” for hippocampus and leukocytes, respectively, and used these signatures for downstream integrative analysis. As illustrated in Fig. 2a, a majority of the hippocampal signatures had increased expression, whereas the opposite is true for leukocyte, suggesting directional differences in the transcriptional response to TBI between the brain and the peripheral. Between the two tissues, 18 common genes were identified (Fig. 2b), including Abca4, Adipor1, Aqp1, Asap1, Cd63, Cdr2, Dennd2c, F5, Fam46a, Gch1, Gulp1, Laptm4b, Lgals3bp, Lgmn, Lrg1, Oasl, Pls1, Vwf. When only considering the gene-level changes, there were 16 shared genes, with 13 (Abca4, Adipor1, Aqp1, Cd63, Cdr2, Dennd2c, Gch1, Gulp1, Laptm4b, Lgals3bp, Lgmn, Oasl, Pls1) showing opposite expression changes and three (Asap1, Lrg1, Vwf) showing the same direction in both tissues. Additionally, we tested the reliability of the differential signals using qPCR experiments on 7 shared genes (F5, Lgmn, Pls1, Cd63, Cdr2, Gch1, Vwf) between hippocampus and leukocytes, and confirmed the differential expression patterns observed in RNA-Seq in both tissues (Table S2 in Dataset S1).
Fig. 2.
Impact of TBI on transcriptome and DNA methylome in hippocampus and leukocytes. A) Heatmaps of differentially expressed genes in both tissues. Blue to red colors indicate low to high expression values. B) Overlap of differential transcriptomic signatures between tissues, and enrichment p value according to Fisher's exact test. C) Overlap of over-represented molecular pathways in the signatures genes between tissues, and enrichment p value according to Fisher's exact test. D) Top shared pathways between tissues. Bars are the fold enrichment of pathways and the values next to the bars are the Bonferroni-corrected enrichment p values according to Fisher's exact test. E) Heatmap of differential methylation loci (DML) in both tissues. Blue to red colors indicate low to high expression values. F) Overlap of DML between tissues, and enrichment p value according to Fisher's exact test.
Among all the hippocampal signatures, there were 28 unique genes showing expression differences at the levels of transcripts or exons but not at gene level, indicative of the importance of TBI-induced alternative splicing in these genes. Similarly, leukocyte signatures also contained 163 unique genes with altered transcriptional activity only at the transcript and exon levels, supporting even broader impact of TBI on the gene alternative splicing events in blood cells. Additionally, many of the signature genes are known splicing regulators, including the hippocampal signature Rbm47 and the leukocyte signatures Bcas2, Clasrp, Lsm6, Luc7l3, Prpf4b, Qk, Rbm25, Rbm38, Rbpms2, Sfrs18, Snrnp40, and Uhmk1.
Increasing evidence indicates that alternative splicing is an inherent mechanism in the etiology of various diseases. Accordingly, TBI affected alternative splicing of genes involved in diverse functions related to neurons (Neurod1, Neurod4, P2ry4, P2ry1, Htr2c, Npy, Agrp), complement and coagulation (C3, F5, Vwf), transcriptions factors (Aff3, Zbtb16), blood pressure (Sgk1, Ace, Angpt1, Gucy1a3, Gucy1b3), inflammation (Tlr7, Ccl9, Cxcl2, Ccr1, Alox15, Alox12), mitochondria (Atp13a4, Atp61f, Atp6v0d1, Cox6a1, Cox8a, Cox20, Ndufa8, Ndufa11, Ndufa12), leptin signaling (Npy, Agrp), insulin signaling (Irs1), and extracellular matrix (ECM) genes (Spp1, Mmp2, Bgn). The alternatively spliced genes F5 and Zbtb16 have been associated with hippocampal atrophy and conduct behavior, respectively in human GWAS.
3.2. Correlation between hippocampal transcriptome signatures of TBI and behavior phenotypes
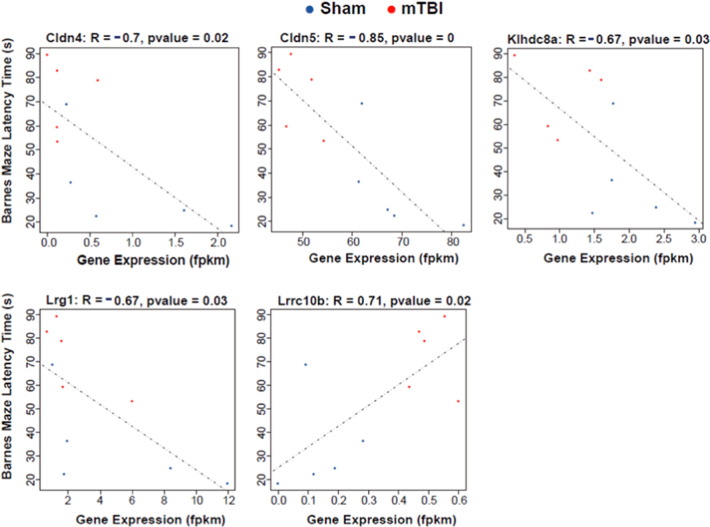
As hippocampus is important for cognitive function, we tested the hippocampal gene expression changes with the effects of TBI on memory. Among the 240 hippocampal signatures with changes at the gene level, 5 (Cldn4, Cldn5, Klhdc8a, Lrg1, and Lrrc10b) showed significant correlation with the latency time detected in the Barnes maze (p < 0.05; Fig. 3).
Fig. 3.
Correlation between TBI hippocampal signature genes and latency time in memory test. Gene expression level is represented by fragments per kilobase of transcript per million RNA-Seq mapped reads (fpkm) on the x-axis and the Barnes maze latency time measured in the matched animals is shown on the y-axis. Correlation strength is indicated by the correlation coefficient R and the p value.
3.3. Functional categorization of hippocampal and leukocyte transcriptomic signatures
We annotated the signature genes based on known pathways or functional categories curated in KEGG, BioCarta, and Reactome, and found enrichment of both tissue-specific and shared biological pathways between hippocampus and leukocytes (Table S3 in Dataset S1). At Bonferroni-corrected p < 0.05, we found 132 and 553 over-represented pathways for hippocampal and leukocyte signatures, respectively, among which 97 were in common when only considering exact matches in pathway names (Fig. 2c). When highly overlapping pathways with non-identical names were considered, the converging pathways between tissues are even greater, with majority of the hippocampal pathways captured in the leukocyte pathways. The shared pathways included axon guidance, neurotransmission, ECM, focal adhesion, lysosome, transmembrane transport of small molecules, hemostasis, platelet activation signaling and aggregation, immune system, vascular smooth muscle contraction, class A1 rhodopsin-like receptors, and various signaling pathways for insulin, glucagon, IGF, FGFR, NGF, neurotrophin, PDGF, and TLR (representative shared pathways are illustrated in Fig. 2d). These results indicate that despite the small overlap in individual signature genes and the largely opposite expression patterns between hippocampus and leukocytes, at pathway level there is substantial overlap.
The main hippocampal-specific pathways included olfactory transduction and growth hormone receptor signaling. In contrast, a larger number of pathways are leukocyte-specific, including cell cycle, oxidative phosphorylation, interferon signaling, cardiomyopathy, PPAR alpha, metabolic pathways for glucose, lipid and lipoproteins, fatty acids and triglycerides, and signaling pathways for NCAM, Notch, TGF-beta, WNT, PARKIN, mTOR, VEGF, and disease related gene sets for AD, PD, depression, and Huntington's disease.
3.4. TBI induced large-scale DNA methylomic changes in hippocampus and leukocytes
To identify epigenomic changes associated with TBI, we examined the DNA methylome using reduced representation bisulfite sequencing (RRBS) that measures millions of potential DNA methylation sites at single base resolution. We found that TBI induced a large-scale switch of the methylation patterns in both tissues: at FDR < 5%, 781 and 758 of differentially methylated loci (DMLs) showed hypomethylation and hypermethylation in hippocampus, respectively. In leukocytes, we identified 915 and 892 DMLs with hypomethylation and hypermethylation, respectively (Fig. 2e). Between the two tissues, there are 269 shared methylation sites, which represent statistically significant overlap (Fig. 2f).
3.5. Relationship between differential methylation and differential expression
To explore the potential relationship between DMLs and gene expression, we mapped the TBI DMLs in each tissue to adjacent genes within 10 kb distance. We found in both hippocampus and leukocytes, ~ 50%, ~ 30%, and 16% of DMLs were located in the intergenic regions, gene body (including 5′-UTR, intron, coding sequence, and 3′-UTR), and the upstream or downstream regions of the genes, respectively (Table S4 in Dataset S1). Assessing the methylome-transcriptome relationship revealed certain co-clustering of DMLs and gene signatures (Fig. S1). For instance, 13 out of the 268 (4.8%) hippocampal signature genes and 65 out of the 1215 (5.3%) leukocyte signature genes co-localized with DMLs within 10 kb distance; 7 hippocampal and 24 leukocyte signature genes were significantly correlated with their local DMLs at p < 0.05 based on Pearson correlation analysis. For example, gene Atp6v0d1 showed a significant positive correlation with its local DML (correlation coefficient r = 0.90, p = 2.18E-3), and gene Igf1r had a significant negative correlation with its local DML (r = − 0.86, p = 6.54E-3) in leukocytes. Additionally, 32 out of the 268 (11.9%) hippocampal signature genes and 130 out of the 1215 (10.7%) leukocyte signature genes co-localized with DMLs within 50 kb distance (Table S5 in Dataset S1), suggesting cis-regulation of gene expression by the local DMLs. Notably, some of the genes containing local DMLs are transcription factors, including Irx6 and Zbtb16 from hippocampus and Dpf3, Foxo3, and Zfp219 from leukocytes, or epigenetic factors such as Gadd45g, Morf4l1, and Rgs1 from leukocytes, which could in turn trans-regulate downstream target genes.
3.6. Identification of key drivers and gene subnetworks mediating the effects of mTBI in hippocampus and leukocytes
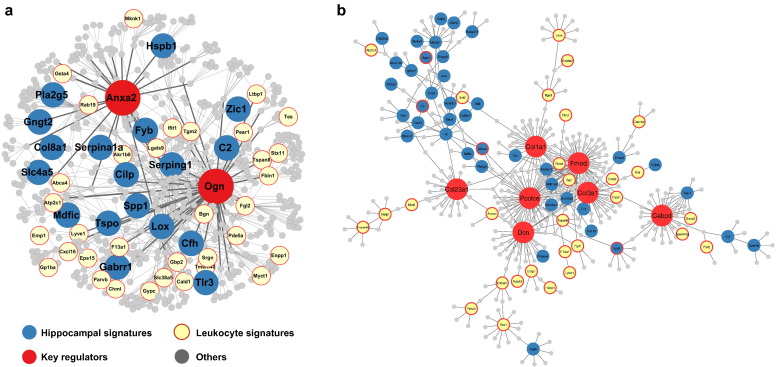
To explore the gene-gene relations among the hippocampus or leukocyte signature genes and to identify potential key perturbation points of TBI that triggered the transcriptomic changes, we employed multiple data-driven Bayesian networks (BNs) (Zhu et al., 2004, Zhu et al., 2008) that elucidate gene-gene regulatory relationships based on brain and blood genomic data of several large independent human and rodent populations (detailed in Materials and methods). Using these networks and a network topology-based key driver analysis (KDA), we identified candidate key driver (KD) genes whose network neighboring genes highly overlapped with the signature genes for hippocampus and leukocytes, among which 127 KDs were common to both tissues (Table S6 in Dataset S1). As illustrated in Fig. 4a, a subnetwork containing two shared KDs, Anxa2 and Ogn, was highly enriched with both hippocampal and leukocyte signatures, although the signature genes themselves did not overlap. Other notable shared KDs between tissues include ECM genes such as Fmod, Dcn, Pcolce, Col1a1, Col3a1, and Col23a1 and transcription factors Tcf7l2 and Cebpd. Some of these KDs also form a subnetwork (Fig. 4b). Both subnetworks coordinate genes primarily functioning in ECM-receptor interaction, complement and coagulation cascades, and focal adhesion. The network-level co-localization of TBI signature genes between the two tissues agrees with the pathway-level overlap observed above, and suggests that TBI affects similar functional processes in both tissues through shared regulatory mechanisms.
Fig. 4.
Shared subnetworks between hippocampal and leukocyte signature genes. A) A subnetwork centered at shared key drivers (KDs) Anax2 and Ogn. B) A subnetwork centered at shared KDs Fmod, collagen genes, and Cebpd. Larger red nodes are the KDs; blue and yellow nodes denote hippocampal and leukocyte signature genes, respectively. Grey nodes are network genes in the neighborhood of KDs that are not affected by TBI.
3.7. Relevance of hippocampal and leukocyte signatures and KDs to human diseases
TBI has been associated with a number of brain disorders. To assess the relevance of our signals from the rodent TBI model to human pathophysiology, we compared the hippocampal and leukocyte signature genes as well as the KDs to human GWAS of brain related diseases or traits. Human GWAS genes of brain disorders most likely play causal roles in disease development, because genetic variations that drive changes in genes or gene products are inherited at birth and precede disease manifestation. If TBI affects these disease genes uncovered in human genetic studies, it is likely that TBI will also impose predisposition to brain disorders. When directly comparing with top candidate genes curated in the GWAS catalog, we found numerous overlapping genes between TBI signature or KD genes and top GWAS hits for a broad range of brain disorders including AD, attention deficient hyperactive disorder, autism, depression, addiction, PTSD, schizophrenia, and PD (representative overlapping genes in hippocampus shown in Table 1; full lists for both tissues in Table S7 in Dataset S1). When using a SNP set enrichment analysis (SSEA) that utilized full sets of GWAS results we had accessed for AD (Heinzen et al., 2010, Lambert et al., 2013, Naj et al., 2011), cognitive traits (Seshadri et al., 2007), psychiatric disorders (Psychiatry Consortium, 2009), and smoking behavior Tobacco and Genetics Consortium, 2010), we found that human orthologs of the TBI signature genes and KDs derived from both hippocampus and leukocytes were significantly enriched for genetic polymorphisms associated with many of these diseases or traits (Table S8 in Dataset S1).
Table 1.
Overlapping genes between TBI hippocampal signatures or KDs and human GWAS genes of brain disorders. Human GWAS candidate genes were derived from the GWAS catalog (p < 1e-5).
| GWAS disease | Overlapping gene in TBI hippocampal signature |
|---|---|
| Alcohol consumption/dependence | CUX2; SLC26A4; ZDHHC21; ESR1; SERINC2 |
| AD | APOE; NMU; PCDH11X; PPP1R3B; PROX1; SCARA3; SMC2; TTLL7; ADAMTS9; ARHGAP20; CAMK4; ELMO1; LIPC; PDE7B; PLEKHG1; SP6 |
| ADHD | BCL11A; CDH13; LRRC7; LYPLAL1; NR4A2; SEMA3A; TCEB1; TGFB2; TSHZ2; ZIC5; PAWR; NDN, ESRRB; ADAMTS2; LRRN3; TYRP1 |
| Bipolar disorder | ANK3; COLEC12; DGKH; DPP10; IRF2BP2; ITIH3; MRPS23; MSI2; NFIA; PDE10A; PGM5; PPM1M; SIAE; SP8; TTC39B; UACA; CGNL1; DMTF1; FAT4 |
| Bipolar disorder and schizophrenia | ARC; ATP6V1B2; BAI1; CACNA2D1; CMYA5; COMMD10; CST7; CTNND2; DDX52; DMD; HACE1; IRX1; KAT2B; MRVI1; MYO1E; PRKCQ; PTPRN2; SIM1; SLC39A12; TMEM212; TTC39B; VPS13C |
| Brain connectivity | CNTN4; EPHA7; NEDD4 |
| Brain structure | BOK; CADPS2; GRIN2B |
| Cognitive function | PTPRO; AFAP1L2; CDH13; FAT4; GABRQ; GRIN2B; HCCS; IMMP2L; IRX1; IRX2; JUN; KIAA1217; KLHL1; LHX2; LIPC; MCTP2; NR2F2; PLCB1; RIT2; TOX; TSHZ3; UNC13C; VANGL2; ZNF788; LMO4 |
| Word reading | NOS1AP; TACSTD2; TYRP1 |
| Working memory | CLDN1; DRD2; LPHN3 |
| Conduct disorder | C1QTNF7; MCTP2; NR2F2; RIT2; ST8SIA4; ZBTB16; KCNA5; PDE10A |
| Eating disorders | CAMK1D; DLGAP1 |
| Hippocampal atrophy/volume | APOE; COL18A1; F5; MAGI2; MAL2; DPP4; MSRB3 |
| Intelligence | CNTN4; COL1A2; GYPC; KIF16B |
| Major depressive disorder | ADCYAP1R1; CCND2; CDH9; EMP1; GRIN2B; HAPLN1; HOMER1; IGFBP3; KCNH5; NDFIP2; NEUROD6; PCLO; RASGEF1B; ATP6V1B2 |
| Parkinson's disease | BMP4; DLG2; GPRIN3; RIT2; RORA; SEMA5A; STK39; VPS13C; WNT3 |
| PTSD | COBL; PCSK2; PRKCA; SLC4A5 |
| Schizophrenia | ANK3; BMP7; CDH13; CXCL12; GNAL; NRGN; PCDH20; PDC; PTGS2; RORA; SNX7; TCF4; VPS13C |
| Smoking behavior | BDNF; CHRNA3; CHRNA5; KCND2; MAOB; PDE1C |
4. Discussion
We utilize systems biology strategies that have yielded breakthroughs in our understanding of mechanisms of complex disorders in recent years (Chen et al., 2008, Huan et al., 2013, Makinen et al., 2014, Meng et al., 2016, Narayanan et al., 2014, Schadt et al., 2005, Wang et al., 2012, Yang et al., 2009, Yang et al., 2010, Zhang et al., 2013) to determine the impact of TBI on fundamental gene regulatory mechanisms and biological pathways underlying pathogenesis. The majority of the genetic causal risks of complex human diseases such as Alzheimer's disease involve alterations of key pathogenic pathways (Lambert et al., 2013, Manolio, 2010). We found that transcriptomic signatures in hippocampus and in leukocytes converge on functional categories of genes modulating important cellular functions such as inflammation, metabolism, and cell communication. The gene regulatory mechanisms uncovered from the current study span from epigenetic regulation and alternative splicing to gene network regulation. Alterations of these regulatory mechanisms studied in our rodent model of TBI could explain how the incidence of TBI alters the course of brain homeostasis and increases the risk of related brain pathologies in humans. This possibility was verified by integrating the results of rodent gene signatures with human GWAS, which catalog putative causal elements in human diseases. We found that key signature genes and their regulators affected by TBI in rodents have been associated with brain disorders with known association to TBI. The overall results of our system biology study provide evidence for the impact of TBI on core aspects of gene regulation, which offers critical mechanistic information at the molecular level and can be used as novel elements of diagnosis and treatment. Compared to previous genomic profiling studies of TBI (Von Gertten et al., 2005, Rojo et al., 2011, Samal et al., 2015, Redell et al., 2013, Lipponen et al., 2016), our study differs in the molecular features assessed (gene expression only in previous studies vs. the combination of epigenome, gene expression, and alternative splicing in the current study), network modeling approaches (literature-based networks in previous studies vs data-driven, tissue-specific networks used here), tissues, time points, and TBI severity, which offer unique mechanistic aspects of the TBI pathology.
DNA methylation is a major epigenetic mechanism responsible for changing the program of genes thus deviating towards disease states (Lardenoije et al., 2015, Szyf et al., 2016). An increasing body of evidence indicates that predisposition to various neurological and psychiatric disorders are saved as epigenetic modifications (Jakovcevski and Akbarian, 2012, Tsankova et al., 2007), and blast exposure results in changes in neuronal DNA methylation (Haghighi et al., 2015). Our results showing the impact of TBI on DNA methylation of multiple genes in both hippocampus and leukocytes provide the opportunity to examine how the effects of TBI can be saved with the potential to regulate gene transcription. Indeed, the action of TBI on the epigenome was revealed on select DNA methylomic alterations that co-localized and correlated with transcriptomic signature genes. It should be noted that correlation is not equal to causality and further validation experiments are warranted to investigate the causal relationship between the epigenome and the transcriptome. Alternative splicing is emerging as another gene regulatory mechanism by which the genome influences the etiology of various diseases such as AD (Xiong et al., 2015). Our results show that TBI affects numerous alternatively spliced genes in the hippocampus and leukocytes involved in functions such as coagulation, blood pressure, inflammation, energy management, and ECM regulation. Further examination of these genes could guide the identification of particularly vulnerable points and mechanisms through which the TBI pathology could deviate to other disorders. For example, many of the signature genes are known splicing regulators such as Rbm47, Sfrs18, Snrnp40, and Uhmk1. Among these, Uhmk1 has been recently related to genetic disposition to cerebral visual impairment (Bosch et al., 2016) while Rbm47 confers genetic risk to high blood pressure and cardiovascular disease (Surendran et al., 2016), and both traits are common aspects of TBI pathology. Our results portray both epigenomic modification and alternative splicing events as putative mechanisms by which TBI impacts gene network programming and disease predisposition.
To date, perturbations in gene network regulation has also been recognized as a key component of the pathogenesis of neurological disorders (Narayanan et al., 2014, Zhang et al., 2013). We identified the impact of TBI on the organization of select networks of genes under regulatory control of key driver genes, which may be responsible for the cascade of cellular events involved in the TBI pathology. We found that TBI affects hippocampal and leukocyte gene networks orchestrated by key driver genes associated with the extracellular matrix (e.g., Fmod, Dcn, Pcolce, collagens), transcription factors (e.g., Tcf7l2, Cebpd), and other functions discussed below (e.g., Anxa2, Ogn). These master genes are located at the center of gene subnetworks surrounded by TBI signature genes, and are likely key regulatory points of these networks. The extracellular matrix genes Fmod and Pcolce have been recently experimentally validated as key regulators of cognitive and metabolic functions (Meng et al., 2016). Tcf7l2 and Cebpd are key transcription factors involved in metabolic processes, diabetes and fat differentiation. The key driver Anxa2 (annexin A2) has been associated with brain tumor formation (Zhai et al., 2011) and the long-term neurological outcomes of focal embolic stroke (Wang et al., 2014). In turn, the fact that Ogn (osteoglycin) is reduced in the amygdala of animals exposed to stress (Jung et al., 2012), suggests that this gene can be an important link between TBI and psychiatric-like disorders such as anxiety and depression (Max, 2014). The fact that TBI-affected genes are tightly connected through key drivers in gene regulatory networks constructed from completely independent studies supports their functional relatedness and their synchronized actions. The identification of key regulatory genes of the entire network spectrum affected by TBI provides a framework for understanding the integrated actions of multiple pathways on the TBI pathology, and offers plausible key interventional targets (Kasarskis et al., 2011, Yang et al., 2012, Schadt et al., 2009).
Although an increasing body of information indicates that TBI is a risk factor for a range of neurological disorders such as CTE, PTSD, AD (Levin and Diaz-Arrastia, 2015, Rabinowitz and Levin, 2014), mechanisms involved remain elusive. In our rodent model, we found that the signature genes Cldn4, Cldn5, Klhdc8a, Lrg1, and Lrrc10b are correlated with the latency time in the Barnes maze. Claudins 4 and 5 belong to a family of genes whose protein products are components of tight junctions and regulate permeability of molecules to the intercellular space. Claudin-5 is a tight junction protein that connects endothelial cells, and is considered important for maintaining permeability of molecules within the extracellular space (Doherty et al., 2016). Therefore, it is possible that genes like Claudins could play a role in the regulation of the BBB, which is a prevalent aspect in the TBI pathology. In turn, the claudin gene CLDN1 has been associated with working memory in human GWAS (Table 1). The actions of klhdc8a, Lrrc10b, and Lrg1 in the brain are poorly understood. However, it has been described that the expression of klhdc8a is elevated in human glioblastomas (Mukasa et al., 2010), and that Lrg1 is associated with regulation of stress (Stankiewicz et al., 2015) and recollection of contextual fear memories (Barnes et al., 2012). Although none of these genes have been studied in the context of TBI, some of their known functions overlap with physiological manifestations of the TBI pathology. By integrating our rodent findings with human GWAS, we found significant overlap between the rodent TBI genes and candidate genes in human GWAS for several neurodegenerative diseases such as AD, cognitive disorders, PTSD and other neuropsychiatric disorders. In particular, we found 16 hippocampal genes including APOE, which are known to provide genetic predisposition to AD (Lupton et al., 2016), and 9 leukocyte genes that overlapped with AD genes. Additionally, four hippocampal genes and one leukocyte gene have known association with human PTSD. Further, we found 14 hippocampal and 5 leukocyte genes are associated with major depressive disorders in humans. These findings provide important genomic leads to elucidate how the TBI pathology can escalate to neurological/psychiatric disorders, and suggest specific interventional targets.
Our system biology approach was key to reveal a genome-scale link between central and peripheral gene regulation as studied in circulating leukocytes in response to TBI. We chose to examine the genomics of blood leukocytes since they are abundant in the circulation and have been lately found to serve as a source of biomarkers for AD (Rezai-Zadeh et al., 2009) and PD (Masliah et al., 2013). The comparative analysis of genomic signatures revealed homology between leukocytes and hippocampus at gene-, epigenome-, pathway-, and network levels related to vascularity, cell integrity, and immune response. These changes are likely a reflection of the early events post-TBI related to breakdown of the BBB and changes in systemic inflammation in response to injury (Kulbe and Geddes, 2016). The 18 common signature genes between the brain and peripheral could point to new leads for leukocyte genomic markers that trace the gene activities in the hippocampus to assess specific functional implications. For example, the share gene F5, involved in coagulation process, has been associated with hippocampal atrophy and conduct behavior in human GWAS. In addition, there was a substantial overlap at pathway and network levels between hippocampus and leukocytes, and the great diversity of common pathways harmonizes with the characteristic broad symptomatology of TBI such a blurred vision (rhodopsin-like receptors), headache, nausea, and dizziness (Levin and Diaz-Arrastia, 2015, Rabinowitz and Levin, 2014). The shared pathways and networks could be used to develop composite genomic biomarkers that aggregate the information from gene expression level, alternative splicing status, and DNA methylation patterns of leukocyte genes within a shared pathway or gene network to accurately predict the pathological state of the brain and link to TBI phenotypes.
In summary, our comprehensive systems investigation shows that concussive injury affects fundamental aspects of gene regulatory mechanisms that maintain brain homeostasis, including epigenomic programming, alternative splicing factors, transcription factors, and novel network regulators. The alterations induced by TBI in all of these gene regulatory components provide molecular support for the emerging clinical reports about the risk posed by TBI on complex human diseases. This information can be used as elements of prognosis as well as a framework to start examining mechanisms by which TBI pathology may contribute to the pathogenesis of select brain disorders. Moreover, our results are unique to identify the effects of TBI on key driver genes that have the capacity to direct main pathways involved in the TBI pathology, and suggest key perturbation points which are plausible interventional targets for TBI. From the biomarker perspective, our results reveal important homologies between central and peripheral gene regulatory mechanisms at transcriptome, epigenome, pathway, and network levels that could serve as basis for novel strategies to identify composite biomarkers of TBI using easily accessible peripheral samples. Lastly, the intersection between numerous genes altered in the rodent model of TBI with human GWAS could account for the risk posed by TBI to neurological and psychiatric disorders in humans. Overall, our results could serve as a platform to investigate the influence of TBI on the etiology of brain disorders.
Funding Sources
XY is supported by NIH grant R01DK104363, American Heart Association Scientist Development Grant 13SDG17290032, Leducq Foundation, Hellman Fellows Award, UCLA Faculty Research Grant, and UCLA CTSI Grant UL1TR000124. FGP is supported by NIH grants R01NS050465, R01DK104363, and The Letten Foundation. None of the funding sources had any role in the design and conduct of the study, the collection, analysis, or interpretation of the data, or the preparation, review, or approval of the manuscript.
Conflict of Interests
The authors have no conflict of interests to declare.
Authors' Contribution
Qingying Meng contributed to collection and analysis of genomic data, and writing and editing of the manuscript; Yumei Zhuang performed the animal lesion experiments and processed tissues for genomic analysis; Zhe Ying coordinated the technical aspect of the study; Rahul Agrawal performed the animal lesion experiments; Xia Yang co-supervised the study, interpreted the data, and wrote the paper; Fernando Gomez-Pinilla co-supervised the study, interpreted the data, and wrote the paper.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.01.046.
Contributor Information
Xia Yang, Email: xyang123@ucla.edu.
Fernando Gomez-Pinilla, Email: fgomezpi@ucla.edu.
Appendix A. Supplementary data
Supplementary material
Co-localization of TBI induced differentially expressed genes (DEG) and differential methylation loci (DML) in hippocampus and leukocytes. Horizontal bars indicate rat chromosomes, and pink and green dots are DEG and DML identified by RNA-Seq and RRBS, respectively.
References
- Agrawal R., Gomez-Pinilla F. ‘Metabolic syndrome’ in the brain: deficiency in omega-3 fatty acid exacerbates dysfunctions in insulin receptor signalling and cognition. J. Physiol. 2012;590:2485–2499. doi: 10.1113/jphysiol.2012.230078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P., Kirtley A., Thomas K.L. Quantitatively and qualitatively different cellular processes are engaged in CA1 during the consolidation and reconsolidation of contextual fear memory. Hippocampus. 2012;22:149–171. doi: 10.1002/hipo.20879. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995:289–300. [Google Scholar]
- Bergsneider M., Hovda D.A., Lee S.M., Kelly D.F., Mcarthur D.L., Vespa P.M., Lee J.H., Huang S.C., Martin N.A., Phelps M.E., Becker D.P. Dissociation of cerebral glucose metabolism and level of consciousness during the period of metabolic depression following human traumatic brain injury. J. Neurotrauma. 2000;17:389–401. doi: 10.1089/neu.2000.17.389. [DOI] [PubMed] [Google Scholar]
- Bosch D.G., Boonstra F.N., De Leeuw N., Pfundt R., Nillesen W.M., De Ligt J., Gilissen C., Jhangiani S., Lupski J.R., Cremers F.P., De Vries B.B. Novel genetic causes for cerebral visual impairment. Eur. J. Hum. Genet. 2016;24:660–665. doi: 10.1038/ejhg.2015.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Y., Cokus S.J., Pellegrini M. BS seeker: precise mapping for bisulfite sequencing. BMC Bioinforma. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., Macneil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., Leonardson A., Castellini L.W., Wang S., Champy M.F., Zhang B., Emilsson V., Doss S., Ghazalpour A., Horvath S., Drake T.A., Lusis A.J., Schadt E.E. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford F., Crynen G., Reed J., Mouzon B., Bishop A., Katz B., Ferguson S., Phillips J., Ganapathi V., Mathura V., Roses A., Mullan M. Identification of plasma biomarkers of TBI outcome using proteomic approaches in an APOE mouse model. J. Neurotrauma. 2012;29:246–260. doi: 10.1089/neu.2011.1789. [DOI] [PubMed] [Google Scholar]
- Derry J.M., Zhong H., Molony C., Macneil D., Guhathakurta D., Zhang B., Mudgett J., Small K., El Fertak L., Guimond A., Selloum M., Zhao W., Champy M.F., Monassier L., Vogt T., Cully D., Kasarskis A., Schadt E.E. Identification of genes and networks driving cardiovascular and metabolic phenotypes in a mouse F2 intercross. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C.P., O'keefe E., Wallace E., Loftus T., Keaney J., Kealy J., Humphries M.M., Molloy M.G., Meaney J.F., Farrell M., Campbell M. Blood-brain barrier dysfunction as a Hallmark pathology in chronic traumatic encephalopathy. J. Neuropathol. Exp. Neurol. 2016;75:656–662. doi: 10.1093/jnen/nlw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., Mouy M., Steinthorsdottir V., Eiriksdottir G.H., Bjornsdottir G., Reynisdottir I., Gudbjartsson D., Helgadottir A., Jonasdottir A., Styrkarsdottir U., Gretarsdottir S., Magnusson K.P., Stefansson H., Fossdal R., Kristjansson K., Gislason H.G., Stefansson T., Leifsson B.G., Thorsteinsdottir U., Lamb J.R., Gulcher J.R., Reitman M.L., Kong A., Schadt E.E., Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Haghighi F., Ge Y., Chen S., Xin Y., Umali M.U., De Gasperi R., Gama Sosa M.A., Ahlers S.T., Elder G.A. Neuronal DNA methylation profiling of blast-related Traumatic Brain Injury (TBI) J Neurotrauma. 2015;32:1200–1209. doi: 10.1089/neu.2014.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen E.L., Need A.C., Hayden K.M., Chiba-Falek O., Roses A.D., Strittmatter W.J., Burke J.R., Hulette C.M., Welsh-Bohmer K.A., Goldstein D.B. Genome-wide scan of copy number variation in late-onset Alzheimer's disease. J. Alzheimers Dis. 2010;19:69–77. doi: 10.3233/JAD-2010-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T., Zhang B., Wang Z., Joehanes R., Zhu J., Johnson A.D., Ying S., Munson P.J., Raghavachari N., Wang R., Liu P., Courchesne P., Hwang S.J., Assimes T.L., Mcpherson R., Samani N.J., Schunkert H., Meng Q., Suver C., O'donnell C.J., Derry J., Yang X., Levy D. A systems biology framework identifies molecular underpinnings of coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2013;33:1427–1434. doi: 10.1161/ATVBAHA.112.300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski M., Akbarian S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Lee Y., Kim G., Son H., Lee D.H., Roh G.S., Kang S.S., Cho G.J., Choi W.S., Kim H.J. Decreased expression of extracellular matrix proteins and trophic factors in the amygdala complex of depressed mice after chronic immobilization stress. BMC Neurosci. 2012;13:58. doi: 10.1186/1471-2202-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasarskis A., Yang X., Schadt E. Integrative genomics strategies to elucidate the complexity of drug response. Pharmacogenomics. 2011;12:1695–1715. doi: 10.2217/pgs.11.115. [DOI] [PubMed] [Google Scholar]
- Kulbe J.R., Geddes J.W. Current status of fluid biomarkers in mild traumatic brain injury. Exp. Neurol. 2016;275(Pt 3):334–352. doi: 10.1016/j.expneurol.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., Destafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thorton-Wells T.A., Jones N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Gerrish A., Schmidt H., Kunkle B., Dunstan M.L., Ruiz A., Bihoreau M.T., Choi S.H., Reitz C., Pasquier F., Cruchaga C., Craig D., Amin N., Berr C., Lopez O.L., De Jager P.L., Deramecourt V., Johnston J.A., Evans D., Lovestone S., Letenneur L., Moron F.J., Rubinsztein D.C., Eiriksdottir G., Sleegers K., Goate A.M., Fievet N., Huentelman M.W., Gill M., Brown K., Kamboh M.I., Keller L., Barberger-Gateau P., Mcguiness B., Larson E.B., Green R., Myers A.J., Dufouil C., Todd S., Wallon D., Love S., Rogaeva E., Gallacher J., St George-Hyslop P., Clarimon J., Lleo A., Bayer A., Tsuang D.W., Yu L., Tsolaki M., Bossu P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fox N.C., Hardy J., Deniz Naranjo M.C., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthews F., European Alzheimer's Disease, I, Genetic, Environmental Risk in Alzheimer's, D, Alzheimer's Disease Genetic, C, Cohorts For, H, Aging Research in Genomic, E, Moebus S., Mecocci P., Del Zompo M., Maier W., Hampel H., Pilotto A., Bullido M., Panza F., Caffarra P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenoije R., Iatrou A., Kenis G., Kompotis K., Steinbusch H.W., Mastroeni D., Coleman P., Lemere C.A., Hof P.R., Van Den Hove D.L., Rutten B.P. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015;131:21–64. doi: 10.1016/j.pneurobio.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H.S., Diaz-Arrastia R.R. Diagnosis, prognosis, and clinical management of mild traumatic brain injury. Lancet Neurol. 2015;14:506–517. doi: 10.1016/S1474-4422(15)00002-2. [DOI] [PubMed] [Google Scholar]
- Levin H.S., Goldstein F.C., High W.M.J., Eisenberg H.M. Disproportionately severe memory deficit in relation to normal intellectual functioning after closed head injury. J. Neurol. Neurosurg. Psychiatry. 1988;51:1294–1301. doi: 10.1136/jnnp.51.10.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipponen A., Paananen J., Puhakka N., Pitkänen A. Analysis of post-traumatic brain injury gene expression signature reveals tubulins, Nfe2l2, Nfkb, Cd44, and S100a4 as treatment targets. Sci. Rep. 2016;6:31570. doi: 10.1038/srep31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton M.K., Strike L., Hansell N.K., Wen W., Mather K.A., Armstrong N.J., Thalamuthu A., Mcmahon K.L., De Zubicaray G.I., Assareh A.A., Simmons A., Proitsi P., Powell J.F., Montgomery G.W., Hibar D.P., Westman E., Tsolaki M., Kloszewska I., Soininen H., Mecocci P., Velas B., Lovestone S., Brodaty H., Ames D., Trollor J.N., Martin N.G., Thompson P.M., Sachdev P.S., Wright M.J., Initiative A.S.D.N. The effect of increased genetic risk for Alzheimer's disease on hippocampal and amygdala volume. Neurobiol. Aging. 2016;40:68–77. doi: 10.1016/j.neurobiolaging.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan D.A.Y.J. Bayesian graphical models for discrete data. Int. Stat. Rev. 1995;63:215–232. [Google Scholar]
- Makinen V.P., Civelek M., Meng Q., Zhang B., Zhu J., Levian C., Huan T., Segre A.V., Ghosh S., Vivar J., Nikpay M., Stewart A.F., Nelson C.P., Willenborg C., Erdmann J., Blakenberg S., O'donnell C.J., Marz W., Laaksonen R., Epstein S.E., Kathiresan S., Shah S.H., Hazen S.L., Reilly M.P., Lusis A.J., Samani N.J., Schunkert H., Quertermous T., Mcpherson R., Yang X., Assimes T.L. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Masliah E., Dumaop W., Galasko D., Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8:1030–1038. doi: 10.4161/epi.25865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max J.E. Neuropsychiatry of pediatric traumatic brain injury. Psychiatr. Clin. N. Am. 2014;37:125–140. doi: 10.1016/j.psc.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Ying Z., Noble E., Zhao Y., Agrawal R., Mikhail A., Zhuang Y., Tyagi E., Zhang Q., Lee J.H., Morselli M., Orozco L., Guo W., Kilts T.M., Zhu J., Zhang B., Pellegrini M., Xiao X., Young M.F., Gomez-Pinilla F., Yang X. Systems nutrigenomics reveals brain gene networks linking metabolic and brain disorders. EBioMedicine. 2016;7:157–166. doi: 10.1016/j.ebiom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J., Winrow C.J., Kasarskis A., Owens J.R., Zhou L., Summa K.C., Fitzpatrick K., Zhang B., Vitaterna M.H., Schadt E.E., Renger J.J., Turek F.W. Identification of causal genes, networks, and transcriptional regulators of REM sleep and wake. Sleep. 2011;34:1469–1477. doi: 10.5665/sleep.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa A., Wykosky J., Ligon K.L., Chin L., Cavenee W.K., Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2616–2621. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., Larson E.B., Bird T.D., Boeve B.F., Graff-Radford N.R., De Jager P.L., Evans D., Schneider J.A., Carrasquillo M.M., Ertekin-Taner N., Younkin S.G., Cruchaga C., Kauwe J.S., Nowotny P., Kramer P., Hardy J., Huentelman M.J., Myers A.J., Barmada M.M., Demirci F.Y., Baldwin C.T., Green R.C., Rogaeva E., St George-Hyslop P., Arnold S.E., Barber R., Beach T., Bigio E.H., Bowen J.D., Boxer A., Burke J.R., Cairns N.J., Carlson C.S., Carney R.M., Carroll S.L., Chui H.C., Clark D.G., Corneveaux J., Cotman C.W., Cummings J.L., Decarli C., Dekosky S.T., Diaz-Arrastia R., Dick M., Dickson D.W., Ellis W.G., Faber K.M., Fallon K.B., Farlow M.R., Ferris S., Frosch M.P., Galasko D.R., Ganguli M., Gearing M., Geschwind D.H., Ghetti B., Gilbert J.R., Gilman S., Giordani B., Glass J.D., Growdon J.H., Hamilton R.L., Harrell L.E., Head E., Honig L.S., Hulette C.M., Hyman B.T., Jicha G.A., Jin L.W., Johnson N., Karlawish J., Karydas A., Kaye J.A., Kim R., Koo E.H., Kowall N.W., Lah J.J., Levey A.I., Lieberman A.P., Lopez O.L., Mack W.J., Marson D.C., Martiniuk F., Mash D.C., Masliah E., Mccormick W.C., Mccurry S.M., Mcdavid A.N., Mckee A.C., Mesulam M., Miller B.L. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., Mcelwee J., Zhang B., Zhang C., Lamb J.R., Xie T., Suver C., Molony C., Melquist S., Johnson A.D., Fan G., Stone D.J., Schadt E.E., Casaccia P., Emilsson V., Zhu J. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014;10:743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Morgan Kaufmann Publishers; San Mateo, Calif.: 1988. Probabilistic reasoning in intelligent systems: networks of plausible inference. [Google Scholar]
- Psychiatry Consortium A framework for interpreting genome-wide association studies of psychiatric disorders. Mol. Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Rabinowitz A.R., Levin H.S. Cognitive sequelae of traumatic brain injury. Psychiatr. Clin. N. Am. 2014;37:1–11. doi: 10.1016/j.psc.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell J.B., Moore A.N., Grill R.J., Johnson D., Zhao J., Liu Y., Dash P.K. Analysis of functional pathways altered after mild traumatic brain injury. J. Neurotrauma. 2013;30:752–764. doi: 10.1089/neu.2012.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezai-Zadeh K., Gate D., Szekely C.A., Town T. Can peripheral leukocytes be used as Alzheimer's disease biomarkers? Expert. Rev. Neurother. 2009;9:1623–1633. doi: 10.1586/ern.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo D.R., Prough D.S., Falduto M.T., Boone D.R., Micci M.A., Kahrig K.M., Crookshanks J.M., Jimenez A., Uchida T., Cowart J.C., Hawkins B.E., Avila M., Dewitt D.S., Hellmich H.L. Influence of stochastic gene expression on the cell survival rheostat after traumatic brain injury. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal B.B., Waites C.K., Almeida-Suhett C., Li Z., Marini A.M., Samal N.R., Elkahloun A., Braga M.F., Eiden L.E. Acute response of the hippocampal transcriptome following mild traumatic brain injury after controlled cortical impact in the rat. J. Mol. Neurosci. 2015;57:282–303. doi: 10.1007/s12031-015-0626-2. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Friend S.H., Shaywitz D.A. A network view of disease and compound screening. Nat. Rev. Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Lamb J., Yang X., Zhu J., Edwards S., Guhathakurta D., Sieberts S.K., Monks S., Reitman M., Zhang C., Lum P.Y., Leonardson A., Thieringer R., Metzger J.M., Yang L., Castle J., Zhu H., Kash S.F., Drake T.A., Sachs A., Lusis A.J. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S., Destefano A.L., Au R., Massaro J.M., Beiser A.S., Kelly-Hayes M., Kase C.S., D'agostino R.B., Sr., Decarli C., Atwood L.D., Wolf P.A. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med. Genet. 2007;8(Suppl. 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz A.M., Goscik J., Majewska A., Swiergiel A.H., Juszczak G.R. The effect of acute and chronic social stress on the hippocampal transcriptome in mice. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran P., Drenos F., Young R., Warren H., Cook J.P., Manning A.K., Grarup N., Sim X., Barnes D.R., Witkowska K., Staley J.R., Tragante V., Tukiainen T., Yaghootkar H., Masca N., Freitag D.F., Ferreira T., Giannakopoulou O., Tinker A., Harakalova M., Mihailov E., Liu C., Kraja A.T., Nielsen S.F., Rasheed A., Samuel M., Zhao W., Bonnycastle L.L., Jackson A.U., Narisu N., Swift A.J., Southam L., Marten J., Huyghe J.R., Stančáková A., Fava C., Ohlsson T., Matchan A., Stirrups K.E., Bork-Jensen J., Gjesing A.P., Kontto J., Perola M., Shaw-Hawkins S., Havulinna A.S., Zhang H., Donnelly L.A., Groves C.J., Rayner N.W., Neville M.J., Robertson N.R., Yiorkas A.M., Herzig K.H., Kajantie E., Zhang W., Willems S.M., Lannfelt L., Malerba G., Soranzo N., Trabetti E., Verweij N., Evangelou E., Moayyeri A., Vergnaud A.C., Nelson C.P., Poveda A., Varga T.V., Caslake M., De Craen A.J., Trompet S., Luan J., Scott R.A., Harris S.E., Liewald D.C., Marioni R., Menni C., Farmaki A.E., Hallmans G., Renström F., Huffman J.E., Hassinen M., Burgess S., Vasan R.S., Felix J.F., Uria-Nickelsen M., Malarstig A., Reilly D.F., Hoek M., Vogt T.F., Lin H., Lieb W., Traylor M., Markus H.S., Highland H.M., Justice A.E., Marouli E., Lindström J., Uusitupa M., Komulainen P., Lakka T.A. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Tang Y.Y., Hill K.G., Musci R. The dynamic epigenome and its implications for behavioral interventions: a role for epigenetics to inform disorder prevention and health promotion. Transl. Behav. Med. 2016;6:55–62. doi: 10.1007/s13142-016-0387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco & Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N., Renthal W., Kumar A., Nestler E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tu Z., Keller M.P., Zhang C., Rabaglia M.E., Greenawalt D.M., Yang X., Wang I.M., Dai H., Bruss M.D., Lum P.Y., Zhou Y.P., Kemp D.M., Kendziorski C., Yandell B.S., Attie A.D., Schadt E.E., Zhu J. Integrative analysis of a cross-loci regulation network identifies app as a gene regulating insulin secretion from pancreatic islets. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Gertten C., Flores Morales A., Holmin S., Mathiesen T., Nordqvist A.C. Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci. 2005;6:69. doi: 10.1186/1471-2202-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Q., Tuominen L.K., Tsai C.J. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics. 2011;27:225–231. doi: 10.1093/bioinformatics/btq650. [DOI] [PubMed] [Google Scholar]
- Wang I.M., Zhang B., Yang X., Zhu J., Stepaniants S., Zhang C., Meng Q., Peters M., He Y., Ni C., Slipetz D., Crackower M.A., Houshyar H., Tan C.M., Asante-Appiah E., O'Neill G., Luo M.J., Thieringer R., Yuan J., Chiu C.S., Lum P.Y., Lamb J., Boie Y., Wilkinson H.A., Schadt E.E., Dai H., Roberts C. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol. Syst. Biol. 2012;8:594. doi: 10.1038/msb.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fan X., Yu Z., Liao Z., Zhao J., Mandeville E., Guo S., Lo E.H. Effects of tissue plasminogen activator and annexin A2 combination therapy on long-term neurological outcomes of rat focal embolic stroke. Stroke. 2014;45:619–622. doi: 10.1161/STROKEAHA.113.003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., Morris Q., Barash Y., Krainer A.R., Jojic N., Scherer S.W., Blencowe B.J., Frey B.J. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki T., Imahori Y., Ohmori Y., Yoshino E., Hohri T., Ebisu T., Ueda S. Cerebral hemodynamics and metabolism of severe diffuse brain injury measured by PET. J. Nucl. Med. 1996;37:1166–1170. [PubMed] [Google Scholar]
- Yang X., Deignan J.L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R.R., Karlsson J., Castellani L.W., Mumick S., Wang K., Xie T., Coon M., Zhang C., Estrada-Smith D., Farber C.R., Wang S.S., Van Nas A., Ghazalpour A., Zhang B., Macneil D.J., Lamb J.R., Dipple K.M., Reitman M.L., Mehrabian M., Lum P.Y., Schadt E.E., Lusis A.J., Drake T.A. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang B., Molony C., Chudin E., Hao K., Zhu J., Gaedigk A., Suver C., Zhong H., Leeder J.S., Guengerich F.P., Strom S.C., Schuetz E., Rushmore T.H., Ulrich R.G., Slatter J.G., Schadt E.E., Kasarskis A., Lum P.Y. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20:1020–1036. doi: 10.1101/gr.103341.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang B., Zhu J. Functional genomics- and network-driven systems biology approaches for pharmacogenomics and toxicogenomics. Curr. Drug Metab. 2012;13:952–967. doi: 10.2174/138920012802138633. [DOI] [PubMed] [Google Scholar]
- Zetterberg H., Blennow K. Fluid biomarkers for mild traumatic brain injury and related conditions. Nat. Rev. Neurol. 2016;12:563–574. doi: 10.1038/nrneurol.2016.127. [DOI] [PubMed] [Google Scholar]
- Zetterberg H., Smith D.H., Blennow K. Biomarkers of mild traumatic brain injury in cerebrospinal fluid and blood. Nat. Rev. Neurol. 2013;9:201–210. doi: 10.1038/nrneurol.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai H., Acharya S., Gravanis I., Mehmood S., Seidman R.J., Shroyer K.R., Hajjar K.A., Tsirka S.E. Annexin A2 promotes glioma cell invasion and tumor progression. J. Neurosci. 2011;31:14346–14360. doi: 10.1523/JNEUROSCI.3299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., Mcelwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., Macdonald M.E., Lamb J.R., Bennett D.A., Molony C., Stone D.J., Gudnason V., Myers A.J., Schadt E.E., Neumann H., Zhu J., Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Lum P.Y., Lamb J., Guhathakurta D., Edwards S.W., Thieringer R., Berger J.P., Wu M.S., Thompson J., Sachs A.B., Schadt E.E. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet. Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- Zhu J., Wiener M.C., Zhang C., Fridman A., Minch E., Lum P.Y., Sachs J.R., Schadt E.E. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput. Biol. 2007;3 doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhang B., Smith E.N., Drees B., Brem R.B., Kruglyak L., Bumgarner R.E., Schadt E.E. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Co-localization of TBI induced differentially expressed genes (DEG) and differential methylation loci (DML) in hippocampus and leukocytes. Horizontal bars indicate rat chromosomes, and pink and green dots are DEG and DML identified by RNA-Seq and RRBS, respectively.