Abstract
Adventitious root formation is essential for the vegetative propagation of perennial woody plants. During the juvenile-to-adult phase change mediated by the microRNA156 (miR156), the adventitious rooting ability decreases dramatically in many species, including apple rootstocks. However, the mechanism underlying how miR156 affects adventitious root formation is unclear. In the present study, we showed that in the presence of the synthetic auxin indole-3-butyric acid (IBA), semi-lignified leafy cuttings from juvenile phase (Mx-J) and rejuvenated (Mx-R) Malus xiaojinensis trees exhibited significantly higher expression of miR156, PIN-FORMED1 (PIN1), PIN10, and rootless concerning crown and seminal roots-like (RTCS-like) genes, thus resulting in higher adventitious rooting ability than those from adult phase (Mx-A) trees. However, the expression of SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE26 (SPL26) and some auxin response factor (ARF) gene family members were substantially higher in Mx-A than in Mx-R cuttings. The expression of NbRTCS-like but not NbPINs and NbARFs varied with miR156 expression in tobacco (Nicotiana benthamiana) plants transformed with 35S:MdMIR156a6 or 35S:MIM156 constructs. Overexpressing the miR156-resistant MxrSPL genes in tobacco confirmed the involvement of MxSPL20, MxSPL21&22, and MxSPL26 in adventitious root formation. Together, high expression of miR156 was necessary for auxin-induced adventitious root formation via MxSPL26, but independent of MxPINs and MxARFs expression in M. xiaojinensis leafy cuttings.
Keywords: adventitious rooting, auxin, leafy cutting, Malus xiaojinensis, miR156
Introduction
Adventitious rooting is a cornerstone of proliferation for most fruit and forest species that are vegetatively propagated from elite genotypes. In the apple rootstocks, Sequoia sempervirens and Pinus radiata for example, propagation via both hard wood and leafy cuttings are constrained by adventitious rooting recalcitrance in the reproductively competent donor trees (Chen et al., 1981; Huang et al., 1992; Sanchez et al., 2007). To date, several techniques have been developed to improve rooting ability in some woody perennials, such as cycles of in vitro culture of apple rootstocks, or the repeated grafting of adult scions onto juvenile rootstocks in English ivy (Hedera helix L.) and chestnut (Castanea sativa; Giovannelli and Giannini, 2000; Xiao et al., 2014). The regulation of adventitious rooting is, however, relatively unexplored.
Adventitious root formation depends on multiple factors, such as genetic background, developmental stage, hormones, and other internal and external cues (Geiss et al., 2009; da Costa et al., 2013). In most tree species, the ability to form adventitious roots decreases during the transition from juvenile to adult development phases. In S. sempervirens, the rooting rate of cuttings from juvenile trees was up to 100% compared to 30% in cuttings from adult trees (Huang et al., 1992). Similarly, the rooting rate of cuttings from 3-year-old juvenile plants was significantly higher (88%) than from 36-year-old adult plants (17%) of Carolina Buckthorn (Rhamnus caroliniana Walt.; Graves, 2002). In our previous study, rooting rates of cuttings from juvenile, juvenile-like, and rejuvenated donor plants were significantly higher (77%) than those of cuttings from adult trees (11%) in Malus xiaojinensis (Xiao et al., 2014).
Numerous genes are differentially expressed between juvenile and adult cuttings prior to root induction. In loblolly pine (Pinus taeda L.), 5NG4, a nodulin-like gene, is highly and specifically induced by auxin in juvenile shoots prior to adventitious root formation, but is then substantially down-regulated in physiologically mature shoots that are adventitious rooting incompetent (Busov et al., 2004). A gene encoding a nitrate reductase involved in nitric oxide production in Eucalyptus grandis is up-regulated in juvenile cuttings as compared to that in mature cuttings, which might lead to increased ability to produce nitric oxide and form adventitious roots (Abu-Abied et al., 2012). Transcriptome data showed that the expression of E. grandis homologs of Peroxidase 72, PIN3, and Aux/IAA 19 (IAA19) were higher at a certain time point in auxin treated juvenile cuttings compared to mature ones (Abu-Abied et al., 2014); however, the insight effect of juvenility on adventitious root formation is not clear.
The juvenile to adult phase change is initiated by a decrease in the expression of the miR156. In Arabidopsis, maize (Zea mays) and various woody plants including Acacia confusa, Acacia colei, E. globulus, H. helix, Quercus acutissima, and Populus × Canadensis, miR156 is highly abundant in seedlings and decreases in adult plants (Wu and Poethig, 2006; Chuck et al., 2007; Wang et al., 2011). In the Congrass1 maize mutant that overexpresses miR156, prop roots are produced at all nodes in the plant, while these roots only grow from shoot-born meristem at the juvenile nodes in wild-type plants (Chuck et al., 2007). Similarly, tomato and tobacco plants overexpressing miR156 exhibit dense aerial roots on their stems, while none appear on the stems of wild type plants (Zhang et al., 2011; Feng et al., 2016). In contrast, Arabidopsis thaliana plants transformed with 35S:MIM156 produce significantly fewer adventitious roots from the base of the hypocotyl than wild-type plants (Xu et al., 2016).
MiR156 acts by repressing the expression of a group of SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) genes. Elevated levels of miR156 promote adventitious root formation in maize, tomato, and tobacco, indicating that SPL proteins inhibit adventitious root formation (Chuck et al., 2007; Zhang et al., 2011; Feng et al., 2016). EgSPL2 and EgSPL5 are up-regulated in mature cuttings compared to juvenile cuttings of E. grandis, and the juvenile cuttings exhibited a higher adventitious rooting percentage (Abu-Abied et al., 2012). These results suggest that miR156 and miR156 putative targeted MxSPL genes may play important roles in adventitious root formation during the juvenile to adult phase change. Twenty-seven SPL gene family members were identified in the apple genome and 13 were predicted to be targets of miR156 using degradome sequencing (Li et al., 2013; Xing et al., 2014). However, which SPL gene members are involved in adventitious root formation is unknown.
Auxin is an effective inducer of adventitious root formation; the dosage and gradient of, and response to, auxin are all important for plant root growth. Synthetic auxins like indole-3-butyric acid (IBA) have been used for almost 80 years to induce adventitious rooting (Zimmerman and Wil-Coxon, 1935). Without IBA treatment, leafy cuttings from juvenile M. xiaojinensis do not form adventitious roots (Xiao et al., 2014). The gradient of auxin controls adventitious root formation; in detached Arabidopsis leaves, an auxin gradient is required in the procambium cells during adventitious root formation (Liu J. C. et al., 2014). Auxin efflux carriers, such as PIN-FORMED (PIN) proteins, establish auxin concentration gradients (Yang and Murphy, 2009; Adamowski and Friml, 2015). Transgenic data suggests that OsPIN1 plays an important role in auxin-dependent adventitious root emergence in rice (Xu et al., 2005). Furthermore, PtoPIN1c is induced and maintained at a 20-fold level during the adventitious root initiation phase in Populus (Liu B. B. et al., 2014). In Arabidopsis, the pin1-1 mutant formed 40% fewer adventitious roots than wild type (Sukumar et al., 2013). All of these results demonstrate that regeneration of adventitious roots requires polar auxin transport.
Beyond auxin gradients and dosage, genes that participated in auxin signaling pathways are also involved in adventitious root formation. The transgenic Arabidopsis line overexpressing the auxin response factor 17 (ARF17) gene develops fewer adventitious roots, while plants overexpressing the ARF6 or ARF8 develop more adventitious roots than wild-type plants (Sorin et al., 2005; Gutierrez et al., 2009). It was previously reported that to promote adventitious root formation, ARF proteins directly regulate LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN (LBD) genes by binding to their promoter sequences (Okushima et al., 2007; Majer et al., 2012). Crown rootless 1 (Crl1) encodes a member of the LBD genes in rice (Oryza sativa), which positively regulate adventitious root formation, and its expression is directly regulated when an ARF binds to the 5′ flanking sequences of Crl1 (Inukai et al., 2005). In maize, rootless concerning crown and seminal root (Rtcs) encodes a LBD protein that positively regulates adventitious root growth, and its expression is regulated by ZmARF34, which binds to an auxin response element in the promoter region of Rtcs (Majer et al., 2012). However, during the adventitious root formation in recalcitrant woody plants, whether and how miR156 interacts with auxin remains unknown.
To evaluate how juvenility mediates adventitious rooting in woody plants and using apple rootstock M. xiaojinensis as an example, we analyzed the transcript level of miR156, miR156 putative target SPL gene expression, and MxPIN and MxARF gene family members during the adventitious rooting process. Then, the function of miR156 in adventitious root formation was validated by generating transgenic tobacco lines. The results elucidate the role of miR156 in adventitious root formation and will be useful in horticultural and forestry industries.
Materials and methods
Plant material
M. xiaojinensis (Mx) was used in these experiments because of its high apomictic rate to ensure stable and robust juvenile materials (Li et al., 2004). The apomictic origin of donor trees used for leafy cutting collection has been confirmed by simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers (Wang et al., 2012). Cuttings were excised from basal suckers (juvenile phase, Mx-J), shoots from the canopy of reproductively mature trees (adult phase, Mx-A), and rejuvenated plants via in vitro apical meristem culture (rejuvenated or juvenile like, Mx-R; Xiao et al., 2014).
Semi-lignified leafy cuttings (8–10 cm in length) were excised from actively growing shoots of donor plants, dipped 1 cm in depth into a solution containing 3000 mg/L indole butyric acid (IBA) (V900325, Sigma-Aldrich, St. Louis, MO, USA) for 1 min, and then the cuttings were plugged into 50 cell trays containing fine sand as a rooting medium. The cuttings were incubated in a solar greenhouse under 90–95% relative humidity (Xiao et al., 2014). Cuttings treated with an IBA-free medium solution were used as controls. The experimental errors were managed by using a complete randomized design for three biological replicates, each including at least 50 leafy cuttings. Rooting parameters were scored at 35 d after application (Xiao et al., 2014).
Plant expression vector construction and transgenic tobacco generation
Nine miR156 precursor genes were previously identified in apple genome (Ma et al., 2014). MdMIR156a6 (Genome location: MDC018927.245), which relative transcript level was higher than the other members, was chosen for overexpression study (Supplementary Figure 1). The apple MdMIR156a6 DNA fragment was amplified from Malus domestica genomic DNA. Artificial target mimics were generated by modifying the sequence of the AtIPS1 gene to knock-down miR156 expression (Franco-Zorrilla et al., 2007). The PCR products were sequenced by BGI (Shenzhen, China). All constructs were cloned behind the constitutive CaMV 35S promoter in the pBI121 vector, and then introduced into Nicotiana benthamiana by Agrobacterium tumefaciens-mediated transformation (Horsch et al., 1985).
MxSPLs coding sequences were amplified from M. xiaojinensis cDNA. The miR156-resistant SPLs (rSPLs) were made by two rounds of mutagenic PCR using KOD-Plus Nero DNA polymerase (KOD-401, TOYOBO LIFE SCIENCE, Japan), and sequencing confirmed the mutations by BGI (Shenzhen, China). The rSPLs variants were driven by the CaMV 35S promoter in the pBI121 vector. These constructs were introduced into 35S:MdMIR156a6 transgenic tobacco leaves by A. tumefaciens-mediated transformation. The infected leaves were selected on MS medium supplemented with 100 mg/L kanamycin and 300 mg/L cefotaxime sodium to generate rSPL and miR156 co-overexpressing transgenic lines. All primers are listed in Supplementary Table 1. Transgenic tobacco plants were proliferated by subculture on hormone-free MS medium. Rooting rate and the number of adventitious roots were recorded at 5, 7, 9, 11, and 13 d after subculture. Plants were incubated at 25°C in long-day light conditions.
Histological analysis
Mx-J, Mx-A, and Mx-R cutting samples were collected at 0, 14, and 28 d after IBA treatment; cuttings from 35S:MdMIR156a6 and 35S:MIM156 transgenic tobacco lines were excised at 0, 3, and 6 d after subculture on hormone-free MS medium. All samples were collected from three biological replicates and n = 10 in each replicate. A one-centimeter section from the bottom of each cutting was excised and fixed in a 5:50:5 (v/v/v) formaldehyde/ethanol/acetic acid (FAA) solution overnight at room temperature. Cuttings were dehydrated in an ethanol series (70, 85, 95, and 100%), infiltrated with xylene, and embedded in paraffin. Then, 15 μm-thick transverse sections were cut with a rotatory microtome (KD-2258, KEDEE, China) and stained with toluidine blue (Rigal et al., 2012).
Gene expression analysis
For relative gene expression assays with M. xiaojinensis, stem bark from 0.5 to 1 cm basal sections of 20 cuttings were frozen in liquid nitrogen at 0, 6, 12, 24, 72, 120, and 168 h after IBA treatment. For gene expression profile analysis in tobacco, stems from 0.5 to 1 cm basal sections of 20 cuttings were sampled at 0, 3, 6, 12, 24, 48, and 72 h after subculture on hormone-free MS medium. Total RNA was isolated from ~500 mg of frozen tissue using a modified cetyltrimethylammonium bromide (CTAB) method (Gasic et al., 2004). The RNA was digested by DNaseI (2313A, Takara, Dalian, China) and reverse-transcribed using oligo-dT18 primers and reverse transcriptase according to the manufacturer's instructions (2641A, Takara, Dalian, China). Semi-quantitative RT-PCR results were quantified by using ImageJ 1.47v (Wayne Rasband, National Institutes of Health, USA) according to the commands in “gels submenu” to analyze one-dimensional electrophoretic gels (https://imagej.nih.gov/ij/docs/menus/analyze.html#gels). Quantitative RT-PCR were performed using SYBR green reagents (RR820A, Takara, Dalian, China) in an Applied Biosystems 7500 real-time PCR system. M. xiaojinensis EF1α or N. benthamiana EF1α was used as the expression control for these experiments. The relative expression level was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). Three independent biological replicates and technical replicates were performed.
MicroRNA was extracted by using the RNAiso for Small RNA kit (9753Q, Takara, Dalian, China) according to the manufacturer's instruction. MiR156 expression level was analyzed by qRT-PCR as described previously (Xiao et al., 2014). The apple PIN and ARF family genes were previously identified (Devoghalaere et al., 2012; Zhang H. et al., 2015). The apple and N. benthamiana RTCS-like gene were selected by using a BLASTP search of known Maize RTCS (GenBank accession number EF051732) gene against the Apple Genome Database (https://www.rosaceae.org/) and Tobacco Genome Database (https://www.solgenomics.net/), respectively. N. benthamiana PIN and ARF genes were selected by using a BLASTP search of known Arabidopsis auxin-related genes against the Tobacco Genome Database (https://www.solgenomics.net/). All primers used for qRT-PCR are listed in Supplementary Tables 2,3.
Statistical analysis
Statistical analysis was performed using the Statistical Product and Service Solutions (SPSS) software (IBM Co., Armonk, USA). All experimental data were tested by Student's t-test or Duncan's multiple-range test.
Results
Adventitious rooting ability of Mx-A, Mx-J, and Mx-R leafy cuttings
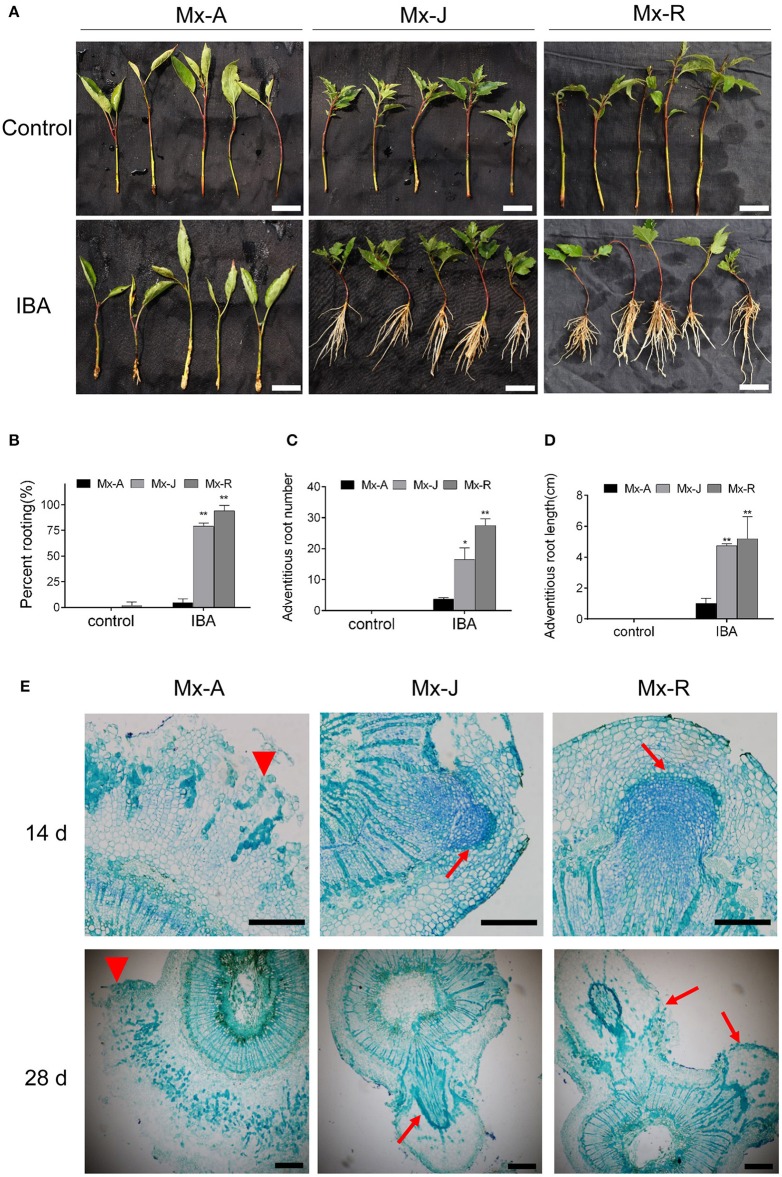
Juvenile cuttings (Mx-J) exhibited a highly adventitious rooting ability in M. xiaojinensis (Figure 1A). Leafy Mx-J cuttings exhibited a 78.63% rooting percentage at day 35 after IBA treatment (Figure 1B). In contrast, the rooting percentage of adult cuttings (Mx-A) was significantly lower (4.38%) than that of Mx-J. The adventitious root number per cutting was also lower in Mx-A cuttings than in Mx-J cuttings (Figure 1C). Similarly, the adventitious root length in Mx-J was significantly longer than that in Mx-A cuttings. Mx-R cuttings exhibited significantly enhanced adventitious rooting percentage and adventitious root number than Mx-A or Mx-J cuttings (Figure 1).
Figure 1.
Adventitious rooting ability of adult phase (Mx-A), juvenile phase (Mx-J) and rejuvenated (Mx-R) leafy cuttings of Malus xiaojinensis. (A) Morphological features, (B) rooting percent, (C) adventitious root number, and (D) adventitious root length of Mx-A, Mx-J, and Mx-R cuttings at 35 d of IBA or control treatments. Scale bars = 20 mm in (A). (B–D) Bars show SD from three biological replicates. n = 50 individuals in each replicate. Asterisks indicate significant difference from the Mx-A (Student's t-test, **P < 0.01, *P < 0.05). (E) Histological features of Mx-A, Mx-J, and Mx-R cuttings during adventitious root formation at 14 days (upper) and 28 days (bottom) after IBA treatment. Toluidine Blue-stained cross section of the stem of Mx-A, Mx-J, and Mx-R at 14 and 28 days after IBA treatment. Arrows indicate emerging adventitious root initials. Scale bars = 200 μm in (E).
To determine whether adult leafy cuttings have defects in the initiation of adventitious root formation primordia, cuttings were processed for histological analysis after IBA treatment. There were no obvious differences in anatomical structure between juvenile and adult softwood cuttings before IBA treatment (Supplementary Figure 2). At 14 d after IBA treatment, root primordia appeared in Mx-J and Mx-R but not Mx-A cuttings. At 28 d, root primordia penetrated the stem cortex and epidermis, and projected well beyond the stem surface in Mx-J and Mx-R (Figure 1E). In contrast, although meristematic cambium cells were observed between the phloem and xylem layers, except for callus, there was almost no differentiated root primordia detected in Mx-A cuttings even after 28 d of auxin treatment (Figure 1E).
The relationship between miR156 expression and adventitious root formation
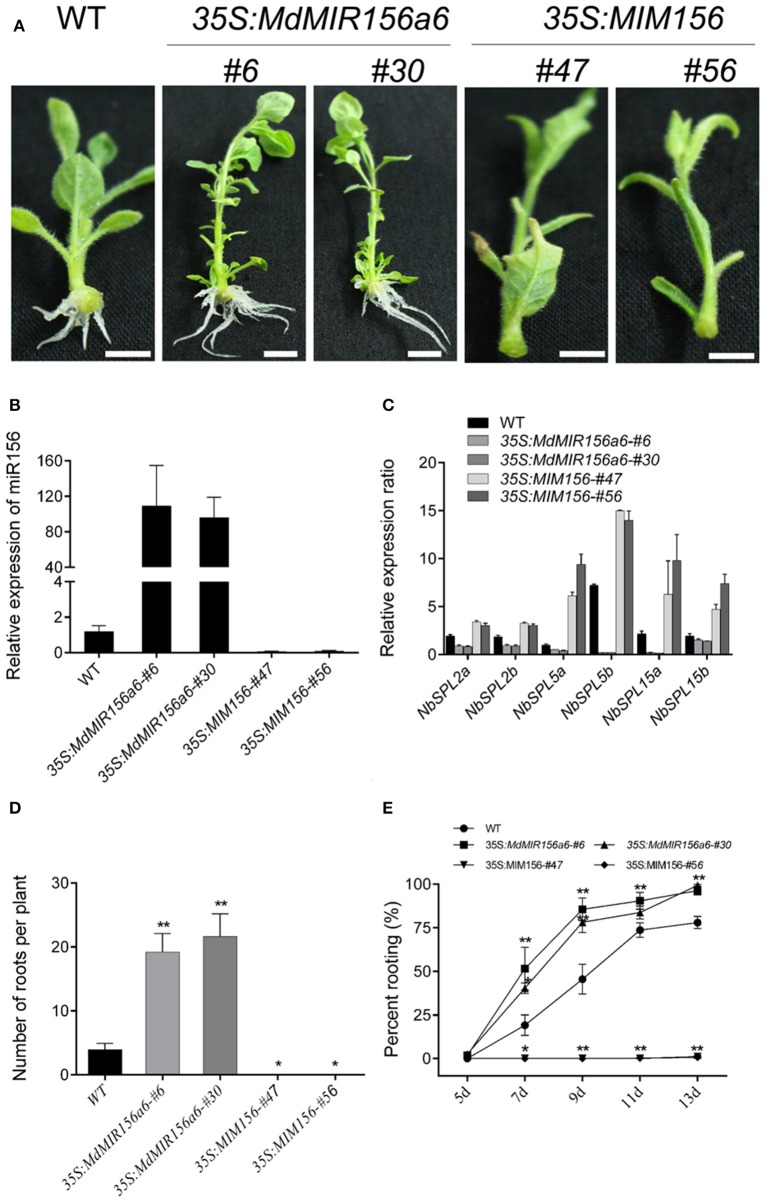
Consistent with the rooting ability, the expression level of miR156 in Mx-J and Mx-R cuttings were significantly higher than that in Mx-A cuttings (Figure 2). To validate if miR156 regulates adventitious root formation, we generated transgenic tobacco plants expressing 35S:MdMIR156a6 or 35S:MIM156 to enhance or inhibit miR156 activity, respectively (Figure 3A). Mature miR156 expression levels were significantly up-regulated in 35S:MdMIR156a6 plants and significantly reduced in 35S:MIM156 plants (Figure 3B). Similarly, the expression levels of some miR156 target genes, including NbSPL2a, NbSPL2b, NbSPL5a, NbSPL5b, NbSPL15a, and NbSPL15b were down-regulated at least 2-fold in 35S:MdMIR156a6 stems, but at least 5-fold up-regulated in 35S:MIM156 stems (Figure 3C). 35S:MdMIR156a6 plants developed almost 20 adventitious roots per cutting, which was 4-fold more than the wild type after 13 days of culture on MS medium. Almost no adventitious roots were observed in 35S:MIM156 cuttings (Figure 3D). In addition to the difference in adventitious root number, the rate of adventitious root development in 35S:MdMIR156a6 plants was significantly faster than that measured in the wild-type plants (Figure 3E).
Figure 2.
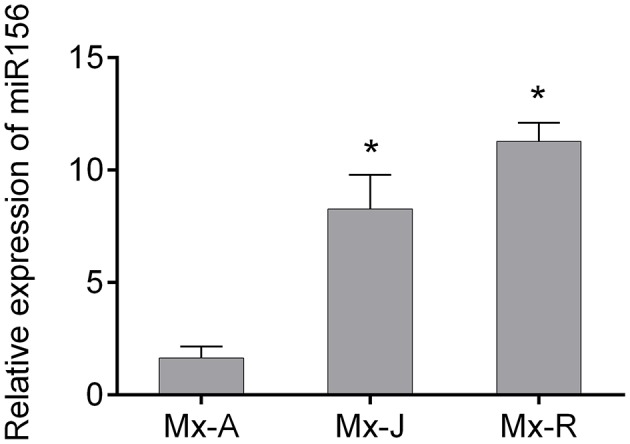
Expression level of miR156 in adult phase (Mx-A), juvenile phase (Mx-J), and rejuvenated (Mx-R) leafy cuttings of Malus xiaojinensis. MicroRNA was extracted from Mx-A, Mx-J, and Mx-R cuttings before IBA treatment. Relative expression was measured with quantitative real time PCR and normalized to that 5S rRNA. Asterisks indicate statistical significance (*P < 0.05) in comparison with Mx-A. Bars show SD from three biological replicates.
Figure 3.
MiR156 affects adventitious rooting capacity in transgenic tobacco plants. (A) Phenotypes of 13-day-old plants (wild-type, 35S:MdMIR156a6, and 35S:MIM156) grown on hormone-free MS medium. Scale bars = 20 mm. (B) Relative expression level of miR156 in transgenic tobacco lines. Bars show SD from three biological replicates. (C) Relative expression levels of NbSPLs in WT, 35S:MdMIR156a6, and 35S:MIM156 plants in stems. (D,E) Rooting ability of tobacco stem cuttings. (D) Adventitious root number per cutting and (E) percent rooting. Adventitious root number was counted after 13 days of growth on MS medium. Bars show SD from three biological replicates; n = 10 in individuals per replicate. Asterisks indicate significant differences from the WT (Student's t-test, **P < 0.01, *P < 0.05).
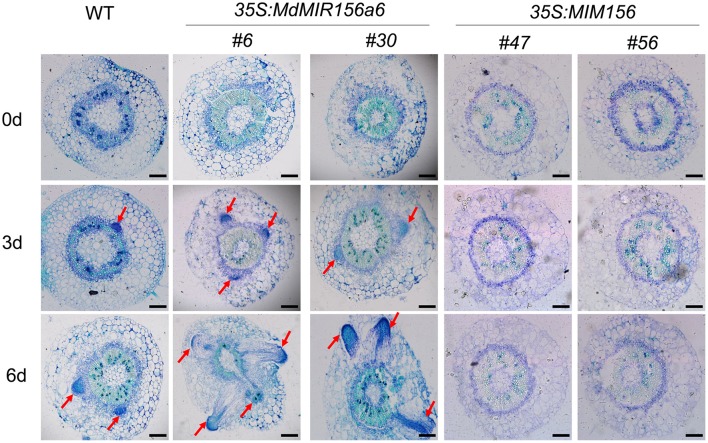
To check whether miR156 affects the initiation of adventitious root primordia, serial cross sections of the stems of wild-type, 35S:MdMIR156a6 expressing transgenic lines, and 35S:MIM156 expressing transgenic tobacco plants were stained with toluidine blue. When compared with the wild-type, the initiation of adventitious root primordia was accelerated and well-developed in 35S:MdMIR156a6 stem cuttings only 3 days after subculture on MS (Figure 4). By contrast, no adventitious root primordia were observed in 35S:MIM156 stem cuttings throughout the experiment (Figure 4).
Figure 4.
Histological features of WT, 35S:MdMIR156a6, and 35S:MIM156 tobacco stems during adventitious root formation. Toluidine Blue-stained cross-section of the stem of WT, 35S:MdMIR156a6, and 35S:MIM156 at 0, 3, and 6 d after subculture on MS medium. Arrows indicate initial emerging adventitious roots and adventitious root primordia. Scale bars = 200 μm.
Interaction between miR156 and auxin during adventitious root formation
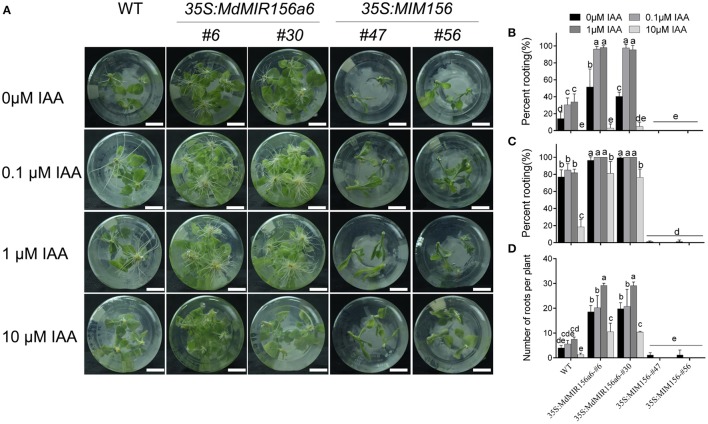
To determine whether miR156 regulates adventitious root formation through modulating endogenous auxin levels or auxin polar transport, we examined adventitious rooting capacity in wild-type, 35S:MdMIR156a6, and 35S:MIM156 stem cuttings grown on MS medium supplemented with different concentrations of indole-3-acetic acid (IAA) or 1-N-naphthylphthalamic acid (NPA). The percent rooting was significantly increased in wild-type and 35S:MdMIR156a6 stem cuttings when cultured in the presence of 0.1 or 1 μM IAA (Figures 5A,B). The 10 μM IAA treatment delayed adventitious root formation in both wild-type and 35S:MdMIR156a6 plants; however, 35S:MdMIR156a6 plants exhibited a significantly higher adventitious rooting percent and adventitious root number than wild-type. In addition, with IAA application, the adventitious root number was clearly increased in both wild-type and 35S:MdMIR156a6 plants (Figures 5A,C) but IAA treatment did not rescue the adventitious rooting capacity defect in 35S:MIM156 plants (Figure 5).
Figure 5.
Effects of IAA on adventitious root formation of 35S:MdMIR156a6 and 35S:MIM156 tobacco plants. (A) Phenotypes of 13-day-old plants (WT, 35S:MdMIR156a6, and 35S:MIM156) grown on MS medium with or without IAA treatment. Scale bars = 15 mm. (B,C) Percent rooting of tobacco stem cuttings grown on MS medium for 7 and 14 d, respectively. (D) Adventitious root number per cutting. Adventitious root number was counted after 14 d on MS medium. Bars show SD with three biological replicates; n = 5 cuttings in each replicate. The statistical analysis was performed by Duncan's multiple range test at level p ≤ 0.05. The same lowercase letter indicate no significant differences.
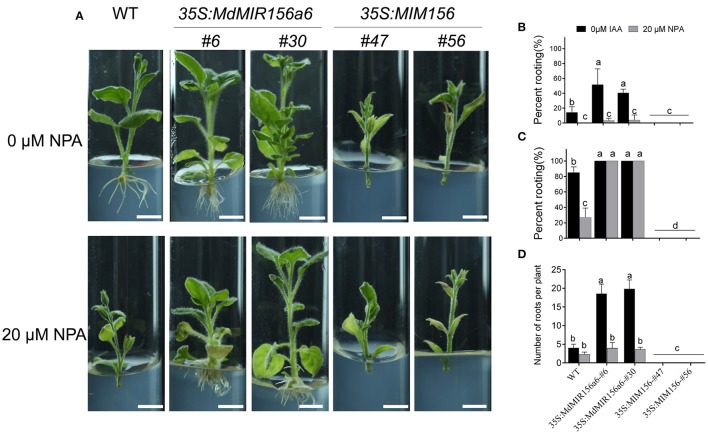
Treatments with 20 μM NPA significantly inhibited the adventitious root formation in wild-type (Supplementary Figure 3); adventitious rooting was obviously delayed and adventitious root number was reduced in both wild-type and 35S:MdMIR156a6 plants treated with 20 μM NPA (Figure 6A). Indeed, rooting percent and root number in 35S:MdMIR156a6 plants were still consistently higher than that in wild-type (Figures 6B,C).
Figure 6.
Effects of NPA on adventitious root formation in 35S:MdMIR156a6 and 35S:MIM156 tobacco plants. (A) Phenotypes of 13-day-old plants (WT, 35S:MdMIR156a6, and 35S:MIM156) grown on MS medium with 20 μM NPA. Scale bars = 15 mm. (B,C) Percent rooting of tobacco stem cuttings grown on MS medium with 20 μM NPA treatment for 7 and 14 d, respectively. (D) Adventitious root numbers per cutting. Bars show SD with three biological replicates; n = 10 cuttings in each replicate. The statistical analysis was performed by Duncan's multiple range test at level p ≤ 0.05. The different lowercase letter indicate significant differences.
PIN genes are not altered by miR156 expression
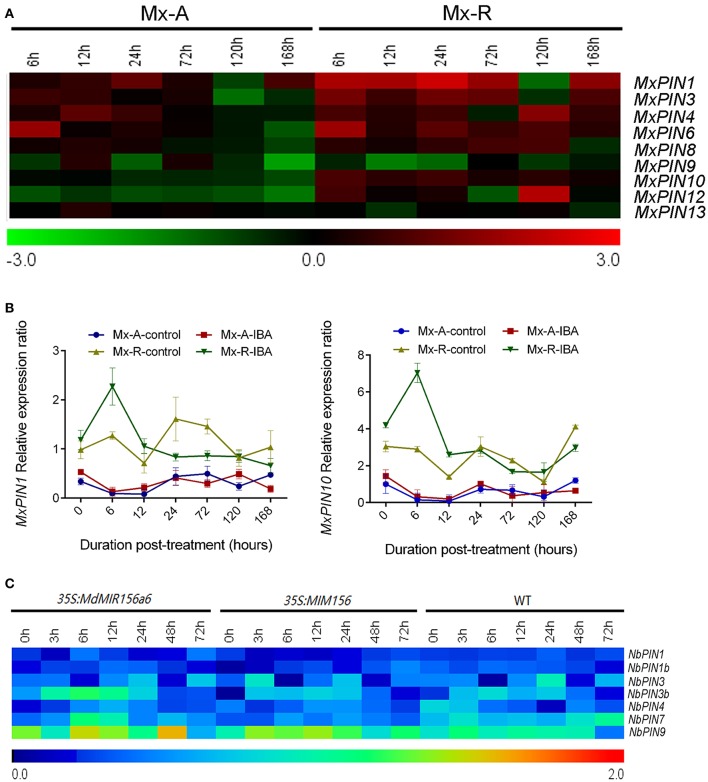
The expression levels of MxPIN3, MxPIN4, MxPIN6, MxPIN8, MxPIN9, MxPIN12, and MxPIN13 did not show obvious differences between Mx-A and Mx-R leafy cuttings during the adventitious root-induction process (Figure 7A and Supplementary Figure 4). The expression of MxPIN2, MxPIN5, MxPIN7, and MxPIN11 was not detected in any of the RNA samples tested. As shown in Supplementary Figure 4, the expression of MxPIN1 and MxPIN10 was higher in Mx-R than in Mx-A with or without IBA treatment. Both MxPIN1 and MxPIN10 expression was induced more than 2-fold after 6 h in the presence of IBA treatment in Mx-R, but not in Mx-A (Figure 7B). The relatively high expression level of MxPIN1 and MxPIN10 may contribute to adventitious rooting competitive in Mx-R cuttings. However, the relationship between miR156 and PIN gene expression was not robust in transgenic tobacco; no distinct changes were detected in the expression of any NbPIN members between wild-type, 35S:MdMIR156a6, and 35S:MIM156 tobacco plants (Figure 7C).
Figure 7.
Expression profiles of PINs during adventitious rooting in Malus xiaojinensis and transgenic Nicotiana benthamiana. All the results of semi-quantitative RT-PCR were quantified using the ImageJ software. Log-transformed values of the relative expression levels of MxPIN family genes under IBA treatment compared to controls were used for hierarchical cluster analysis with MeV 4.8.1. The color scale represents relative expression levels with red denoting up-regulation and green denoting down-regulation. The relative expression levels of NbPIN genes compared to NbEF1α were used for hierarchical cluster analysis with MeV 4.8.1. Sampling times are indicated at top of the figure. (A) Expression profiles of MxPIN genes in stem bark of Mx-A and Mx-R during adventitious root formation process after IBA treatment (original results show in Supplementary Figure 4). (B) Quantitative RT-PCR analysis of the expression dynamics of MxPIN1 and MxPIN10 in stem bark from Mx-A and Mx-R during adventitious root formation after IBA treatment. Bars show SD from three biological replicates. (C) NbPIN genes expression patterns in WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic tobacco stems during the adventitious rooting process (original results show in Supplementary Figure 5).
ARF genes expression did not change with miR156 levels
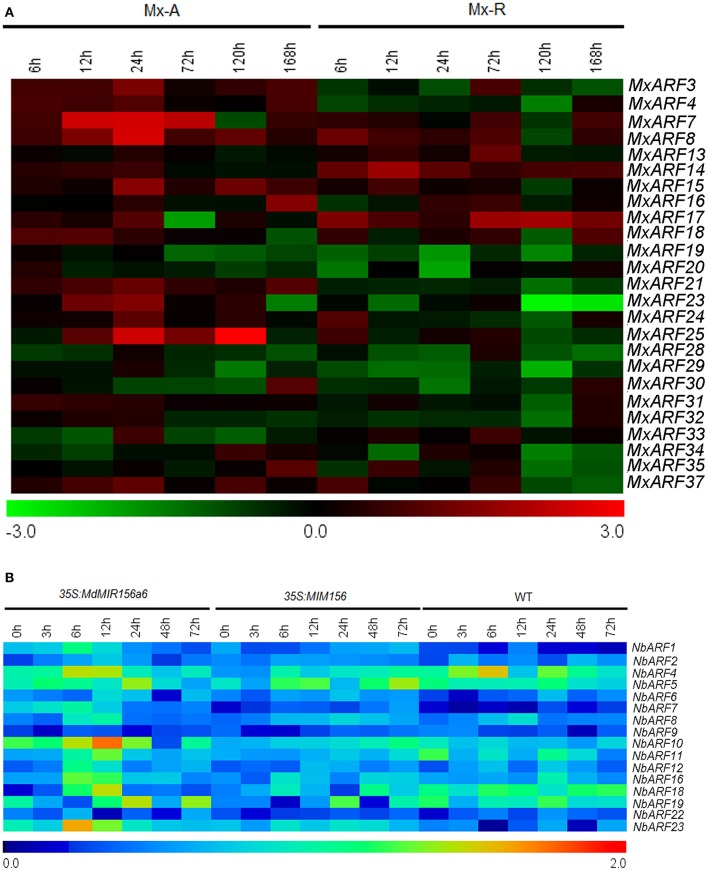
The MxARF4, MxARF7, MxARF14, MxARF15, MxARF16, MxARF17, MxARF19, MxARF20, and MxARF28 genes showed higher expression in Mx-A than in Mx-R cuttings with or without IBA treatment during the adventitious root formation process (Supplementary Figure 6). In contrast, expression of MxARF3 and MxARF8 was higher in Mx-R than in Mx-A cuttings (Supplementary Figure 6). MxARF21, MxARF23 and MxARF25 were down regulated in plants treated with IBA in Mx-R but not Mx-A cuttings (Figure 8A). However, no distinct changes were detected in 16 putative NbARF gene family members between wild-type, 35S:MdMIR156a6, and 35S:MIM156 tobacco plants during the adventitious rooting process (Figure 8B). This indicates that miR156 may not be associated with the expression of ARF transcription factors during the regulation of adventitious root formation.
Figure 8.
Expression profiles of ARFs during adventitious rooting in Malus xiaojinensis and transgenic Nicotiana benthamiana. All the results of semi-quantitative RT-PCR were quantified using the ImageJ software. Log-transformed values of the relative expression levels of MxARF family genes under IBA treatment compared to controls were used for hierarchical cluster analysis with MeV 4.8.1. The color scale represents relative expression levels with red denoting up-regulation and green denoting down-regulation. The relative expression levels of NbARFs genes compared to NbEF1α were used for hierarchical cluster analysis with MeV 4.8.1. Sampling times are indicated at top of the figure. (A) Expression profiles of MxARF genes in stem bark of Mx-A and Mx-R during adventitious root formation process after IBA treatment (original results show in Supplementary Figure 6). (B) NbARF genes expression patterns in WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic tobacco stems during the adventitious rooting process (original results show in Supplementary Figure 7).
RTCS-like gene expression varied with miR156 levels
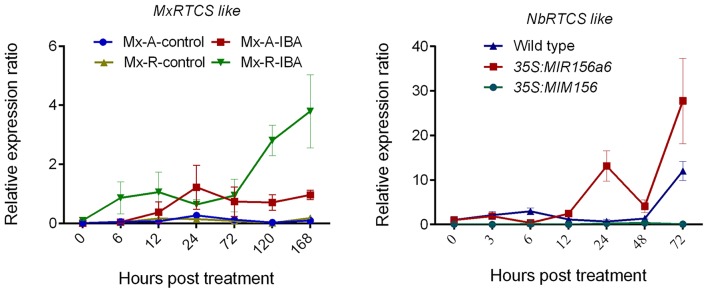
In M. xiaojinensis, the MxRTCS-like gene was up-regulated 6–24 h after IBA treatment in both Mx-A and Mx-R cuttings, but the maximum induction of MxRTCS was observed in Mx-R cuttings at 120–168 h, which was 4-fold higher than that measured in Mx-A cuttings (Figure 9). Similarly, the expression of NbRTCS was also significantly induced 24–72 h after subculture in hormone-free medium in 35S:MdMIR156a6 plants, but was delayed 72 h subculture in hormone-free medium in wild-type. However, the expression of NbRTCS did not change throughout the experiment in 35S:MIM156 plants (Figure 9).
Figure 9.
Quantitative RT-PCR analysis the expression profiles of MxRTCS-like and NbRTCS-like genes during adventitious root formation in Malus xiaojinensis and transgenic Nicotiana benthamiana. Bars show SD from three biological replicates.
Response in MxSPL gene expression to miR156 levels
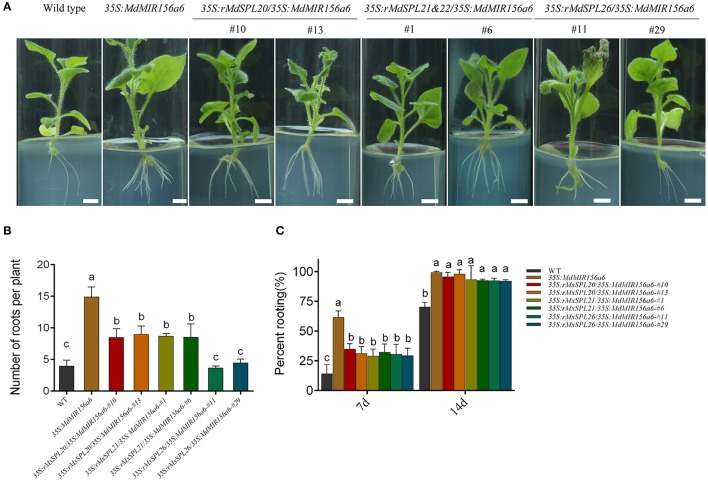
Of the 13 putative miR156-regulated SPL gene family members in apple genome, nine were actively expressed in both Mx-J and Mx-A plant cuttings, but did not include MxSPL3, MxSPL10, MxSPL11, and MxSPL12. To investigate which M. xiaojinensis SPL gene family member was involved in adventitious root formation, mutants of SPL genes, indicated here as resistant SPLs (rSPLs), were designed to no longer be targeted by miR156 (Supplementary Figure 8; Schwab et al., 2005). MxrSPL4a&4b, MxrSPL18, MxrSPL19, MxrSPL20, MxrSPL21&22, MxrSPL24, and MxrSPL26, driven by CaMV 35S promoter, were transformed into 35S:MdMIR156a6 transgenic tobacco plants (Supplementary Figure 9). In independent bivalent transgenic lines, 35S:rSPL4a&4b/35S:MdMIR156a6, 35S:rSPL18/35S:MdMIR156a6, 35S:rSPL19/35S:MdMIR156a6, and 35S:rSPL24/35S:MdMIR156a6 the adventitious rooting rate and adventitious root number did not significantly differ from that of plants expressing 35S:MdMIR156a6 (Supplementary Figure 10), indicating that MxSPL4a&4b, MxSPL18, MxSPL19, and MxSPL24 are not involved in adventitious rooting. In contrast, the bivalent transformants 35S:rSPL20/35S:MdMIR156a6, 35S:rSPL21&22/35S:MdMIR156a6, and 35S:rSPL26/35S:MdMIR156a6 exhibited reduced adventitious rooting ability (Figure 10A). 35S:rSPL20/35S:MdMIR156a6 and 35S:rSPL21&22/35S:MdMIR156a6 produced significantly fewer adventitious roots than 35S:MdMIR156a6 plants, but still more than wild-type plants (Figure 10B). The adventitious root number in 35S:rSPL26/35S:MdMIR156a6 plants was significantly fewer than that in 35S:MdMIR156a6 plants (Figure 10B). In addition, the adventitious root development rate in 35S:rSPL20/35S:MdMIR156a6, 35S:rSPL21&22/35S:MdMIR156a6, and 35S:rSPL26/35S:MdMIR156a6 plants were similar as that measured in wild-type, but slower than that observed in 35S:MdMIR156a6 plants 7 d after culture on MS medium (Figure 10C). These results indicate that MxSPL20, MxSPL21&22, and MxSPL26 not only affected adventitious root number but also delayed rooting rate.
Figure 10.
Role of MxSPL20, 21&22, and 26 genes during adventitious root formation in transgenic Nicotiana benthamiana. (A) Adventitious root formation of 35S:rMxSPL/35S:MdMIR156a6 plants. Scale bars = 1 cm. (B,C) Quantitative analysis of rooting ability of tobacco stem cuttings. (B) Adventitious root numbers per cutting were counted after 14 d on MS medium. (C) Percent rooting was investigated after 7 and 14 d of growth on MS medium. Bars show SD with three biological replicates; n = 5 for each replicate. The statistical analysis was performed by Duncan's multiple range test at level p ≤ 0.05. The same lowercase letter indicate no significant differences.
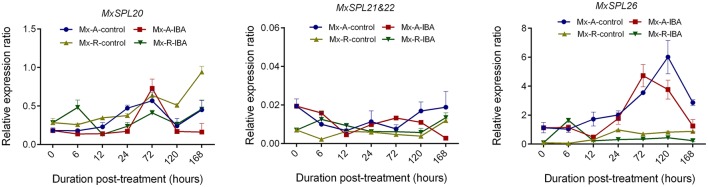
The MxSPL20, MxSPL21&22, and MxSPL26 expression profile was detected at 0, 6, 12, 24, 72, 120, and 168 h after IBA treatment (Figure 11). There were no obviously differences in the expression of MxSPL20 and MxSPL21&22 between Mx-A and Mx-R cuttings; however, the expression of MxSPL26 was significantly higher in Mx-A than in Mx-R cuttings, indicating MxSPL26 could be a key SPL family member involved in adventitious rooting of leafy cuttings.
Figure 11.
Expression profile of MxSPL20, 21&22, and 26 genes during adventitious root formation in Malus xiaojinensis leafy cuttings. RNA was extracted from Mx-A and Mx-R stem bark after IBA treatment at serial time-points. Relative expression was measured using quantitative real time PCR normalized to MxEF1α. Bars show SD from three biological replicates.
Discussion
Although, it appears that juvenile phase softwood cuttings are much easier to root than the adult ones, there is little data detailing the mechanism of these differences. The expression level of miR156 was decreased during juvenile to adult phase change (Du et al., 2015; Ji et al., 2016). However, the effect of miR156 on adventitious root formation barely rated a mention in previous reports (Zhang et al., 2011; Feng et al., 2016; Massoumi et al., 2017). Although, miR156-targeted SPLs are known to control varied physiological and developmental processes (Wang et al., 2008; Shikata et al., 2009; Yu et al., 2010, 2015; Gou et al., 2011), with which the miR156-targeted SPL gene member is associated with adventitious rooting and how miR156 interacts with other rooting regulatory factors such as IAA are so far not fully understood to date. In the present study, we found that the high miR156 expression was required for adventitious roots formation in an apple rootstock, M. xiaojinensis. We also confirmed the involvement of MxSPL26 in inhibiting adventitious root formation.
Auxin and high miR156 expression level are both necessary for adventitious rooting
For some rooting recalcitrant woody plants, juvenility is necessary for efficient adventitious rooting. In general, cuttings from juvenile nodes root more readily than cuttings from adult nodes in woody plants (Greenwood, 1995). Indeed, miR156 expression levels were significantly higher in Mx-J and Mx-R cuttings than Mx-A cuttings (Figure 2), and the rooting ability of Mx-R and Mx-J cuttings were consistently higher than that of Mx-A cuttings (Figure 1). A significant decrease in miR156 expression was observed in shoots taken from 1.4 m trunk above ground compared to shoots taken from below 1.4 m in M. xiaojinesis seedlings (Ji et al., 2016). In our previous data, the micro-shoots from adult phase M. xiaojinnesis explants were rejuvenated successfully after 15 passages of in vitro subculture, marked by the elevated expression of miR156 expression in leaves of the micro-shoots, and coupled with recovered adventitious rooting ability and leaf lobes (Xiao et al., 2014).
When miR156 level was manipulated via transformation with a 35S:MIR156 construct in tomato, tobacco, or Arabidopsis, the adventitious rooting increased (Zhang et al., 2011; Feng et al., 2016; Massoumi et al., 2017). In the present study, the miR156 expression level was manipulated in transgenic tobacco to analyze how miR156/SPL modules are involved in adventitious rooting. Consistent with the previous reports, the adventitious rooting capacity also varied with 35S:MIM156 and 35S:MdMIR156a6 overexpressing transgenic tobacco plants (Figure 3). However, the uncoupling of miR156 expression and adventitious root formation was reported in E. grandis (Levy et al., 2014); therefore, miR156 may be necessary but not sufficient for adventitious rooting in woody plants. Except for high expression of miR156, auxin was also integrant for adventitious root formation. In absence of IBA treatment, even Mx-J and Mx-R leafy cuttings exhibit adventitious root defects (Figure 1). In agreement to this result, the adventitious rooting ability was reduced in all tobacco lines upon treatment with the polar auxin transport inhibitor NPA (Figure 6).
miR156 affects adventitious root formation independently from PIN and ARF genes expression
Auxin and miR156 are both involved in adventitious root development (Zhang et al., 2011; Feng et al., 2016; Steffens and Rasmussen, 2016; Massoumi et al., 2017). However, the interaction of miR156 and auxin signaling pathway is unexplored during adventitious root development. In comparison to the wild-type samples, NbPINs and NbARFs expressions was not substantially modulated in 35S:MdMIR156a6 and 35S:MIM156 plants (Figures 7, 8), indicating that miR156 promoted adventitious root formation independently from changing PIN and ARF genes expression. These results are supported by the transcriptome data, which show that PIN and ARF genes in Medicago sativa are not significantly differentially expressed between miR156 overexpression and wild-type plants (Gao et al., 2016). Similarly, the auxin response was not changed in either Pro35S:MIR156 or Pro35S:MIM156 Arabidopsis, indicating that miR156 does not modulate the auxin response during regulating shoot regeneration (Zhang T. Q. et al., 2015). Collectively, these findings suggest that miR156 does not modulate the early auxin response.
Conversely, auxin can induce the expressions of two MIR156 genes and two SPL genes during lateral root development in transgenic Arabidopsis (Yu et al., 2015). GUS staining revealed that MIR156B was specifically expressed in primary and lateral root primordia, MIR156D was especially active in primary and lateral root tip, and two SPL genes were also highly or specifically expressed in root (Yu et al., 2015). Hence, auxin inducing MIR156 and SPL expression may be tissue/organ specific during lateral root development. Our results revealed that the expression level of MxSPL26 or MxSPL20 or MxSPL21&22 was not obviously affected under IBA treatment (Figure 11). Similarly, miR156 expression was not affected by IBA application during the induction of adventitious root development in both juvenile and adult E. grandis stem cuttings (Levy et al., 2014). Thus, miR156/SPL modules were not downstream targets of auxin during adventitious root formation.
Rtcs and Rtcl in maize are orthologs of CRL1 in rice (O. sativa) and of AtLBD29 in A. thaliana, and all of these genes are involved in the initiation of adventitious root formation and are targets of the ARF gene family (Inukai et al., 2005; Taramino et al., 2007; Liu J. C. et al., 2014). Although MxRTCS-like gene expression was induced in both Mx-A and Mx-R cuttings after IBA treatment, the distinct fold-change in expression occurred only in Mx-R cuttings (Figure 9). In agreement with these finding, the NbRTCS expression was significantly induced in 35S:MdMIR156a6 transgenic tobacco plants, but no substantial changes were detected in 35S:MIM156 transformants (Figure 9). These data suggest that high expression level of miR156 is required for auxin inducing expression of RTCS-like during adventitious root formation in M. xiaojinesis and transgenic tobacco. Therefore, we speculate that decline of miR156 expression in adult phase leafy cuttings may inhibit transcript abundance of RTCS-like gene induced by auxin, thereby reducing the adventitious rooting capacity (Figure 12). The histological features showed that Mx-A leafy cuttings and 35S:MIM156 transgenic tobacco plants exhibited defect in adventitious root primordia initiation (Figures 1E,4), which provided an evidence for above mentioned speculation. In agreement with our observations, maize mutant rtcs and rice mutant rtcl both failed in adventitious root primordia initiation (Inukai et al., 2005; Muthreich et al., 2013). However, the molecular mechanism underlying miR156 modulate auxin induced RTCS-like gene expression remains unknown.
Figure 12.
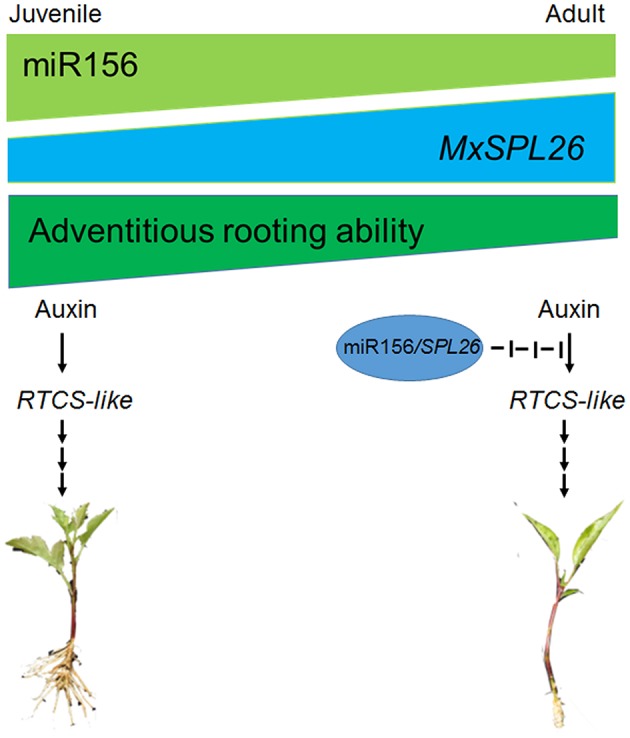
Illustration of role of juvenility and auxin in triggering adventitious rooting. In adult leafy cuttings, decline of miR156 caused high expression level of SPL26. This modules may inhibit transcript abundances of RTCS-like gene induced by auxin, thereby reducing the adventitious rooting capacity in adult plants.
Although, the expressions of PIN and ARF genes were independently from the miR156 expression level, the response of MxPIN1 and MxPIN10 gene expression to auxin treatment differed in Mx-R and Mx-A samples. The different responses of PIN family members to auxin treatment between Mx-R and Mx-A cuttings may be caused by other developmental signals, such as glutathione content. In apple seedlings, the glutathione content and glutathione/glutathione disulfide ratio were much higher in the juvenile phase than in the adult phase, and modulating glutathione content caused concomitant changes in miR156 expression levels (Du et al., 2015). It has been proved that the reduction of glutathione availability by L-buthionine-(S,R)-sulfoximine (BSO) treatment reduced the expression of PIN1, PIN2, and PIN3 in Arabidopsis (Bashandy et al., 2010).
In addition, MxARF4, MxARF7, MxARF14, MxARF15, MxARF16, MxARF17, MxARF19, MxARF20, and MxARF28 showed relative higher expression levels in Mx-A than in Mx-R cuttings during adventitious root formation (Supplementary Figure 6). Interestingly, the peptide sequences of these ARFs, except for ARF20, are enriched in serine, proline, and leucine in their middle regions (Supplementary Figure 11). Arabidopsis ARFs with this middle region are usually transcriptional repressors (Guilfoyle and Hagen, 2007). The differential expression of MxPIN and MxARF might partly cause Mx-A recalcitrant to root formation.
MxSPL26 is the key target gene in miR156 regulation of adventitious rooting in M. xiaojinensis
MiR156 and its targeted SPL genes were demonstrated to control plant phase transition and related traits such as cell size and number, trichome development, anthocyanin synthesis, leafy morphology, flowering time, and lateral root development (Wang et al., 2008; Shikata et al., 2009; Yu et al., 2010, 2015; Gou et al., 2011). Yet, the function of SPL genes involving in adventitious rooting are largely unexplored. It was reported that hypocotyls of spl2/9/11/13/15 mutants, as well as plants transformed with 35S:MIR156A, produced the same number of adventitious roots as wild-type plants (Xu et al., 2016). However, it is not clear which SPL gene involved in regulating adventitious rooting in Arabidopsis, because the expression level of miR156 targeted SPLs had no significantly difference among spl2/9/11/13/15 mutants, 35S:MIR156A plants and wild-type young seedlings (Xu et al., 2016). Here, MxSPL26 was experimentally confirmed to be involved in inhibiting adventitious root formation most possibly in function redundancy with SPL20 and SPL21&22, but SPL26 seemed to play a major role. The expression of MxSPL26 was constantly higher than that of MxSPL20 and MxSPL21&22 in M. xiaojinensis cuttings. MxSPL26 relative expression exhibited a 5- to 10-fold increase in stem bark from Mx-A cuttings than in that from Mx-R after 72~120 h from plugging into the media, but no obvious differences in the expression of MxSPL20 and MxSPL21&22 genes were found between Mx-A and Mx-R (Figure 11). The number of roots per plant was significantly reduced in all the 35S:rMxSPLs/35S:MdMIR156a6 lines but MxSPL26 resistant form transgenic tobacco lines produced the least number of adventitious roots compared with rMxSPL20, rMxSPL21&22 and 35S:MdMIR156a6 transgenic lines (Figure 10 and Supplementary Figure 10). According to the phylogenetic tree, MxSPL20 and MxSPL21&22 exhibited close evolutionary, but quite far phylogenetic relationship from MxSPL26 (Supplementary Figure 12). Notably, although SPL3, SPL9, and SPL10 were all involved in repressing Arabidopsis lateral root development, SPL10 seemed to play a major role (Yu et al., 2015). Similarly, AtSPL3 and AtSPL9 exhibited close evolutionary, but quite far phylogenetic relationship from AtSPL10 (Supplementary Figure 12).
In conclusion, in semi-lignified leafy cuttings of M. xiaojinensis, a relatively higher expression of the miR156 is necessary for adventitious root formation. MiR156 functions via its target gene MxSPL26 and rooting-related genes such as MxRTCS-like; but acts independently of MxPIN or MxARF family members in response to auxin. These results provided a potential strategy for the improvement of the adventitious rooting ability of perennial woody plants via manipulating miR156 and SPL gene expression.
Author contributions
XZ, YW, TW, XzX, and ZH prepared the plant materials and designed the experiments. XzX, XL, and XH conducted the experiments. XzX took the photographs. XzX and XZ analyzed the data and XzX wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by Special Fund for Agro-scientific Research in the Public Interest (201203075); the Modern Agricultural Industry Technology System (CARS-28); the National Natural Science Foundation of China (NSFC) (Grant no. 31372020; 31672107); Key Laboratory of Biology and Genetic Improvement of Horticulture Crop (Nutrition and Physiology), Ministry of Agriculture, and Beijing Collaborative innovation center for eco-environmental improvement with forestry and fruit trees.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01059/full#supplementary-material
Expression profiles of nine miR156 precursors were analyzed using semi-quantitative RT-PCR in Malus xiaojinensis stem bark.
Histological features of Mx-A, Mx-J, and Mx-R leafy cuttings of Malus xiaojinensis before IBA treatment. Cross sections of the stems were stained with toluidine blue. Scale bars = 200 μm.
NPA (1-N-naphthylphthalamic acid) concentration selected for wild type tobacco plant treatment. Tobacco plants were transferred to MS medium with 5, 20, or 80 μM NPA for 14 days. (A) The phenotype and (B) percent rooting were evaluated. Bars show SD from three biological replicates; n = 10 individuals in each replicate. Scale bars = 15 mm.
Semi-quantitative RT-PCR analysis of the expression dynamics of MxPINs in stem barks from Mx-A and Mx-R during adventitious root formation in Malus xiaojinensis under IBA treatment. MxEF1α was used as an internal control. The upper and lower bands represent treatment and control, respectively.
NbPIN genes expression pattern in tobacco stems from WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic lines growing on MS medium during the adventitious rooting process. NbEF1α was used as an internal control.
Semi-quantitative RT-PCR analysis of the expression dynamics of MxARFs in stem barks from Mx-A and Mx-R during adventitious root formation in Malus xiaojinensis under IBA treatment. MxEF1α was used as an internal control. The upper and lower bands represent treatment and control, respectively.
NbARF genes expression pattern in tobacco stems from WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic lines growing on MS medium during the adventitious rooting process. NbEF1α was used as an internal control.
Diagram of the miR156 target sites of the WT and modified version of MxSPLs. Capital letters indicate the amino acid sequences in Malus xiaojinesis.
Molecular characterization of transgenic tobacco plants. (A) Semi-quantitative PCR analysis of rMxSPLs DNA and mRNA levels and (B) miR156 expression levels in 10-day old seedling leaves.
Role of MxSPL4a&4b, 18, 19, and 24 genes during adventitious root formation in transgenic Nicotiana benthamiana. (A) Adventitious root formation in 35S:rMxSPL/35S:MdMIR156a6 plants. Scale bars = 1 cm. (B,C) Quantitative analysis of the rooting ability in tobacco stem cuttings. (B) Percent rooting was investigated after 7 and 14 d on MS medium. (C) Adventitious root numbers per cutting were counted after 14 d on MS medium. Bars show SD from three biological replicates; n = 5 plants in each individual replicate. The statistical analysis was performed by Duncan's multiple range test at level p ≤ 0.05; means with different letters are significantly different from each other.
The MxARF family of transcription factors in Malus domestic. MdARF2, 11, 12, 20, 30, 33, and 36–39 have an activation domain (AD) that is enriched in glutamine (Q), serine (S), and leucine (L). The remainder of the ARFs consist of transcriptional repressors with a repression domain (RD) that is enriched in serine (S) and in some cases proline (P). All ARFs contain a conserved DNA binding domain (DBD).
Phylogenetic analysis of miR156-targeted SPL between apple and Arabidopsis. Phylogenetic tree was constructed with SBP domain protein sequences. Phylogenetic tree was constructed using MEGA 4.0 software with the neighbor-joining (NJ) method and the bootstrap test replicated 1,000 times.
Primers for constructing vector.
PCR primers of genes in Malus xiaojinesis.
PCR primers of genes in Nicotiana benthamiana.
References
- Abu-Abied M., Szwerdszarf D., Mordehaev I., Levy A., Rogovoy O., Belausov E., et al. (2012). Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of Eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. Plant J. 71, 787–799. 10.1111/j.1365-313X.2012.05032.x [DOI] [PubMed] [Google Scholar]
- Abu-Abied M., Szwerdszarf D., Mordehaev I., Yaniv Y., Levinkron S., Rubinstein M., et al. (2014). Gene expression profiling in juvenile and mature cuttings of Eucalyptus grandis reveals the importance of microtubule remodeling during adventitious root formation. BMC Genomics 15:826. 10.1186/1471-2164-15-826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T., Guilleminot J., Vernoux T., Caparros-Ruiz D., Ljung K., Meyer Y., et al. (2010). Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22, 376–391. 10.1105/tpc.109.071225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V. B., Johannes E., Whetten R. W., Sederoff R. R., Spiker S. L., Lanz-Garcia C., et al. (2004). An auxin-inducible gene from loblolly pine (Pinus taeda L.) is differentially expressed in mature and juvenile-phase shoots and encodes a putative transmembrane protein. Planta 218, 916–927. 10.1007/s00425-003-1175-4 [DOI] [PubMed] [Google Scholar]
- Chen S. W., Zha X. J., Ma B. K., Li Z. Z., Yuan X. L. (1981). A preliminary study on propagation of several fruit trees and rootstocks from hardwood cuttings. J. Agric. Univ. Hebei. 4, 14–21 (in chinese: ). [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544–549. 10.1038/ng2001 [DOI] [PubMed] [Google Scholar]
- da Costa C. T., de Almeida M. R., Ruedell C. M., Schwambach J., Maraschin F. S., Fett-Neto A. G. (2013). When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 4:133. 10.3389/fpls.2013.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoghalaere F., Doucen T., Guitton B., Keeling J., Payne W., Ling T. J., et al. (2012). A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol. 12:7. 10.1186/1471-2229-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Jia X. L., Wang Y., Wu T., Han Z. H., Zhang X. Z. (2015). Redox homeostasis and reactive oxygen species scavengers shift during ontogenetic phase changes in apple. Plant Sci. 236, 283–294. 10.1016/j.plantsci.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Feng S. J., Xu Y. M., Guo C. K., Zheng J. R., Zhou B. Y., Zhang Y. T., et al. (2016). Modulation of miR156 to identify traits associated with vegetative phase change in tobacco (Nicotiana tabacum). J. Exp. Bot. 67, 1493–1504. 10.1093/jxb/erv551 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Gao R., Austin R. S., Amyot L., Hannoufa A. (2016). Comparative transcriptome investigation of global gene expression changes caused by miR156 overexpression in Medicago sativa. BMC Genomics 17:658. 10.1186/s12864-016-3014-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K., Hernandez A., Korban S. S. (2004). RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 22, 437–438. 10.1007/BF02772687 [DOI] [Google Scholar]
- Geiss G., Gutierrez L., Bellini C. (2009). Adventitious root formation: new insights and perspectives, in Annual Plant Reviews Vol 37: Root Development, ed Beeckman T. (Ames, IA: Wiley-Blackwell; ), 127–156. [Google Scholar]
- Giovannelli A., Giannini R. (2000). Reinvigoration of mature chestnut (Castanea sativa) by repeated graftings and micropropagation. Tree Physiol. 20, 1243–1248. 10.1093/treephys/20.18.1243 [DOI] [PubMed] [Google Scholar]
- Gou J. Y., Felippes F. F., Liu C. J., Weigel D., Wang J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. 10.1105/tpc.111.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves W. R. (2002). IBA, juvenility, and position on ortets influence propagation of Carolina buckthorn from softwood cuttings. J. Environ. Hortic. 20, 57–61. 10.24266/0738-2898-20.1.57 [DOI] [Google Scholar]
- Greenwood M. S. (1995). Juvenility and maturation in conifers: current concepts. Tree Physiol. 15, 433–438. 10.1093/treephys/15.7-8.433 [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J. D., Pacurar D. I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of auxin response factor transcripts and microRNA abundance. Plant Cell 21, 3119–3132. 10.1105/tpc.108.064758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B., Fry J. E., Hoffmann N. L., Eichholtz D., Rogers S. G., Fraley R. T. (1985). A simple and general-method for transferring genes into plants. Science 227, 1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Huang L. C., Lius S., Huang B. L., Murashige T., Mahdi el F. M., Van Gundy R. (1992). Rejuvenation of Sequoia sempervirens by repeated grafting of shoot tips onto juvenile rootstocks in vitro: model for phase reversal of trees. Plant Physiol. 98, 166–173. 10.1104/pp.98.1.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., et al. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell 17, 1387–1396. 10.1105/tpc.105.030981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N., Wang Y., Wu T., Zhang X. Z., Han Z. H. (2016). Correlation analysis of miR156 expression and redox status during the phase change of Malus xiaojinensis seedlings. J. China. Agric. Univ. 21, 59–64. 10.11841/j.issn.1007-4333 [DOI] [Google Scholar]
- Levy A., Szwerdszarf D., Abu-Abied M., Mordehaev I., Yaniv Y., Riov J., et al. (2014). Profiling microRNAs in Eucalyptus grandis reveals no mutual relationship between alterations in miR156 and miR172 expression and adventitious root induction during development. BMC Genomics 15:524. 10.1186/1471-2164-15-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hou H. M., Li X. Q., Xiang J., Yin X. J., Gao H., et al. (2013). Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 70, 100–114. 10.1016/j.plaphy.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Li Y. H., Han Z. H., Xu X. (2004). Segregation patterns of AFLP markers in F-1 hybrids of a cross between tetraploid and diploid species in the genus Malus. Plant Breed. 123, 316–320. 10.1111/j.1439-0523.2004.00994.x [DOI] [Google Scholar]
- Liu B. B., Zhang J., Wang L., Li J. B., Zheng H. Q., Chen J., et al. (2014). A survey of Populus PIN-formed family genes reveals their diversified expression patterns. J. Exp. Bot. 65, 2437–2448. 10.1093/jxb/eru129 [DOI] [PubMed] [Google Scholar]
- Liu J. C., Sheng L. H., Xu Y. Q., Li J. Q., Yang Z. N., Huang H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. 10.1105/tpc.114.122887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma C., Lu Y., Bai S., Zhang W., Duan X., Meng D., et al. (2014). Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS–LRR protein class gene in apple (Golden Delicious). Mol. Plant 7, 218–230. 10.1093/mp/sst101 [DOI] [PubMed] [Google Scholar]
- Majer C., Xu C. Z., Berendzen K. W., Hochholdinger F. (2012). Molecular interactions of rootless concerning crown and seminal roots, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. B. 367, 1542–1551. 10.1098/rstb.2011.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi M., Krens F. A., Visser R. G., De Klerk G. M. (2017). Azacytidine and miR156 promote rooting in adult but not in juvenile Arabidopsis tissues. J. Plant Physiol. 208, 52–60. 10.1016/j.jplph.2016.10.010 [DOI] [PubMed] [Google Scholar]
- Muthreich N., Majer C., Beatty M., Paschold A., Schutzenmeister A., Fu Y., et al. (2013). Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiol. 163, 419–430. 10.1104/pp.113.221481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal A., Yordanov Y. S., Perrone I., Karlberg A., Tisserant E., Bellini C., et al. (2012). The aintegumenta like1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in Poplar. Plant Physiol. 160, 1996–2006. 10.1104/pp.112.204453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C., Vielba J. M., Ferro E., Covelo G., Sole A., Abarca D., et al. (2007). Two Scarecrow-like genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 27, 1459–1470. 10.1093/treephys/27.10.1459 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. 10.1016/j.devcel.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Shikata M., Koyama T., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. 10.1093/pcp/pcp148 [DOI] [PubMed] [Google Scholar]
- Sorin C., Bussell J. D., Camus I., Ljung K., Kowalczyk M., Geiss G., et al. (2005). Auxin and light control of adventitious rooting in Arabidopsis require agronaute1. Plant Cell 17, 1343–1359. 10.1105/tpc.105.031625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B., Rasmussen A. (2016). The physiology of adventitious roots. Plant Physiol. 170, 603–617. 10.1104/pp.15.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P., Maloney G. S., Muday G. K. (2013). Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 162, 1392–1405. 10.1104/pp.113.217174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G., Sauer M., Stauffer J. L., Multani D., Niu X. M., Sakai H., et al. (2007). The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50, 649–659. 10.1111/j.1365-313X.2007.03075.x [DOI] [PubMed] [Google Scholar]
- Wang J. W., Park M. Y., Wang L. J., Koo Y. J., Chen X. Y., Weigel D., et al. (2011). MiRNA control of vgetative phase change in trees. PLoS Genet. 7:e1002012. 10.1371/journal.pgen.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. 10.1105/tpc.108.058180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Han D. G., Gao C., Wang Y., Zhang X. Z., Xu X. F., et al. (2012). Paternity and ploidy segregation of progenies derived from tetraploid Malus xiaojinensis. Tree Genet. Genomes 8, 1469–1476. 10.1007/s11295-012-0535-2 [DOI] [Google Scholar]
- Wu G., Poethig R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. 10.1242/dev.02521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z. F., Ji N., Zhang X. Z., Zhang Y. Z., Wang Y., Wu T., et al. (2014). The lose of juvenility elicits adventitious rooting recalcitrance in apple rootstocks. Plant Cell Tissue Organ. 119, 51–63. 10.1007/s11240-014-0513-5 [DOI] [Google Scholar]
- Xing L. B., Zhang D., Li Y. M., Zhao C. P., Zhang S. W., Shen Y. W., et al. (2014). Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. BMC Genomics 15:1125. 10.1186/1471-2164-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. L., Hu T. Q., Zhao J. F., Park M. Y., Earley K. W., Wu G., et al. (2016). Developmental functions of miR156-Regulated Squamosa promoter binding protein-like (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. 10.1371/journal.pgen.1006263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zhu L., Shou H. X., Wu P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. 10.1093/pcp/pci183 [DOI] [PubMed] [Google Scholar]
- Yang H. B., Murphy A. S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59, 179–191. 10.1111/j.1365-313X.2009.03856.x [DOI] [PubMed] [Google Scholar]
- Yu N., Cai W. J., Wang S., Shan C. M., Wang L. J., Chen X. Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322–2335. 10.1105/tpc.109.072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Niu Q. W., Ng K. H., Chua N. H. (2015). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685. 10.1111/tpj.12919 [DOI] [PubMed] [Google Scholar]
- Zhang H., An H. S., Wang Y., Zhang X. Z., Han Z. H. (2015). Low expression of PIN gene family members is involved in triggering the dwarfing effect in M9 interstem but not in M9 rootstock apple trees. Acta Physiol. Plant. 37:104 10.1007/s11738-015-1851-6 [DOI] [Google Scholar]
- Zhang T. Q., Lian H., Tang H. B., Dolezal K., Zhou E. M., Yu S., et al. (2015). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 27, 349–360. 10.1105/tpc.114.135186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. H., Zou Z., Zhang J. H., Zhang Y. Y., Han Q. Q., Hu T. X., et al. (2011). Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 585, 435–439. 10.1016/j.febslet.2010.12.036 [DOI] [PubMed] [Google Scholar]
- Zimmerman P. W., Wil-Coxon F. (1935). Several chemical growth substances which cause initiation of roots and other responses in plants. Contr. Boyce Thompson Inst. 7, 209–229. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of nine miR156 precursors were analyzed using semi-quantitative RT-PCR in Malus xiaojinensis stem bark.
Histological features of Mx-A, Mx-J, and Mx-R leafy cuttings of Malus xiaojinensis before IBA treatment. Cross sections of the stems were stained with toluidine blue. Scale bars = 200 μm.
NPA (1-N-naphthylphthalamic acid) concentration selected for wild type tobacco plant treatment. Tobacco plants were transferred to MS medium with 5, 20, or 80 μM NPA for 14 days. (A) The phenotype and (B) percent rooting were evaluated. Bars show SD from three biological replicates; n = 10 individuals in each replicate. Scale bars = 15 mm.
Semi-quantitative RT-PCR analysis of the expression dynamics of MxPINs in stem barks from Mx-A and Mx-R during adventitious root formation in Malus xiaojinensis under IBA treatment. MxEF1α was used as an internal control. The upper and lower bands represent treatment and control, respectively.
NbPIN genes expression pattern in tobacco stems from WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic lines growing on MS medium during the adventitious rooting process. NbEF1α was used as an internal control.
Semi-quantitative RT-PCR analysis of the expression dynamics of MxARFs in stem barks from Mx-A and Mx-R during adventitious root formation in Malus xiaojinensis under IBA treatment. MxEF1α was used as an internal control. The upper and lower bands represent treatment and control, respectively.
NbARF genes expression pattern in tobacco stems from WT, 35S:MdMIR156a6, and 35S:MIM156 transgenic lines growing on MS medium during the adventitious rooting process. NbEF1α was used as an internal control.
Diagram of the miR156 target sites of the WT and modified version of MxSPLs. Capital letters indicate the amino acid sequences in Malus xiaojinesis.
Molecular characterization of transgenic tobacco plants. (A) Semi-quantitative PCR analysis of rMxSPLs DNA and mRNA levels and (B) miR156 expression levels in 10-day old seedling leaves.
Role of MxSPL4a&4b, 18, 19, and 24 genes during adventitious root formation in transgenic Nicotiana benthamiana. (A) Adventitious root formation in 35S:rMxSPL/35S:MdMIR156a6 plants. Scale bars = 1 cm. (B,C) Quantitative analysis of the rooting ability in tobacco stem cuttings. (B) Percent rooting was investigated after 7 and 14 d on MS medium. (C) Adventitious root numbers per cutting were counted after 14 d on MS medium. Bars show SD from three biological replicates; n = 5 plants in each individual replicate. The statistical analysis was performed by Duncan's multiple range test at level p ≤ 0.05; means with different letters are significantly different from each other.
The MxARF family of transcription factors in Malus domestic. MdARF2, 11, 12, 20, 30, 33, and 36–39 have an activation domain (AD) that is enriched in glutamine (Q), serine (S), and leucine (L). The remainder of the ARFs consist of transcriptional repressors with a repression domain (RD) that is enriched in serine (S) and in some cases proline (P). All ARFs contain a conserved DNA binding domain (DBD).
Phylogenetic analysis of miR156-targeted SPL between apple and Arabidopsis. Phylogenetic tree was constructed with SBP domain protein sequences. Phylogenetic tree was constructed using MEGA 4.0 software with the neighbor-joining (NJ) method and the bootstrap test replicated 1,000 times.
Primers for constructing vector.
PCR primers of genes in Malus xiaojinesis.
PCR primers of genes in Nicotiana benthamiana.