Abstract

Fluorescent proteins (FPs) are indispensable markers for two-photon imaging of live tissue, especially in the brains of small model organisms. The quantity of physiologically relevant data collected, however, is limited by heat-induced damage of the tissue due to the high intensities of the excitation laser. We seek to minimize this damage by developing FPs with improved brightness. Among FPs with the same chromophore structure, the spectral properties can vary widely due to differences in the local protein environment. Using a physical model that describes the spectra of FPs containing the anionic green FP (GFP) chromophore, we predict that those that are blue-shifted in one-photon absorption will have stronger peak two-photon absorption cross sections. Following this prediction, we present 12 blue-shifted GFP homologues and demonstrate that they are up to 2.5 times brighter than the commonly used enhanced GFP (EGFP).
Neuroscience research needs better tools for large-scale study of neural circuits.1 These tools include genetically encoded fluorescent proteins (FPs),2 which can be incorporated into biosensors,3 and hardware such as two-photon (2P) microscopes.4 Two-photon laser microscopy (2PLM) enables recording from large populations of neurons at greater sampling rates and depths than are attainable with one-photon excitation microscopy.5,6 The high intensities of near-infrared lasers (700–1300 nm) required for 2PLM, however, constrain the illumination time due to inevitable tissue damage through multiphoton-induced photochemistry of intrinsic cellular chromophores7−10 or heating, which is more important than the former for in vivo imaging of the mouse brain.11 Heating of the tissue is linearly proportional to the average laser intensity, and the rate of damage depends exponentially on the elevated steady-state temperature above a certain threshold.12 By improving the 2P brightness (also known as the action cross section, defined as the cross section multiplied by the fluorescence quantum yield) of a FP, an equivalent fluorescence signal can be acquired with a lower laser power and thus less heating. Therefore, the use of brighter FPs would exponentially increase the time available for imaging before damaging the tissue and make it possible to extract more information from the living brain.
How can we find 2P brighter FPs? Previously, we established unified structure–property relationships for the one-photon absorption (1PA) and two-photon absorption (2PA) properties for a series of FPs with the same chromophore structure but different local protein surroundings.13,14 Here we use these relationships to predict a correlation between the 1PA peak wavelength and the 2PA maximum cross section for FPs with the anionic green FP (GFP)-type chromophore. Specifically, we will show that FPs with 1PA spectra shifted to shorter wavelengths tend to have stronger 2PA.
The primary factor responsible for FP to FP variation of both the 1PA and 2PA properties is the internal electric field E created by the protein environment surrounding the chromophore. Any changes in the field E (projected onto the chromophore axis) translate into changes in the difference between the permanent dipole moments in the excited (μe) and ground (μg) states, Δμ = μe – μg. This difference represents the amount of charge transfer from one part of the molecule to another upon electronic excitation. In the case of the S0 → S1 transition in the anionic GFP chromophore, the electron density shifts from the phenolate (P) group to the imidazolinone (I) group;14−18 thus, Δμ points from I → P, which we take as the positive direction of axis x. In FPs, Δμ depends on E because the field-induced part Δμind must be considered in addition to the vacuum part, Δμ0.14,19 In a one-dimensional approximation, Δμind = ΔαE, where Δα is the difference of polarizabilities along the x direction between the excited and ground states and Δμind and E are the projections of the Δμind and E vectors on the x axis. The total Δμ value along the x axis is therefore a linear function of E(20)
| 1 |
Importantly, changes in the electric field due to mutation-induced alterations in the protein environment and the consequential changes in Δμ will cause variations in the 1PA and 2PA properties.
In the case of 1PA, the chromophore transition frequency undergoes a field-dependent Stark shift that is unique for each FP, and because of non-negligible Δμind, it contains both linear and quadratic terms of E(20)
| 2 |
where ν̅ and ν̅0 are the transition frequencies in cm–1 (i.e., 1PA peak positions) in the presence and absence of a field, respectively, c is the speed of light, and h is the Planck constant. Solving eq 1 for E and substituting it into eq 2, we obtain the dependence of the transition frequency on Δμ
| 3 |
Defining A = −(2hcΔα)−1 and C = ν̅0 + Δμ02/2hcΔα, we can rewrite eq 3 as follows:
| 4 |
The quadratic dependence of ν̅ on Δμ with positive curvature (A = 72 cm–1 D–2 with Δα = −35 Å3) and y-intercept C = 19300 cm–1 was demonstrated experimentally for numerous FP variants with the anionic GFP chromophore.14 Because Δα is negative, we can predict that applying the field along the P → I direction (i.e., opposite to x) will result in the increase of Δμ. An example could involve concentrating positive charge closer to P or negative charge closer to I. This, in turn, will correspond to a higher-frequency (blue) shift of the 1PA maximum.
In terms of 2PA, it has been observed experimentally that in FPs with anionic chromophores, the peak transition frequency of the 2PA spectrum is systematically higher by 1000–2000 cm–1 with respect to the 1PA peak frequency.13 This effect was explained by an intensification of the vibronic 0–1 transition in the 2PA spectrum from a Herzberg–Teller-type interaction of Δμ with Qa, a bond-length-alternating vibrational coordinate whose equilibrium position shifts due to charge transfer upon excitation.13 Quantum mechanical calculations of the 2PA spectrum of the anionic GFP chromophore substantiate this general idea21,22 and even specify the vibrations responsible for the effect (i.e., the in-plane stretching mode of the exocyclic C–C=C bond and, to some extent, the stretching mode of the C=O bond).21
This model predicts the following expression for the peak 2PA cross section, assuming a two-level 2PA system
| 5 |
where a is a constant independent of the local field, λ2PA is the transition wavelength (i.e., one-half of the laser wavelength) corresponding to the 2PA maximum, ε(λ1PA) is the peak 1PA extinction coefficient, ε(λ2PA) is the extinction coefficient at λ2PA, and HT is a Herzberg–Teller parameter13 assumed to be independent of the local field. (The two-level approximation for the S0 → S1 2P transition was validated with quantum chemical calculations using the sum-over-states formula for the anionic GFP chromophore in vacuum and in 30 different types of local surroundings corresponding to the broad spectral range of GFP homologues. Including more than two levels in these calculations affected the σ2 value by less than 5%.14) To the first approximation, all ε(λ1PA), ε(λ2PA), and λ2PA can also be considered nearly constants over the series of FPs under investigation.
Combining eqs 4 and 5, we finally get the explicit dependence of the peak 2PA cross section on the 1PA transition frequency
| 6 |
where the new constants are
Note that eq 6 predicts a monotonic (almost linear) increase of σ2,max as a function of ν̅. In other words, GFP homologues with blue-shifted (i.e., higher-frequency, shorter-wavelength) 1PA transitions are expected to have higher 2PA cross sections.
To test this theoretical correlation between the 1PA wavelength and 2PA strength, we measured the 2PA properties of 12 FPs with the anionic GFP chromophore, including 8 previously described,23−25 1 unpublished but deposited in GenBank (Accession AF545829), and 3 previously undescribed FPs. These FPs peak in 1PA at shorter wavelengths (≤465 nm) than the commonly used enhanced GFP (EGFP) (489 nm). For comparison, we also measured the 2PA spectra of EGFP (Clontech) and mNeonGreen (Allele Biotechnology), a GFP homologue with extremely bright fluorescence under 1P excitation.26 These results, along with the 1PA and fluorescence properties, are summarized in Table 1 and SI Figure 1.
Table 1. 1PA, Fluorescence, and 2PA Properties of FPs Described in this Letter, Ordered by 2P Brightness (last column)a.
| protein | 1PA peak (nm) | emission peak (nm) | φb (±10%) | εc (M–1 cm–1) (±1%) | 2PA peak (nm) | σ2,maxd (GM) (±13%) | σ2,maxφe (GM) (±16%) |
|---|---|---|---|---|---|---|---|
| eqFP486 | 445 | 486 | 0.81 | 49 900 | 856 | 125 | 100 |
| Rosmarinus | 437 | 482 | 0.85 | 45 000 | 852 | 110 | 95 |
| amFP486/K68M | 458 | 489 | 0.93 | 51 500 | 862 | 94 | 87 |
| dTFP0.2 | 461 | 489 | 0.83 (0.68)28 | 45 000 (60 000)28 | 869 | 100 | 85 |
| meleCFP | 453 | 486 | 0.86 (0.74)24 | 47 400 (47 400)24 | 857 | 90 | 77 |
| meffCFP | 465 | 490 | 0.80 (0.55)24 | 61 000 (88 600)24 | 872 | 92 | 74 |
| Tam1 | 452 | 486 | 0.82 | 46 300 | 866 | 81 | 66 |
| efasCFP | 462 | 490 | 0.88 (0.77)24 | 57 000 (40 333)24 | 867 | 73 | 64 |
| EG-4 | 438 | 485 | 0.88 | 46 000 | 853 | 70 | 62 |
| dsFP483 | 439 | 483 | 0.76 (0.46,23 0.7841) | 46 000 (23 900)23 | 856 | 78 | 59 |
| KCyG4219 | 457 | 488 | 0.83 (0.80)25 | 38 000 (21 100)25 | 862 | 69 | 57 |
| amFP486 | 455 | 486 | 0.75 (0.24,23 0.7127) | 49 200 (40 000)23 | 861 | 75 | 56 |
| EGFP | 489 | 510 | 0.76 (0.61)42 | 58 300 (55 000)42 | 911 | 54 | 41f |
| mNeonGreen | 506 | 517 | 0.78 (0.80)26 | 116 000 (116 000)26 | 944 | 29 | 23 |
All photophysical parameters are presented per single mature chromophore.
Fluorescence quantum yield. Relative errors of measurements are shown in parentheses.
Extinction coefficient.
2PA maximum cross section.
2P brightness.
The value of 41 GM presented here for the 2P brightness of EGFP corresponds to that reported by Blab et al.43 but does not match the value of 30 GM previously obtained by Drobizhev et al.29 This is likely due to the difference in the measured extinction coefficient used to evaluate the chromophore concentration in the 2PA cross section measurement. The value of 46 000 M–1 cm–1 published in Drobizhev et al.29 was based on fluorescence lifetime measurements with 400 nm excitation and the Strickler–Berg equation relating the extinction coefficient and radiative lifetime. However, 400 nm light causes fluorescence of the transient anionic I* state, not the steady anionic B* state, whose lifetimes differ by a factor of 1.26, that is, 3.4 vs 2.7 ns.44 This led to an underestimation of the extinction coefficient and 2PA cross section by the same factor in Drobizhev et al.29
All of the newly investigated blue-shifted GFPs emit cyan fluorescence (peaking at 476–500 nm), have 1PA peak positions between 438 and 465 nm, range in maximum 2PA cross section (σ2,max) from 69 to 125 GM, and have high fluorescence quantum yields (φ = 0.75–0.95). The peak 2P brightness values (σ2,maxφ) range from 57 to 100 GM, which are higher than those of EGFP (41 GM) and mNeonGreen (23 GM). As expected for blue-shifted FPs, the position of the maximum 2PA has shifted to shorter wavelengths (837–873 nm) relative to EGFP (911 nm). Interestingly, the blue shifts in amFP486, the TFP series (including dTFP0.2), and KCy4219 were proposed to be related to the presence of a cationic histidine residue (H199, H197, and H193, respectively) in close proximity to the phenolate group of the chromophore.25,27,28 This is in agreement with our model that predicts both the high-frequency shift of the 1PA transition and enhancement of 2PA due to an increase of the Δμ value when a positive charge is concentrated near the chromophore phenolate.
Plotting the new data together with published data29,30 as the cross section (σ2) versus transition frequency (ν̅) reveals that the model prediction is qualitatively accurate: a more blue-shifted 1PA is associated with a higher 2PA maximum cross section (Figure 1). The fitted curve is based on eq 6, keeping the C value constant at 19300 cm–114 and using β and γ as fitting parameters. The quantitative discrepancies of some data points with the model function (especially at large frequencies) can be explained by experimental errors and some simplifying assumptions in our model. For example, we have not considered the field-related broadening of spectral bands nor the changes of Franck–Condon factors. Because of this, the model presented here serves as a qualitative guideline to find brighter 2P GFP homologues.
Figure 1.

Correlation between σ2,max and the peak 1PA position. The fitted curve is based on eq 6 in the text. Error bars are at ±13%.
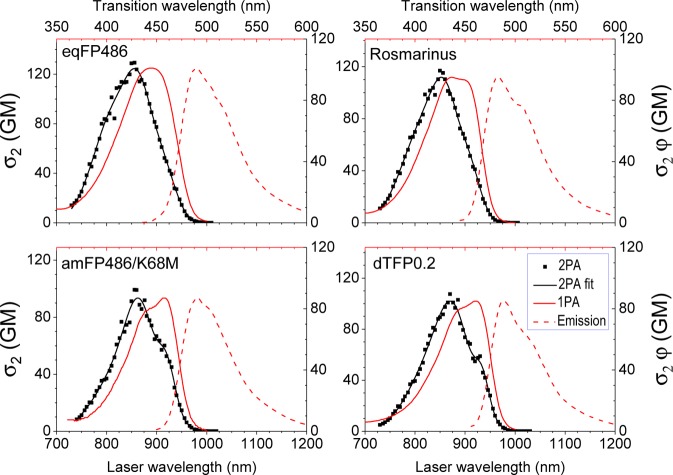
Four proteins stand out in terms of their 2PA cross sections and brightness: eqFP486 (GenBank Accession AF545829), dTFP0.2,28 amFP486/K68M,27,31 and a new hybrid mutant that we call Rosmarinus (GenBank Accession KY931461). Their respective 2PA, 1PA, and emission spectra are presented in Figure 2. Notably, the 2PA spectrum of each of them is blue-shifted from double the wavelength of the 1PA spectrum due to enhancement of the vibronic 0–1 transition, as described in our model. This shift, as well as alkaline titration experiments (showing no change of the absorption spectrum before the onset of denaturation), suggests that the chromophore is present in the anionic state in these proteins. For the neutral chromophore, the 1PA and 2PA spectra coincide.29,32 While eqFP486 is the brightest (σ2,maxφ = 100 GM), Rosmarinus is a more efficient folder (see the Methods section) and is similarly bright (σ2,maxφ = 95 GM). The protein dTFP0.2 is a dimer precursor to mTFP1 with a slightly blue-shifted 1PA spectrum. A single-point mutation of the wild-type protein amFP486, K68M, confers it with increased brightness under both 1P31 and 2P excitation. These four proteins are highlighted as potential markers for 2P imaging, but they have yet to be fully optimized for in vivo work.
Figure 2.
eqFP486, Rosmarinus, amFP486/K68M, and dTFP0.2 spectra: 2PA, 1PA, and emission. The top axis shows the transition wavelengths for 1PA and emission spectra, and the bottom axis shows the laser wavelengths for the 2PA spectra. The left axis is the 2PA cross section, and the right axis is the 2P brightness, which is scaled equally to show differences between FPs. The 2PA fit is displayed as a guide to the eye. The 1PA and the fluorescence emission spectra (excitation 450 nm) are normalized to the intensity of the 2PA peak.
Depending on the imaging application, the oligomerization state of FP probes can be important.33−35 To evaluate the oligomerization state of Rosmarinus, eqFP486, and amFP486/K68M, we determined their rotational correlation time and hydrodynamic volume by measuring the dependence of fluorescence anisotropy on solution viscosity with Perrin plots (see SI Figures S2–S5 and the SI). At low micromolar concentrations (5–6 μM) in buffer solution, eqFP486 is a dimer while amFP486/K68M and Rosmarinus are tetramers. At lower concentrations (∼1.2 μM), amFP486/K68M and eqFP486 become monomeric, but Rosmarinus remains tetrameric and becomes dimeric only at nanomolar concentrations (∼70 nM). Thus, these FPs may be useful for imaging applications that do not require monomers.36,37
An important implication of our findings is that FPs that are very bright under 1P excitation are not necessarily bright under 2P excitation. This is clearly illustrated by comparing the 1P and 2P brightness of Rosmarinus, EGFP, and mNeonGreen (Figure 3). While mNeonGreen is about two times brighter than the other two proteins in 1P, it is nearly two times dimmer than EGFP and four times dimmer than Rosmarinus in 2P. According to our model, this can be attributed to its red-shifted 1PA spectrum. Although the model as presented solely applies to the anionic GFP chromophore, much of the background comes from studies of the red FP chromophore,13,19 and a similar analysis to find 2P-brighter red FP variants is warranted.
Figure 3.

Absolute 1P (top) and 2P (bottom) brightness of Rosmarinus, EGFP, and mNeonGreen plotted versus the excitation wavelength. ε, extinction coefficient; φ, fluorescence quantum yield; σ2, 2PA cross section.
Our findings suggest that efforts to optimize FPs for 1P and 2P imaging should proceed independently because the desirable properties for each type of excitation may be incompatible. Blue-shifted forms of GFP have been used for some applications (e.g., FRET),38 but they have not been widely adopted in imaging. This is largely because a blue-shifted absorbance is poorly suited for 1P imaging due to increased phototoxicity, tissue absorbance, and scattering. The surprising result presented here is that these obscure proteins may be important tools for 2P imaging: a FP with a more blue-shifted 1PA is likely to have a stronger 2PA peak. While this will also be blue-shifted to some degree, it will still fall in the range of ∼850–870 nm, which is well within the near-IR optical window where tissue is most transparent to light and illumination-dependent heating is minimized. Water absorption decreases at shorter wavelengths, and lipid absorption has a peak at 930 nm with much weaker absorption at 850–870 nm.39 Additionally, these wavelengths are ideal for the conventional Ti-Sapphire laser that is often used for 2P imaging. One can assume that blue-shifted fluorescence would not cause major issues in 2P imaging because, although the scattering is higher, 2P microscope objectives with large numerical aperture and low magnification will still gather most of the photons.40 With a brighter 2P probe, less laser power is needed for the same fluorescence signal, and more imaging can be completed without causing damage to the tissue. Our model relating the 1PA peak position and 2PA cross section led us to blue-shifted GFP homologues up to 2.5 times brighter than EGFP. These FPs are highly promising starting points for further optimization of their 2PA properties.
Methods
Protein Purification. Proteins were expressed and purified as described in Barnett et al.45 All measurements were carried out in elution buffer, 1× PBS with imidazole at pH ≈ 8, except for dTFP0.2 and Tam1, which were in 1× TBS at pH ≈ 7.5.
Folding Rates. Relative folding rates were determined by qualitative observation of E. coli colonies expressing the protein of interest after 16 h of incubation at 37 °C and the subsequent time at room temperature that it took for them to appear fluorescent.
Fluorescence Quantum Yield. To measure the fluorescence quantum yield (φ), emission spectra were collected with the PC1 spectrofluorimeter (ISS), using fluorescein in 1 M NaOH as a reference standard. A correction function (created in the ISS software with quinine sulfate in 1 M H2SO4) was applied to the spectra to adjust for spectral sensitivity of the detection system, and the integral under the curve was divided by the OD at the excitation wavelength to find the quantum yield relative to that of fluorescein (φ = 0.95).46 All samples had a maximum OD < 0.1 to avoid reabsorption effects.
Extinction Coefficients. Alkaline titrations were performed for each protein with 1 M NaOH, with absorption spectra collected at every titration step until the peak at ∼447 nm of the denatured chromophore reached its maximum. The peak extinction coefficient of the protein was calculated by dividing the initial maximum OD over the final maximum OD (after correcting for the volume of added NaOH) and multiplying that by the extinction coefficient of the denatured chromophore (44 100 M–1 cm–1).47
For several of the proteins (meffCFP, efasCFP, EG-4, eqFP486, and dsFP483), it was not clear when the denatured form peaked; therefore, in addition to the aforementioned method, fluorescence detection with an alkaline titration was used. The sample was excited at 450 nm, and emission was collected at 490 nm; the titration was stopped when the fluorescence signal dropped to 1000 times below its initial value and leveled off, an indication that the protein had fully denatured. The OD was measured before and after denaturation, and the same calculations as before were applied to determine the extinction coefficient.
Two-Photon Measurements. Two-photon (2P) characterization was performed as in Barnett et al.45 Briefly, 2P excitation spectra and 2PA cross sections were measured using femtosecond excitation (with the tunable femtosecond laser InSight DeepSee, Newport) and fluorescein in 1 M NaOH as a reference standard.48,49 The fluorescence signal was collected with the 2P microscope setup (MOM, Sutter Instruments) from a 1 mm thick spectroscopic cell containing the sample. For the 2P excitation spectra, a 480/30 filter (Semrock) was used in the emission channel. 2PA cross sections at 840 and 900 nm were measured with collection of fluorescence through a 535/10 nm narrow bandpass filter (custom-made). The 2P excitation spectra were then normalized to those cross sections, and the average of the two are presented. OriginLab was used to fit 2–3 Gaussian peaks to the averaged spectrum of each protein, with the peak value of the summed curve presented in Table 1 as the 2PA maximum cross section. The 2P excitation spectrum of EGFP measured with this setup was compared to published data,29 and the shape and the absolute value coincided within experimental errors.
Development of New FP Variants, Rosmarinus, EG-4, and Tam1. Rosmarinus was made using PCR and the In-Fusion cloning technique (Clontech) to fuse the front half of the coding region for mc550 to the back half of meleCFP24 at a five amino acid identical stretch in their coding sequences. EG-4 is a tetrameric cyan FP cloned from the large polyp stony coral Euphyllia glabrescens (full description in preparation). Tam1 was discovered by colony-based screening of libraries of randomly mutated dTFP0.2.28 The gene encoding dTFP0.2 was used as the template for error-prone PCR.35 The forward primer contained an XhoI, site and the reverse primer contained an EcoRI site, and following digestion, the resulting product was ligated into the corresponding sites of pBAD/His B (ThermoFisher Scientific). The fluorescence of colonies (∼10 000 total) on Petri dishes was imaged using a custom imaging system.35 The brightest (excitation at 470/40 nm and emission at 510/20 nm) and most blue-shifted (ratio of emission at 470/40 to 510/20 nm, with excitation at 436/20 nm) colonies were picked and cultured overnight. Proteins were extracted using BPER (ThermoFisher Scientific), and spectra were collected with a SafireII platereader (Tecan). The brightest and most blue-shifted variant was designated as Tam1 and is equivalent to dTFP0.2 Q66L.
Acknowledgments
This work was supported by NIH Grant R01 GM121944 and the BRAIN initiative Grant U01 NS094246. We thank Lauren Barnett, Yan Li, Landon Zarowny, Dr. Matthew Wiens, and the Molecular Biology Service Unit for technical support.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.7b00960.
Method on evaluating oligomerization state of FPs; SI Figure 1. 2PA, 1PA, and emission spectra of all FPs presented in the paper; SI Figure 2. Perrin plots for EGFP and dTFP0.2; SI Figure 3. Perrin plots for amFP486/K68 M at different concentrations; SI Figure 4. Perrin plots for eqFP486 at different concentrations; SI Figure 5. Perrin plots for Rosmarinus at different concentrations; and SI Table 1. Molecular parameters used for evaluation of the FP aggregation state in solution (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Alivisatos A. P.; Chun M.; Church G. M.; Deisseroth K.; Donoghue J. P.; Greenspan R. J.; McEuen P. L.; Roukes M. L.; Sejnowski T. J.; Weiss P. S.; et al. Neuroscience. The Brain Activity Map. Science 2013, 339, 1284–1285. 10.1126/science.1236939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E. A.; Campbell R. E.; Lin J. Y.; Lin M. Z.; Miyawaki A.; Palmer A. E.; Shu X.; Zhang J.; Tsien R. Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. 10.1016/j.tibs.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem A.; Campbell R. E. Designs and Applications of Fluorescent Protein-Based Biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Sofroniew N. J.; Flickinger D.; King J.; Svoboda K.. A Large Field of View Two-Photon Mesoscope with Subcellular Resolution for in Vivo Imaging. eLife 2016, 5. 10.7554/eLife.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel W. R.; Williams R. M.; Webb W. W. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- Helmchen F.; Denk W. Deep Tissue Two-Photon Microscopy. Nat. Methods 2005, 2, 932–940. 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Koester H. J.; Baur D.; Uhl R.; Hell S. W. Ca2+ Fluorescence Imaging with Pico- and Femtosecond Two-Photon Excitation: Signal and Photodamage. Biophys. J. 1999, 77, 2226–2236. 10.1016/S0006-3495(99)77063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopt A.; Neher E. Highly Nonlinear Photodamage in Two-Photon Fluorescence Microscopy. Biophys. J. 2001, 80, 2029–2036. 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N.; Magee J. C.; Betzig E. High-Speed, Low-Photodamage Nonlinear Imaging Using Passive Pulse Splitters. Nat. Methods 2008, 5, 197–202. 10.1038/nmeth.1175. [DOI] [PubMed] [Google Scholar]
- Akerboom J.; Chen T.-W.; Wardill T. J.; Tian L.; Marvin J. S.; Mutlu S.; Calderón N. C.; Esposti F.; Borghuis B. G.; Sun X. R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgorski K.; Ranganathan G. Brain Heating Induced by near-Infrared Lasers during Multiphoton Microscopy. J. Neurophysiol. 2016, 116, 1012–1023. 10.1152/jn.00275.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey W. C. Arrhenius Relationships from the Molecule and Cell to the Clinic. Int. J. Hyperthermia 2009, 25, 3–20. 10.1080/02656730902747919. [DOI] [PubMed] [Google Scholar]
- Drobizhev M.; Makarov N. S.; Tillo S. E.; Hughes T. E.; Rebane A. Describing Two-Photon Absorptivity of Fluorescent Proteins with a New Vibronic Coupling Mechanism. J. Phys. Chem. B 2012, 116, 1736–1744. 10.1021/jp211020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobizhev M.; Callis P. R.; Nifosì R.; Wicks G.; Stoltzfus C.; Barnett L.; Hughes T. E.; Sullivan P.; Rebane A. Long- and Short-Range Electrostatic Fields in GFP Mutants: Implications for Spectral Tuning. Sci. Rep. 2015, 5, 13223. 10.1038/srep13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altoe’ P.; Bernardi F.; Garavelli M.; Orlandi G.; Negri F. Solvent Effects on the Vibrational Activity and Photodynamics of the Green Fluorescent Protein Chromophore: A Quantum-Chemical Study. J. Am. Chem. Soc. 2005, 127, 3952–3963. 10.1021/ja0451517. [DOI] [PubMed] [Google Scholar]
- Sinicropi A.; Andruniow T.; Ferré N.; Basosi R.; Olivucci M. Properties of the Emitting State of the Green Fluorescent Protein Resolved at the CASPT2//CASSCF/CHARMM Level. J. Am. Chem. Soc. 2005, 127, 11534–11535. 10.1021/ja045269n. [DOI] [PubMed] [Google Scholar]
- Filippi C.; Buda F.; Guidoni L.; Sinicropi A. Bathochromic Shift in Green Fluorescent Protein: A Puzzle for QM/MM Approaches. J. Chem. Theory Comput. 2012, 8, 112–124. 10.1021/ct200704k. [DOI] [PubMed] [Google Scholar]
- Wanko M.; García-Risueño P.; Rubio A. Excited States of the Green Fluorescent Protein Chromophore: Performance of Ab Initio and Semi-Empirical Methods. Phys. Status Solidi B 2012, 249, 392–400. 10.1002/pssb.201100536. [DOI] [Google Scholar]
- Drobizhev M.; Tillo S.; Makarov N. S.; Hughes T. E.; Rebane A. Color Hues in Red Fluorescent Proteins Are Due to Internal Quadratic Stark Effect. J. Phys. Chem. B 2009, 113, 12860–12864. 10.1021/jp907085p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins P. W.; Friedman R. S.. Molecular Quantum Mechanics; Oxford University Press: Oxford, U.K., 1997. [Google Scholar]
- Ai Y.; Tian G.; Luo Y. Role of Non-Condon Vibronic Coupling and Conformation Change on Two-Photon Absorption Spectra of Green Fluorescent Protein. Mol. Phys. 2013, 111, 1316–1321. 10.1080/00268976.2013.771800. [DOI] [Google Scholar]
- Ma H.; Zhao Y.; Liang W. Assessment of Mode-Mixing and Herzberg-Teller Effects on Two-Photon Absorption and Resonance Hyper-Raman Spectra from a Time-Dependent Approach. J. Chem. Phys. 2014, 140, 094107. 10.1063/1.4867273. [DOI] [PubMed] [Google Scholar]
- Matz M. V.; Fradkov A. F.; Labas Y. A.; Savitsky A. P.; Zaraisky A. G.; Markelov M. L.; Lukyanov S. A. Fluorescent Proteins from Nonbioluminescent Anthozoa Species. Nat. Biotechnol. 1999, 17, 969–973. 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Alieva N. O.; Konzen K. A.; Field S. F.; Meleshkevitch E. A.; Hunt M. E.; Beltran-Ramirez V.; Miller D. J.; Wiedenmann J.; Salih A.; Matz M. V. Diversity and Evolution of Coral Fluorescent Proteins. PLoS One 2008, 3, e2680. 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A.; Fukumura E.; Karasawa S.; Shiro Y.; Miyawaki A. Crystal Structure of a New Cyan Fluorescent Protein and Its Hue-Shifted Variants. Biochemistry 2009, 48, 5276–5283. 10.1021/bi801658p. [DOI] [PubMed] [Google Scholar]
- Shaner N. C.; Lambert G. G.; Chammas A.; Ni Y.; Cranfill P. J.; Baird M. A.; Sell B. R.; Allen J. R.; Day R. N.; Israelsson M.; et al. A Bright Monomeric Green Fluorescent Protein Derived from Branchiostoma Lanceolatum. Nat. Methods 2013, 10, 407–409. 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J. N.; Remington S. J. Crystal Structures and Mutational Analysis of amFP486, a Cyan Fluorescent Protein from Anemonia Majano. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 12712–12717. 10.1073/pnas.0502250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H.-W.; Henderson J. N.; Remington S. J.; Campbell R. E. Directed Evolution of a Monomeric, Bright and Photostable Version of Clavularia Cyan Fluorescent Protein: Structural Characterization and Applications in Fluorescence Imaging. Biochem. J. 2006, 400, 531–540. 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobizhev M.; Makarov N. S.; Tillo S. E.; Hughes T. E.; Rebane A. Two-Photon Absorption Properties of Fluorescent Proteins. Nat. Methods 2011, 8, 393–399. 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. R.; Barnett L. M.; Drobizhev M.; Wicks G.; Mikhaylov A.; Hughes T. E.; Rebane A. Two-Photon Directed Evolution of Green Fluorescent Proteins. Sci. Rep. 2015, 5, 11968. 10.1038/srep11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanushevich Y. G.; Staroverov D. B.; Savitsky A. P.; Fradkov A. F.; Gurskaya N. G.; Bulina M. E.; Lukyanov K. A.; Lukyanov S. A. A Strategy for the Generation of Non-Aggregating Mutants of Anthozoa Fluorescent Proteins. FEBS Lett. 2002, 511, 11–14. 10.1016/S0014-5793(01)03263-X. [DOI] [PubMed] [Google Scholar]
- Sarkisyan K. S.; Goryashchenko A. S.; Lidsky P. V.; Gorbachev D. A.; Bozhanova N. G.; Gorokhovatsky A. Y.; Pereverzeva A. R.; Ryumina A. P.; Zherdeva V. V.; Savitsky A. P.; et al. Green Fluorescent Protein with Anionic Tryptophan-Based Chromophore and Long Fluorescence Lifetime. Biophys. J. 2015, 109, 380–389. 10.1016/j.bpj.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C.; Steinbach P. A.; Tsien R. Y. A Guide to Choosing Fluorescent Proteins. Nat. Methods 2005, 2, 905–909. 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Prescott M.; Battad J. M.; Wilmann P. G.; Rossjohn J.; Devenish R. J. Recent Advances in All-Protein Chromophore Technology. Biotechnol. Annu. Rev. 2006, 12, 31–66. 10.1016/S1387-2656(06)12002-5. [DOI] [PubMed] [Google Scholar]
- Ai H.-W.; Baird M. A.; Shen Y.; Davidson M. W.; Campbell R. E. Engineering and Characterizing Monomeric Fluorescent Proteins for Live-Cell Imaging Applications. Nat. Protoc. 2014, 9, 910–928. 10.1038/nprot.2014.054. [DOI] [PubMed] [Google Scholar]
- Fei Y.; Hughes T. E. Transgenic Expression of the Jellyfish Green Fluorescent Protein in the Cone Photoreceptors of the Mouse. Vis. Neurosci. 2001, 18, 615–623. 10.1017/S0952523801184117. [DOI] [PubMed] [Google Scholar]
- Heintz N. Gene Expression Nervous System Atlas (GENSAT). Nat. Neurosci. 2004, 7, 483–483. 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Day R. N.; Booker C. F.; Periasamy A. Characterization of an Improved Donor Fluorescent Protein for Forster Resonance Energy Transfer Microscopy. J. Biomed. Opt. 2008, 13, 031203. 10.1117/1.2939094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S. L. Optical Properties of Biological Tissues: A Review. Phys. Med. Biol. 2013, 58, R37–R61. 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- Zinter J. P.; Levene M. J. Maximizing Fluorescence Collection Efficiency in Multiphoton Microscopy. Opt. Express 2011, 19, 15348–15362. 10.1364/OE.19.015348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo G. D.; Wang M.; Wu D.; Stelling A. L.; Tonge P. J.; Wachter R. M. Crystal Structure and Raman Studies of dsFP483, a Cyan Fluorescent Protein from Discosoma Striata. J. Mol. Biol. 2008, 378, 871–886. 10.1016/j.jmb.2008.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.; Day R. N.; Piston D. Fluorescent Protein Spectra. J. Cell Sci. 2001, 114, 837–838. [DOI] [PubMed] [Google Scholar]
- Blab G. A.; Lommerse P. H. M.; Cognet L.; Harms G. S.; Schmidt T. Two-Photon Excitation Action Cross-Sections of the Autofluorescent Proteins. Chem. Phys. Lett. 2001, 350, 71–77. 10.1016/S0009-2614(01)01282-9. [DOI] [Google Scholar]
- Cotlet M.; Hofkens J.; Maus M.; Gensch T.; Van der Auweraer M.; Michiels J.; Dirix G.; Van Guyse M.; Vanderleyden J.; Visser A. J. W. G.; et al. Excited-State Dynamics in the Enhanced Green Fluorescent Protein Mutant Probed by Picosecond Time-Resolved Single Photon Counting Spectroscopy. J. Phys. Chem. B 2001, 105, 4999–5006. 10.1021/jp003813i. [DOI] [Google Scholar]
- Barnett L. M.; Hughes T. E.; Drobizhev M. Deciphering the Molecular Mechanism Responsible for GCaMP6m’s Ca2+-Dependent Change in Fluorescence. PLoS One 2017, 12, e0170934. 10.1371/journal.pone.0170934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy; Springer: New York, 2006. [Google Scholar]
- Ward W. W.Biochemical and Physical Properties of Green Fluorescent Protein. Green Fluorescent Protein: Properties, Applications and Protocols; John Wiley & Sons, Inc., 2005; pp 39–65. [Google Scholar]
- Xu C.; Webb W. W. Measurement of Two-Photon Excitation Cross Sections of Molecular Fluorophores with Data from 690 to 1050 Nm. J. Opt. Soc. Am. B 1996, 13, 481–491. 10.1364/JOSAB.13.000481. [DOI] [Google Scholar]
- Makarov N. S.; Drobizhev M.; Rebane A. Two-Photon Absorption Standards in the 550–1600 Nm Excitation Wavelength Range. Opt. Express 2008, 16, 4029–4047. 10.1364/OE.16.004029. [DOI] [PubMed] [Google Scholar]
- Kelmanson I. V.; Matz M. V. Molecular Basis and Evolutionary Origins of Color Diversity in Great Star Coral Montastraea Cavernosa (Scleractinia: Faviida). Mol. Biol. Evol. 2003, 20, 1125–1133. 10.1093/molbev/msg130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.