Abstract
Enzyme IIC (EIIC) is a membrane-embedded sugar transport protein that is part of the phosphoenolpyruvate-dependent phosphotransferases. Crystal structures of two members of the glucose EIIC superfamily, bcChbC in the inward-facing conformation and bcMalT in the outward-facing conformation, were previously solved. Comparing the two structures led us to the hypothesis that sugar translocation could be achieved by an elevator-type transport mechanism in which a transport domain binds to the substrate and, through rigid body motions, transports it across the membrane. To test this hypothesis and to obtain more accurate descriptions of alternate conformations of the two proteins, we first performed collective variable-based steered molecular dynamics (CVSMD) simulations starting with the two crystal structures embedded in model lipid bilayers, and steered their transport domain toward their own alternative conformation. Our simulations show that large rigid-body motions of the transport domain (55° in rotation and 8 Å in translation) lead to access of the substrate binding site to the alternate side of the membrane. H-bonding interactions between the sugar and the protein are intact, although the side chains of the binding-site residues were not restrained in the simulation. Pairs of residues in bcMalT that are far apart in the crystal structure become close to each other in the simulated model. Some of these pairs can be cross-linked by a mercury ion when mutated to cysteines, providing further support for the CVSMD-generated model. In addition, bcMalT binds to maltose with similar affinities before and after the cross-linking, suggesting that the binding site is preserved after the conformational change. In combination, these results support an elevator-type transport mechanism in EIIC.
Main Text
The phosphoenolpyruvate-dependent phosphotransferase system (PTS) is ubiquitous in bacteria and crucial for bacterial growth (1, 2, 3). Each PTS has five components: enzyme I (EI), histidine containing phospho-carrier protein (HPr), enzyme IIA (EIIA), enzyme IIB (EIIB), and enzyme IIC (EIIC). EIIC is the only component that is embedded in the bacterial plasma membrane and transports a sugar substrate across the membrane. A phosphate group from a phosphoenolpyruvate is transferred sequentially through EI, HPr, EIIA, EIIB, and eventually to the incoming sugar while it is still on EIIC. Since PTS is absent in eukaryotes (4, 5), it could be a potential target for novel antibiotics.
Currently, there are two crystal structures of the glucose EIIC superfamily: a chitobiose-specific (bcChbC) (6) and a maltose-specific (bcMalT) EIIC (7), and both are from Bacillus cereus. The amino acid sequences of bcChbC and bcMalT are ∼50% similar and ∼19% identical, and they share the same structural fold. Both proteins form a homodimer, and each protomer has 10 transmembrane (TM) helices, two reentrant loops, and two amphipathic helices (Fig. S1 A–D). When the two structures are aligned, it becomes obvious that each protomer has two structurally conserved domains and that the two domains have a different position relative to each other in the two structures. The interface domain, which is composed of TM1–5 and the first amphipathic helix (AH1), forms the interface between the dimers. The transport domain, which is composed of TM6–10 and the two reentrant loops (HP1–2), contains the substrate binding site. The two domains are connected by the second amphipathic helix (AH2). Although the interface and the transport domains from the two proteins can individually be aligned (Fig. S1, E and F), the two structures represent different conformations. The substrate binding site of bcChbC is closer to the intracellular side, so it is likely in an inward-facing (IF) conformation (Fig. 1 A), whereas that of bcMalT is closer to the extracellular side and thus in the outward-facing (OF) conformation (Fig. 1 B).
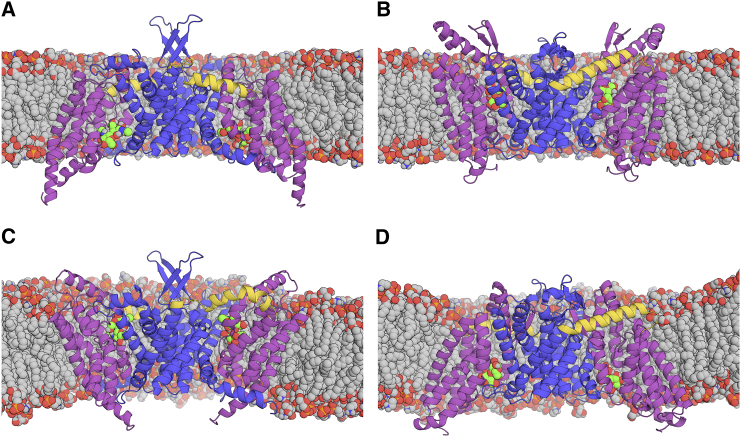
Figure 1.
Snapshots of (A and B) initial and (C and D) final simulation systems for (A) bcChbC-IF, (B) bcMalT-OF, (C) bcChbC-OF CVSMD model, and (D) bcMalT-IF CVSMD model structures in mixed bilayers of 3:1 POPE/POPG. The transport domain, the interface domain, and the linker between the two domains are represented by purple, blue, and yellow cartoons, respectively. Sugar and lipid molecules are represented by green and gray spheres. Water and ions are omitted for clarity. To see this figure in color, go online.
When the interface domains from the two crystal structures are aligned, the two structures can be superposed by a rigid-body motion of rotating and translating the transport domain by 44° and 9 Å, respectively. This rigid-body motion could bring the substrate from one side of the membrane to the other, a mechanism commonly known as the elevator-type transport mechanism (8, 9, 10, 11, 12). Since this prediction is based on the structures of two different proteins, how conformational changes occur in each of the proteins remains unknown. In this study, we try to bridge this knowledge gap by using both computational and experimental approaches. Since the conformational change of transporters generally occurs on micro-/millisecond timescales (13), it is difficult to observe such natural transition with the conventional MD simulation. Therefore, we used the collective variable-based steered molecular dynamics (CVSMD) simulation technique to accelerate the conformational transition.
The initial simulation systems were prepared using CHARMM-GUI Membrane Builder (14, 15, 16) with a 3:1 mixture of POPE/POPG lipids (Fig. 1, A and B); see Section 1 of the Supporting Material for the computational details. The rotational and translational harmonic restraints were applied to the Cα atoms of the transport domain, and the transport domain was smoothly steered toward the target rotation angle and the target z translation distance (Fig. S2; Movie S1). Note that the “elevator transport mechanism” is already implied by the simulation protocol, and we tried to see if the simulation result could capture the “elevator”-type transporter properties. Although the target rotation angle and translation distance of the transport domain could be identified by comparing the bcChbC-IF structure with the bcMalT-OF structure, these values could not be directly used without testing, because there are detailed interactions that need to be considered during the CVSMD simulation. Thus, to optimize the target rotation angle and translation distance, a set of CV pairs was first examined with 2-ns CVSMD. Based on these simulation results, the target rotation angle of 55° and target z translation distance of 8 Å were selected and used for slower 22-ns CVSMD simulations. Three independent CVSMD simulations were performed for each system using NAMD (17), and 50-ns MD simulations with no restraints were performed to equilibrate the final CVSMD structures.
From the CVSMD simulations, we were able to obtain the bcChbC-OF and bcMalT-IF model structures (Fig. 1, C and D). Each model structure has remarkably similar quaternary structural features against its counter crystal structure (Fig. S3). In this study, the structure similarity was examined by the TM-score (18) with a value between 0 and 1, with 1 representing the identical structures. The TM-score of the bcChbC crystal structure against the bcMalT crystal structure is 0.52, and it is dramatically increased to 0.65 after the CVSMD. Note that the TM-score of each domain between bcChbC and bcMalT is 0.63 (interface) and 0.65 (transport), reflecting the intrinsic structural differences between bcChbC and bcMalT even in the same state.
The sugar molecules in the binding site are spontaneously transported to the opposite side of the membrane during the CVSMD simulations (Fig. S4; Movie S1) while maintaining H-bonding interactions with the transport domain (Fig. S5). In bcChbC, the sugar molecules interact with W245, H250, D290, S298, N333, and E334, and the sugar molecules in bcMalT interact with E231, R232, H240, H241, K307, T354, and E355, which are all H-bonding interactions observed in the crystal structures. Since the sugar molecules are transported to the opposite side without losing the key interactions, our simulations indicate that the sugar binding site in the transport domain is not perturbed during the translocation.
To investigate how the interactions between the interface and transport domains change during the transportation, a Cα-Cα close-contact frequency was measured with a 10-Å cutoff. Movie S2 shows the changes in the Cα contact frequency as a function of simulation time, and Fig. S6 shows the difference (Δf) between the contact frequencies of the initial (0–1 ns; finit) and final (21–22 ns; ffin) stages: Δf = ffin – finit. In both the bcChbC and bcMalT simulations, the interaction pattern changes occurred mostly in HP1 (Fig. S6, orange) and TM7 (Fig. S6, green; TM7-PH1 for bcMalT). In the bcChbC CVSMD simulation, the C-terminus of TM7 provides the most favorable interactions with the interface domain initially, but the interface interactions are moved to its N-terminus or replaced by HP1 at the final stage. Considering that the TM7 C-terminus of bcChbC is replaced by β5/β6/PH1 in bcMalT, the bcMalT systems show the exact opposite interaction pattern changes compared to the bcChbC system. The interactions between the transport domain of one protomer and the interface domain of the other (Fig. S6) were not significantly changed during the simulations, indicating that the dimer formation may provide a scaffold for sugar transportation.
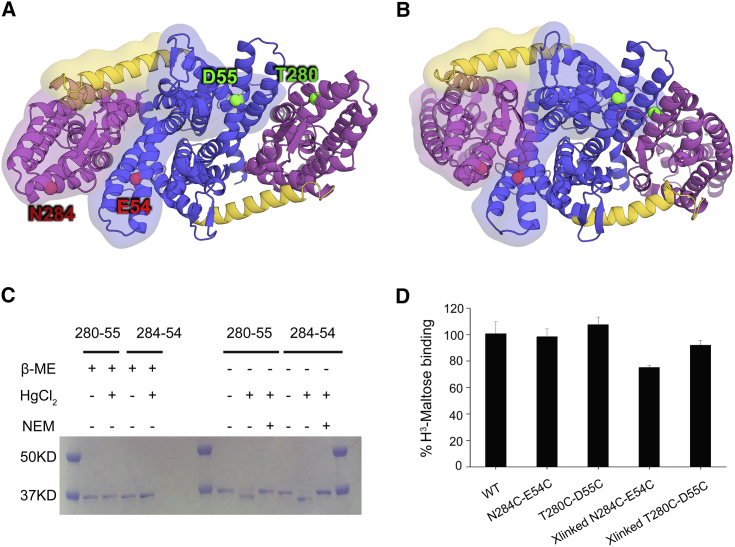
We then evaluated the CVSMD-generated model by testing its prediction of proximity between the transport and interface domains. We identified pairs of residues, one from the interface domain and one from the transport domain, that are 15–30 Å away in the bcMalT crystal structure but are in close proximity in the CVSMD-derived model (Table S1 and see Section 2 of the Supporting Material for the experimental details). Each pair of residues was then mutated to cysteines and tested with cross-linking reagents. Among the pairs, N284-E54 and T280-D55 (Fig. 2, A and B) showed acceptable Cα-Cα distances during the simulation (Fig. S7), and their cysteine mutants were cross-linked in the presence of Hg (II), as indicated by faster migration on an SDS-PAGE gel (Fig. 2 C). In both cross-linking reactions, the mercury concentration was ∼15 μM, close to the protein concentration. The cross-linking reaches completion within seconds, and maltose does not affect the rate of cross-linking (Fig. S8). The cross-linking was reversed in the presence of a thioreducing reagent, β-mercaptoethonal (β-ME), and blocked when the protein was pretreated with N-ethylmaleimide (NEM), which eliminates free thiogroups (Fig. 2 C). In addition, no cross-linking was observed in single cysteine mutations (Fig. S9).
Figure 2.
Cross-linking the transport and interface domains. (A and B) The dimeric bcMalT-OF crystal structure (A) and the bcMalT-IF computational model (B) are shown viewed from the extracellular side. The same color scheme as in Fig. 1 is used for each domain. The E54-N284 pair is represented by red spheres and the D55-T280 pair by green spheres. (C) SDS-PAGE of bcMalT double-cysteine mutants under various conditions. The molecular weight marker is shown on the left. (D) Maltose binding in bcMalT before and after cross-linking. To see this figure in color, go online.
We then tested whether the cross-linked MalT is capable of binding to the substrate maltose. In both double cysteine mutations, maltose binding was mildly affected compared to that of the wild-type, and after cross-linking, the binding was largely preserved (Fig. 2 D; Fig. S10). Assuming that bcMalT has both the IF and OF conformations in the solution, these results are consistent with the computational results showing that the transport domain moves as a rigid body and that the protein-substrate interactions are preserved.
In conclusion, we have studied structural changes required for sugar translocation using the CVSMD technique, and then validated the results with cross-linking and binding experiments. Our observations support an elevator-type transportation mechanism in EIIC, showing that the sugar molecules are stabilized during the transportation by maintaining the key H-bonding interactions with the transport domain. The contact analysis provides insight into the structural characteristics of each conformational state. For example, favorable interactions between the interface and the transport domains are mediated by AH1-TM1 and TM7 in the IF EIIC and by AH1-TM1 and HP1 in the OF EIIC. In addition, we were able to obtain two stable cross-linked proteins with the selected pairs from the close-contact analysis. The cross-linked proteins provide candidates for structural studies to further visualize the IF bcMalT, and eventually to calculate the free-energy landscape associated with the conformational changes during sugar transport in EIIC.
Author Contributions
J.L., Z.R., M.Z., and W.I. conceived the study and wrote the manuscript; J.L. and Z.R. performed research. J.L., Z.R., M.Z., and W.I. analyzed data and edited the manuscript.
Acknowledgments
This work was supported by National Science Foundation grant NSF DBI-1707207, XSEDE MCB070009 (to W.I.), National Institutes of Health grants U54GM087519 (to W.I. and M.Z.), GM098878, DK088057, and HL086392, 12EIA8850017 from the American Heart Association, and Cancer Prevention and Research Institute of Texas grants R12MZ (to M.Z.).
Editor: Bert de Groot.
Footnotes
Jumin Lee and Zhenning Ren contributed equally to this work.
Supporting Materials and Methods, twelve figures, two tables, two movies, and two structure data files are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30456-3.
Contributor Information
Ming Zhou, Email: mzhou@bcm.edu.
Wonpil Im, Email: wonpil@lehigh.edu.
Supporting Material
Trajectory Smoothing Window Size of 2 is used to remove the thermal fluctuation effect.
The close contact frequencies (f) are calculated in between the intra-molecular (left) and inter-molecular (right) proteins for bcChbC (top) and bcMalT (bottom). The data points having |f| < 0.5 are not shown for clarity.
References
- 1.Steinsiek S., Bettenbrock K. Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J. Bacteriol. 2012;194:5897–5908. doi: 10.1128/JB.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang Q., Zhang F., Qi Q. Comparison of individual component deletions in a glucose-specific phosphotransferase system revealed their different applications. Sci. Rep. 2015;5:13200. doi: 10.1038/srep13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picon A., Teixeira de Mattos M.J., Postma P.W. Reducing the glucose uptake rate in Escherichia coli affects growth rate but not protein production. Biotechnol. Bioeng. 2005;90:191–200. doi: 10.1002/bit.20387. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher J., Aké F.M., Joyet P. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siebold C., Flükiger K., Erni B. Carbohydrate transporters of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) FEBS Lett. 2001;504:104–111. doi: 10.1016/s0014-5793(01)02705-3. [DOI] [PubMed] [Google Scholar]
- 6.Cao Y., Jin X., Zhou M. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011;473:50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCoy J.G., Ren Z., Zhou M. The structure of a sugar transporter of the glucose EIIC superfamily provides insight into the elevator mechanism of membrane transport. Structure. 2016;24:956–964. doi: 10.1016/j.str.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes N., Ginter C., Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C., Kang H.J., Drew D. A two-domain elevator mechanism for sodium/proton antiport. Nature. 2013;501:573–577. doi: 10.1038/nature12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy J.G., Levin E.J., Zhou M. Structural insight into the PTS sugar transporter EIIC. Biochim. Biophys. Acta. 2015;1850:577–585. doi: 10.1016/j.bbagen.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulligan C., Fenollar-Ferrer C., Mindell J.A. The bacterial dicarboxylate transporter VcINDY uses a two-domain elevator-type mechanism. Nat. Struct. Mol. Biol. 2016;23:256–263. doi: 10.1038/nsmb.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machtens J.P., Kortzak D., Fahlke C. Mechanisms of anion conduction by coupled glutamate transporters. Cell. 2015;160:542–553. doi: 10.1016/j.cell.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Bahar I., Lezon T.R., Shrivastava I.H. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 2010;110:1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 15.Wu E.L., Cheng X., Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trajectory Smoothing Window Size of 2 is used to remove the thermal fluctuation effect.
The close contact frequencies (f) are calculated in between the intra-molecular (left) and inter-molecular (right) proteins for bcChbC (top) and bcMalT (bottom). The data points having |f| < 0.5 are not shown for clarity.