Significance
Cellular chemical reactions are slow, and to make them compatible with biological life, enzymes have evolved to accelerate their associated rate constants. Enzymatic catalysis is a complex process where the increase of rate constants predominantly depends on a reduction of the free energy barrier for product formation. It is now established that transient, so-called high-energy, enzyme states are indispensable entities that contribute to lowering of free energy barriers. Such states are inherently difficult to study. Here, we have been able to arrest a catalytically indispensable high-energy state of the enzyme adenylate kinase. A detailed characterization of its structure, dynamics, and function has revealed several aspects that together increase the understanding of how enzymes can perform their spectacular function.
Keywords: enzymatic catalysis, ligand binding, structural biology, adenylate kinase
Abstract
Proteins can bind target molecules through either induced fit or conformational selection pathways. In the conformational selection model, a protein samples a scarcely populated high-energy state that resembles a target-bound conformation. In enzymatic catalysis, such high-energy states have been identified as crucial entities for activity and the dynamic interconversion between ground states and high-energy states can constitute the rate-limiting step for catalytic turnover. The transient nature of these states has precluded direct observation of their properties. Here, we present a molecular description of a high-energy enzyme state in a conformational selection pathway by an experimental strategy centered on NMR spectroscopy, protein engineering, and X-ray crystallography. Through the introduction of a disulfide bond, we succeeded in arresting the enzyme adenylate kinase in a closed high-energy conformation that is on-pathway for catalysis. A 1.9-Å X-ray structure of the arrested enzyme in complex with a transition state analog shows that catalytic sidechains are properly aligned for catalysis. We discovered that the structural sampling of the substrate free enzyme corresponds to the complete amplitude that is associated with formation of the closed and catalytically active state. In addition, we found that the trapped high-energy state displayed improved ligand binding affinity, compared with the wild-type enzyme, demonstrating that substrate binding to the high-energy state is not occluded by steric hindrance. Finally, we show that quenching of fast time scale motions observed upon ligand binding to adenylate kinase is dominated by enzyme–substrate interactions and not by intramolecular interactions resulting from the conformational change.
Substantial proportions of cellular processes depend on chemical reactions that in aqueous solution often are several orders of magnitude too slow to support biological life (1). This difference between “chemical” and “biological” time scales is bridged by acceleration of the rates of chemical reactions by enzymes (2). The catalytic power of an enzyme depends on a significant reduction of the free energy barrier for the chemical reaction (2). Several factors collectively contribute to an enzyme’s efficiency as catalysts, including balanced substrate-binding affinity to ensure selectivity but at the same time avoid kinetic traps, optimal alignment of substrates, activation of a substrate’s functional groups, and dehydration of active sites. All of these factors to some degree depend on conformational dynamics of the enzyme (3), where dynamics is defined as the time-dependent displacement of atomic coordinates. Structural excursions from enzymatic ground states to high-energy states have been observed with NMR spectroscopy, and in a few cases, were shown to be rate limiting for catalytic turnover (3–7). Furthermore, it has been proposed that the conformational dynamics required for substrate binding can be present already in substrate-free enzymes (5, 8) such that the free energy landscapes are inherently primed for catalysis.
Both NMR (5, 9, 10) and single-molecule fluorescence resonance energy transfer (smFRET) (10–12) experiments have indicated that substrate-free enzymes can transiently populate “active-like” structural ensembles. The catalytically active state of adenylate kinase (AdK), the enzyme in focus here, is a closed conformation, for which the structure (with bound ligand) has been determined by X-ray crystallography (13). Sampling of a closed conformation in a ligand-free “apo enzyme” is one of the prerequisites for the conformational selection model (14), in which functionally active states are populated in the absence of substrate. In this model, substrate binding redistributes the statistical weights in a manner that favors active conformations. Critics of the conformational selection model pointed out that substrate binding to a closed conformational state may be hampered by steric hindrance (15). It is inherently difficult to address the function of high-energy states directly because they are, first, transient and, second, in a dynamic equilibrium with more stable ground states. To enable molecular studies of such states, we have developed a chemical approach whereby a high-energy state of apo AdK was arrested with a covalent disulfide bond. The relevance of cross-linking proteins through disulfide bonding (16–18) has been shown, e.g., by inhibition of fibril nucleation and elongation of α-synuclein, amyloid-β peptide, and islet amyloid polypeptide (19). The disulfide-linked AdK variant could be purified to homogeneity in the arrested high-energy state enabeling subsequent comprehensive biophysical characterization of the structure, dynamics, and activity of this functionally crucial conformation.
Results
Approach for Arresting AdK in a High-Energy Conformation.
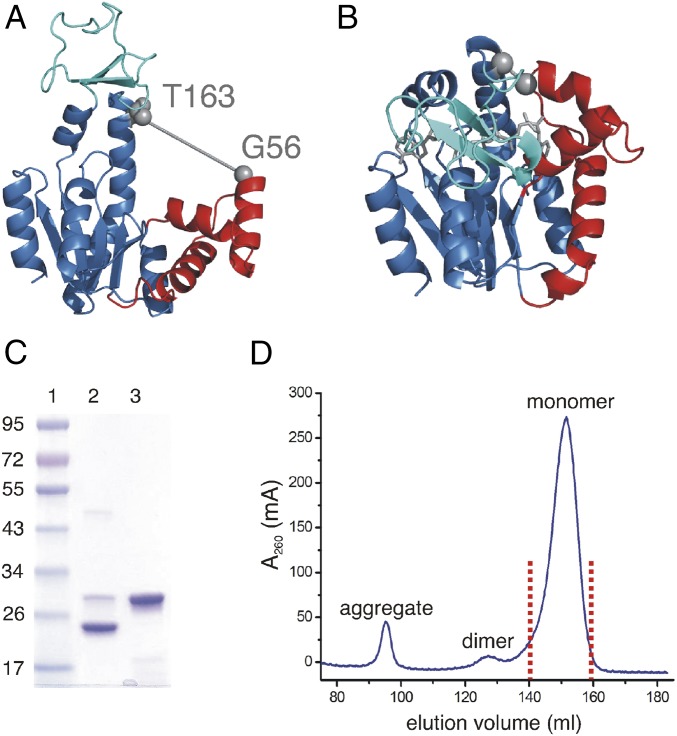
Cysteine pairs were designed by using the computer program Disulfide by Design (20) with the boundary conditions that the positions should be separated as much as possible in the open apo structure (Fig. 1A, PDB ID code: 4AKE; ref. 21) but in close proximity in the closed structure (Fig. 1B, PDB ID code: 1AKE; ref. 13) of AdK. A successful disulfide bond formation is thus designed to occur only if the substrate-free enzyme fluctuates on the open-to-closed conformational coordinate. Thus, if the apo enzyme meeting these distance conditions samples the fully closed conformation then, over time, formation of a disulfide bond will arrest the enzyme in a high-energy state even in the absence of bound substrate. Introducing cysteins at positions Gly56 (G56C) and Thr163 (T163C) (Fig. 1 A and B) promoted the formation of a disulfide bond in the substrate-free enzyme (see below). The successful disulfide bond formation in this double cysteine variant is enabling isolation of a substrate-free and at the same time closed AdK state. This AdK variant (denoted AdKcc) could be purified to homogeneity (Fig. 1 C and D) in amounts sufficient for subsequent biophysical studies, including NMR spectroscopy and X-ray crystallography.
Fig. 1.
Design of, and biochemical evidence for, a disulfide bond in monomeric apo-AdKcc,ox. (A) Wild-type apo AdK in the open conformation (PDB ID code: 4AKE) with AMPbd (red), ATPlid (cyan), and core domains (light blue). The positions of Gly56 and Thr163 used for cysteine mutations are indicated with gray spheres. The distance between the Cα atoms of Thr163 and Gly56 in the open form is 23.6 Å and is indicated by a straight line. (B) Wild-type AdK in complex with Ap5a (gray sticks) in the closed conformation (PDB ID code: 1AKE) with AMPbd (red), ATPlid (cyan), and core domains (light blue). The two sites used for cysteine mutation are indicated by gray spheres. The distance between the Cα atom of Gly56 and Thr163 in the closed form is 4.9 Å and is indicated with a straight line. (C) SDS/PAGE analysis of AdKcc from the monomeric fraction as indicated in D. Lane 1, molecular mass is indicated in kilodaltons; lane 2, AdKcc under oxidative conditions; lane 3, AdKcc under reduced conditions obtained with β-mercaptoethanol. (D) Size-exclusion chromatogram of AdKcc. Fractions containing monomers, dimers, and higher-order aggregates are indicated. The pool used for subsequent analysis of monomeric AdKcc,ox is marked by red dots.
Biochemical Evidence for Formation of an Intramolecular Disulfide Bond.
Initial evidence for formation of a disulfide bond in substrate-free AdKcc was obtained from SDS/PAGE analysis. The migration through the polyacrylamide matrix of a protein containing a disulfide bond depends on the proteins’ oxidation state, i.e., the absence or presence of the disulfide bond (22). The migration behavior of AdKcc under SDS/PAGE conditions (Fig. 1C) differs in oxidizing and reducing conditions, as expected for a protein containing a disulfide bond (22, 23). These findings confirm that AdKcc can be purified in an oxidized state with a disulfide bond present (designated AdKcc,ox, whereas the reduced form is designated AdKcc,red). By using size exclusion chromatography, it could be further shown that AdKcc,ox elutes as a monomeric enzyme (Fig. 1D). The kinetics of disulfide bond reduction in AdKcc,ox was quantified by following the change in intensity of a selected methyl group in real time. This time-dependent process could be accurately fitted to an exponential function yielding a half-life time for reduction of 54 ± 15 min at 25 °C (SI Appendix, Fig. S1 A and B).
Structure of Disulfide Arrested Adenylate Kinase.
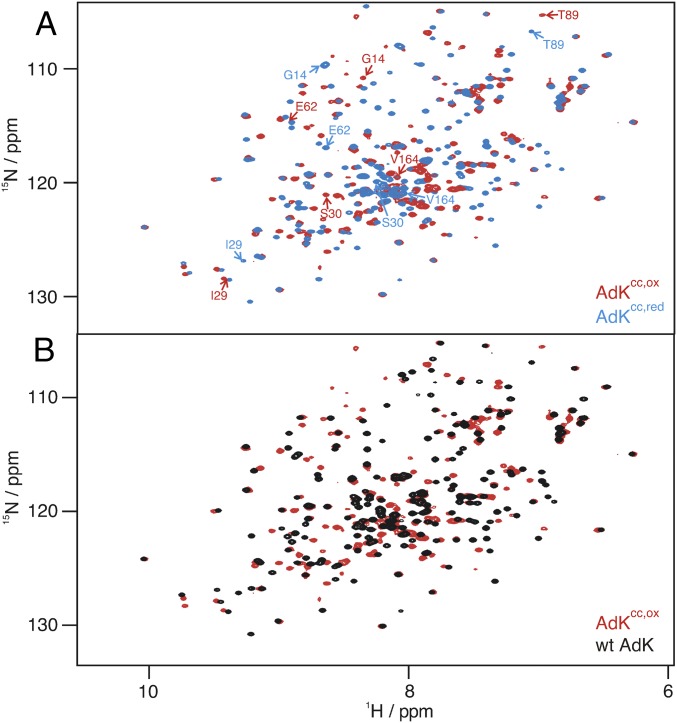
Substrate-free AdKcc,ox is a well-folded protein as judged by the chemical shift dispersion in a 1H-15N 2D heteronuclear single quantum correlation (HSQC) NMR spectrum (Fig. 2 A and B). We obtained a high degree of assignment for backbone 1HN, 15N, CO, and Cα resonances of this enzyme variant: 76% (1HN, 15N, and CO) and 89% (Cα) completeness for AdKcc,ox; 95% (1HN, 15N, and CO), and 99% (Cα) completeness for AdKcc,red. These numbers are on the same level as the degree of assignments of 1HN and 15N chemical shifts for wild-type AdK (97% for apo and 95% for Ap5a-bound state). The assignments enabled quantitative analysis of secondary structure by conducting analysis of torsion angles (ψ and ϕ) with the empirical program TALOS+ (24) (SI Appendix, Fig. S2). Profound spectral differences were observed in the NMR spectra following reduction of AdKcc,ox with the reducing agent Tris(2-carboxyethyl)phosphine (TCEP) (Fig. 2A). These spectral differences between AdKcc in the oxidized and reduced states were analyzed on basis of changes of 1H-15N chemical shifts (SI Appendix, Table S1) and mapped onto the structure of AdK in Fig. 3A. The overall chemical shift perturbation pattern between the oxidized and reduced states of AdKcc is similar to the pattern observed for the open-to-closed transition of wild-type AdK following binding to the tight binding inhibitor P1,P5-di(adenosine-5′)pentaphosphate (Ap5a) (25) (Fig. 3B and SI Appendix, Table S2). Note that Ap5a is a well-characterized bisubstrate analog for AdK, that binds to AdK with nanomolar affinity (3, 26, 27). Ap5a consists of one ATP and one AMP molecule bridged by an additional phosphate group. The convergence of the chemical shift perturbation patterns for reduction of AdKcc,ox and Ap5a binding to wild-type AdK is suggesting that substrate-free AdKcc,ox populates a closed-like conformation. Note that the amplitude of the overall perturbation of 1HN and 15N chemical shifts for the open-to-closed transition of wild-type AdK triggered by adding Ap5a (Fig. 3B) is larger than for the transition between the reduced and oxidized states of AdKcc (Fig. 3A). This effect depends on additional contributions to the chemical shifts from the presence of the strongly negatively charged Ap5a molecule because Ap5a alters both the overall protein conformation and the local chemical environment close to the interaction site. Analysis of the backbone torsion angles ψ and ϕ provides additional support for the closed nature of apo AdKcc,ox. Similar correlations for the open-to-closed transitions were found by either monitoring the open-to-closed transition by addition of Ap5a to wild-type AdK or by reducing AdKcc,ox into AdKcc,red (SI Appendix, Fig. S2).
Fig. 2.
High-resolution NMR spectra of AdKcc. (A) Overlay of 1H-15N HSQC spectra acquired for AdKcc,ox (red) and with its TCEP-reduced form AdKcc,red (blue). Residues that show largest changes in chemical shifts by comparing AdKcc,ox to AdKcc,red are indicated (compare Fig. 3A). (B) Overlay of 1H-15N HSQC spectra acquired for apo AdKcc,ox (red) and apo wild-type AdK (black).
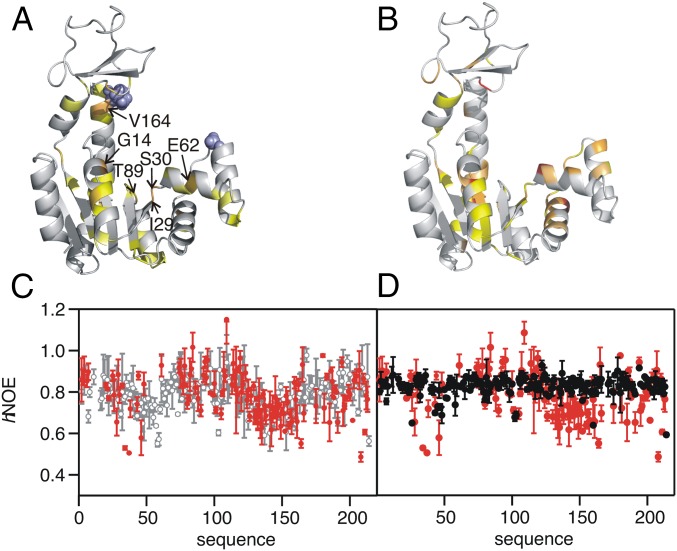
Fig. 3.
Structural and dynamic features of AdKcc under oxidative and reducing conditions. (A) Change of weighted 1H-15N chemical shifts, Δω, induced by adding TCEP to AdKcc,ox. Cysteine mutation sites are shown by blue spheres. Color coding: Δω > 0.2 ppm, light orange; 0.15 < Δω < 0.2 ppm, yellow orange; 0.1 < Δω < 0.15 ppm, yellow; 0.07 < Δω < 0.1 ppm, pale yellow. Residues with most substantial changes in chemical shifts are indicated (compare Fig. 2A) (B) Change of Δω induced by adding Ap5a to wild-type AdK. Color coding: Δω > 0.8 ppm, red; 0.6 < Δω < 0.8 ppm, bright orange; 0.4 < Δω < 0.6 ppm, light orange; 0.3 < Δω < 0.4 ppm, dark yellow. (C) hNOE for 15N nuclei of AdKcc under oxidative (red) and reducing conditions (gray). (D) hNOE for 15N nuclei of AdKcc,ox (red) and wild-type AdK in complex with Ap5a (black).
Chemical shifts of carbons are known to be highly sensitive to local molecular topology (28–30), and we exploited this feature to further characterize the structural state of AdKcc,ox. Analysis of changes of chemical shifts for backbone Cα and CO chemical shifts for the transitions: (i) wild-type open apo AdK to the Ap5a-bound closed state and (ii) the structural transition occurring upon reducing AdKcc,ox to AdKcc,red confirms that AdKcc,ox adopts a closed conformation (SI Appendix, Fig. S3 and Tables S3 and S4). Specifically, the chemical shift differences for these two transitions cluster on the same spatial location of the structure of AdK (SI Appendix, Fig. S3) mainly in the core of the central α-helix (T175, A176) and in the AMP binding domain (AMPbd) (around residues M34, K40, L45, A55). Note that the importance of the central helix in the enzyme’s opening-to-closing dynamics has been established by all-atom molecular dynamic simulations (31) and a comprehensive NMR study (3). The Cα and CO chemical shift changes were further analyzed quantitatively for residues in the AMPbd and the central helix. A striking linear correlation exists for these two residue clusters when the chemical shifts, corresponding to the transitions resulting from closure of wild-type AdK and reduction of AdKcc,ox, are plotted against each other (SI Appendix, Fig. S4). These correlations reinforce the notion that AdKcc,ox is populating a closed conformational state in solution.
The finding that AdKcc,ox is occupying a closed conformational state was further supported by analysis of residual dipolar couplings (RDCs). RDCs are NMR parameters that carry information about the angle formed between a specific bond vector with the external magnetic field. Hence, RDCs provide additional structural information of a protein’s topology (32). In total, 1H-15N RDC values were obtained for 90 residues of AdKcc,ox. For subsequent analysis, the RDCs were grouped into three classes corresponding to: AMPbd (Fig. 1A, colored in red), ATP binding domain (ATPlid) (Fig. 1A, cyan), and core domain residues (Fig. 1A, light blue). Quantitative analysis was performed by comparing the experimental RDCs with calculated RDCs from either the closed Ap5a-bound structure (1AKE) or the open apo structure (4AKE). For residues in both the AMPbd domain (SI Appendix, Fig. S5 A and B and Table S5) and the ATPlid domain (SI Appendix, Fig. S5 C and D and Table S5), the correlation between experimental and calculated RDCs is stronger when the closed structure is considered. Measured RDC values for residues belonging to the core domain of AdKcc,ox show no dependency on the structure chosen for RDC calculation (SI Appendix, Fig. S5 E and F and Table S5). Hence, the structural changes in the core domain are likely to be too small to generate a measurable perturbation to the RDCs. Taken together, the RDC analysis confirms the hypothesis that both the AMPbd and ATPlid domains are populating a closed-like conformation in the apo AdKcc,ox structure, which is fully consistent with changes in chemical shifts observed for apo AdKcc (Fig. 3 A and B and SI Appendix, Fig. S3).
The structural analyses carried out with different, independent NMR methods demonstrate that formation of the disulfide bond between positions Cys56 and Cys163 traps AdKcc,ox in a closed-like conformation. This conformation corresponds to a high-energy enzyme state that would be anticipated in a model where substrate binding occurs by conformational selection (33).
Dynamic Signature of Disulfide-Arrested Adenylate Kinase.
Next, we have investigated the intrinsic dynamics of AdKcc on a picosecond-to-nanosecond time scale. Overall, AdKcc,ox has similar picosecond-to-nanosecond dynamics compared with AdKcc,red and wild-type AdK when quantified at the level of amino acid residues with heteronuclear nuclear Overhauser effect (hNOE) experiments (Fig. 3C). This comparison indicates that the enzyme’s arrest in a closed-like state does not affect its fast time scale dynamics. In contrast, there is significant quenching of wild-type AdK’s picosecond-to-nanosecond dynamics when the closed state is generated by binding to Ap5a (Fig. 3D). Further evidence for this property is provided from analysis of crystallographic B factors, as described in the section below where the X-ray structure of Ap5a-bound AdKcc,ox is presented. Thus, the fast time scale motions are predominantly affected by enzyme-to-substrate interactions and not by intramolecular interactions resulting from the conformational change.
Crystallographic Structure of Inhibitor-Bound Cross-Linked Adenylate Kinase.
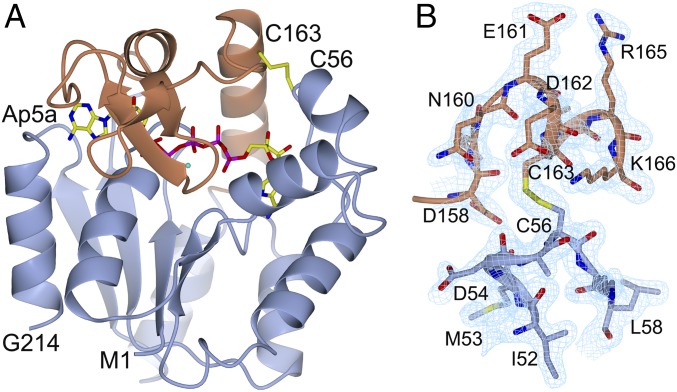
It has been demonstrated that Ap5a can form a complex with AdK, which adequately mimics the ternary complex formed by AdK, ATP, and AMP (3). In the study reported here, diffracting crystals of AdKcc,ox in complex with Ap5a were obtained by adding an excess of Ap5a to purified AdKcc,ox. A complete dataset to 1.9 Å was obtained, and the phases for 3D structure determination were obtained by using molecular replacement with Ap5a-bound wild-type AdK used as a search model (Fig. 4A and SI Appendix, Table S6). The electron density between C56 and C163 clearly shows the presence of a disulfide bond in AdKcc,ox with bound Ap5a (Fig. 4 A and B). Overall, the AdKcc,ox structure in complex with Ap5a presented here is similar to the wild-type structure in complex with Ap5a (1AKE; ref. 13) with an overall rmsd of 0.40 Å for 214 aligned Cα atoms. Notably, the orientation of catalytic side chains are in optimal positions for catalysis as deduced from a comparison with a catalytically competent cocrystal structure of AdK in complex with the nonhydrolyzable ATP analog AMPPNP and AMP (34) (SI Appendix, Fig. S6). Apparently, the Ap5a bound and cross-linked structure is in a conformation that is poised for catalysis and it can be concluded that the conformation of the closed and active state of AdK is not disturbed by the introduction of the disulfide bond. A clear correlation between motions on a picosecond-to-nanosecond time scale determined from NMR spectroscopy and crystallographic B factors has been established (35). Qualitatively, the quenching of fast time scale motions in AdK following binding to Ap5a is reinforced by analysis of crystallographic B factors in apo wild-type AdK and Ap5a-bound Adkcc,ox. As can be seen in SI Appendix, Fig. S7 A and B, there exist a significant redistribution of B factors when comparing these two states. Whereas the B factors are large in the flexible ATPlid and AMPbd of apo wild-type AdK (SI Appendix, Fig. S7A), the corresponding values in Ap5a-bound Adkcc,ox are substantially attenuated (SI Appendix, Fig. S7B). These observations indicate that the motions that affect the B factors are quenched in AdK as a result of binding to the transition state mimic Ap5a.
Fig. 4.
Crystal structure of AdKcc,ox in complex with Ap5a. (A) Ribbon drawing of the closed Ap5a-bound complex with the ATPlid (residues 113–176) colored in brown. The Ap5a molecule and the disulfide bond connecting C56 and C163 (yellow) are highlighted. (B) Close-up of the disulfide bond and neighboring residues. The σA-weighted (2m|Fo|-D|Fc|) electron density map is contoured at 1.0 times the root-mean-square value of the electron density in the asymmetric unit.
We were unable to crystallize apo AdKcc,ox. One can think of two reasons that may prevent crystallization of the trapped high-energy state: (i) structural heterogeneity caused by isomerization of the disulfide bond or (ii) energetic frustration (36) in apo AdKcc,ox. The isomerization of disulfide-bonded cysteines plays a vital role in protein folding (37) and has been shown to occur, e.g., as rotamer transitions in bovine pancreatic trypsin inhibitor on a microsecond-to-millisecond time scale (38, 39). However, relaxation dispersion experiments (40) on apo AdKcc,ox could not show contributions to chemical exchange on this time scale (SI Appendix, Fig. S8). For this reason, it is more likely that the disulfide bond between C56 and C163 forces AdKcc,ox into an energetically frustrated state, thus preventing protein crystallization. Indeed, calculation of the local mutational frustration index (36) suggested that the closed AdK structure is energetically more frustrated compared with the open conformation (SI Appendix, Fig. S9).
The Trapped High-Energy State Is On-Pathway for Catalysis.
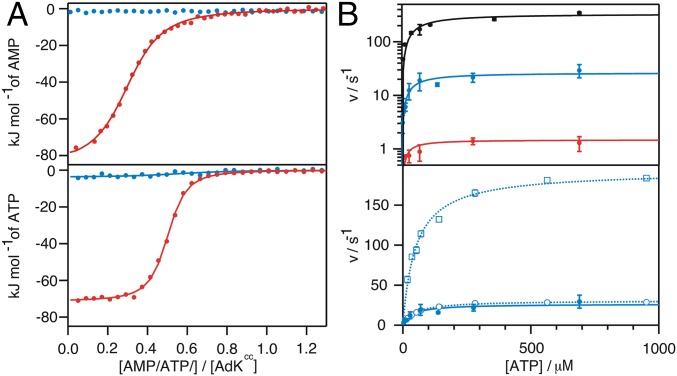
The structural analysis above suggests that AdKcc,ox is arrested in a closed-like structure that resembles the conformation of the catalytically active state. From a thermodynamic point of view, arrest of AdKcc,ox in an on-pathway state for substrate binding should result in significantly stronger substrate binding affinities compared with those of wild-type enzyme (16, 33). This argument will hold true if substrate binding is not occluded through steric hindrance in the arrested state (15). Binding of the natural substrates ATP and AMP to AdKcc,ox was quantified with isothermal titration calorimetry (ITC). The binding affinities (KD) of ATP and AMP to AdKcc,ox were found to be 200 times stronger relative to the wild-type enzyme (Fig. 5A and SI Appendix, Table S7). Obviously there is a strong convergence in the improvement of binding of both ATP and AMP to arrested AdKcc,ox. Note that confining apo AdKcc,ox in a closed-like conformation gives rise to a large entropic penalty seen for both titrations (SI Appendix, Table S7). Because AdK is known to possess a strong entropy-enthalpy compensation upon ligand binding (3), the enthalpic component due to AMP and ATP interaction with AdKcc,ox is for this reason expected to be significantly reduced in comparison with wild-type AdK. That is indeed the case (SI Appendix, Table S7). However, addition of ATP or AMP to AdKcc,red and to both single mutation variants AdK G56C and AdK T163C shows enthalpic and entropic components on the same order of magnitude as seen for ligand interaction with wild-type AdK (SI Appendix, Table S7). Taken together, the 200-fold increase in both ATP and AMP binding establishes that arrested AdKcc,ox is on-pathway for ligand binding and, therefore, also for catalysis. Binding of the transition state mimic Ap5a to AdKcc,ox is also significantly stronger compared with wild-type AdK as shown by ITC (SI Appendix, Fig. S10A). Comparing NMR spectra between apo AdK variants and related Ap5a-bound states structurally underlines that profound interaction (SI Appendix, Fig. S10 B–H).
Fig. 5.
Substrate affinity and catalytic activity of AdKcc. (A) Interactions of AMP (Upper) and ATP (Lower) with AdKcc under oxidative (red) and reduced conditions (blue) as probed by ITC. Corresponding thermodynamic parameters are listed in SI Appendix, Table S7. (B) Catalytic activity of wild-type AdK (black) and AdKcc under reducing (blue) and oxidative conditions (red) probed using a coupled ATPase assay and shown with a logarithmic scale (Upper). The catalytic activity of AdKcc under reducing conditions (presented as closed circles) is shown in Lower on a linear scale and relative to activities of the two variants with single-mutations, AdK G56C (open circles) and AdK T163C (open rectangles). Corresponding Michaelis–Menten parameters are listed in Table 1.
Next, the enzymatic activity of arrested AdKcc,ox was quantified by using an established coupled kinetic assay (41). The catalytic model for AdK involves a rate limiting conformational change from a substrate-bound closed state (4) to a substrate-bound open state (3). From this model it is predicted that AdKcc,ox should have a significantly lower catalytic efficiency than wild-type AdK, because of restriction of the opening process caused by the disulfide bond. Accordingly, AdKcc,ox showed only weak residual activity, with a kcat of 1.5 ± 0.1 s−1, but with preserved Michaelis–Menten characteristics and a KM of 18 ± 8 μM (Fig. 5B and Table 1). In control experiments, activities of the single mutation variants AdK G56C and AdK T163C were quantified (Fig. 5B and Table 1). Their kcat values were found to be 31 ± 1 s−1 and 195 ± 5 s−1, respectively, confirming that the orders of magnitude slower catalytic turnover of AdKcc,ox is primarily due to conformational restriction of the enzyme because of the disulfide bond and not a consequence of the selected two mutation sites.
Table 1.
Enzymatic parameters of AdK variants
| AdK variant | kcat, s−1 | KM, μM |
| Wild-type | 340 ± 30 | 53 ± 17 |
| AdKcc,ox | 1.5 ± 0.1 | 18 ± 8 |
| AdKcc,red | 26 ± 2 | 37 ± 12 |
| AdK G56C | 31 ± 1 | 46 ± 2 |
| AdK T163C | 195 ± 5 | 50 ± 5 |
Discussion
The key contribution to enzymatic rate enhancement is lowering of the transition state barrier for product formation. On the microscopic scale, this phenomenon depends on a large number of events that collectively conspire to enable lowering of the transition state energy. It has been established that enzymatic high-energy states are conformational ensembles that contribute to lowering the transition state energy barrier. These states are only transiently populated, and have so far only been detected and characterized experimentally by indirect methods such as smFRET (10–12) and NMR spectroscopy (5, 6, 9, 10). An emerging property of proteins in general (42, 43) including enzymes is that their substrate-free states may sample high-energy states that resemble the target-bound conformation (5, 6). In the case of adenylate kinase, such dynamics have been inferred from NMR spectroscopy with paramagnetic relaxation enhancement (10) and chemical shift analysis (9), and with smFRET methodology (10, 11). A missing piece in our understanding of the contribution to enzymatic catalysis from the sampling of high-energy states is a detailed understanding of their structure, dynamics, and function. We have succeeded in arresting a high-energy state that is on-pathway for ligand binding and catalysis for adenylate kinase. The approach was to introduce a pair of cysteine residues that are distant in the open and inactive apo structure while in close proximity in the closed and active conformation. Successful disulfide bond formation could then only occur for the substrate-free state if the enzyme fluctuates on the open-to-closed conformational coordinate. Our data conclusively show that the innate structural dynamics of apo AdK enables sampling of the fully closed and catalytically active state. This property indicates that AdK can, in fact, bind to substrates with both induced fit and conformational selection pathways. We have previously proposed that ATP binding to AdK occurs with a hybrid mechanism that uses aspects of both these general models (44). On the microscopic level, this hybrid mechanism corresponds to an initial induced-fit recognition of ATP occurring with modest structural rearrangements of the p-loop segment followed by the large conformational change occurring with a thermally driven conformational selection event. The data presented here establish that AdK has the inherent properties required for this hybrid model. Inside cells, the scenario is more complex and the actual pathway with dominant flux depends on both enzyme and ligand concentration as outlined in ref. 33.
For several cases, it has been shown that ligand binding to proteins is accompanied by a loss of flexibility of the proteins polypeptide backbone, which is manifested by increased order parameters as quantified from fast (picosecond-to-nanosecond) time scale motions probed by NMR spectroscopy. For AdK, a similar scenario was described by comparisons of the dynamics of the substrate-free apo state and an inhibitor (Ap5a) bound state (45). An open question is whether the change in fast time scale (picosecond-to-nanosecond) dynamics depends on enzyme–ligand interactions or on intramolecular interactions resulting from the conformational change. Our data clarifies this question for AdK, because we show that the fast time scale dynamics of the trapped closed state are similar to that of the apo wild-type enzyme. Hence, the changes in fast time scale dynamics for AdK primarily depend on the interactions formed between the enzyme and the ligand. There exist at least one notable exception to the general trend that ligand binding to proteins is inducing quenching of fast time scale motions. For the carbohydrate binding protein Galectin-3, an increase in fast time scale motions has been observed for the ligand-bound state in comparison with the substrate-free state (46). This effect points toward a substantial contribution from increased polypeptide backbone entropy in ligand recognition.
Using a steric occlusion argument, it has been suggested that closed states of enzymes should generally not be binding competent states (15). However, our results contradict this suggestion, at least for the closed and active state of AdK designed and explored in the present study. This arrested high-energy state of AdK has a 200-fold higher binding affinity for the natural substrates AMP and ATP than the wild-type enzyme. According to the model of AdK catalysis (4), substrate release, which depends on the slow reopening of the ATPlid and AMPbd substrate-binding subdomains, is the rate limiting step of catalytic turnover. This model predicts that the enzyme’s activity should be significantly reduced in the disulfide bond arrested state compared with the wild-type state. Our measurements validated the prediction, because enzymatic activity was 100-fold lower in the disulfide-locked state. Thus, the increase in binding affinity of the trapped high-energy state is accompanied by a significant decrease of its catalytic activity.
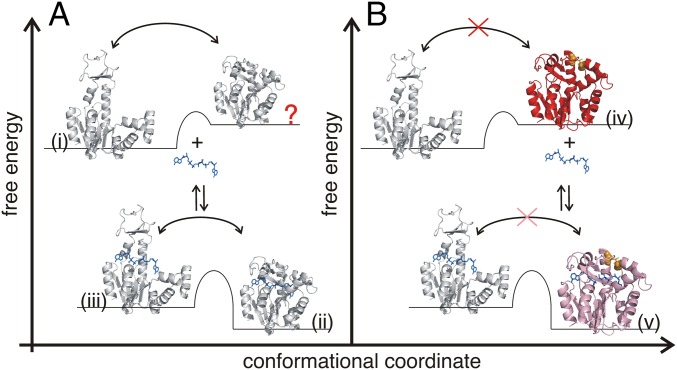
The results presented here establish—in the context of previous findings (6, 8, 10)—that the energy landscape (47) of enzymes evolved to allow sampling of catalytically active states even in the absence of substrates (Fig. 6). Thus, substrates are not per se required to activate enzymes but to reshape the free energy landscape in favor of the catalytically active conformation. This feature of the free energy landscape was originally postulated in the Monod–Wyman–Changeux (MWC) model of cooperative binding of oxygen to hemoglobin (48). The work presented here reveals that the sampling of binding competent high-energy states is of substantial functional relevance. Nevertheless, the benefit of the significant increase in substrate affinity by trapping AdK into a high-energy structural state has to be paid by a decrease in enzymatic turnover. Thus, an artificially designed network of disulfide bonds can be used to control both the affinity and the turnover of an enzyme. This basic principle may open an avenue for the targeted design of enzymes and other functional biomolecules.
Fig. 6.
Free energy landscapes of substrate binding for wild-type AdK and disulfide-trapped AdKcc,ox. (A) Wild-type apo AdK fluctuates between the open- (i, 4AKE; ref. 21) and closed-like structural states (indicated with a red question mark). Substrate (blue) binding lowers the free energy and stabilizes the closed and active state (ii, 1AKE; ref. 13), which undergoes conformational exchange dynamics with an open-like and bound structure (3). (B) Trapping of apo AdK in the high-energy state (iv, AdKcc,ox, red) by introducing a disulfide bond between C56 and C163 (orange) excludes transitions from the closed to open-like conformations in the absence of substrate. Substrate binding decreases the free energy of AdKcc,ox but the conformational exchange with an open-like bound state is prohibited. The disulfide bond effectively forces AdKcc,ox to bind substrates with a conformational selection mechanism, whereas substrate binding to wild-type AdK likely occurs with a mixed induced fit and conformational selection mechanism. The structural states presented in (iii), (iv), and marked by “?” have been modeled whereas the structure presented in v has been solved in the present study (Fig. 4). The left–right arrows indicate equilibria between open and closed structural states while the up–down arrows indicate substrate-binding events. The red crosses indicate reaction trajectories that are inaccessible for the AdKcc,ox variant.
Methods
Detailed experimental procedures describing site-specific mutagenesis, protein production, crystallization of AdKcc, X-ray data collection, phasing and refinement, the kinetic assay used for probing enzyme activity, isothermal titration calorimetry, and NMR spectroscopy are described in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank the staff of beamline 911-2 at Max II laboratory for assistance with data collection. The Umeå Protein Expertise Platform is acknowledged for providing reagents for protein overexpression and purification. This work was supported by the Swedish Research Council (M.W.-W. and A.E.S.-E.) and Deutsche Forschungsgemeinschaft Grant KO 4687/1-1 (to M.K.); Helge Magnussen, Per Rogne, and Jörgen Ådén regarding the AdK variants G56C and T163C; The Kempe Foundation for supporting the NMR and X-ray crystallography infrastructure at Umeå University; and The Knut and Alice Wallenberg Foundation for funding of the NMR infrastructure (NMR4life).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank, www.rcsb.org (PDB ID code 5EJE). NMR resonance assignments have been deposited in the Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu (accession nos. 25628 and 25629).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700919114/-/DCSupplemental.
References
- 1.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Fersht A. Structure and Mechanism in Protein Science. W.H. Freeman and Company; New York: 2000. [Google Scholar]
- 3.Kovermann M, et al. Structural basis for catalytically restrictive dynamics of a high-energy enzyme state. Nat Commun. 2015;6:7644. doi: 10.1038/ncomms8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf-Watz M, et al. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 5.Beach H, Cole R, Gill ML, Loria JP. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 6.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 7.Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341:899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 9.Ådén J, Verma A, Schug A, Wolf-Watz M. Modulation of a pre-existing conformational equilibrium tunes adenylate kinase activity. J Am Chem Soc. 2012;134:16562–16570. doi: 10.1021/ja3032482. [DOI] [PubMed] [Google Scholar]
- 10.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 11.Hanson JA, et al. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc Natl Acad Sci USA. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahra D, et al. Conformational plasticity and dynamics in the generic protein folding catalyst SlyD unraveled by single-molecule FRET. J Mol Biol. 2011;411:781–790. doi: 10.1016/j.jmb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Müller CW, Schulz GE. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. J Mol Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- 14.Gianni S, Dogan J, Jemth P. Distinguishing induced fit from conformational selection. Biophys Chem. 2014;189:33–39. doi: 10.1016/j.bpc.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan SM, Holyoak T. Enzymes with lid-gated active sites must operate by an induced fit mechanism instead of conformational selection. Proc Natl Acad Sci USA. 2008;105:13829–13834. doi: 10.1073/pnas.0805364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moleschi KJ, Akimoto M, Melacini G. Measurement of state-specific association constants in allosteric sensors through molecular stapling and NMR. J Am Chem Soc. 2015;137:10777–10785. doi: 10.1021/jacs.5b06557. [DOI] [PubMed] [Google Scholar]
- 17.Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. Pathways of allosteric regulation in Hsp70 chaperones. Nat Commun. 2015;6:8308. doi: 10.1038/ncomms9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lendel C, et al. A hexameric peptide barrel as building block of amyloid-β protofibrils. Angew Chem Int Ed Engl. 2014;53:12756–12760. doi: 10.1002/anie.201406357. [DOI] [PubMed] [Google Scholar]
- 19.Shaykhalishahi H, et al. Contact between the b1 and b2 segments of a-synuclein that inhibits amyloid formation. Angew Chem Int Ed. 2015;54:8837–8840. doi: 10.1002/anie.201503018. [DOI] [PubMed] [Google Scholar]
- 20.Dombkowski AA. Disulfide by Design: A computational method for the rational design of disulfide bonds in proteins. Bioinformatics. 2003;19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 21.Müller CW, Schlauderer GJ, Reinstein J, Schulz GE. Adenylate kinase motions during catalysis: An energetic counterweight balancing substrate binding. Structure. 1996;4:147–156. doi: 10.1016/s0969-2126(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 22.Kiritsi MN, Fragoulis EG, Sideris DC. Essential cysteine residues for human RNase κ catalytic activity. FEBS J. 2012;279:1318–1326. doi: 10.1111/j.1742-4658.2012.08526.x. [DOI] [PubMed] [Google Scholar]
- 23.Owen GR, et al. Human CD4 metastability is a function of the allosteric disulfide bond in domain 2. Biochemistry. 2016;55:2227–2237. doi: 10.1021/acs.biochem.6b00154. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lienhard GE, Secemski II. P 1, P 5 -Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973;248:1121–1123. [PubMed] [Google Scholar]
- 26.Schrank TP, Bolen DW, Hilser VJ. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc Natl Acad Sci USA. 2009;106:16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsson U, Wolf-Watz M. Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nat Commun. 2010;1:111. doi: 10.1038/ncomms1106. [DOI] [PubMed] [Google Scholar]
- 28.Barraud P, Schubert M, Allain FH-T. A strong 13C chemical shift signature provides the coordination mode of histidines in zinc-binding proteins. J Biomol NMR. 2012;53:93–101. doi: 10.1007/s10858-012-9625-6. [DOI] [PubMed] [Google Scholar]
- 29.Mulder FAA, Filatov M. NMR chemical shift data and ab initio shielding calculations: Emerging tools for protein structure determination. Chem Soc Rev. 2010;39:578–590. doi: 10.1039/b811366c. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 31.Brokaw JB, Chu JW. On the roles of substrate binding and hinge unfolding in conformational changes of adenylate kinase. Biophys J. 2010;99:3420–3429. doi: 10.1016/j.bpj.2010.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: Information for structure determination in solution. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammes GG, Chang Y-C, Oas TG. Conformational selection or induced fit: A flux description of reaction mechanism. Proc Natl Acad Sci USA. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry MB, et al. The closed conformation of a highly flexible protein: The structure of E. coli adenylate kinase with bound AMP and AMPPNP. Proteins. 1994;19:183–198. doi: 10.1002/prot.340190304. [DOI] [PubMed] [Google Scholar]
- 35.Powers R, Clore GM, Garrett DS, Gronenborn AM. Relationships between the precision of high-resolution protein NMR structures, solution-order parameters, and crystallographic B factors. J Magn Reson B. 1993;101:325–327. [Google Scholar]
- 36.Jenik M, et al. Protein frustratometer: A tool to localize energetic frustration in protein molecules. Nucleic Acids Res. 2012;40:W348–W351. doi: 10.1093/nar/gks447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bošnjak I, Bojović V, Šegvić-Bubić T, Bielen A. Occurrence of protein disulfide bonds in different domains of life: A comparison of proteins from the Protein Data Bank. Protein Eng Des Sel. 2014;27:65–72. doi: 10.1093/protein/gzt063. [DOI] [PubMed] [Google Scholar]
- 38.Grey MJ, Wang C, Palmer AG., 3rd Disulfide bond isomerization in basic pancreatic trypsin inhibitor: Multisite chemical exchange quantified by CPMG relaxation dispersion and chemical shift modeling. J Am Chem Soc. 2003;125:14324–14335. doi: 10.1021/ja0367389. [DOI] [PubMed] [Google Scholar]
- 39.Takeda M, Miyanoiri Y, Terauchi T, Kainosho M. (13)C-NMR studies on disulfide bond isomerization in bovine pancreatic trypsin inhibitor (BPTI) J Biomol NMR. 2016;66:37–53. doi: 10.1007/s10858-016-0055-8. [DOI] [PubMed] [Google Scholar]
- 40.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for charactherizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 41.Rhoads DG, Lowenstein JM. Initial velocity and equilibrium kinetics of myokinase. J Biol Chem. 1968;243:3963–3972. [PubMed] [Google Scholar]
- 42.Lange OF, et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 43.Chakrabarti KS, et al. Conformational selection in a protein-protein interaction revealed by dynamic pathway analysis. Cell Reports. 2016;14:32–42. doi: 10.1016/j.celrep.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ådén J, Weise CF, Brännström K, Olofsson A, Wolf-Watz M. Structural topology and activation of an initial adenylate kinase-substrate complex. Biochemistry. 2013;52:1055–1061. doi: 10.1021/bi301460k. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro YE, Sinev MA, Sineva EV, Tugarinov V, Meirovitch E. Backbone dynamics of escherichia coli adenylate kinase at the extreme stages of the catalytic cycle studied by (15)N NMR relaxation. Biochemistry. 2000;39:6634–6644. doi: 10.1021/bi992076h. [DOI] [PubMed] [Google Scholar]
- 46.Diehl C, et al. Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J Am Chem Soc. 2010;132:14577–14589. doi: 10.1021/ja105852y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 48.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.